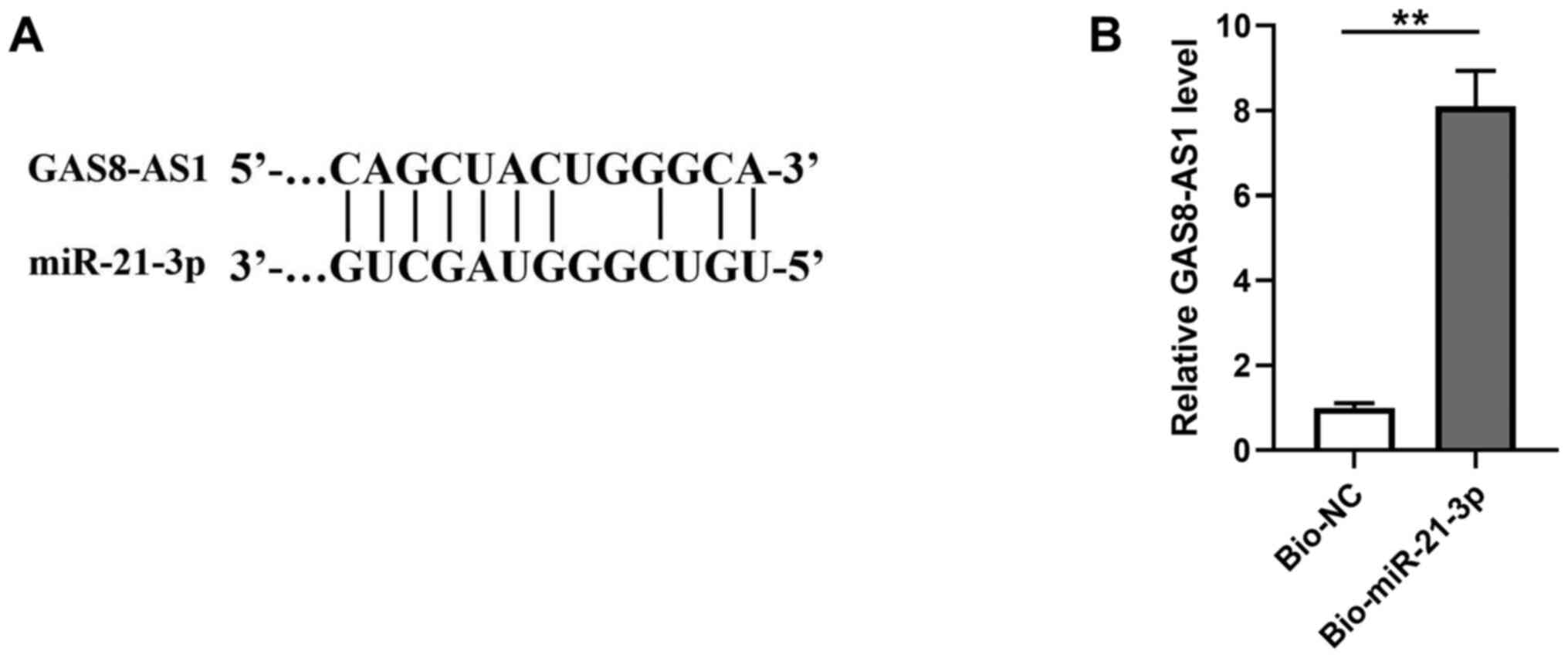Introduction
Gastric cancer is a frequently diagnosed cancer
worldwide, with the highest incidence rates in regions such as
Central and South America, Eastern Europe and East Asia (1). Early-stage gastric cancer is
radically treated with surgery; however, effective therapies for
advanced gastric cancer are still lacking (2). A considerable number of tumor
suppressors and oncogenes have been identified as critical players
in gastric cancer. For instance, microRNA-223 (miRNA/miR-223) and
long non-coding RNA (lncRNA) UCA1 have been found to have oncogenic
roles in gastric cancer (3,4),
while miR-4317 and lncRNA MEG3 display anticancer functions in
gastric cancer (5,6). However, the pathogenesis of this
disease remains to be fully elucidated (7), leading to difficulties in clinical
treatment.
The functions of lncRNAs, which are >200
nucleotides in length, in cancer occurrence and progression, such
as carcinogenesis, metastasis and chemoresistance have recently
been investigated (8). In effect,
regulation of the expression of specific lncRNAs has demonstrated
potential in cancer treatment and prevention (9,10).
Various lncRNAs have been characterized as critical regulators in
gastric cancer. For example, lncRNA H19, UCA1 and HOXA11-AS have
been found to enhance the carcinogenesis and metastasis of gastric
cancer (4,11,12),
while lncRNA MEG3 and MIR22HG have been found to inhibit gastric
cancer progression (6,13). Recent studies have reported that
lncRNA GAS8-AS1 (chromosome 16) is a tumor suppressor in multiple
types of cancer. For example, GAS8-AS1 has demonstrated inhibitory
effects during cell proliferation in papillary thyroid carcinoma
(14), cancer cell migration and
invasion in osteosarcoma (15),
and cancer cell proliferation in glioblastoma (16). Mechanistically, GAS8-AS1 may
function by regulating downstream miRNAs and target genes, such as
the miR-187-3p/autophagy protein 5 axis and the
miR-135b-5p/G1/S-specific cycin-D2 axis (14,17).
However, the role of GAS8-AS1 in gastric cancer remains to be fully
elucidated.
miR-21-3p has been identified as an oncogenic miRNA
and is frequently upregulated in a variety of malignancies such as
colorectal and breast cancer, with roles in multiple processes of
cancer development, such as cancer cell proliferation, migration
and invasion (18,19). Previous studies have revealed that
miR-21-3p plays a pivotal role in the pathogenesis and progression
of gastric cancer (20,21). Therefore, the present study aimed
to investigate the role of GAS8-AS1 in gastric cancer and determine
whether its effects are mediated by regulating miR-21-3p. A total
of 70 patients with gastric adenocarcinoma were involved in the
study. GAS8-AS1 and miR-21-3p levels in cancer and non-cancerous
tissues were detected, and a survival analysis was performed. In
gastric cell lines, RNA pull-down and reverse
transcription-quantitative (RT-q)PCR assays were performed to
explore the interaction between GAS8-AS1 and miR-21-3p. Finally,
CCK-8 and Transwell assays were performed to assess the effects of
GAS8-AS1 and miR-21-3p on cell proliferation, migration and
invasion.
Materials and methods
Patients
The present study included 70 patients with gastric
adenocarcinoma (40 male and 30 female patients; age, 48.4±7.1
years; range, 30–66 years) selected from 168 patients who were
admitted at the Affiliated Hospital of Xuzhou Medical University
(Xuzhou, China) between March 2010 and March 2013.
The inclusion criteria were as follows: i)
First-time diagnosis; ii) no previous history of cancer; and iii)
completed 5-year follow-up after admission. The exclusion criteria
were as follows: i) Diagnosis of other clinical conditions; and ii)
patients who failed to complete the 5-year follow-up. There were
12, 18, 22 and 18 cases with American Joint Committee on Cancer
clinical stage I, II, III and IV, respectively (22). These 70 patients included 43 cases
of high-grade (poorly differentiated) and 27 cases of low-grade
(highly differentiated) cancer.
All the patients provided written informed consent
before involved in the present study. The present study was
approved by the Ethics Committee of the Affiliated Hospital of
Xuzhou Medical University prior to patient admission (approval no.
XZNU-2010-032).
Tissues and cells
Gastric cancer and non-cancerous adjacent tissues
were collected through gastric biopsies. SNU-1 and AGS (American
Type Culture Collection) gastric cancer cell lines and a normal
gastric cell line Hs 738.St/Int (American Type Culture Collection)
were used. The cells were authenticated using the STR method and
mycoplasma contamination was excluded using the
MycAway™-Color One-Step Mycoplasma Detection kit (cat.
no. 40611ES25; Shanghai Yeasen Biotechnology Co., Ltd.). Cells were
cultivated in 90% RPMI-1640 medium (Invitrogen; Thermo Fischer
Scientific, Inc.) supplemented with 10% FBS (Thermo Fisher
Scientific, Inc.) at 37°C for 72 h before the experiments.
Follow-up
From the day of admission, all 70 patients were
followed up monthly using telephone communication for 5 years to
record survival rates.
RT-qPCR
Total RNA from the cancer and non-cancerous adjacent
tissues, as well as from the SNU-1, AGS and Hs 738.St/Int cell
lines, was extracted using RNAzol® reagent
(Sigma-Aldrich; Merck KGaA). Total RNA was reverse transcribed
using SuperScript™ IV First-Strand Synthesis System
(Invitrogen; Thermo Fisher Scientific, Inc.) to synthesize cDNA
according to the manufacturer's protocol. PowerUp™
SYBR™ Green Master Mix (Applied Biosystems; Thermo
Fisher Scientific, Inc.) was used for the preparation of qPCR
reactions. The expression levels of GAS8-AS1 were determined with
18S rRNA as the endogenous control. To measure the expression of
miR-21-3p, miRNA extraction was performed using miRNeasy kit
(Qiagen, Inc.), and U6 was used as the internal control of
miR-21-3p. The All-in-One™ miRNA qRT-PCR Detection kit
(GeneCopoeia, Inc.) was used to perform RT-qPCR. The PCR thermal
cycling conditions were as follows: 95°C for 1 min, followed by 40
cycles of 95°C for 15 sec, 58°C for 20 sec and a final extension of
72°C for 45 sec. Cq values of target genes were normalized to
internal controls using the 2−∆∆Cq method (23). The primer sequences are listed in
Table I.
 | Table I.Primers used for reverse
transcription-quantitative PCR. |
Table I.
Primers used for reverse
transcription-quantitative PCR.
| Gene name | Forward, 5′-3′ | Reverse, 5′-3′ |
|---|
| GAS8-AS1 |
CAACGAGCAAACAAGAAGGA |
TGAGCCAAACAGACCAGTCA |
| PTEN |
AAGACCATAACCCACCACAGC |
CACCAGTTCGTCCCTTTCCA |
| 18S rRNA |
CTACCACATCCAAGGAAGCA |
TTTTTCGTCACTACCTCCCCG |
| miR-21 |
GCCCGCTAGCTTATCAGACTGATG | GTGCAGGGTCCGAGGT |
| U6 |
ATTGGAACGATACAGAGAAGATT |
GGAACGCTTCACGAATTTG |
Transient transfection
Transient transfection was applied to investigate
the effects of short-term overexpression of GAS8-AS1 and miR-21-3p
on gastric cancer cell lines. The overexpression of GAS8-AS1 and
miR-21-3p was achieved by transfecting 10 nM GAS8-AS1 expression
vector or miR-21-3p mimic into in vitro cultivated cells
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at room temperature for 24 h. Backbone pcDNA3.1
vector (Invitrogen; Thermo Fisher Scientific, Inc.) was used to
construct the expression vector of GAS8-AS1. Negative control (NC)
miRNA (scrambled mimic; 5′-CCUUCCGAGAGAAGAGCC-3′) and miR-21-3p
mimic (5′-CGGGUAGCUGACCACAAC-3′) were purchased from Invitrogen
(Thermo Fisher Scientific, Inc.). NC experiments were performed by
transfecting cells with 10 nM empty vector or NC miRNA. Control (C)
cells were untransfected cells. The subsequent experiments were
performed at 24 h post-transfection.
RNA pull-down assay
The interaction between GAS8-AS1 and miR-21-3p was
determined by RNA pull-down assay using the Pierce Magnetic
RNA-Protein Pull-Down kit (Thermo Fisher Scientific, Inc.) in
accordance with the manufacturer's protocol. The
desthiobiotin-labeled DNA probes targeting back-splicing sequence
of miR-21-3p and NC miRNA, named Bio-miR-21-3p or Bio-NC, were
purchased from Invitrogen; Thermo Fisher Scientific, Inc. As
aforementioned, SNU-1 cells were co-transfected with 10 nM GAS8-AS1
expression vector and 10 nM Bio-miR-21-3p or Bio-NC at 37°C using
Lipofectamine 2000. At 48 h post-transfection, 400 µl
TRIzol® lysis buffer (Thermo Fisher Scientific, Inc.)
was prepared and RNA pull-down was performed using 50 µl
streptavidin agarose magnetic beads (Thermo Fisher Scientific,
Inc.) at room temperature. After incubation at 4°C for 1.5 h with
rotation streptavidin magnetic beads were washed three times with
the washing buffer from the Pierce Magnetic RNA-Protein Pull-Down
kit, and then incubated with 50 µl Elution buffer at 37°C for 15
min with agitation. Afterwards, the mixture was centrifuged at 800
× g for 15 min at 4°C. Total RNA was extracted from pull-down
samples, which was followed by RT-qPCR, as aforementioned, to
determine the expression of GAS8-AS1.
Cell Counting Kit-8 (CCK-8) assay
Cells were seeded into a 96-well plate (5,000 cells
per well in 0.1 ml medium) and the cells were cultured under the
aforementioned conditions, followed by cell collection every 24 h
until 96 h. CCK-8 (Sigma-Aldrich; Merck KGaA) was added to reach
10% final concentration at 4 h before cell collection. Finally, OD
values (460 nm) were measured.
Transwell assay
Cell migration and invasion were assessed by using
Transwell assays. The migration assay was performed the same as the
invasion assay but without the Matrigel precoating. For the
invasion assay, the Transwell inserts were incubated with 100 µl
Matrigel (MilliporeSigma) at 37°C for 6 h. Then, 1×105
cells were transferred into the upper chamber with 1 ml serum-free
RPMI-1640 medium, and 1 ml RPMI-1640 medium containing 20% FBS was
added into the lower chamber. After 48 h of culture at 37°C, 0.5%
crystal violet (Sigma-Aldrich; Merck KGaA) was used to stain
membranes for 20 min at room temperature. Finally, an optical
microscope (Olympus Corporation) was used to observe the migratory
cells (×200 magnification).
Statistical analysis
Each experiment was repeated three times to
calculate the mean values, and the experimental data are presented
as the mean ± standard deviation. The statistical analysis was
performed in SPSS ver. 22.0 (IBM Corp.). A paired Student's t-test
was used to compare gastric cancer and non-cancerous tissues.
One-way ANOVA followed by Tukey's post hoc test was used to compare
different clinical stages or different cell treatment groups.
Survival analysis was performed by dividing patients into low
(n=38) and high (n=32) expression levels of GAS8-AS1 (cancer
tissue) groups according to Youden's index (24). Kaplan-Meier plotter and log-rank
test were used to plot and compare survival curves. Associations
between clinical factors and the expression levels of GAS8-AS1 were
analyzed using a χ2 test. Pearson's correlation coefficient
analysis was performed to analyze the correlation between GAS8-AS1
level and miR-21-3p. StarBase Database (http://starbase.sysu.edu.cn/index.php) was used to
predict the potential targets of GAS8-AS1. P<0.05 was considered
to indicate a statistically significant difference.
Results
GAS8-AS1 is downregulated in patients
with gastric cancer
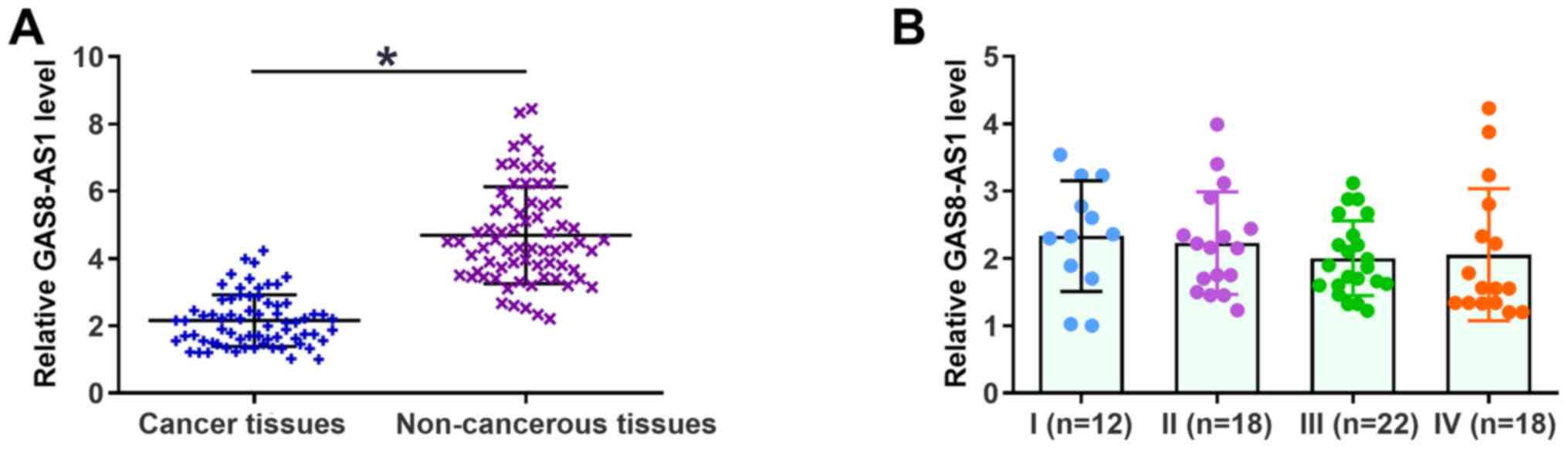
The expression levels of GAS8-AS1 in gastric cancer
and non-cancerous tissues were detected via RT-qPCR. As shown in
Fig. 1A, the expression levels of
GAS8-AS1 were significantly lower in gastric cancer tissues
compared with those in non-cancerous tissues. However, the
expression levels of GAS8-AS1 in cancerous tissues at different
clinical stages revealed no significant difference (P>0.05;
Fig. 1B). The χ2 test
revealed that the expression of GAS8-AS1 had no association with
patients' age, sex, BMI, history of drinking and smoking, tumor
grade or clinical stage (data not shown).
Reduced expression levels of GAS8-AS1
in gastric cancer tissues predict poor survival
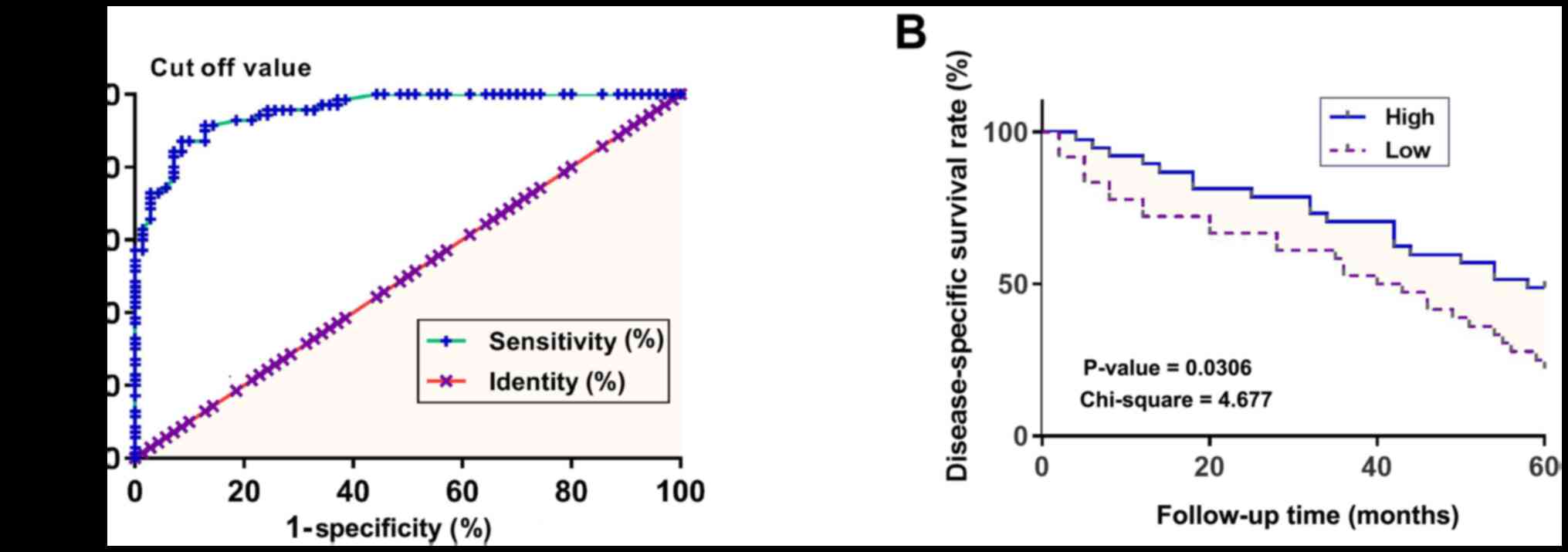
Patients with gastric cancer were divided into high
and low GAS8-AS1 expression groups according to Youden's index
(Fig. 2A). Results of a survival
curve analysis demonstrated that patients with low expression
levels of GAS8-AS1 exhibited a markedly lower disease-specific
survival rate within the 5-year follow-up compared to those with
high expression levels (Fig.
2B).
miR-21-3p is upregulated in gastric
cancer tissues and is inversely correlated with GAS8-AS1
expression
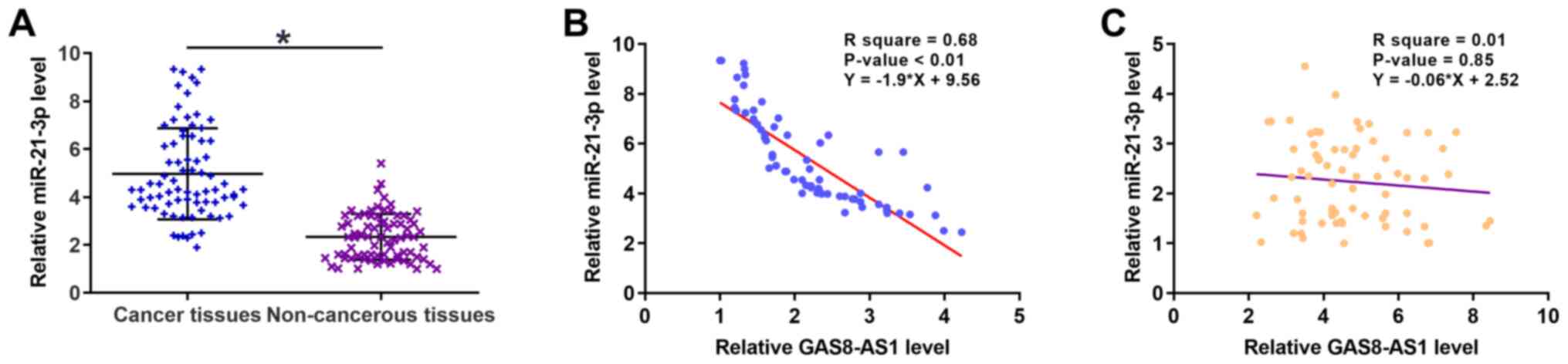
The expression levels of miR-21-3p in gastric cancer
and non-cancerous tissues were also detected via RT-qPCR. As is
evident in Fig. 3A (P<0.05),
the expression levels of miR-21-3p in cancer tissues were
significantly higher compared with those in adjacent non-cancerous
tissues. Pearson's correlation coefficient analysis demonstrated
that the expression levels of GAS8-AS1 and miR-21-3p were inversely
correlated in cancer tissues, while they exhibited no correlation
in the non-cancerous tissues (Fig. 3B
and C).
Overexpression of GAS8-AS1 reduces the
expression levels of miR-21-3p in gastric cancer cells
As shown in Fig.
4A, miR-21-3p was predicted to be a potential target of
GAS8-AS1 using the starBase database. The RNA pull-down assay
revealed that, compared with the Bio-NC pull-down sample, the
Bio-miR-21-3p pull-down sample exhibited significantly higher
expression levels of GAS8-AS1 (Fig.
4B; P<0.01), confirming the direct interaction between
GAS8-AS1 and miR-21-3p. In addition, SNU-1 and AGS cells were
transfected with GAS8-AS1 expression vector or miR-21-3p mimic and
their corresponding controls.
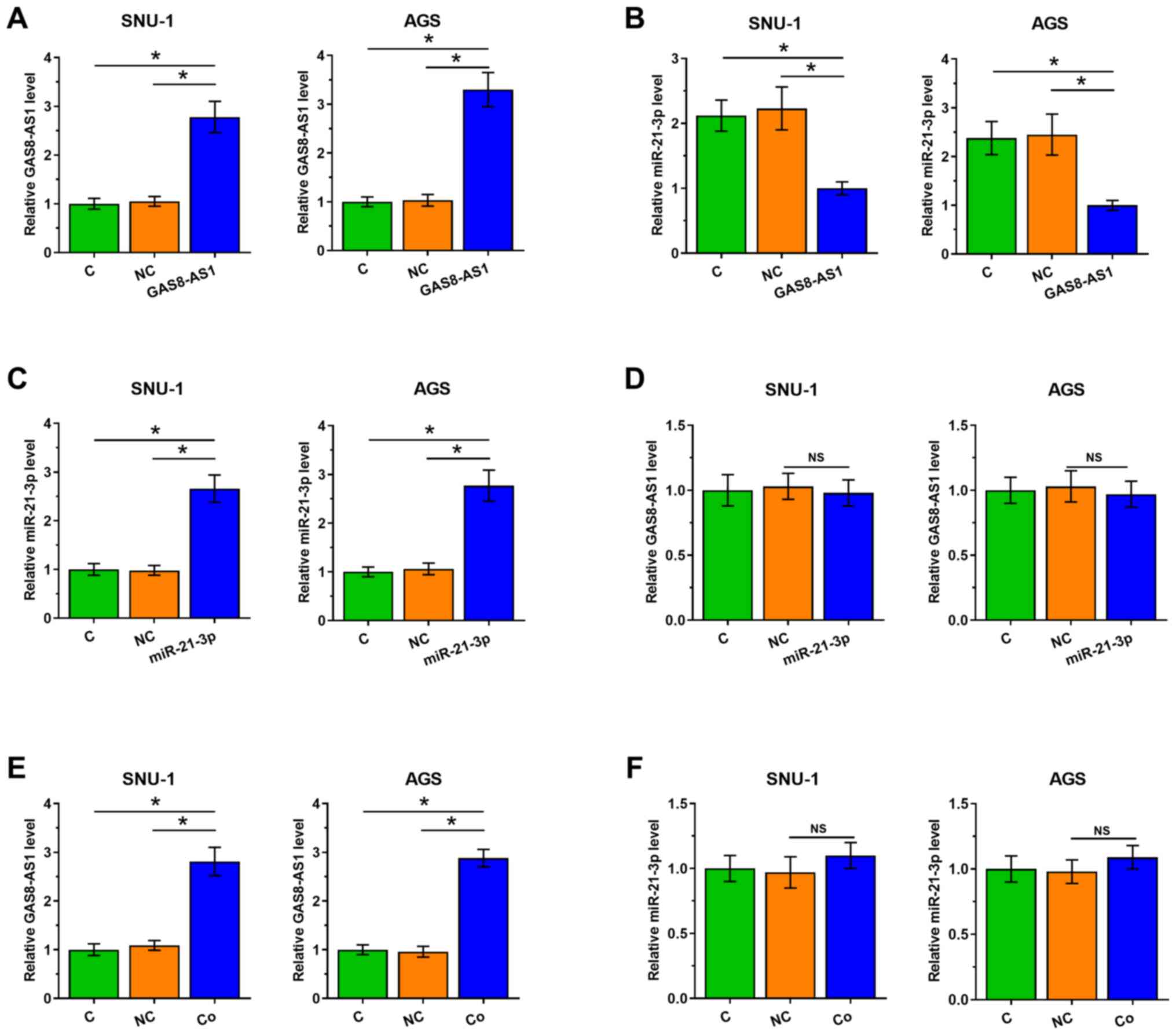
As demonstrated in Fig.
5A, the expression level of GAS8-AS1 was significantly
increased in cells transfected with the GAS8-AS1 expression vector
compared with that in cells transfected with NC (P<0.05).
Overexpression of GAS8-AS1 significantly reduced the expression
levels of miR-21-3p (Fig. 5B;
P<0.05) compared with the NC group, while the overexpression of
miR-21-3p did not affect the expression of GAS8-AS1 (Fig. 5C and D; P<0.05) compared with
the NC group. In addition, SNU-1 and AGS cells were also
co-transfected with GAS8-AS1 expression vector and miR-21-3p mimic.
The results demonstrated that the expression levels of GAS8-AS1
were significantly increased in the co-transfected cells compared
with those in the NC group, while the expression levels of
miR-21-3p in the co-transfected cells indicated no notable changes
(Fig. 5E and F; P<0.05)
compared with those in the NC group. These results indicated that
GAS8-AS1 directly targeted miR-21-3p and inhibited its
expression.
GAS8-AS1 regulates cancer cell
proliferation via miR-21-3p
Subsequently, the effects of GAS8-AS1 on the
proliferation, migration and invasion of gastric cancer cells were
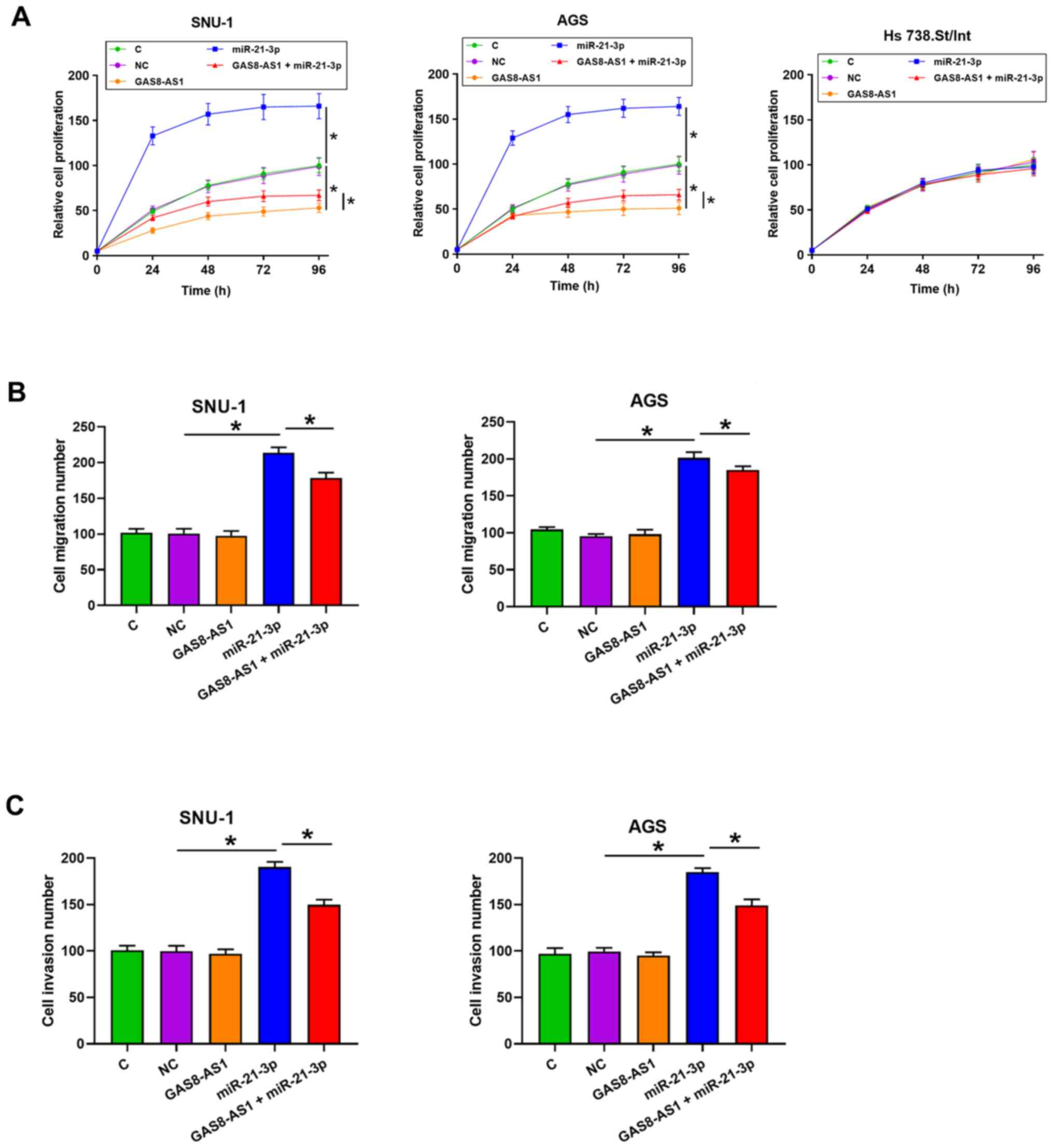
investigated. As shown in Fig. 6A,
the overexpression of GAS8-AS1 significantly inhibited cancer cell
proliferation, while the overexpression of miR-21-3p inhibited
cancer cell proliferation (P<0.05). In addition, the cells
transfected with GAS8-AS1 and miR-21-3p had enhanced cell
proliferation compared with the cells transfected with the GAS8-AS1
expression vector. In addition, the overexpression of GAS8-AS1 and
miR-21-3p did not affect the proliferation of the normal Hs
738.St/Int cell line. Furthermore, the overexpression of miR-21-3p
markedly enhanced cancer cell migration and invasion compared with
the NC group, while the overexpression of GAS8-AS1 did not affect
the migration and invasion of gastric cancer cells compared with
the NC group (Figs. 6B and C, and
S1; P<0.05). However, the
overexpression of GAS8-AS1 partly alleviated the effect of
miR-21-3p overexpression on cancer cell migration and invasion
compared with the cells transfected with miR-21-3p mimic.
Discussion
The survival rate of patients with gastric cancer
currently remains poor, particularly for patients at stages III and
IV (25). Therefore, accurate
prognosis is critical for the development of effective treatments.
A considerable number of prognostic biomarkers have been identified
for gastric cancer. For example, lncRNA MALAT1 has been found to be
significantly upregulated in gastric cancer and serves as a
predictor of poor survival (26).
Furthermore, lncRNA LINC-PINT was demonstrated to be downregulated
in patients with gastric cancer and predicted poor survival
(27). In the present study,
downregulation of GAS8-AS1 in patients with gastric cancer was
observed. In addition, low expression levels of GAS8-AS1 predicted
poor survival of patients with gastric cancer compared with high
expression levels. Therefore, GAS8-AS1 may serve as a potential
prognostic marker for gastric cancer. Of note is that no
significant difference was found in the expression levels of
GAS8-AS1 among the four clinical stages.
Studies have revealed that GAS8-AS1 is a tumor
suppressor in papillary thyroid carcinoma, osteosarcoma and
glioblastoma (14–16). In the present study, it was
revealed that the overexpression of GAS8-AS1 significantly
inhibited gastric cancer cell proliferation compared with the NC,
suggesting an anticancer role of GAS8-AS1 in gastric cancer. To the
best of our knowledge, the present study was the first to report
the role of GAS8-AS1 in gastric cancer.
miR-21-3p has been identified as a critical
regulator in gastric cancer (20).
In the present study, miR-21-3p expression was markedly upregulated
in the gastric cancer tissues compared with non-cancerous tissues,
and its expression was negatively correlated with the expression of
GAS8-AS1 in gastric cancer tissues. In addition, miR-21-3p was
predicted to be a potential target of GAS8-AS1, and this
interaction was confirmed using an RNA pull-down assay. Moreover,
it was observed that GAS8-AS1 inhibited the expression of
miR-21-3p, while overexpression of miR-21-3p exhibited no effect on
the expression level of GAS8-AS1. Therefore, we hypothesized that
GAS8-AS1 may inhibit the expression of miR-21-3p by directly
sponging it. To the best of our knowledge, this is the first study
to report the association between GAS8-AS1 and miR-21-3p. In
addition, overexpression of miR-21-3p promoted cancer cell
proliferation compared with the NC, and alleviated
GAS8-AS1-mediated inhibition of cancer cell proliferation in two
cancer cell lines compared with that in GAS8-AS1 overexpression
group. Collectively, the results of the present study indicate that
GAS8-AS1 may play a protective role in gastric cancer by regulating
miR-21-3p.
However, the present study has some limitations.
Firstly, the sample size of patients with gastric cancer was small.
Future studies will collect more samples to strengthen the
findings. In addition, in vivo experiments were not performed in
the study, but will be performed in future studies to further
confirm the role of GAS8-AS1 in gastric cancer.
In conclusion, GAS8-AS1 was found to be
significantly downregulated in cancer tissues compared with
non-cancerous tissues in patients with gastric cancer, and low
expression levels of GAS8-AS1 predicted poor survival. Furthermore,
the results showed that GAS8-AS1 may inhibit gastric cancer cell
proliferation by downregulating miR-21-3p. Future experiments will
focus on the detailed mechanisms by which GAS8-AS1 exerts its
anticancer function in gastric cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CL was responsible for writing the manuscript,
performing the literature search, data analysis and statistical
analysis. HW conducted the data and statistical analyses. SM, JH,
LY and YS performed the literature search, and were responsible for
data collection and sample collection. XZ was responsible for
designing the study and revising the manuscript. CL and XZ confirm
the authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study received ethics approval from the
Ethics Committee of the Affiliated Hospital of Xuzhou Medical
University (Xuzhou, China) prior to patient admission. Written
informed consent was obtained from all individual participants
included in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rawla P and Barsouk A: Epidemiology of
gastric cancer: Global trends, risk factors and prevention. Prz
Gastroenterol. 14:26–38. 2019.PubMed/NCBI
|
|
2
|
Sumiyama K: Past and current trends in
endoscopic diagnosis for early stage gastric cancer in Japan.
Gastric Cancer. 20 (Suppl 1):20–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li X, Zhang Y, Zhang H, Liu X, Gong T, Li
M, Sun L, Ji G, Shi Y, Han Z, et al: miRNA-223 promotes gastric
cancer invasion and metastasis by targeting tumor suppressor
EPB41L3. Mol Cancer Res. 9:824–833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY,
Liu Q, Zhao G and Zhang ZZ: The lncRNA UCA1 promotes proliferation,
migration, immune escape and inhibits apoptosis in gastric cancer
by sponging anti-tumor miRNAs. Mol Cancer. 18:1152019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hu X, Zhang M, Miao J, Wang X and Huang C:
miRNA-4317 suppresses human gastric cancer cell proliferation by
targeting ZNF322. Cell Biol Int. 42:923–930. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wei GH and Wang X: lncRNA MEG3 inhibit
proliferation and metastasis of gastric cancer via p53 signaling
pathway. Eur Rev Med Pharmacol Sci. 21:3850–3856. 2017.PubMed/NCBI
|
|
7
|
Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B
and Guo J: Global circular RNA expression profile of human gastric
cancer and its clinical significance. Cancer Med. 6:1173–1180.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Poursheikhani A, Abbaszadegan MR,
Nokhandani N and Kerachian MA: Integration analysis of long
non-coding RNA (lncRNA) role in tumorigenesis of colon
adenocarcinoma. BMC Med Genomics. 13:1082020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma Z, Gu G, Pan W and Chen X: Chen X and
therapy: LncRNA PCAT6 accelerates the progression and
chemoresistance of cervical cancer through up-regulating ZEB1 by
sponging miR-543. OncoTargets Ther. 13:1159–1170. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang L, Wang L, Wang Y, Chen T, Liu R,
Yang W, Liu Q and Tu K: LncRNA KTN1-AS1 promotes tumor growth of
hepatocellular carcinoma by targeting miR-23c/ERBB2IP axis. Biomed
Pharmacother. 109:1140–1147. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun M, Nie F, Wang Y, Zhang Z, Hou J, He
D, Xie M, Xu L, De W, Wang Z, et al: LncRNA HOXA11-AS Promotes
Proliferation and Invasion of Gastric Cancer by Scaffolding the
Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res.
76:6299–6310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li H, Yu B, Li J, Su L, Yan M, Zhu Z and
Liu B: Overexpression of lncRNA H19 enhances carcinogenesis and
metastasis of gastric cancer. Oncotarget. 5:2318–2329. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li H and Wang Y: Long Noncoding RNA
(lncRNA) MIR22HG Suppresses Gastric Cancer Progression through
Attenuating NOTCH2 Signaling. Med Sci Monit. 25:656–665. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen N, Yin D, Lun B and Guo X: LncRNA
GAS8-AS1 suppresses papillary thyroid carcinoma cell growth through
the miR-135b-5p/CCND2 axis. Biosci Rep. 9:BSR201814402019.
View Article : Google Scholar
|
|
15
|
Zha Z, Han Q, Liu W and Huo S: lncRNA
GAS8-AS1 downregulates lncRNA UCA1 to inhibit osteosarcoma cell
migration and invasion. J Orthop Surg Res. 15:382020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu X, Jiang T, Huang R and Xiao X: LncRNA
GAS8-AS1 downregulates lncRNA NEAT1 to inhibit glioblastoma cell
proliferation. Brain Behav. 11:e021282021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qin Y, Sun W, Wang Z, Dong W, He L, Zhang
T, Shao L and Zhang H: ATF2-Induced lncRNA GAS8-AS1 Promotes
Autophagy of Thyroid Cancer Cells by Targeting the miR-187-3p/ATG5
and miR-1343-3p/ATG7 Axes. Mol Ther Nucleic Acids. 22:584–600.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu Y, Song Y, Xiong Y, Wang X, Xu K, Han
B, Bai Y, Li L, Zhang Y and Zhou L: MicroRNA-21 (Mir-21) Promotes
Cell Growth and Invasion by Repressing Tumor Suppressor PTEN in
Colorectal Cancer. Cell Physiol Biochem. 43:945–958. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang H, Tan Z, Hu H, Liu H, Wu T, Zheng C,
Wang X, Luo Z, Wang J, Liu S, et al: microRNA-21 promotes breast
cancer proliferation and metastasis by targeting LZTFL1. BMC
Cancer. 19:7382019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Q, Li B, Li Q, Wei S, He Z, Huang X,
Wang L, Xia Y, Xu Z, Li Z, et al: Exosomal miR-21-5p derived from
gastric cancer promotes peritoneal metastasis via
mesothelial-to-mesenchymal transition. Cell Death Dis. 9:8542018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan J, Liu T, Zhou X, Dang Y, Yin C and
Zhang G: FZD6, targeted by miR-21, represses gastric cancer cell
proliferation and migration via activating non-canonical wnt
pathway. Am J Transl Res. 8:2354–2364. 2016.PubMed/NCBI
|
|
22
|
Amin MB, Edge SB, Greene FL, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, et al: AJCC Cancer Staging Manual. 8th edition.
Springer; Cham, Switzerland: pp. 3077–3079. 2017
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Δ Δ C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fluss R, Faraggi D and Reiser B:
Estimation of the Youden Index and its associated cutoff point.
Biom J. 47:458–472. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song Z, Wu Y, Yang J, Yang D and Fang X:
Progress in the treatment of advanced gastric cancer. Tumour Biol.
39:10104283177146262017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dai Q, Zhang T and Li C: LncRNA MALAT1
regulates the cell proliferation and cisplatin resistance in
gastric cancer via PI3K/AKT pathway. Cancer Manag Res.
12:1929–1939. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Feng H, Zhang J, Shi Y, Wang L, Zhang C
and Wu L: Long noncoding RNA LINC-PINT is inhibited in gastric
cancer and predicts poor survival. J Cell Biochem. 120:9594–9600.
2019. View Article : Google Scholar : PubMed/NCBI
|