Introduction
The global incidence of oral squamous cell carcinoma
(OSCC) has increased to more than 355,000 new patients and 177,000
related deaths annually (1–3). The
most common sites for intraoral cancer are the tongue, gingiva, and
floor of the mouth, which account for more than half of all OSCCs
(1). The oral cavity is central to
several essential functions such as speech, breathing, and feeding,
and cancers in this region can severely affect several functions
essential for day-to-day life (4).
Early detection and treatment of OSCC can improve quality of life.
Most OSCCs arise from oral epithelial dysplasia (OED). Several
studies have focused on OSCC detection; however, candidate
biomarkers for the early detection of malignant transformation have
not yet been identified (5). This
may be attributed to the lack of an experimental model that allows
continuous observation of molecular changes occurring in the
dysplasia-carcinoma sequence in the same organism (6).
Liquid-based cytology (LBC) is a non-invasive
technique that can be used for the early diagnosis of OSCC. LBC has
higher diagnostic accuracy than conventional cytology; moreover,
gene expression analysis and immunocytochemistry (ICC) can be
performed on the residual LBC specimens (7–11).
There are several reports on ICC and gene expression analysis;
however, none of the studies have reported the continuous changes
in morphology and the gene and protein expression during
carcinogenesis. Most cytology specimens are collected from
superficial or keratinized cell layers, and the changes occurring
in this layer during carcinogenesis need to be observed. Therefore,
the focus should be on identifying biomarkers expressed in the
keratinized cell layers.
Animal experiments are useful for observing changes
in molecular expression during carcinogenesis. We previously
reported that Dark-Agouti (DA) rats are highly susceptible to
4-nitroquinoline 1-oxide (4NQO)-induced tongue cancer (TC)
(12–14). DA rats with 4NQO-induced TC are
considered an ideal experimental model for human TC because tumors
in both share several morphological, molecular, and biological
properties (14). Using LBC
techniques on this model, it is theoretically possible to
continuously observe malignant transformation events in the same
organism. Previous studies analyzed quantitative trait loci, loss
of heterozygosity, and microarray results in 4NQO-induced TC and
identified that genes encoding c-Myc and p53 are associated with
the promotion and progression of OSCC (12,14–16).
Therefore, we focused on these genes in this study. Furthermore, we
analyzed the role of bromodomain protein 4 (Brd4) because Brd4
transcriptionally regulates Myc expression and is expressed in the
superficial or keratinized cell layers during the early stages of
carcinogenesis (17,18).
Brd4 is a member of the bromodomain and
extraterminal domain (BET) family of transcriptional regulatory
proteins. It plays a key role in carcinogenesis and cancer
progression (19). Brd4 expression
increases during 4NQO-induced carcinogenesis in animal models, and
it modulates tumorigenicity and tumor overgrowth (18). Significantly higher levels of Brd4
mRNA and protein expression are observed in the head and neck
squamous cell carcinoma (SCC) tissues, compared to that in normal
tissues (18,20,21).
Brd4 expression correlates with matrix metallopeptidase 2
expression in OSCC. Reportedly, Brd4 is overexpressed in OSCC
patients with lymph node metastasis than in patients without
metastasis (22,23). Furthermore, Brd4 binds and
activates the c-Myc promoter to induce the c-Myc overexpression
(24). c-Myc plays a key role in
cell proliferation, metabolism, differentiation, promotion of p53
expression, and apoptosis. However, when Tp53 is mutated, cell
death is avoided, leading to the development of OSCC (25,26).
Reportedly, c-Myc expression levels are associated with the
advanced stages of OED and OSCC (18,27,28),
wherein c-Myc and p53 are overexpressed in the early stages of oral
carcinogenesis. Therefore, these are considered useful markers for
the early detection of premalignant lesions using
immunohistochemistry (IHC) (29).
However, none of the studies have explored the usefulness and
reliability of these markers for ICC-based cancer detection.
The aim of this study was to establish an
experimental model, wherein the sequential mRNA and protein
expression patterns of Brd4, c-Myc, and p53 could be observed in
the same organism using LBC in a 4NQO-induced rat model of TC and
verify whether these biomarkers are useful for the early detection
of OSCC when combined with cytological diagnosis.
Materials and methods
Animals
Inbred DA rats (DA/Slc) were purchased from the
Shizuoka Laboratory Animal Center (Hamamatsu, Japan). All rats were
housed in a controlled environment with a 12 h light/dark cycle and
temperature of 22±2˚C and fed a commercial pellet-based feed
(Nosan, Yokohama, Japan). Fifty-one 4-weeks old male DA rats were
used for the cytological analysis, and twelve rats were used for
histological experiments. This study was reviewed by the Committee
of the Ethics on Animal Experiments in Niigata University and
carried out according to the Guidelines for Animal Experiments in
Niigata University, Niigata, Japan (SA00507). All experiments were
conducted in accordance with relevant national legislation on the
use of animals for research.
TC induction
A stock solution of 4NQO (200 mg/l in 5% ethanol;
Nacalai Tesque Inc., Kyoto, Japan) was prepared and stored at 4˚C
until use. Starting 6 weeks of age, all rats were given drinking
water containing 0.001% 4NQO ad libitum. At week 21 of the
experiment, all rats were sacrificed by sodium pentobarbital (150
mg/kg body wt, i.p.), followed by full necropsy and
histopathological examinations.
LBC specimens
LBC specimens were collected from the tongue region
of rats at 2, 5, 8, 11, 14, 17, and 21 weeks of the experiment. An
Orcellex brush (Rovers Medical Devices B.V., Oss, Netherlands) or
interdental brush (Dentalpro Co., Osaka, Japan) was rotated on the
lesion surface 20 times. The collected contents were transferred
into a methanol-based preservative solution (SurePath, BD
Diagnostics, Franklin Lakes, NJ, USA) or RNAlater reagent (Ambion,
Austin, TX, USA). LBC preparations were processed according to the
manufacturer's protocol (30).
Fixed specimens were rehydrated with distilled water and
subsequently, subjected to nuclear staining with hematoxylin
solution and cytoplasmic staining with Orange G solution (Muto Pure
Chemicals Co., Ltd, Tokyo, Japan) and Eosin Azure solution (Muto
Pure Chemicals Co., Ltd.). Cytological diagnosis was based on the
oral Bethesda system (31).
Histopathological specimens
We performed another carcinogenesis experiment using
12 DA rats; three rats without 4NQO treatment were analyzed as
controls. We sacrificed three rats each (a total of nine DA rats)
at the time point of the low-grade squamous intraepithelial lesion
(LSIL), high-grade squamous intraepithelial lesion (HSIL), and SCC
formation. The tongue specimens were fixed in buffered 10% formalin
and embedded in paraffin. Serial 4-µm sections were cut, dewaxed in
xylene, and rehydrated in graded ethanol. The normal mucosal
epithelium, hyperplasia, OED, and SCC sections were stained with
hematoxylin and eosin to confirm the histological diagnosis, while
the other sections were used for immunohistochemical analyses.
Immunostaining of Brd4, c-Myc, and p53
in the cytological and the histological specimens
Immunostaining was performed to observe the
expression patterns of Brd4, c-Myc, and p53 in the cytological and
histological specimens. All slides were subjected to antigen
retrieval in a microwave oven, with a maximum strength of 1000 W,
using EDTA (pH 8.0), for 20 min followed by incubation with either
a rabbit polyclonal anti-p53 antibody (1:50 dilution at room
temperature (RT, 24±2˚C) for 2 h; clone ab131442; Abcam, Cambridge,
MA, USA), a rabbit monoclonal anti-c-Myc antibody (1:100 dilution
at RT for 2 h; clone ab32072; Abcam), or a rabbit monoclonal
anti-BRD4 antibody (1:100 dilution at RT for 2 h; clone ab128874;
Abcam). The slides were then washed and incubated with the
Envision+/HRP system (Dako, Glostrup, Denmark). Immunoreactive
cells were visualized using DAB (Dojindo, Kumamoto, Japan) and
counterstained with hematoxylin.
Labeling index analysis of ICC
The percentages of positively stained nuclei in
specimens stained with each of the antibodies (anti-Brd4,
anti-c-Myc, and anti-p53) were calculated as the labeling index
(LI) using the images captured at ×200 magnification and analyzed
using e-Count2 cell counting software (e-Path, Kanagawa, Japan).
Six random fields containing an average of 200 cells each were
selected for analysis, and the average value for LI was
determined.
RNA isolation and quantitative
real-time PCR (qRT-PCR)
The LBC specimens in the RNAlater reagent were
processed using the NucleoSpin RNA XS-kit (Macherey-Nagel, Düren,
Germany) according to the manufacturer's instructions. Reverse
transcription was carried out using the High-Capacity cDNA Reverse
Transcription Kit with RNase inhibitor (Applied Biosystems, Foster
City, CA, USA) according to the manufacturer's protocol. cDNA was
amplified using a TaqMan Universal PCR Master Mix (Applied
Biosystems) and TaqMan Gene Expression Assays (Applied Biosystems)
for Brd4 (Rn01535560_m1), Myc (Rn07310910_m1), and Tp53
(Rn00755717_m1); 18S mRNA (Hs99999901_s1) was used as the internal
standard (32). To compare the
levels of target gene transcripts in LSIL, HSIL, and SCC samples
with those in negative for intraepithelial lesion or malignancy
(NILM) samples, the 2−ΔΔCq method was used to calculate
relative mRNA levels (33).
Statistical analyses
The Shapiro-Wilk tests showed that the Brd4, c-Myc,
and p53-LI and the Brd4 and Tp53 mRNA levels were normally
distributed; therefore, these data were analyzed parametrically. We
performed ANOVA with a post-hoc Tukey test for comparisons between
multiple groups. In contrast, the c-Myc mRNA levels were not
normally distributed; therefore, the data were assessed with a
Kruskal-Wallis followed by a Dunn's post hoc test. The correlation
between mRNA expression and LI was assessed using the Spearman rank
correlation test. We evaluated the diagnostic ability of these
biomarkers using receiver operating characteristic (ROC) curves. As
a global measure for the accuracy of diagnosis, we also calculated
the area under the ROC curve (AUC) for each biomarker. The optimal
cut-off values for the LIs for Brd4, c-Myc, and p53 were determined
using ‘closest-topleft’ (34). In
addition, sample size calculations showed that 12 samples were
necessary for each group to reach 80% power at a 5% significance
level. All statistical comparisons were performed using GraphPad
Prism for Windows version 6.00 (GraphPad Software Inc., San Diego,
CA, USA) and R version 4.0.2 (R Foundation, Vienna, Austria). A
p-value of <0.05 was considered significant, and all statistical
tests were two-sided.
Results
Sample collection
The cytology specimens from the tongues of 51 rats
were classified into four groups, NILM, LSIL, HSIL, and SCC,
according to the oral Bethesda system. LSIL was recognized after 11
weeks of treatment with 4NQO. At 17 weeks, all the diagnostic
classifications, from NILM to SCC, could be recognized. Finally,
all the samples were evaluated as SCC at 21 weeks. The results of
the cytopathological examination are summarized in Table I. Furthermore, the mean incidence
periods of LSIL, HSIL, and SCC were 14.3±2.1, 16.8±0.7, and
20.8±0.8 weeks after the beginning of the carcinogenesis
experiments, respectively.
 | Table I.Cytological diagnoses of 51 rats at
each week. |
Table I.
Cytological diagnoses of 51 rats at
each week.
|
| 4NQO treatment
period (weeks) |
|---|
|
|
|
|---|
| Cytological
diagnosis | 2 | 5 | 8 | 11 | 14 | 17 | 21 |
|---|
| NILM | 51 (100.0) | 51 (100.0) | 51 (100.0) | 44 (86.3) | 25 (49.0) | 7 (13.7) | 0 (0.0) |
| LSIL | 0 (0.0) | 0 (0.0) | 0 (0.0) | 7 (13.7) | 25 (49.0) | 24 (47.1) | 0 (0.0) |
| HSIL | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (2.0) | 18 (35.3) | 0 (0.0) |
| SCC | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (3.9) | 51 (100.0) |
Expression analysis of Brd4, c-Myc,
and p53 using ICC and IHC
Following Papanicolaou staining, NILM specimens
showed orangeophilic keratinized cells without atypical or higher
brightness (Fig. 1A). The LSIL
specimens had mild atypical high-bright orangeophilic keratinized
cells (Fig. 1B), whereas the HSIL
specimens had moderate atypical high-bright orangeophilic
keratinized cells (Fig. 1C). In
addition, several atypical parabasal/basal cell clusters were
detected in SCC specimens (Fig.
1D). NILM, LSIL, HSIL, and SCC were further evaluated by
performing ICC staining for Brd4, c-Myc, and p53. NILM specimens
tested mostly negative for these markers (Fig. 1E, I and M). In LSIL and HSIL
specimens, positive staining for Brd4 and c-Myc was observed along
with the atypically enlarged nuclei of the superficial and
intermediate cells (Fig. 1F, G, J and
K). In SCC specimens, Brd4 and c-Myc expression were detected
in the parabasal/basal cells that appeared small and rounded and
showed clear evidence of nuclear changes, such as nuclear
enlargement, nuclear shape abnormalities, and increased nuclear to
cytoplasmic ratios (Fig. 1H and
L). In contrast, p53 expression was observed only in SCC
specimens, localized in the atypical nuclei of parabasal/basal
cells (Fig. 1N-P).
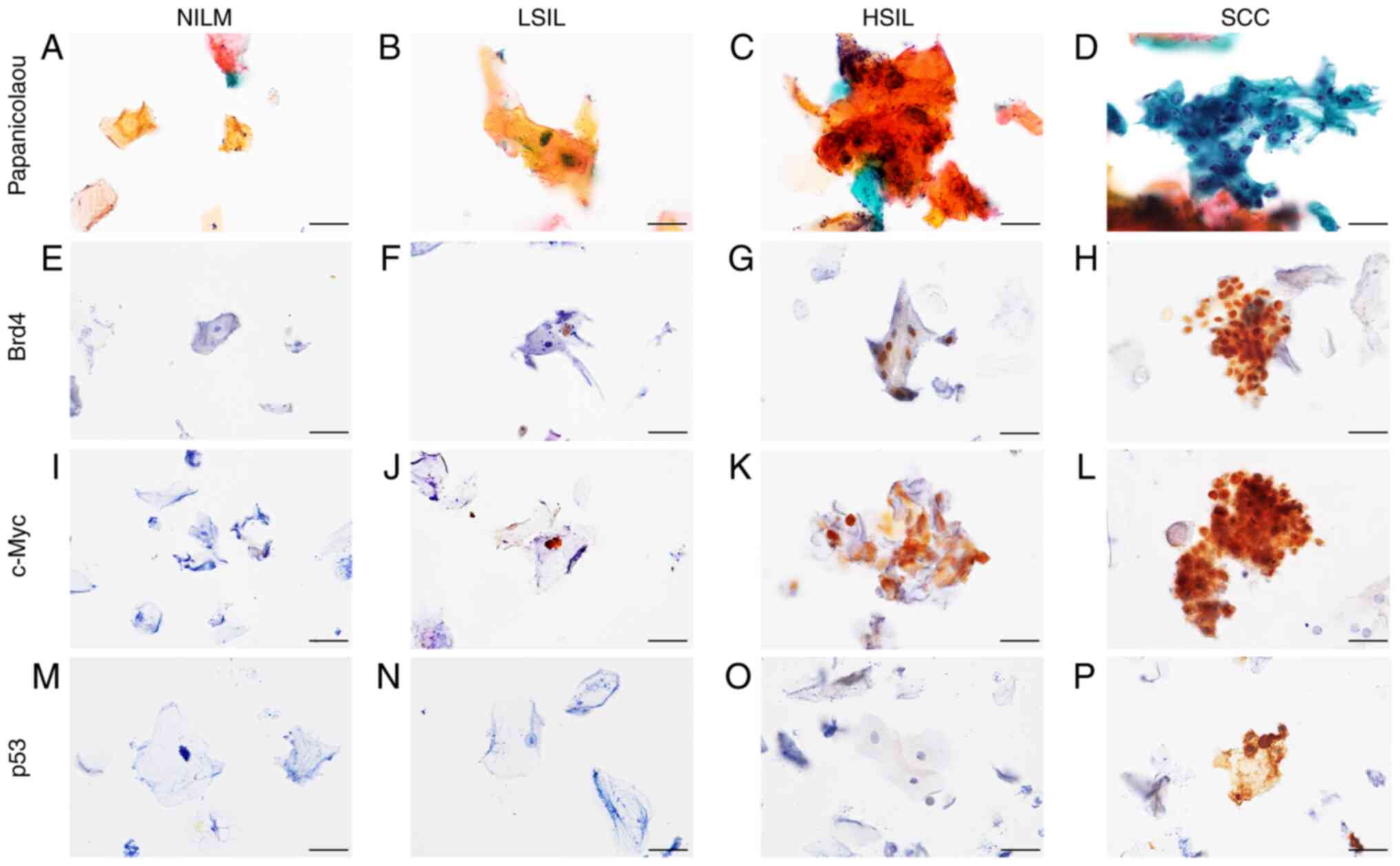 | Figure 1.Immunoexpression of Brd4, c-Myc and
p53 in cytological specimens. Representative cytological findings
based on Papanicolaou staining of liquid-based cytology specimens:
(A) NILM, (B) LSIL, (C) HSIL and (D) SCC. (E-H) Brd4
immunocytochemical staining of oral smears. Although Brd4 staining
was generally negative in (E) NILM specimens, (F) positive staining
in the nuclei was observed in LSIL, (G) HSIL and (H) SCC specimens.
(I-L) c-Myc immunocytochemical staining of oral smears. Although
c-Myc staining was generally negative in (I) NILM specimens,
positive staining in the nuclei was observed in (J) LSIL, (K) HSIL
and (L) SCC specimens. (M-P) p53 immunocytochemical staining of
oral smears. Although p53 staining was generally negative in (M)
NILM, (N) LSIL and (O) HSIL specimens, positive staining in the
nuclei was observed in (P) SCC specimens. Original magnification,
×600. Scale bars, 20 µm. NILM, negative for intraepithelial lesion
or malignancy; LSIL, low-grade squamous intraepithelial lesion;
HSIL, high-grade squamous intraepithelial lesion; SCC, squamous
cell carcinoma; Brd4, bromodomain protein 4. |
After confirming the histological diagnosis of the
normal mucosal epithelium (Fig.
2A), hyperplasia (Fig. 2B),
OED (Fig. 2C), and SCC (Fig. 2D) by hematoxylin and eosin
staining, the expression of Brd4, c-Myc, and p53 was further
compared by IHC staining. Brd4 expression was observed from the
basal layer to the superficial layer in the hyperplasia, OED, and
OSCC tissue specimens (Fig. 2E-H),
while the c-Myc expression was limited to the basal and parabasal
layers of the hyperplasia. c-Myc positive cells were observed from
the basal layer to the superficial layers in the OED and OSCC
tissue specimens (Fig. 2I-L).
Diffuse and intensive p53 nuclear positivity was detected in OSCC
tissue specimens but not overexpression was detected in the normal
mucosal epithelium, hyperplasia, and OED tissue specimens (Fig. 2M-P). Thus, consistent expression
patterns were observed using ICC and IHC.
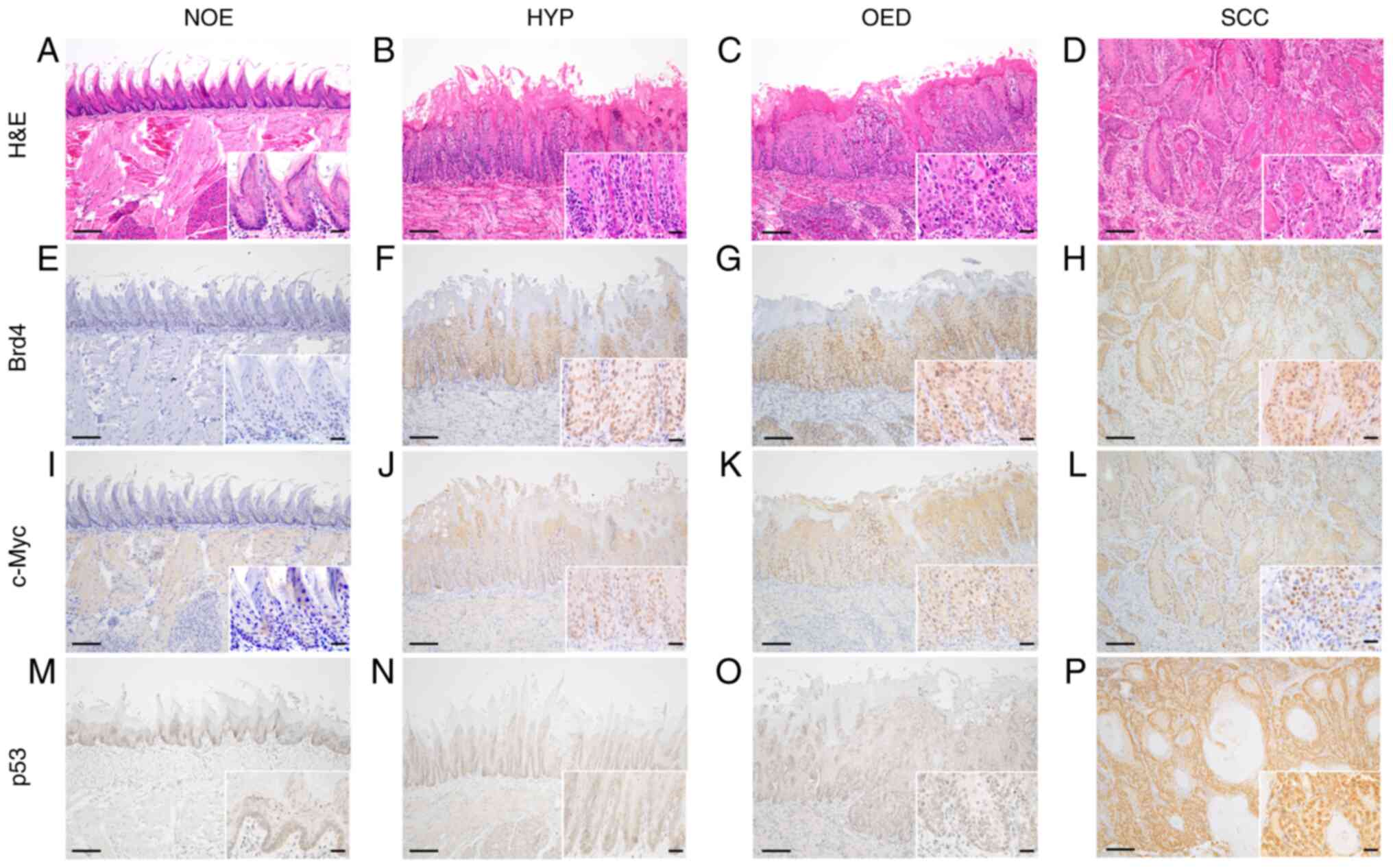 | Figure 2.Representative histopathological and
immunohistochemical findings for Brd4, c-Myc and p53 expression
patterns in control and 4NQO-treated rats at 21 weeks. (A, E, I, M)
NOE, (B, F, J, N) HYP, (C, G, K, O) OED and (D, H, L, P) SCC. (A-D)
H&E, (E-H) Brd4, (I-L) c-Myc and (M-P) p53. Original
magnification, ×100 and 400 (inset). Scale bars, 100 and 20 µm
(inset). 4NQO, 4-nitroquinoline 1-oxide; NOE, normal epithelium;
HYP, hyperplasia; OED, oral epithelial dysplasia; SCC, squamous
cell carcinoma; H&E, hematoxylin and eosin; Brd4, bromodomain
protein 4. |
mRNA expression levels of candidate
markers in LBC specimens
Results of qRT-PCR for Brd4, c-Myc, and Tp53 mRNA
expression revealed a statistically significant difference in the
expression of each marker among the oral Bethesda categories
(P<0.01; Tukey's multiple comparisons test for Brd4 and Tp53,
Kruskal-Wallis test for c-Myc). In addition, the levels of Brd4,
c-Myc, and Tp53 mRNAs were significantly higher in SCC specimens
than that in the NILM, LSIL, and HSIL specimens (P<0.01;
Fig. 3A-C). These results showed
that expression levels of the Brd4, c-Myc, and Tp53 mRNAs were
upregulated during the carcinogenesis in the 4NQO-induced TC
model.
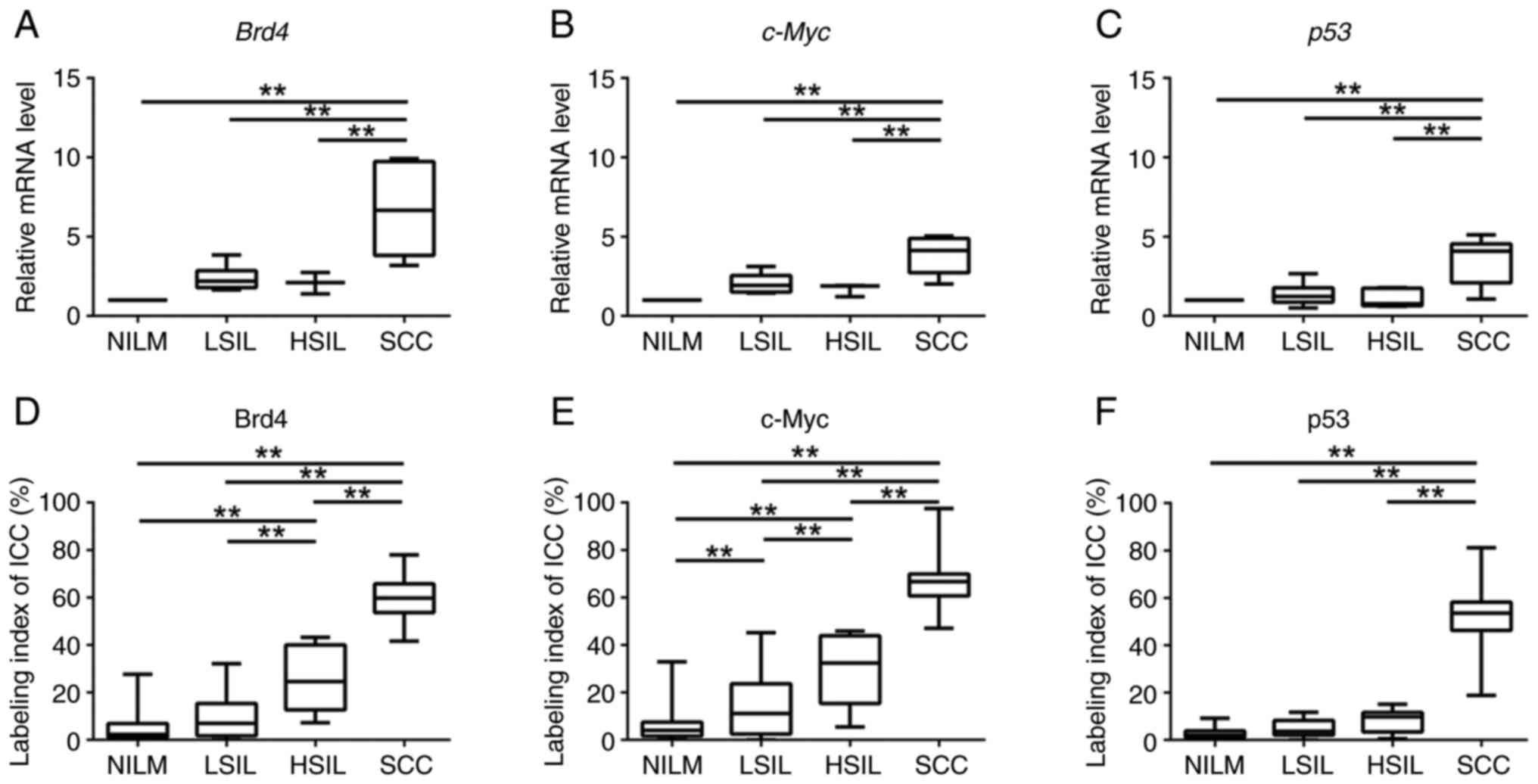 | Figure 3.Box plots showing normalized
expression of (A) Brd4, (B) c-Myc and (C) p53 in NILM, LSIL, HSIL
and SCC specimens. The box represents 50% quartiles (>25% and
<75%), and the solid line within each box is the median gene
expression value. ANOVA followed by Tukey's multiple comparisons
test and the Kruskal-Wallis test followed by Dunn's post hoc test
were used to determine statistical significance. Boxplots for
labeling indices from ICC of (D) Brd4, (E) c-Myc and (F) p53 in
NILM, LSIL, HSIL and SCC specimens. ANOVA followed by Tukey's
multiple comparisons test was used to determine statistical
significance. **P<0.01, as indicated. NILM, negative for
intraepithelial lesion or malignancy; LSIL, low-grade squamous
intraepithelial lesion; HSIL, high-grade squamous intraepithelial
lesion; SCC, squamous cell carcinoma; ICC, immunocytochemistry;
Brd4, bromodomain protein 4. |
Protein levels of candidate markers in
LBC specimens
To identify Brd4, c-Myc, and p53 protein expression
patterns in the different grades within the oral Bethesda system,
we analyzed the expression of Brd4, c-Myc, and p53 proteins in
NILM, LSIL, HSIL, and SCC specimens using ICC. Tukey's multiple
comparisons test showed significant differences in the LI for Brd4
(BRD4-LI), c-Myc (c-Myc-LI), and p53 (p53-LI) among the oral
Bethesda categories (P<0.01). The BRD4-LI was significantly
higher in HSIL and SCC specimens than that in the NILM and LSIL
specimens, based on the ICC results (P<0.01; Tukey's multiple
comparisons test) (Fig. 3D). In
line with Brd4 expression, c-Myc-LI was significantly higher in the
HSIL and SCC specimens than that in the NILM and LSIL specimens
(P<0.01; Tukey's multiple comparisons test) (Fig. 3E). Similarly, p53-LI was
significantly higher in the SCC specimens compared to that in the
NILM, LSIL, and HSIL specimens (P<0.01; Tukey's multiple
comparisons test) (Fig. 3F). These
results showed that the expression of Brd4, c-Myc, and p53 proteins
increased with disease severity. In particular, the levels of Brd4
and c-Myc were higher in the LSIL and HSIL specimens.
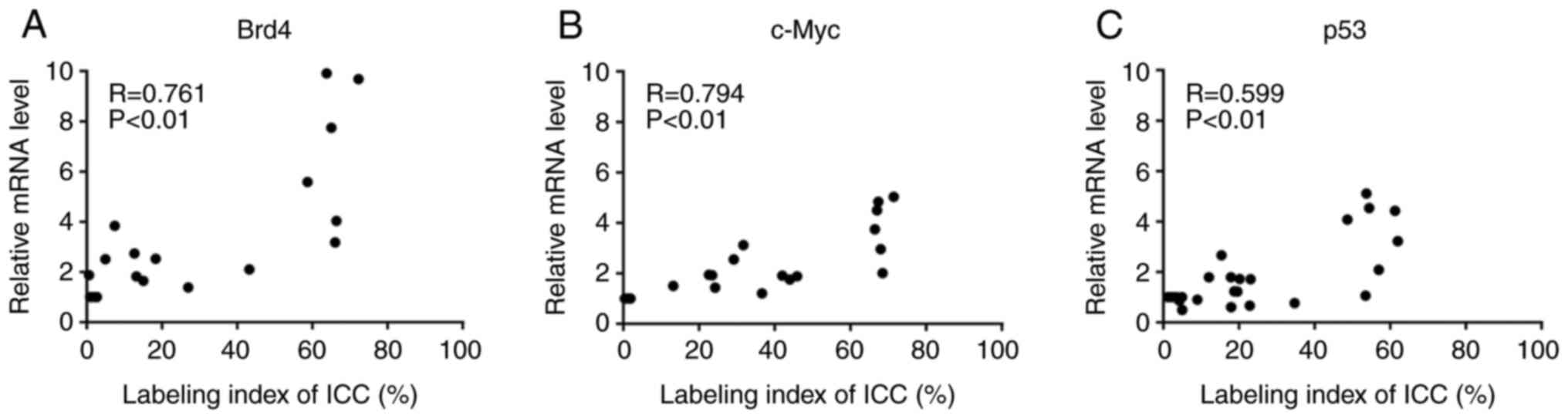
Relationship between mRNA expression
levels and LI for Brd4, c-Myc, and p53
To analyze the relationship between mRNA expression
levels and the LI for ICC, we performed the correlation analysis
for each candidate marker. Fig. 4
shows the relationship between Brd4, c-Myc, and p53 mRNA expression
levels. Significant correlations were detected for Brd4 (R=0.761,
P<0.01; Fig. 4A), c-Myc
(R=0.794, P<0.01; Fig. 4B) and
p53 (R=0.599; P<0.01; Fig. 4C)
expression levels.
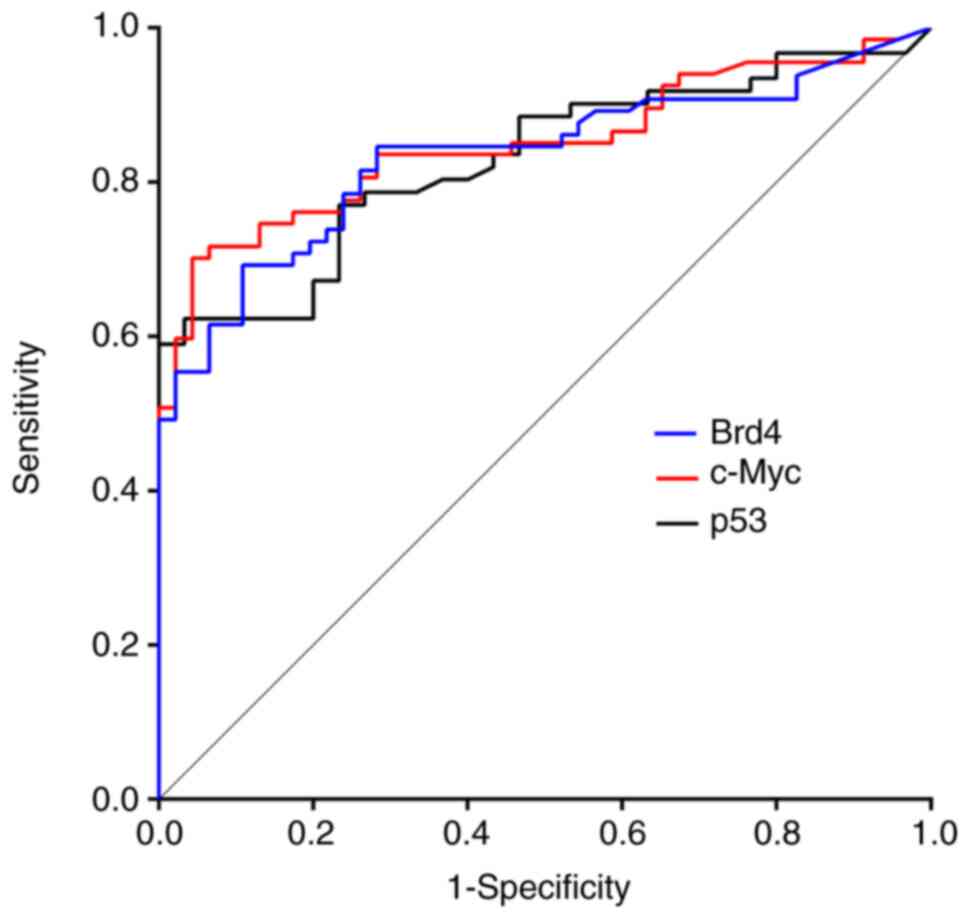
Diagnostic accuracy of candidate
markers
To evaluate the diagnostic accuracy of the increased
LI for individual candidate markers, we stratified the LI of each
marker as positive (LI below cut-off) or negative (LI above the
cut-off). We calculated the sensitivities and specificities to
detect LSIL or a higher category by performing ROC analysis
(Fig. 5). The AUCs were 0.833
(standard error [SE]: 0.039; 95% CI: 0.758–0.909) for Brd4, 0.849
(SE: 0.036; 95% CI: 0.778–0.921) for c-Myc, and 0.829 (SE: 0.042;
95% CI: 0.747–0.911) for p53. When we classified into negative
(NILM) and positive (LSIL, HSIL, and SCC), Table II shows the sensitivity,
specificity, false-negative rate (FNR), negative predictive value
(NPV), positive predictive value (PPV), and diagnostic accuracy of
all markers. Using the oral Bethesda system, cut-off BRD4-LI values
greater than 6.0% were observed in 60.0% of LSIL and 100% of HSIL
specimens; likewise, for c-Myc, cut-off c-Myc-LI values greater
than 12.0% were observed in 50.0% of LSIL and 78.0% of HSIL
specimens. In contrast, a cut-off p53-LI greater than 3.6% was
observed in only 20% of LSIL and 58% of HSIL specimens.
 | Table II.Cut-off values, sensitivity,
specificity, FNRs, NPVs and PPVs for BRD4-LI, c-Myc-LI and
p53-LI. |
Table II.
Cut-off values, sensitivity,
specificity, FNRs, NPVs and PPVs for BRD4-LI, c-Myc-LI and
p53-LI.
| Markers | Cut-off value,
% | Sensitivity, % | Specificity, % | FNR, % | PPV, % | NPV, % | Accuracy, % |
|---|
| Brd4 | 6.0 | 81.5 | 73.9 | 18.5 | 81.5 | 73.9 | 78.3 |
| c-Myc | 12.0 | 74.6 | 87.0 | 25.3 | 89.3 | 70.2 | 79.6 |
| p53 | 3.6 | 59.0 | 100.0 | 41.0 | 100.0 | 54.4 | 72.5 |
Discussion
To identify biomarkers useful for the early
diagnosis of OSCC, we established a novel experimental rat model
that would enable evaluating the biological changes occurring
during the multi-step carcinogenesis process in the same animal. In
this result, Papanicolaou smears showed sequential progression from
NILM to LSIL/HSIL to SCC, and the morphological changes were
similar to that observed during the progression of human oral
carcinogenesis (35). We also
succeeded, for the first time, in performing ICC and qRT-PCR on
mRNA isolated from cytology specimens in a 4NQO-induced rat model
of TC. Several studies have suggested that the 4NQO-induced rat
model of TC can be used to visualize histological as well as
molecular changes in human oral carcinogenesis (14,36,37).
In addition, Arduino et al (38) reported that the mean age of the
patient diagnosed with OED is 63.8 years (SD±10.7 years), and the
mean period for malignant transformation of OED is 29.8 months
(range: 9–120 months). These results suggest that in humans, the
time for OED to develop (approximately 60 years) is longer with a
short period of transformation from OED to OSCC, and are consistent
with our results in the rat model. Taken together, these findings
support that our 4NQO-induced rat model of TC may be a suitable
model of human OSCC. Furthermore, to the best of our knowledge, the
morphological and molecular changes using cytology specimens have
not been explored in previous studies. Therefore, we believe this
is the first study to demonstrate the changes occurring in
morphology and mRNA and protein expression during carcinogenesis,
using the cytology specimens obtained from the same organism.
As most of the cytological specimens are collected
from the superficial and keratinized cell layers, the biomarkers
expressed in these layers could be useful. A previous study has
shown that immunohistochemical staining for cytokeratin 13 (CK13),
CK17, and p53 is useful to adjunct the histological diagnosis of
OED and OSCC. However, the expression patterns of CK13 and CK17
vary in the reactive lesions and are not specific for the
neoplastic changes (39). Similar
results have also been reported in other studies. For example, Noda
et al (40) investigated
the immunocytochemical expression patterns of CK13 and CK17 and
reported that these markers could not distinguish non-neoplastic
and neoplastic lesions in the oral cytology specimens. In this
study, we selected p53 as a candidate marker because p53
overexpression is reportedly involved in OSCC development, and Tp53
mutations are observed in OED and OSCC (41). However, in this study, the
immunohistochemical results revealed the absence of p53
overexpression in the normal mucosal epithelium, hyperplasia, and
OED specimens; therefore, it was difficult to detect the
overexpression of p53 in the cytology specimens from the
premalignant lesions. In contrast, hyperplasia, OED, and OSCC
specimens showed Brd4 expression from the basal to superficial
layers. c-Myc was expressed in the basal to spinous cell layer in
the hyperplasia specimens and in the basal to superficial layer in
the OED and SCC specimens; therefore, we considered Brd4, and c-Myc
as candidate markers that were detected by ICC in the hyperplasia
and OED specimens and examined their usefulness and reliability in
this novel experimental model.
Results of the qRT-PCR analysis showed that Brd4,
c-Myc, and Tp53 mRNA expression increased with the progression from
NILM to SCC, and each marker was significantly overexpressed in SCC
specimens than in the NILM, LSIL, and HSIL specimens. These
observations are consistent with those reported in the previous
studies in OSCC (18,42,43).
The level of Brd4 mRNA is aberrantly upregulated in head and neck
SCC samples and a 4NQO-induced animal model of OSCC (18). c-Myc and p53 are overexpressed in
OSCC tissues and TSCCA and CAL-27 cell lines than in normal tissues
and normal human oral keratinocyte cells (37,42).
The LI values for Brd4, c-Myc, and p53 increased
with the progression from NILM to SCC. Although c-Myc
immunostaining patterns can be divided into three types-nuclear,
granular perinuclear, and diffuse cytoplasmic-we analyzed only the
nuclear-positive patterns. In frozen specimens of normal tissues,
Loke et al (44) found a
predominantly nuclear localization of the c-Myc protein in the
liver, spleen, kidney, lungs, and so on; however, when tissues were
fixed in formalin, this pattern was altered, suggesting possible
protein translocation to the cytoplasm during fixation. c-Myc is
ubiquitinated in the nucleus and exported to the cytoplasm for
degradation (45). c-Myc is a
nuclear transcription factor, and therefore, it is most active in
the nucleus (46). The c-Myc
expression pattern in OSCC remains debatable; however,
collectively, these studies indicate that the cytoplasmic
localization of c-Myc immunoreactivity is unlikely to be considered
positive, and it may be reasonable to evaluate the nuclear
localization of c-Myc in OSCC. Moreover, the methods used for
calculating LI were adequate because a significant correlation was
observed between the LI and mRNA expression level for each marker
(Fig. 4). Also, ICC was more
effective in detecting positive cells than qRT-PCR because ICC
calculates the LI for the hot spots in the cytology specimens,
whereas qRT-PCR evaluates entire specimens. Consequently, ICC is a
more useful method than qRT-PCR for the early detection of OSCC
from cytology specimens.
LI for Brd4 and c-Myc increased significantly during
the early stages of the carcinogenesis, and therefore, they may be
good predictors of OSCC. Cytologic diagnoses based on morphologic
changes are frequently difficult because of the occurrence of false
negatives. In particular, differentiating between
reactive/regenerative or neoplastic changes, namely NILM and LSIL,
is challenging (40). Our analysis
showed that the cells positive for Brd4 or c-Myc expression were
more frequently detected in LSIL specimens than the cells positive
for p53 expression. Therefore, assessing BRD4-LI and c-Myc-LI by
ICC, in addition to cytological diagnosis, can improve the
diagnostic accuracy of the cytology of OSCC. c-Myc is a well-known
oncogene and a downstream target of Brd4 (18). c-Myc promotes p53 expression; it
also induces the expression of cyclins D/E and cyclin-dependent
protein kinases 2/4/6 and represses p21CIP1 and
p27KIP1 levels, leading to cell cycle progression
(47–52). Brd4 and c-Myc expressions are
usually upregulated in OSCC and significantly associated with
aggressive clinicopathological features and poor survival (18,53).
However, their usefulness as markers for the early detection of
OSCC remains unknown. Combining cytology and ICC to increase
diagnostic accuracy is widely reported for other cancers, but just
a few studies have been conducted for evaluating the pathological
conditions of the oral cavity (8,54–59).
Therefore, this study offers a new cytological diagnosis tool for
OSCC.
In conclusion, the novel experimental model reported
in this study allowed us to observe the sequential morphologic
changes and expression patterns of mRNAs and proteins in the same
animal during carcinogenesis. Our data suggested that ICC-based
detection of Brd4 or c-Myc expression from cytology specimens could
improve the diagnostic accuracy. Therefore, in combination with
cytology, immunocytochemistry can improve the accuracy of OSCC
diagnosis.
Acknowledgements
Not applicable.
Funding
This research was funded by the Grants-in-Aid for Scientific
Research from the Japan Society for the Promotion of Science (JSPS
KAKENHI grant no. 19K10069).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MK was responsible for data acquisition (management
of patient samples), analysis and interpretation, development of
methodology, and the writing, editing/reviewing and intellectual
content of the manuscript. MY designed the methodology, and was
responsible for the writing, editing/reviewing and intellectual
content of the manuscript. SM was responsible for the analysis and
interpretation of data, and the writing, editing/reviewing and
intellectual content of the manuscript. TA, TKo and TM were
responsible for the analysis and interpretation of data, and the
writing, editing/reviewing and intellectual content of the
manuscript. NC and TKi were responsible for acquisition of data
(management of patient samples). JT was responsible for the study
concept and design, development of methodology, analysis and
interpretation of data, writing, editing/reviewing and intellectual
content of the manuscript, and study supervision. MK and JT confirm
the authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Animal Care
Guidelines of Niigata University (Niigata, Japan; approval no.
SA00507).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
El-Naggar AK, Chan JKC, Grandis JR, Takata
T and Slootweg PJ: World Health Organization classification of Head
and Neck Tumours. 9. 4th edition. IARC Press; Lyon: pp. 109–111.
2017
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miranda-Filho A and Bray F: Global
patterns and trends in cancers of the lip, tongue and mouth. Oral
Oncol. 102:1045512020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tirelli G, Gatto A, Bonini P, Tofanelli M,
Arnež ZM and Piccinato A: Prognostic indicators of improved
survival and quality of life in surgically treated oral cancer.
Oral Surg Oral Med Oral Pathol Oral Radiol. Jan 31–2018.(Epub ahead
of print). doi: 10.1016/j.oooo.2018.01.016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Blatt S, Krüger M, Ziebart T, Sagheb K,
Schiegnitz E, Goetze E, Al-Nawas B and Pabst AM: Biomarkers in
diagnosis and therapy of oral squamous cell carcinoma: A review of
the literature. J Craniomaxillofac Surg. 45:722–730. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Omar E: Current concepts and future of
noninvasive procedures for diagnosing oral squamous cell
carcinoma-a systematic review. Head Face Med. 11:62015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Osaka R, Hayashi K, Onda T, Shibahara T
and Matsuzaka K: Evaluation of liquid based cytology for tongue
squamous cell carcinoma: Comparison with conventional cytology.
Bull Tokyo Dent Coll. 60:29–37. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kujan O, Huang G, Ravindran A, Vijayan M
and Farah CS: CDK4, CDK6, cyclin D1 and Notch1 immunocytochemical
expression of oral brush liquid-based cytology for the diagnosis of
oral leukoplakia and oral cancer. J Oral Pathol Med. 48:566–573.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sivakumar N, Narwal A, Kumar S, Kamboj M,
Devi A, Pandiar D and Bhardwaj R: Application of the Bethesda
system of reporting for cervical cytology to evaluate human
papilloma virus induced changes in oral leukoplakia, oral squamous
cell carcinoma, and oropharyngeal squamous cell carcinoma: A
cytomorphological and genetic study. Diagn Cytopathol.
49:1036–1044. 2021. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rossi ED, Bizzarro T, Schmitt F and
Longatto-Filho A: The role of liquid-based cytology and ancillary
techniques in pleural and pericardic effusions: An institutional
experience. Cancer Cytopathol. 123:258–266. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mehrotra R: The role of cytology in oral
lesions: A review of recent improvements. Diagn Cytopathol.
40:73–83. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tanuma JI, Shisa H, Hiai H, Higashi S,
Yamada Y, Kamoto T, Hirayama Y, Matsuuchi H and Kitano M:
Quantitative trait loci affecting 4-nitroquinoline 1-oxide-induced
tongue carcinogenesis in the rat. Cancer Res. 58:1660–1664.
1998.PubMed/NCBI
|
|
13
|
Tanuma JI, Kitano M, Shisa H and Hiai H:
Polygenetic susceptibility and resistance to 4-nitroquinoline
1-oxide-induced tongue carcinomas in the rat. J Exp Anim Sci.
41:68–77. 2000. View Article : Google Scholar
|
|
14
|
Tanuma JI, Hiai H, Shisa H, Hirano M,
Semba I, Nagaoka S and Kitano M: Carcinogenesis modifier loci in
rat tongue are subject to frequent loss of heterozygosity. Int J
Cancer. 102:638–642. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tanuma JI, Fujii K, Hirano M, Matsuuchi H,
Shisa H, Hiai H and Kitano M: Five quantitative trait loci
affecting 4-nitroquinoline 1-oxide-induced tongue cancer in the
rat. Jpn J Cancer Res. 92:610–616. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Suwa H, Hirano M, Kawarada K, Nagayama M,
Ehara M, Muraki T, Shisa H, Sugiyama A, Sugimoto M, Hiai H, et al:
Pthlh, a promising cancer modifier gene in rat tongue
carcinogenesis. Oncol Rep. 31:3–12. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zuber J, Shi J, Wang E, Rappaport AR,
Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al:
RNAi screen identifies Brd4 as a therapeutic target in acute
myeloid leukaemia. Nature. 478:524–528. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu Y, Wang Y, Diao P, Zhang W, Li J, Ge H,
Song Y, Li Z, Wang D, Liu L, et al: Therapeutic targeting of BRD4
in head neck squamous cell carcinoma. Theranostics. 9:1777–1793.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lovén J, Hoke HA, Lin CY, Lau A, Orlando
DA, Vakoc CR, Bradner JE, Lee TI and Young RA: Selective inhibition
of tumor oncogenes by disruption of super-enhancers. Cell.
153:320–334. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu X, Li Q, Huang P, Tong D, Wu H and
Zhang F: EGFR-mediated signaling pathway influences the sensitivity
of oral squamous cell carcinoma to JQ1. J Cell Biochem.
119:8368–8377. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao L, Li P, Zhao L, Wang M, Tong D, Meng
Z, Zhang Q, Li Q and Zhang F: Expression and clinical value of
PD-L1 which is regulated by BRD4 in tongue squamous cell carcinoma.
J Cell Biochem. 121:1855–1869. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cho HY, Lee SW, Jeon YH, Lee DH, Kim GW,
Yoo J, Kim SY and Kwon SH: Combination of ACY-241 and JQ1
synergistically suppresses metastasis of HNSCC via regulation of
MMP-2 and MMP-9. Int J Mol Sci. 21:68732020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yamamoto T, Hirosue A, Nakamoto M, Yoshida
R, Sakata J, Matsuoka Y, Kawahara K, Nagao Y, Nagata M, Takahashi
N, et al: BRD4 promotes metastatic potential in oral squamous cell
carcinoma through the epigenetic regulation of the MMP2 gene. Br J
Cancer. 123:580–590. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Delmore JE, Issa GC, Lemieux ME, Rahl PB,
Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Chesi M,
et al: BET bromodomain inhibition as a therapeutic strategy to
target c-Myc. Cell. 146:904–917. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Poter JR, Fisher BE, Baranello L, Liu JC,
Kambach DM, Nie Z, Koh WS, Luo J, Stommel JM, Levens D and
Batchelor E: Global inhibition with specific activation: How p53
and MYC redistribute the transcriptome in the DNA double-strand
break response. Mol Cell. 67:1013–1025. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ries JC, Schreiner D, Steininger H and
Girod SC: p53 mutation and detection of p53 protein expression in
oral leukoplakia and oral squamous cell carcinoma. Anticancer Res.
18:2031–2036. 1998.PubMed/NCBI
|
|
27
|
Pallavi N, Nalabolu GRK and Hiremath SKS:
Bcl-2 and c-Myc expression in oral dysplasia and oral squamous cell
carcinoma: An immunohistochemical study to assess tumor
progression. J Oral Maxillofac Pathol. 22:325–331. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pérez-Sayáns M, Suárez-Peñaranda JM, Pilar
G, Barros-Angueira F, Gándara-Rey JM and García-garcía A: What real
influence does the proto-oncogene c-myc have in OSCC behavior? Oral
Oncol. 47:688–692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Papakosta V, Vairaktaris E, Vylliotis A,
Derka S, Nkenke E, Vassiliou S, Lazaris A, Mourouzis C, Rallis G,
Spyridonidou S, et al: The co-expression of c-myc and p53 increases
and reaches a plateau early in oral oncogenesis. Anticancer Res.
26:2957–2962. 2006.PubMed/NCBI
|
|
30
|
Norimatsu Y, Yamaguchi T, Taira T, Abe H,
Sakamoto H, Takenaka M, Yanoh K, Yoshinobu M, Irino S, Hirai Y and
Kobayashi TK: Inter-observer reproducibility of endometrial
cytology by the osaki study group method: utilising the becton
dickinson surepath™ liquid-based cytology. Cytopathology.
27:472–478. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Suzuki T, Isaka E, Hiraga C, Akiyama Y,
Okamura M, Oomura Y, Hashimoto K, Sato K, Tanaka Y and Nomura T:
New oral cytodiagnostic criteria predict change to oral epithelial
dysplasia (OED) and cancerization. Oral Sci Int. 18:203–208. 2021.
View Article : Google Scholar
|
|
32
|
Ogawa K, Tanuma JI, Hirano M, Hirayama Y,
Semba I, Shisa H and Kitano M: Selective loss of resistant alleles
at p15INK4B and p16INK4A genes in chemically-induced rat tongue
cancers. Oral Oncol. 42:710–717. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12:772011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kitano M: Host genes controlling the
susceptibility and resistance to squamous cell carcinoma of the
tongue in a rat model. Pathol Int. 50:353–362. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kanojia D and Vaidya MM:
4-Nitroquinoline-1-oxide induced experimental oral carcinogenesis.
Oral Oncol. 42:655–667. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Moon SM, Ahn MY, Kwon SM, Kim SA, Ahn SG
and Yoon JH: Homeobox C5 expression is associated with the
progression of 4-nitroquinoline 1-oxide-induced rat tongue
carcinogenesis. J Oral Pathol Med. 41:470–476. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Arduino PG, Surace A, Carbone M, Elia A,
Massolini G, Gandolfo S and Broccoletti R: Outcome of oral
dysplasia: A retrospective hospital-based study of 207 patients
with a long follow-up. J Oral Pathol Med. 38:540–544. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ikeda M, Shima K, Kondo T and Semba I:
Atypical immunohistochemical patterns can complement the
histopathological diagnosis of oral premalignant lesions. J Oral
Biosci. 62:93–98. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Noda Y, Kondo Y, Sakai M, Sato S and
Kishino M: Galectin-1 is a useful marker for detecting neoplastic
squamous cells in oral cytology smears. Hum Pathol. 52:101–109.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Osugi Y: p53 expression in various stages
of 4-nitroquinoline 1-oxide induced carcinoma in the rat tongue. J
Osaka Dent Univ. 30:29–35. 1996.PubMed/NCBI
|
|
42
|
Li S, Zhang S and Chen J: c-Myc induced
upregulation of long non-coding RNA SNHG16 enhances progression and
carcinogenesis in oral squamous cell carcinoma. Cancer Gene Ther.
26:400–410. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ota K, Fujimori H, Ueda M, Jono H,
Shinriki S, Ota T, Sueyoshi T, Taura M, Taguma A, Kai H, et al:
Midkine expression is correlated with an adverse prognosis and is
down-regulated by p53 in oral squamous cell carcinoma. Int J Oncol.
37:797–804. 2010.PubMed/NCBI
|
|
44
|
Loke SL, Neckers LM, Schwab G and Jaffe
ES: c-myc protein in normal tissue. Effects of fixation on its
apparent subcellular distribution. Am J Pathol. 131:29–37.
1988.PubMed/NCBI
|
|
45
|
Lee CM: Transport of c-MYC by Kinesin-1
for proteasomal degradation in the cytoplasm. Biochim Biophys Acta.
1843:2027–2036. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pérez-Sayáns M, Suárez-Peñaranda JM,
Padín-Iruegas E, Gayoso-Diz P, Reis-De Almeida M, Barros-Angueira
F, Gándara-Vila P, Blanco-Carrión A and García-García A:
Quantitative determination of c-myc facilitates the assessment of
prognosis of OSCC patients. Oncol Rep. 31:1677–1682. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bouchard C, Dittrich O, Kiermaier A,
Dohmann K, Menkel A, Eilers M and Lüscher B: Regulation of cyclin
D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP
recruitment and histone acetylation at the cyclin D2 promoter.
Genes Dev. 15:2042–2047. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tumbarello DA and Turner CE: Hic-5
contributes to epithelial-mesenchymal transformation through a
RhoA/ROCK-dependent pathway. J Cell Physiol. 211:736–747. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hermeking H, Rago C, Schuhmacher M, Li Q,
Barrett JF, Obaya AJ, O'Connell BC, Mateyak MK, Tam W, Kohlhuber F,
et al: Identification of CDK4 as a target of c-MYC. Proc Natl Acad
Sci USA. 97:2229–2234. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yap CS, Peterson AL, Castellani G, Sedivy
JM and Neretti N: Kinetic profiling of the c-Myc transcriptome and
bioinformatic analysis of repressed gene promoters. Cell Cycle.
10:2184–2196. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Claassen GF and Hann SR: A role for
transcriptional repression of p21CIP1 by c-Myc in overcoming
transforming growth factor β-induced cell-cycle arrest. Proc Natl
Acad Sci USA. 97:9498–9503. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Acosta JC, Ferrándiz N, Bretones G,
Torrano V, Blanco R, Richard C, O'Connell B, Sedivy J, Delgado MD
and León J: Myc inhibits p27-induced erythroid differentiation of
leukemia cells by repressing erythroid master genes without
reversing p27-mediated cell cycle arrest. Mol Cell Biol.
28:7286–7295. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Waitzberg AF, Nonogaki S, Nishimoto IN,
Kowalski LP, Miguel RE, Brentani RR and Brentani MM: Clinical
significance of c-myc and p53 expression in head and neck squamous
cell carcinomas. Cancer Detect Prev. 28:178–186. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ribeiro DA, Kitakawa D, Aparecida M,
Domingues C, Cabral LAG, Marques MEA and Salvadori DMF: Survivin
and inducible nitric oxide synthase production during 4NQO-induced
rat tongue carcinogenesis: A possible relationship. Exp Mol Pathol.
83:131–137. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Vlajnic T, Savic S, Barascud A, Baschiera
B, Bihl M, Grilli B, Herzog M, Rebetez J and Bubendorf L: Detection
of ROS1-positive non-small cell lung cancer on cytological
specimens using immunocytochemistry. Cancer Cytopathol.
126:421–429. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Nakra T, Nambirajan A, Guleria P, Phulware
RH and Jain D: Insulinoma-associated protein 1 is a robust nuclear
immunostain for the diagnosis of small cell lung carcinoma in
cytology smears. Cancer Cytopathol. 127:539–548. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Jain D, Nambirajan A, Borczuk A, Chen G,
Minami Y, Moreira AL, Motoi N, Papotti M, Rekhtman N, Russell PA,
et al: Immunocytochemistry for predictive biomarker testing in lung
cancer cytology. Cancer Cytopathol. 127:325–339. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Metovic J, Righi L, Delsedime L, Volante M
and Papotti M: Role of immunocytochemistry in the cytological
diagnosis of pulmonary tumors. Acta Cytol. 64:16–29. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Tone K, Ohno S, Honda M, Notsu A, Sasaki K
and Sugino T: Application of enhancer of zeste homolog 2
immunocytochemistry to bile cytology. Cancer Cytopathol.
129:612–621. 2021. View Article : Google Scholar : PubMed/NCBI
|