Introduction
Endometrial cancer (EC) accounts for around 76,000
deaths in females worldwide each year (1,2).
Abnormal gene expression regulation results in numerous types of
human diseases, including cancer. Non-coding RNA (ncRNA) represents
a large proportion of human genome transcripts (3). Long non-coding RNA (lncRNA) is a
large family of ncRNAs that accounts for 80% of all RNA
transcripts. Emerging evidence has indicated that lncRNAs serve
vital roles in carcinogenesis and cancer progression (4,5).
Multiple studies have also proposed different mechanisms of action
for lncRNAs, the key mechanism of which is named the competing
endogenous RNAs (ceRNAs) theory (6).
Down syndrome cell adhesion molecule antisense 1
(DSCAM-AS1) was reported to serve as an oncogenic lncRNA in
different cancer types (7–9). High DSCAM-AS1 levels were identified
as a predictor for the overall survival of patients with colorectal
cancer (7). Moreover, DSCAM-AS1
was determined to stimulate non-small cell lung cancer progression
by regulating high mobility group box protein 1 expression via
sponging microRNA (miR)-577 (8).
Another study indicated that DSCAM-AS1 promoted breast cancer
progression by regulating the miR-204-5p/ribonucleotide reductase
M2 axis (9). However, it is
unclear whether DSCAM-AS1 serves a role in affecting EC
carcinogenesis and progression.
The aim of the present study was to analyze
DSCAM-AS1 expression level in EC tissues and cells and to explore
its biological roles in regulating EC progression. In addition, the
detailed acting mechanism of DSCAM-AS1 in EC was analyzed using
both in vitro and in vivo experiments.
Materials and methods
Patient tissues
EC tissues and adjacent non-cancerous tissues were
collected from 34 patients (mean age, 59.3 years; age range, 47–68
years) who underwent treatment at The Second Affiliated Hospital of
Fujian Medical University (Fujian, China) between January 2015 and
December 2015. Non-cancerous tissues were collected at 2 cm distant
from tumor tissues. The study protocol was approved by the Ethics
Committee of The Second Affiliated Hospital of Fujian Medical
University. Inclusion criteria for patients were as follows: i)
Diagnosed as EC by medical examination; ii) did not receive any
anticancer treatments; and iii) without other malignancies or
chronic diseases. Additionally, patients who did not have complete
clinical information were excluded. The treatment protocols of
patients with EC were in accordance with the Diagnosis and Therapy
Guideline for Endometrial cancer 4th edition (10). After surgery, patients were
treatment with radiotherapy, chemotherapy, or combination
therapies. Estrogen receptor (ER), Lymph-vascular space invasion
(LVSI) and histology type were classified by pathologists at our
hospital in a blinded manner according to the criteria of previous
literature (1–13). Written informed consent was
obtained from all enrolled patients.
Cell culture and treatment
The EC cells (HEC-1-B, HEC-1-A and KLE) used in the
present study were purchased from the American Type Culture
Collection, while normal uterine endometrial epithelial cells
(NUEEC) were obtained from Chi Scientific, Inc. RPMI-1640
(Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10%
fetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc.),
100 U/ml penicillin, and 0.1 mg/ml streptomycin were used to
incubate cells. Cell culture was conducted at 37°C in an incubator
filled with 5% CO2.
Cell transfection
Small interfering (si)RNA against DSCAM1-AS1
(si-DSCAM1-AS1, 5′-GUUCUGGUCUCAUCAUGAUTT−3′), control siRNA
(si-con, 5′-AUAGACUCGCUUGUUGUUCTT−3′), miR-136-5p mimic
(5′-CAUCAUCGUCUCAAAUGAGUCU-3′), miR-136-5p inhibitor
(5′-AGACUCAUUUGAGACGAUGAUG-3′) and control miRNA (mimic-con,
5′-ACAUUAACGUAUCGUCACUCUG-3′; or inhibitor-con,
5′-GCGAGAUCGCUGAUAUGAAUAU-3′) were synthesized by Shanghai
GenePharma Co., Ltd. miRNA or siRNA (50 nM) transfection was
conducted using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C according to the manufacturer's
instructions. After 48 h transfection, cells were collected for
subsequent analyses.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation rate was measured using a CCK-8
assay (Sigma-Aldrich; Merck KGaA). In brief, cells were seeded into
96-well plate at a density of 4×103 cells/well. Plates
were maintained at 37°C in the incubator, as aforementioned. A
total of 10 µl CCK-8 reagent was added after 24, 48 and 72 h
incubation and then incubated for a further 2 h at 37°C. Optical
density at the wavelength of 450 nm was measured using microplate
reader.
Flow cytometry assay
Cell apoptosis was measured using the
Annexin-V-FITC/PI cell apoptosis kit (Beyotime Institute of
Biotechnology). After treatment with 0.25% trypsin at 37°C for 2
min, 5×106 cells were collected, suspended in binding
buffer and stained with 5 µl Annexin-V-FITC at 4°C for 15 min in
the dark. Then, 5 µl PI was added to cells and incubated at 4°C for
5 min. Finally, cell apoptosis rate was measured using LSRFortessa™
(BD Biosciences) and analyzed with FlowJo 10.7 software (BD
Biosciences). The cells with the Annexin-FITC label were regarded
as apoptotic cells.
Reverse transcription-quantitative
(RT-qPCR) analysis
RNA samples of tissues and cultured cells were
isolated with TRIzol® kit (Invitrogen; Thermo Fisher
Scientific, Inc.) and reverse transcribed into complementary DNA
using the PrimeScript® RT Reagent kit (Takara
Biotechnology Co., Ltd.) according to the provided protocols.
RT-qPCR was performed using an ABI 7500 PCR instrument (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using SYBR Green
(Takara Biotechnology Co., Ltd.). Primers were synthesized by
Sangon Biotech Co., Ltd. and the sequences were as follows:
DSCAM-AS1 forward, 5′-GTGACACAGCAAGACTCCCT-3′ and reverse,
5′-GATCCGTCGTCCATCTCTGT-3′; GAPDH forward,
5′-AAGGTGAAGGTCGGAGTCAA-3′ and reverse, 5′-AATGAAGGGGTCATTGATGG-3′;
miR-136-5p forward, 5′-ACACTCCAGCTGGGACTCCATTTGTTTT-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGT-3′; and U6 small nuclear (sn)RNA (U6 snRNA)
forward, 5′-TCCGATCGTGAAGCGTTC−3′ and reverse,
5′-GTGCAGGGTCCGAGGT-3′. Gene expression levels were calculated with
the 2−∆∆Cq method (14). The thermocycling conditions used
were as follows: 95°C for 30 sec (1 cycle), 95°C for 5 sec; and
60°C for 30 sec (40 cycles).
Target prediction
miRNA targets for DSCAM-AS1 were analyzed using the
Encyclopedia of RNA Interactomes (ENCORI) (http://starbase.sysu.edu.cn/agoClipRNA.php?source=lncRNA&flag=target&clade=mammal&genome=human&assembly=hg19&miRNA=all&clipNum=1&deNum=0&panNum=0&target=DSCAM-AS1).
Among all predicted targets results in ENCORI, miR-136-5p ranked
first and therefore was selected for subsequent analyses.
Dual-luciferase activity reporter
assay
Chemically synthesized wild-type (WT)
3′-untranslated region sequence of DSCAM-AS1 was inserted into a
pMIR-reporter (Promega Corporation) to generate a WT-DSCAM-AS1
construct. A site-direct mutagenesis kit (Takara Biotechnology Co.,
Ltd.) was used to generate MT-DSCAM-AS1 construct from
WT-DSCAM-AS1. WT-DSCAM-AS1 or MT-DSCAM-AS1 and the aforementioned
miRNAs were co-transfected into EC cells using
Lipofectamine® 2000 according to the manufacturer's
instructions. After 48 h, cells were collected to measure relative
luciferase activity using the Dual-luciferase activity system
(Promega Corporation) with Renilla luciferase activity used
as the internal control.
DSCAM-AS1 expression level analysis in
EC using online database
DSCAM-AS1 expression level in EC tissues and normal
tissues was measured using ENCORI.
RNA immunoprecipitation (RIP)
assay
A Magna RIP RNA-Binding Protein Immunoprecipitation
kit (EMD Millipore) was utilized to detect the potential
interactions of DSCAM-AS1 and miR-136-5p based on supplier's
instructions. Cells were lysed with RIP buffer and then incubated
with anti-Argonaute 2 (anti-Ago2, MA5-23515, Thermo Fisher
Scientific, Inc.) or normal immunoglobulin G (IgG)-conjugated
magnetic beads. RNA samples were then extracted using
TRIzol® and subjected to RT-qPCR analysis (as described
in the RT-qPCR section) to detect relative DSCAM-AS1 and miR-136-5p
expression levels using the protocols described above.
In vivo tumorigenesis assay
The animal experiment protocol was approved by the
Ethics Committee of The Second Affiliated Hospital of Fujian
Medical University. BALB/C nude female mice (4 weeks old; n=5 for
each group, 10 in total; 18–20 g) were purchased from the National
Laboratory Animal Center and cultured in specific pathogen-free
conditions with controlled temperature (23±3°C) and humidity
(40±10%), and under a 12 h light/dark cycle. Mice had free access
to laboratory food and water. Sample size calculation was
accompanied using an online calculation tool (https://clincalc.com/). sh-DSCAM-AS1
(5′-CCGGGCTGCAGTGAGCTGAGATCATCTCGAGATGATCTCAGCTCACTGCAGCTTTTTG−3′)
or sh-con
5′-CCGGGCTGTTAAGCCGTGAGAGCTACTCGAGTAGCTCTCACGGCTTAACAGCTTTTTG−3;
both Sangon Biotech Co., Ltd. were cloned into pLKO.1 and packaged
into lentivirus particles together with psPAX2 and pMD2.G through
co-transfecting 293T cells with Polyethylenimine (PEI, 1 mg/ml,
Sigma-Aldrich; Merck KGaA) in DMEM supplemented with 10% fetal
bovine serum w at 37°C incubator filled with 5% CO2.
After 48 h, lentivirus particles were collected from supernatant
and then transduced into HEC-1-B cells. Stably infected
DSCAM-AS1-silenced cells were selected via puromycin (0.5 mg/ml)
for 7 days. The successful transduction of sh-DSCAM-AS1 was
confirmed by RT-qPCR. Then, 1×107 cells in PBS were
injected into the right flank of each mouse. On the 7th day after
injection, tumor width and length were measured weekly to calculate
tumor volume using the following formula: length ×
width2 × 0.5. After 4 weeks, mice were sacrificed via
cervical dislocation. In accordance with the IACUC guidelines, the
tumor diameter was maintained at <1.5 cm throughout the
experiment period. Finally, tumor tissues were collected and
weighed.
Statistical analysis
Data obtained from three independent experiments
were analyzed using SPSS 21.0 software (IBM Corp.) and presented as
the mean ± SD. Paired Student's t-test was conducted to analyze
differences between two groups, while one-way ANOVA followed by
Dunnett's post hoc test was conducted to analyze differences among
three or more groups. Log-rank test was used to analyze survival
difference between high- and low-DSCAM-AS1 groups using the mean
expression value as cut-off value. The χ2 test was used
to calculate associations between DSCAM-AS1 expression and
clinicopathological features of patients with EC. Spearman's
correlation co-efficient for DSCAM-AS1 and miR-136-5p was
calculated. P<0.05 was considered to indicate a statistically
significant difference.
Results
DSCAM-AS1 is highly expressed in
EC
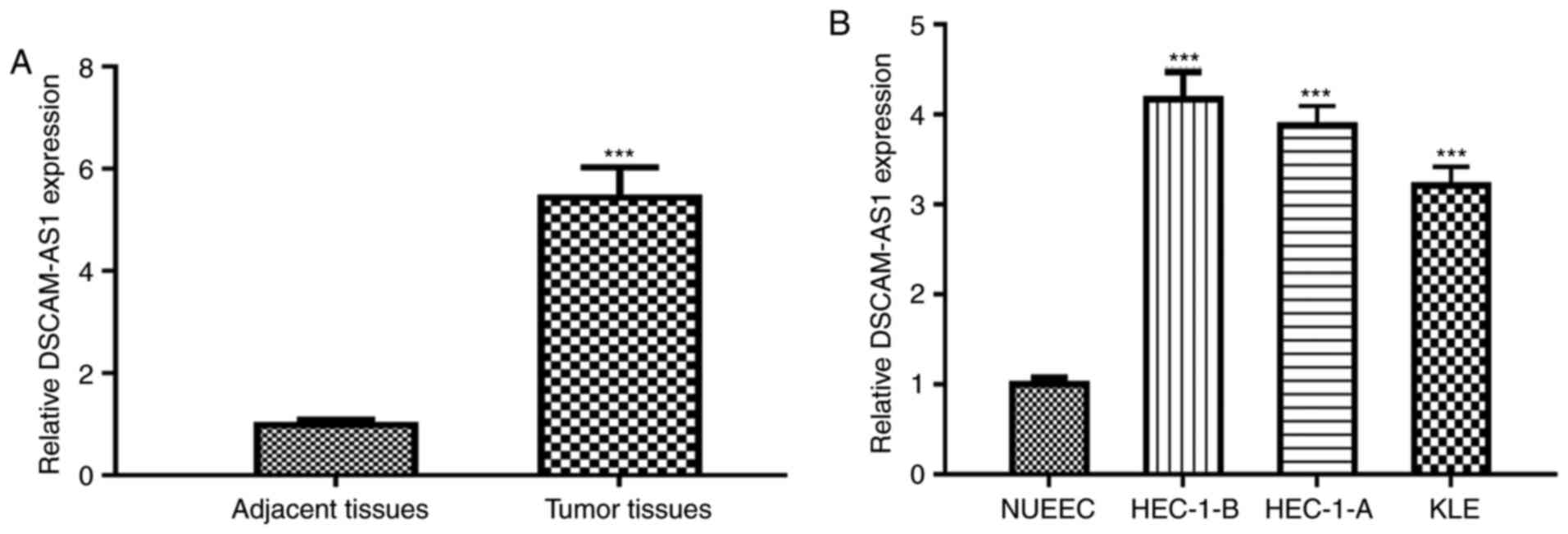
DSCAM-AS1 expression levels were analyzed in EC
tissues. The results revealed that DSCAM-AS1 was highly expressed
in EC tissues compared with normal tissues (Fig. 1A). Additionally, analysis of TCGA
data revealed that DSCAM-AS1 expression was also elevated in
clinical EC samples (Fig. S1).
Moreover, the DSCAM-AS1 expression level was higher in EC cells
(HEC-1-B, HEC-1-A and KLE) compared with in NUEEC cells (Fig. 1B).
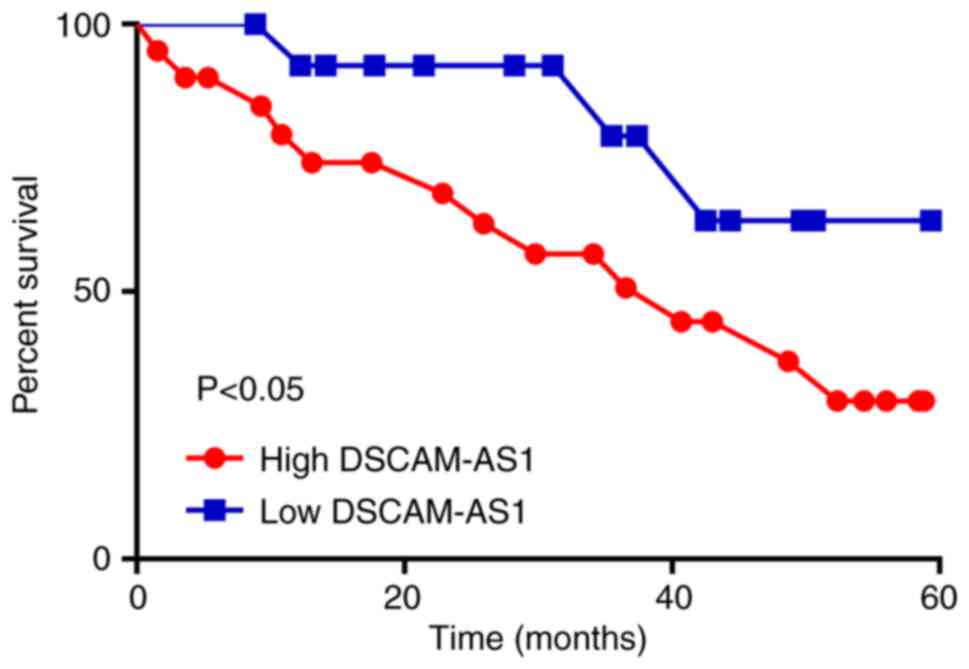
High DSCAM-AS1 expression is
associated with poor overall survival of patients with EC
Patients were classified into two groups based on
mean DSCAM-AS1 expression level (2.78). Patients with low DSCAM-AS1
expression levels exhibited better overall survival than those with
high expression (Fig. 2). In
addition, it was demonstrated that high DSCAM-AS1 levels were
significantly associated with FIGO stage and lymph node metastasis,
but were not associated with other clinical features (Table I).
 | Table I.Association of DSCAM-AS1 expression
and clinicopathological features of EC patients. |
Table I.
Association of DSCAM-AS1 expression
and clinicopathological features of EC patients.
|
|
| DSCAM-AS1
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Variable | n | High | Low |
χ2-value | P-value |
|---|
| Age (years) |
|
|
| 2.835 | 0.092 |
| ≥50 | 18 | 13 | 5 |
|
|
|
<50 | 16 | 7 | 9 |
|
|
| FIGO stage |
|
|
| 5.247 | 0.022a |
| I–II | 14 | 5 | 9 |
|
|
| III | 20 | 15 | 5 |
|
|
| Grade |
|
|
| 0.486 | 0.485 |
|
G1/G2 | 17 | 11 | 6 |
|
|
| G3 | 17 | 9 | 8 |
|
|
| Lymph node
metastasis |
|
|
| 3.927 | 0.048a |
| No | 15 | 6 | 9 |
|
|
| Yes | 19 | 14 | 5 |
|
|
| Myometrial
invasion |
|
|
| 0.064 | 0.800 |
|
<1/2 | 21 | 12 | 9 |
|
|
|
<1/2 | 13 | 8 | 5 |
|
|
| Estrogen
receptor |
|
|
| 0.105 | 0.746 |
|
Positive | 16 | 8 | 8 |
|
|
|
Negative | 18 | 8 | 10 |
|
|
| Lymph-vascular
space invasion |
|
|
| 0.007 | 0.933 |
|
Positive | 13 | 7 | 6 |
|
|
|
Negative | 21 | 10 | 11 |
|
|
| Histological
type |
|
|
| 1.574 | 0.455 |
|
Serous | 12 | 5 | 7 |
|
|
|
Endometrioid | 15 | 8 | 7 |
|
|
| Mixed
serous and endometrioid | 7 | 5 | 2 |
|
|
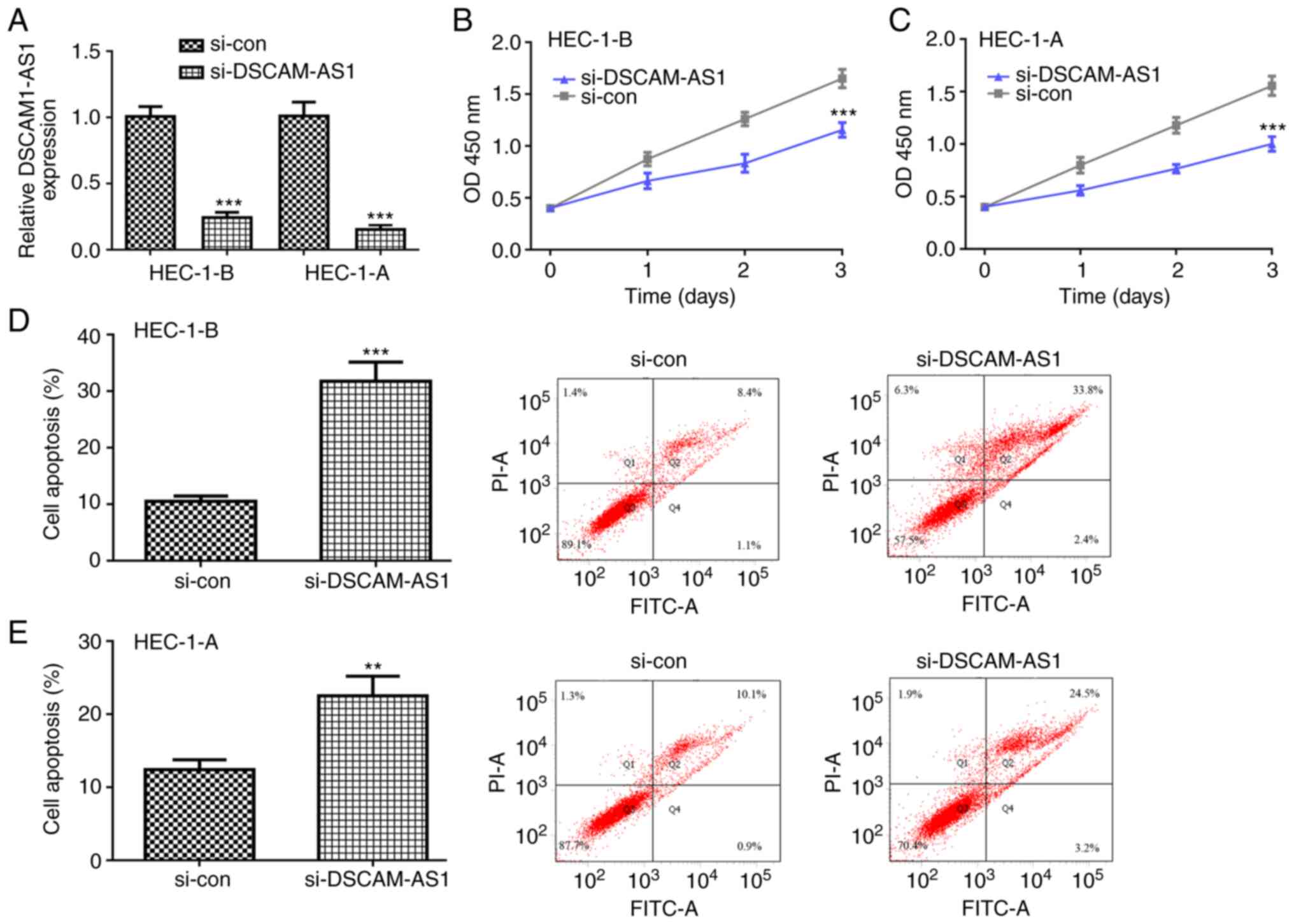
Knockdown of DSCAM-AS1 suppresses EC
cell growth
Subsequently, si-DSCAM-AS1 was transfected into
HEC-1-B and HEC-1-A cells and it was revealed that DSCAM-AS1 levels
were significantly decreased by si-DSCAM-AS1 in both cells
(Fig. 3A). The CCK-8 assay
revealed that the proliferation rate of HEC-1-B and HEC-1-A cells
was decreased following si-DSCAM-AS1 transfection (Fig. 3B and C). Furthermore, flow
cytometry revealed that knockdown of DSCAM-AS1 stimulated HEC-1-B
and HEC-1-A cell apoptosis (Fig. 3D
and E).
DSCAM-AS1 directly interacts with
miR-136-5p
miR-136-5p was identified as the top DSCAM-AS1
potential target (Fig. 4A). The
transfection of the miR-136-5p mimic significantly increased, while
miR-136-5p inhibitor decreased miR-136-5p levels in both cell lines
(Fig. 4B). It was also revealed
that miR-136-5p overexpression significantly decreased relative
luciferase activity in WT-DSCAM-AS1-transfected HEC-1-B and HEC-1-A
cells (Fig. 4C and D). Moreover,
miR-136-5p exhibited a decreased expression in EC samples and cells
compared with the normal counterparts (Fig. 4E and F). The results of the RIP
assay revealed that DSCAM-AS1 and miR-136-5p can directly interact
with each other (Fig. 4G and H).
Subsequently, DSCAM-AS1 was enriched in the miR-136-5p
mimic-transfected cells following anti-Ago2 treatment (Fig. 4I). In addition, si-DSCAM-AS1
transfection significantly increased miR-136-5p levels in HEC-1-B
and HEC-1-A cells (Fig. 4J and K).
Notably, miR-136-5p levels were negatively correlated with
DSCAM-AS1 levels in EC tissues (Fig.
4L).
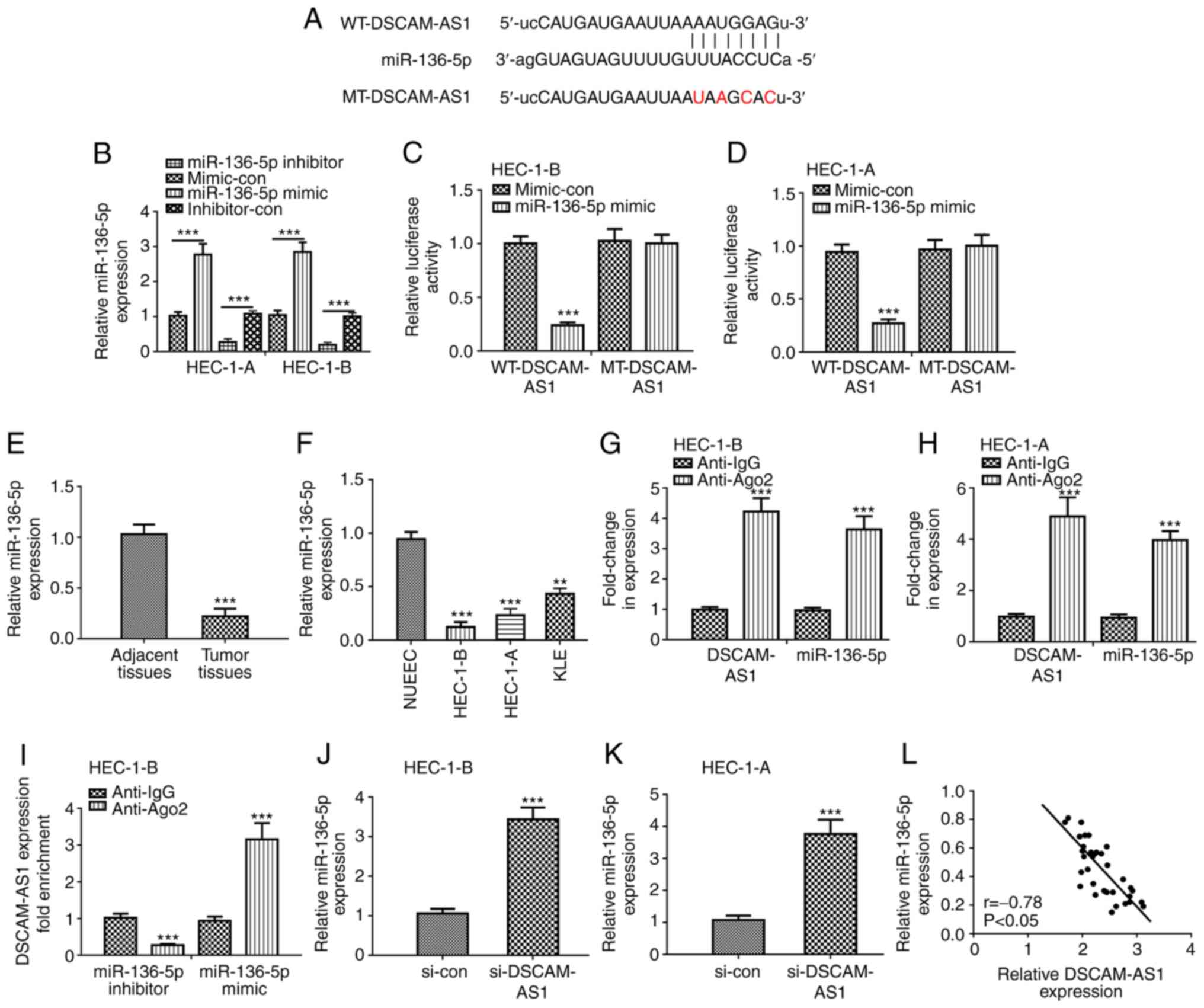 | Figure 4.DSCAM-AS1 specifically binds
miR-136-5p and regulates miR-136-5p expression. (A) The putative
binding sites between DSCAM-AS1 and miR-136-5p and the mutant
sequences of DSCAM-AS1 are presented. (B) miR-136-5p levels were
assessed in HEC-1-B and HEC-1-A cells after miR-136-5p mimic or
inhibitor transfection. Luciferase activity was detected in (C)
HEC-1-B and (D) HEC-1-A cells co-transfected with WT-DSCAM-AS1 or
MT-DSCAM-AS1 and miR-136-5p mimic or mi-con. ***P<0.001 vs.
mi-con. DSCAM-AS1 expression was measured in (E) 34 pairs of EC
tissues and adjacent normal tissues. ***P<0.001 vs. normal
tissues. (F) DSCAM-AS1 expression was measured in EC cells
(HEC-1-B, HEC-1-A and KLE) and NUEECs by RT-qPCR assay. **P<0.01
and ***P<0.001 vs. NUEEC. Relative enrichment of DSCAM-AS1 and
miR-136-5p in (G) HEC-1-B and (H) HEC-1-A cells was detected by RIP
assay. ***P<0.001 vs. anti-IgG. (I) Fold enrichment of DSCAM-AS1
in miR-136-5p mimic or miR-136-5p inhibitor group. ***P<0.001
vs. anti-IgG. The expression of miR-136-5p in (J) HEC-1-B and (K)
HEC-1-A cells after si-DSCAM-AS1 transfection was detected by
RT-qPCR. ***P<0.001 vs. si-con. (L) Correlation analysis between
DSCAM-AS1 and miR-136-5p expression was conducted by Pearson
analysis in EC tissues. DSCAM-AS1, down syndrome cell adhesion
molecule antisense 1; EC, endometrial cancer; RT-qPCR, reverse
transcription-quantitative PCR; si-DSCAM-AS1, small interfering RNA
against DSCAM-AS1; si-con, negative control siRNA; mimic-con,
negative control miRNA for miR-136-5p mimic; inhibitor-con,
negative control for miR-136-5p inhibitor; miR-136-5p,
microRNA-136-5p; RIP, RNA immunoprecipitation; NUEEC, normal
uterine endometrial epithelial cell. |
DSCAM-AS1 regulates EC cell growth by
targeting miR-136-5p
To validate the functional associations of DSCAM-AS1
and miR-136-5p in EC, rescue experiments were conducted. RT-qPCR
analysis revealed that miR-136-5p levels were elevated following
si-DSCAM-AS1 transfection, which was partially abrogated by
miR-136-5p knockdown (Fig. 5A).
The CCK-8 assay revealed that the inhibitory effects of DSCAM-AS1
knockdown on HEC-1-B cell proliferation were attenuated by
miR-136-5p knockdown (Fig. 5B).
Meanwhile, the flow cytometry assay indicated that inhibition of
miR-136-5p partially reversed the stimulatory effects of DSCAM-AS1
knockdown on regulating cell apoptosis (Fig. 5C).
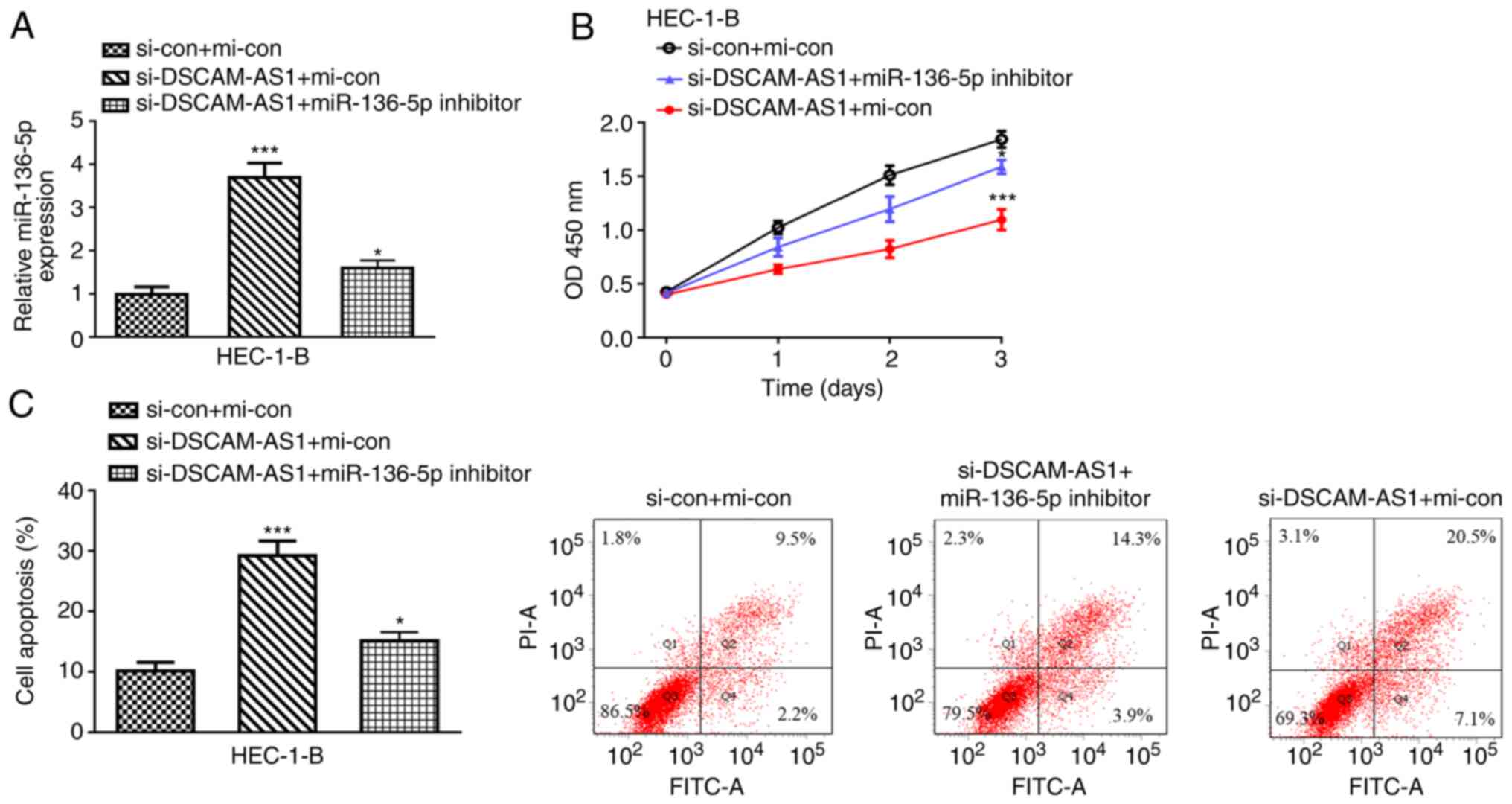 | Figure 5.DSCAM-AS1 regulates EC cell growth via
miR-136-5p. (A) The transfection efficacy of si-con + mi-con,
si-DSCAM-AS1 + mi-con, si-DSCAM-AS1 + miR-136-5p inhibitor was
measured by reverse transcription-quantitative PCR assay. (B) Cell
viability was evaluated in HEC-1-B after si-con + mi-con,
si-DSCAM-AS1 + mi-con, si-DSCAM-AS1 + miR-136-5p inhibitor
transfection by Cell Counting Kit-8 assay. (C) Cell apoptosis
percentage was evaluated in HEC-1-B after si-con + mi-con,
si-DSCAM-AS1 + mi-con, si-DSCAM-AS1 + miR-136-5p inhibitor
transfection by flow cytometry assay. *P<0.05 and ***P<0.001
vs. si-con + mi-con. DSCAM-AS1, down syndrome cell adhesion
molecule antisense 1; EC, endometrial cancer; si-DSCAM-AS1, small
interfering RNA against DSCAM-AS1; si-con, negative control siRNA;
mi-con, negative control miRNA. |
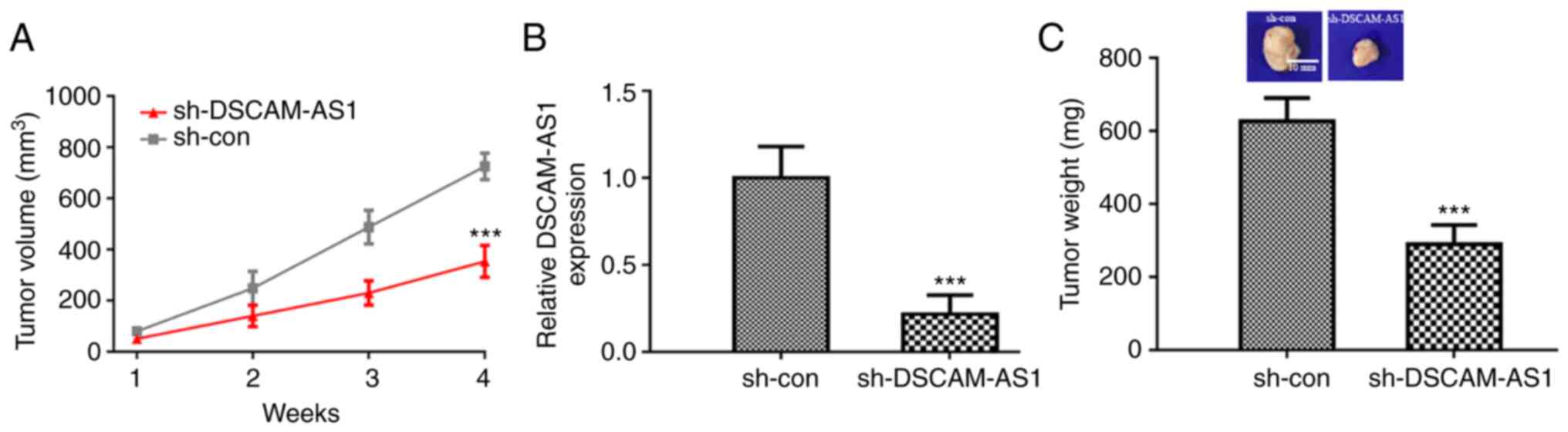
Silencing of DSCAM-AS1 suppresses EC
tumor growth
Finally, the effects of DSCAM-AS1 on EC tumor growth
were explored. It was revealed that mice injected with EC cells
with stable knockdown of DSCAM-AS1 had smaller tumors compared to
the sh-con group (Fig. 6A). The
decreased expression of DSCAM-AS1 following sh-DSCAM-AS1
transfection was also confirmed by RT-qPCR (Fig. 6B). In addition, it was demonstrated
that the weight of tumors from the sh-DSCAM-AS1 group were
significantly lower than those from the sh-con group (Fig. 6C).
Discussion
In the present study, DSCAM-AS1 was found to be
expressed at a high level in EC samples, and strongly associated
with a less favorable overall survival, higher FIGO stage and
poorer lymph node metastasis in patients with cancer. EC is prone
to metastasis and this typically results in a poor prognosis;
therefore, it is necessary to better understand the mechanisms
underlying EC progression. Notably, recent studies have revealed
that lncRNAs can affect the malignant behavior of cells (15,16).
In addition, numerous studies have revealed that DSCAM-AS1 can
affect tumorigenesis, but its role in EC remains unclear (7–9,17,18).
Therefore, the aim of the present study was to investigate the
biological functions of DSCAM-AS1 in EC.
Functional experiments were conducted in the present
study, the results of which revealed that DSCAM-AS1 silencing
inhibits EC cell proliferation via promoting cell apoptosis in
vitro. In addition, in vivo animal experiments revealed
that knockdown of DSCAM-AS1 suppressed EC tumor growth. Previous
studies have reported that DSCAM-AS1 serves as oncogenic lncRNA in
cancers including colorectal cancer, non-small cell lung cancer,
breast cancer, cervical cancer and hepatocellular carcinoma
(7–9,17,18).
Consistent with these findings, the present study also indicated
that DSCAM-AS1 could stimulate EC progression.
The ceRNA theory helps explain the underlying
mechanisms of ncRNAs, with the hypothesis that lncRNAs compete with
miRNAs to bind mRNA (19).
Notably, this theory connects ncRNAs and mRNAs and results in a
better understanding of the role of gene regulatory networks in
regulating normal development and disease progression (19). Based on this theory, emerging
studies have indicated that DSCAM-AS1 can sponge miRNAs to affect
tumorigenesis and tumor progression (7–9,17,18).
miRNAs including miR-384, miR-577, miR-204-5p, miR-877-5p and
miR-338-3p were previously identified as DSCAM-AS1 targets in
various types of cancer (7–9,17).
However, whether DSCAM-AS1 sponges miR-136-5p remains unclear.
miR-136-5p was identified as a tumor suppressive miRNA in numerous
types of cancer (20–23). Notably, miR-136-5p was found to be
regulated by several ncRNAs, including circular RNA TLK1, forkhead
box P4 antisense RNA, non-coding RNA activated by DNA damage and
family with sequence similarity 83 member H antisense RNA 1 in
renal cell carcinoma, cervical cancer, retinoblastoma, and
triple-negative breast cancer (20–23).
In the current study, it was demonstrated that miR-136-5p
expression was regulated by DSCAM-AS1 in EC. Rescue experiments
demonstrated that the effects of DSCAM-AS1 silencing on EC cell
proliferation can be partially abolished by miR-136-5p knockdown.
These results implied that DSCAM-AS1 regulates EC progression by
affecting miR-136-5p.
Collectively, the current study revealed that
DSCAM-AS1 stimulates EC progression by regulating miR-136-5p. A
limitation of the present work was that the detailed mechanisms of
DSCAM-AS1 in EC were not fully investigated. Another limitation is
that the enrolled population size was small in the present study
and the background of these patients varied. In the present study
only the prognostic value of DSCAM-AS1 in all patients with EC were
collected and the effect of different clinicopathological
characteristics was not investigated. In the future, a large cohort
study should be conducted by co-operating with other research
groups to further validate the conclusions of the present
study.
In conclusion, it was demonstrated that DSCAM-AS1
expression was upregulated in EC tissues and cell lines. Moreover,
DSCAM-AS1 may bind with miR-136-5p to affect EC progression. To the
best of the authors' knowledge, the present study provided the
first evidence to highlight the importance of the
DSCAM-AS1/miR-136-5p axis in the regulation of cancer
progression.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding
The present study was supported by the Quanzhou
Science and Technology Project Grant (grant no. 2018N021S).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LHL designed the study. LHL, PPC, BBH and PYC
performed the experiments and collected the data. LHL wrote the
manuscript. LHL and PPC confirm the authenticity of all the raw
data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of The Second Affiliated Hospital of Fujian Medical
University. Written informed consent was obtained from all patients
prior to enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Raffone A, Travaglino A, Saccone G, Di
Maio A, Mollo A, Mascolo M, De Rosa R, De Placido G, Insabato L and
Zullo F: Diabetes mellitus and responsiveness of endometrial
hyperplasia and early endometrial cancer to conservative treatment.
Gynecol Endocrinol. 35:932–937. 2019. View Article : Google Scholar
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar
|
|
3
|
ENCODE Project Consortium: An integrated
encyclopedia of DNA elements in the human genome. Nature.
489:57–74. 2012. View Article : Google Scholar
|
|
4
|
Zhao X, Tang DY, Zuo X, Zhang TD and Wang
C: Identification of lncRNA-miRNA-mRNA regulatory network
associated with epithelial ovarian cancer cisplatin-resistant. J
Cell Physiol. 234:19886–19894. 2019. View Article : Google Scholar
|
|
5
|
Tang D, Zhao X, Zhang L and Wang C:
Comprehensive analysis of pseudogene HSPB1P1 and its potential
roles in hepatocellular carcinoma. J Cell Physiol. 235:6515–6527.
2020. View Article : Google Scholar
|
|
6
|
Kondo Y, Shinjo K and Katsushima K: Long
non-coding RNAs as an epigenetic regulator in human cancers. Cancer
Sci. 108:1927–1933. 2017. View Article : Google Scholar
|
|
7
|
Li B, Sun H and Zhang J: lncRNA DSCAM-AS1
promotes colorectal cancer progression by acting as a molecular
sponge of miR-384 to modulate AKT3 expression. Aging (Albany NY).
12:9781–9792. 2020. View Article : Google Scholar
|
|
8
|
Qiu Z, Pan XX and You DY: lncRNA DSCAM-AS1
promotes non-small cell lung cancer progression via regulating
miR-577/HMGB1 axis. Neoplasma. 67:871–879. 2020. View Article : Google Scholar
|
|
9
|
Liang WH, Li N, Yuan ZQ, Qian XL and Wang
ZH: DSCAM-AS1 promotes tumor growth of breast cancer by reducing
miR-204-5p and up-regulating RRM2. Mol Carcinog. 58:461–473. 2019.
View Article : Google Scholar
|
|
10
|
Specialized Committee of Gynecological
Tumors of China Anticancer Association, . Guidelines for Diagnosis
and Treatment of Endometrial Cancer (Fourth Edition). Chin J Pract
Gynecol Obstet. 34:880–886. 2018.(In Chinese).
|
|
11
|
Matias-Guiu X and Prat J: Molecular
pathology of endometrial carcinoma. Histopathology. 62:111–123.
2013. View Article : Google Scholar
|
|
12
|
Ryu YJ, Kang SJ, Cho JS, Yoon JH and Park
MH: Lymphovascular invasion can be better than pathologic complete
response to predict prognosis in breast cancer treated with
neoadjuvant chemotherapy. Medicine (Baltimore). 97:e116472018.
View Article : Google Scholar
|
|
13
|
Smith D, Stewart CJR, Clarke EM, Lose F,
Davies C, Armes J, Obermair A, Brennan D, Webb PM, Nagle CM and
Spurdle AB: ER and PR expression and survival after endometrial
cancer. Gynecol Oncol. 148:258–266. 2018. View Article : Google Scholar
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
Qu Y, Tan HY, Chan YT, Jiang H, Wang N and
Wang D: The functional role of long noncoding RNA in resistance to
anticancer treatment. Ther Adv Med Oncol. 12:17588359209278502020.
View Article : Google Scholar
|
|
16
|
Grixti JM and Ayers D: Long noncoding RNAs
and their link to cancer. Noncoding RNA Res. 5:77–82. 2020.
View Article : Google Scholar
|
|
17
|
Liang J, Zhang S, Wang W, Xu Y, Kawuli A,
Lu J and Xiu X: Long non-coding RNA DSCAM-AS1 contributes to the
tumorigenesis of cervical cancer by targeting miR-877-5p/ATXN7L3
axis. Biosci Rep. 40:BSR201920612020. View Article : Google Scholar
|
|
18
|
Ji D, Hu G, Zhang X, Yu T and Yang J: Long
non-coding RNA DSCAM-AS1 accelerates the progression of
hepatocellular carcinoma via sponging miR-338-3p. Am J Transl Res.
11:4290–4302. 2019.
|
|
19
|
Karreth FA and Pandolfi PP: ceRNA
cross-talk in cancer: When ce-bling rivalries go awry. Cancer
Discov. 3:1113–1121. 2013. View Article : Google Scholar
|
|
20
|
Li J, Huang C, Zou Y, Ye J, Yu J and Gui
Y: CircTLK1 promotes the proliferation and metastasis of renal cell
carcinoma by sponging miR-136-5p. Mol Cancer. 19:1032020.
View Article : Google Scholar
|
|
21
|
Zhao J, Yang T and Li L: lncRNA FOXP4-AS1
is involved in cervical cancer progression via regulating
miR-136-5p/CBX4 axis. Onco Targets Ther. 13:2347–2355. 2020.
View Article : Google Scholar
|
|
22
|
Yang XL, Hao YJ, Wang B, Gu XL, Wang XX
and Sun JF: Long noncoding RNA NORAD promotes the progression of
retinoblastoma by sponging miR-136-5p/PBX3 axis. Eur Rev Med
Pharmacol Sci. 24:40552020.
|
|
23
|
Han C, Fu Y, Zeng N, Yin J and Li Q:
lncRNA FAM83H-AS1 promotes triple-negative breast cancer
progression by regulating the miR-136-5p/metadherin axis. Aging
(Albany NY). 12:3594–3616. 2020. View Article : Google Scholar
|