The occurrence and development of tumors is a
complicated process involving numerous factors. Cancer stem cells
(CSCs) are considered to be the seed of the tumor and are
characterized by self-renewal and differentiation (1). CSCs serve an important role in
maintaining the proliferation, invasion, drug resistance,
metastasis and recurrence of malignant tumors (2). In recent years, the CSC model has
received increasing attention.
Energy metabolism serves a significant role in the
process of substance metabolism. Metabolic reprogramming is one of
the most important hallmarks of cancer (3). Tumor-initiating cells (TICs) and CSCs
exhibit different biology behaviors compared with non-stem cancer
cells (4). The metabolic
regulation of ATP synthesis and biological building block formation
in CSCs is different compared with that in differential non-stem
cancer cells, but similar to that in normal tissue-derived stem
cells (5). In the past decade, the
role of metabolism in CSC biology has developed into an active area
of research, particularly in lipid metabolism. Previous studies
have shown that lipid metabolism serves an important role in
maintaining the stemness of CSCs (6) and meeting their energy needs,
ultimately leading to cancer growth and invasion. In the present
review, the origin and evolution of the CSC model are summarized.
In addition, the characteristics and mechanisms of lipid metabolism
of CSCs, as well as their role in radiotherapy and chemotherapy
resistance are discussed.
Rudolf Virchow observed similarities between tumor
tissues and embryonic tissues ~150 years ago, establishing the
Embryonic-Rest hypothesis of tumor formation (7). Subsequently, his student Julius
Cohnheim extended this theory, suggesting that tumors originate
from stem cells remaining from embryonic development and sustained
in the tissues (7). Since the 19th
century, significant progress has been achieved in understanding
CSC biology and the existence of CSCs has also been confirmed in a
variety of solid tumor types, including breast carcinoma (8), ovarian carcinoma (9), colon carcinoma (10), pancreatic carcinoma (11) and liver cancer (12).
CSCs, also termed tumor-initiating cells, display
significant self-renewing abilities (13). As well as being responsible for the
origin and development of tumors (14), CSCs are resistant to chemotherapy
and radiotherapy (15), which
allows for recurrence and metastasis (16).
Epigenetic regulation of the genome is associated
with tumor progression. A variety of epigenetic pathways can
contribute to the development and progression of tumors, especially
in CSC maintenance and survival. Abnormal epigenetic changes may
transform normal stem cells into CSCs. DNA methylation and histone
modifications are two key factors involved in the developmental
programming of stem cells to specific cell and tissue
differentiation lineages (17).
The important role of DNA methylation in maintaining CSC properties
has been reported in leukemia, lung and colon stem cells (18,19).
DNA methylation serves a critical role in this transformation
process in the presence of DNA methyltransferases (20). Increasing evidence indicates that
the silencing of tumor suppressor genes and activation of various
cancer genes, which contribute to the formation of CSCs, are
closely related to DNA hypermethylation (21).
In addition, epigenetic mechanisms regulate a number
of key CSC pathways, including the Wnt/β-catenin, Hedgehog (Hh) and
Notch signaling pathways. Specifically, the Wnt/β-catenin signaling
pathway serves an important role in normal tissue development and
maintenance, as well as in the self-renewal and differentiation of
CSCs (22,23). The Hh signaling pathway also
regulates the proliferation and maintains the stemness of
progenitor cells and CSCs in several tissues (24). Notch signaling is an evolutionarily
conserved pathway that regulates proliferation and differentiation
in a wide range of cell types and different stages of cell lineage
progression, as well as in CSC differentiation and self-renewal
(25).
CSCs are considered to be the seed in tumor
initiation, angiogenesis and maintenance. As aforementioned, CSCs
are also an important factor in tumor therapy resistance and
metastasis (26). Compared with
non-stem cancer cells, CSCs are more resistant to radiotherapy and
chemotherapy (27). The following
characteristics contribute to CSC chemotherapy or radiotherapy
resistance (28): Quiescent
phenotype, efficient DNA repair, high expression of drug efflux
pumps and antiapoptotic protein expression.
The target of radiotherapy and/or chemotherapy is
primarily focused on fast growing cells (29). CSCs and normal stem cells are
quiescent, thus CSCs may be insensitive to traditional radiotherapy
and/or chemotherapy (15). CSCs
express a high level of ATP-binding cassette transporters (ABC
transporters), which contributes to the efflux of chemotherapeutic
agents, leading to multidrug resistance (15,30).
CSCs are inherently resistant to DNA damage. The
innate defense system of CSCs protects them against DNA-targeted
chemicals and radiotherapy (31).
Moreover, even under a radiation dose that causes DNA damage, CSCs
can repair the damaged DNA more quickly (26). Indeed, checkpoint kinase (Chk)1 and
Chk2, DNA damage and replication Chks, become activated on
genotoxic stress to initiate cell cycle arrest and attempt repair
or induce apoptosis if the damage is too great (32,33).
Chk1 and Chk2 are highly expressed in CSCs. Inhibition of the
Chk1/2 kinases with a small molecule inhibitor disrupted the
radioresistance of CSCs (34).
Moreover, overamplifying apoptotic inhibitor proteins also
contributes to CSC treatment resistance. Various CSCs express
higher levels of apoptosis protein inhibitors, including X-linked
inhibitor of apoptosis protein (XIAP) isoform, which are associated
with poor therapeutic responses (35). XIAP can alleviate the
radioresistance of CSCs by promoting apoptosis (36).
Numerous other mechanisms also account for CSC
resistance to therapy, including the increased production of
free-radical scavengers and molecular metabolism mediators
(37). Furthermore, certain
mutated genes, including tumor suppressors P53, can help to rescue
CSCs under stress conditions, including radiation therapy, tissue
damage and exposure to toxins (38).
Therefore, to target CSCs more effectively, the
molecular mechanisms of CSCs in proliferation and survival require
further investigation.
Due to the heterogeneity of tumor cells, the energy
metabolism of cancer cells is distinct from that of normal cells
and they are heavily dependent on glucose and aerobic oxidation for
their energy supply (39).
Metabolic reprogramming is one of the hallmarks of cancer cells
(3). Cancer cells display a
disrupted metabolism; even under oxygen-rich conditions, these
cells still depend on glycolysis for energy and survival, a
phenomenon termed the Warburg effect (40). Compared to oxidative
phosphorylation (OXPHOS), glycolysis results in rapid production of
ATP and an increase in metabolic intermediates for anabolic
reactions (41). Although the
metabolic characteristics of CSCs have been researched in recent
years, the exact metabolism of CSCs remains to be elucidated.
A number of studies have reported that CSCs
preferentially utilize glycolysis for survival, while others have
shown that CSCs may also rely on OXPHOS (42,43).
Ciavardelli et al (42)
demonstrate that inhibiting the glycolysis of CD44+
CD24− breast CSCs reduces their proliferation,
indicating that this population is glycolytic (42). Nasopharyngeal carcinoma, ovarian
cancer, osteosarcoma, glioblastoma (GBM) and colon cancer are
primarily dependent on mitochondrial OXPHOS for energy (44–48).
Increasing evidence has indicated that non-stem
cancer cell population dedifferentiation to CSCs is accompanied by
the transformation of metabolic pathways from mitochondrial OXPHOS
to glycolysis (49). Therefore,
CSCs display highly metabolic heterogeneity and plasticity
abilities that allow them to adapt to the changing tumor
microenvironment.
Despite the dependence of cancer on glycolysis,
glycolytic inhibitors, such as 2-deoxyglucose, exhibit minimal
effects on tumor growth inhibition. Therefore, other metabolic
pathways are also critical to cancer cell survival, such as lipid
metabolism (50). In addition to
providing and storing energy as nutrients, lipids also function as
the major component of the cell and signal molecules.
Phospholipids, including glycerophospholipids and sphingolipids and
cholesterol are the main components of the cell membrane (51). Changes in lipid metabolism could
directly affect cell membrane synthesis and proliferation.
In addition, various lipid molecules and their
metabolic intermediates participate in cell signal transduction,
proliferation, cell adhesion and movement, inflammation and
vascular regulation (51). The
unlimited proliferation of cancer cells requires more fatty acids
(FAs) and increased lipid droplet metabolism (52).
Unlike normal cells that preferentially utilize free
FAs, cancer cells are dependent on reconstituted FAs, thus display
enhanced de novo synthesis of FAs. A series of lipid
synthesis enzymes are upregulated in cancer cells, including
sterol-regulatory element binding proteins (SREBPs), acetyl-CoA
carboxylase (ACC), FA synthase (FASN) and stearoyl-CoA desaturase 1
(SCD1) (53–60). In addition, citrate derived from
the citrate (TCA) cycle can be used to produce acetyl-groups for FA
synthesis (61). Therefore, lipid
metabolism is also critical for the maintenance of cancer cell
malignant biological behaviors.
In contrast to the dedifferentiation of non-stem
cancer cells, increasing studies have reported that lipid
metabolism is highly related to the stemness of CSCs (62). In addition to energy generation,
biosynthesis and redox homeostasis, FA metabolism has a vital role
in determining the fate of CSCs (63–65).
FA synthesis and oxidation are essential for the maintenance of
CSCs. For example, NANOG, a critical regulator of CSCs, can promote
mitochondrial FA oxidation (FAO) to satisfy energy requirements for
TICs (66). NANOG promotes the
self-renewal abilities, tumor-initiation properties and generation
of stem-like TICs, as well as the hepatocellular carcinoma (HCC)
oncogenesis of TICs through metabolic reprogramming from OXPHOS to
FAO (66). Peroxisomal
proliferation-activated receptors (PPARδ) have significant effects
on lipid metabolism and are strongly associated with NANOG
expression (67). Overexpression
of PPARδ or NANOG in TICs increases the probability of FAO
occurring (66).
FAs are strictly regulated by CSCs to maintain their
self-renewal ability and therapy resistance (65). As a critical intracellular
organelle for the storage of excess lipids (68), the content of lipid droplets (LDs)
is significantly increased in several solid tumor CSCs, including
colorectal, breast, prostate (69–71)
and ovarian CSCs (64). Of note,
de novo lipogenesis is more active in GBM CSCs compared with
that in non-stem cancer cells (72).
Sterol regulatory element binding protein 1 (SREBP1)
belongs to the SREBP transcription factor family and serves an
important role in the biosynthesis of FAs and cholesterol (81). SREBP1 is the major transcriptional
regulator of lipogenesis and directly regulates several lipogenic
enzymes, including ATP citrate lyase (ACLY), ACC1 and FASN
(57,81). Overexpression of SREBP1 can promote
the growth of various tumors and maintain the stemness of CSCs
(81). On the other hand, SREBP1
can also induce the expression of SCD1, which further induces CSC
generation and stemness maintenance (82).
It has been reported that elevated FAO could help
nutrient-deficient and hypoxic cancer cells survival, especially
those with glycolytic deficiency (83,84).
On the one hand, FAO plays a key role in meeting the heightened
energy demands of CSCs. On the other hand, FAO could reduce
intracellular reactive oxygen species production and maintain an
internal steady state (84).
3-hydroxy-3-methylglutharyl-coenzyme A reductase is
the rate-limiting enzyme in the mevalonate pathway and the
molecular target of statins (85).
The mevalonate pathway represents a metabolic pathway leading to
the production of steroid hormones, cholesterol and non-sterol
isoprenoids. The mevalonate cascade culminates in the production of
farnesyl pyrophosphate and geranylgeranyl pyrophosphate, which are
essential for correct membrane anchoring of Rho family of small
guanosine triphosphatases (GTPases). A number of oncogenic receptor
tyrosine kinases require Ras proteins for signaling and EGFR
signaling is particularly important for CSC maintenance.
Furthermore, Rho GTPases maintain stemness by activating the Hippo
transducers yes1 associated transcriptional regulator/tafazzin and
by promoting the degradation of P27kip, leading to
inhibition of retinoblastoma protein activation, which is
ultimately conducive to the differentiation of CSCs (86).
Liver cancer is the second leading cause of cancer
mortality in the world. Among primary liver cancer, HCC is the
major histological subtype (87).
HCC CSCs are currently considered as a specific subpopulation with
significant tumorigenic potential and contribute to the development
and recurrence of HCC (88). At
present, several special markers have been identified for HCC CSCs,
including CD133. Studies have demonstrated that CD133+
HCC CSCs show a significant enhancement of FAO rate and glycolysis,
as well as a significant decrease in mitochondrial OXPHOS capacity
(66,89).
In addition, inhibiting the prolongation of FAs in
HCC CSCs results in a reduction in the content of polyunsaturated
FAs, which is critical for HCC CSC ferroptosis resistance (66). NANOG is a critical regulator of HCC
CSC lipid metabolic reprogramming. Knockdown of NANOG in HCC CSCs
promotes the expression of FASN and ACLY, which is accompanied by
increased OXPHOS and inhibition of glycolysis (66). Overexpression of NANOG induces
CD133− HCC cancer cell dedifferentiation to
CD133+ HCC CSCs and enhanced FAO activity, indicating
that NANOG serves a vital role in regulating FA metabolism in liver
CSCs (66). Overall, these studies
show that liver CSCs suppress OXPHOS. In general, HCC CSCs favor
FAO to support their stemness, self-renewal ability and therapy
resistance.
CRC is the third most frequently diagnosed cancer
and one of the most lethal types of cancer in both men and women
worldwide (90). CRC CSCs induce
tumorigenesis, proliferation, migration and metastasis of CRC. CRC
CSCs are defined by a group of cell-surface markers, including
CD44, CD133, CD24, epithelial cell adhesion factor molecule,
leucine rich repeat containing G protein-coupled receptor 5 and
Lin-28 homolog A (91). CRC CSCs
also display a special lipid metabolism pathway. Genes associated
with FA biosynthesis are downregulated, whereas genes involved in
glycolysis, the TCA cycle and one-carbon metabolism pathway are
upregulated in CD133+ CRC CSCs, suggesting a strong
preference to glycolysis but suppression of FA biosynthesis
(92). Tirinato et al
(69) found that CD133+
CSCs contain more lipids. The lipid content in cancer cells is also
positively correlated with the expression level of CD133 and
Wnt/β-catenin pathway activity, which are markers of CSCs (69). CD133high cells possess
more LDs compared with CD133low cells. Using the
label-free Raman spectroscopy technology, researchers reported that
the number of LDs in CRC CSCs was related to their tumorigenicity.
The higher the number of LDs, the stronger the tumorigenicity
(69). LDs may potentially open a
new horizon for more specific ex vivo CRC CSCs
diagnostics.
As the most common primary malignant brain tumor,
GBM is extremely aggressive, with a median overall survival of
<15 months (93). GBM CSCs
contribute to the aggressive behaviors of GBM. GBM CSCs are
considered to locate in a special environment, including
perivascular, hypoxic and necrotic niches, as well as tumor border
regions. Identified by the incorporation of 14
[C]-glucose and 14 [C]-acetate into the lipids, GBM CSCs
are reported to have a higher rate of de novo lipogenesis
compared with differentiated non-stem cancer cells (72). Increased de novo lipogenesis
and extracellular lipid uptake result in LD accumulation in GBM
CSCs (94). Moreover, GBM CSCs
express a higher level of FASN protein, which facilitates the
synthesis of FAs. Increased de novo lipogenesis may also
contribute to the upregulation of FASN, thus maintaining the
stemness of GBM CSCs (72).
Inhibition of FASN expression decreased the expression of stemness
markers and inhibited the proliferation and migration of GBM CSCs
(72).
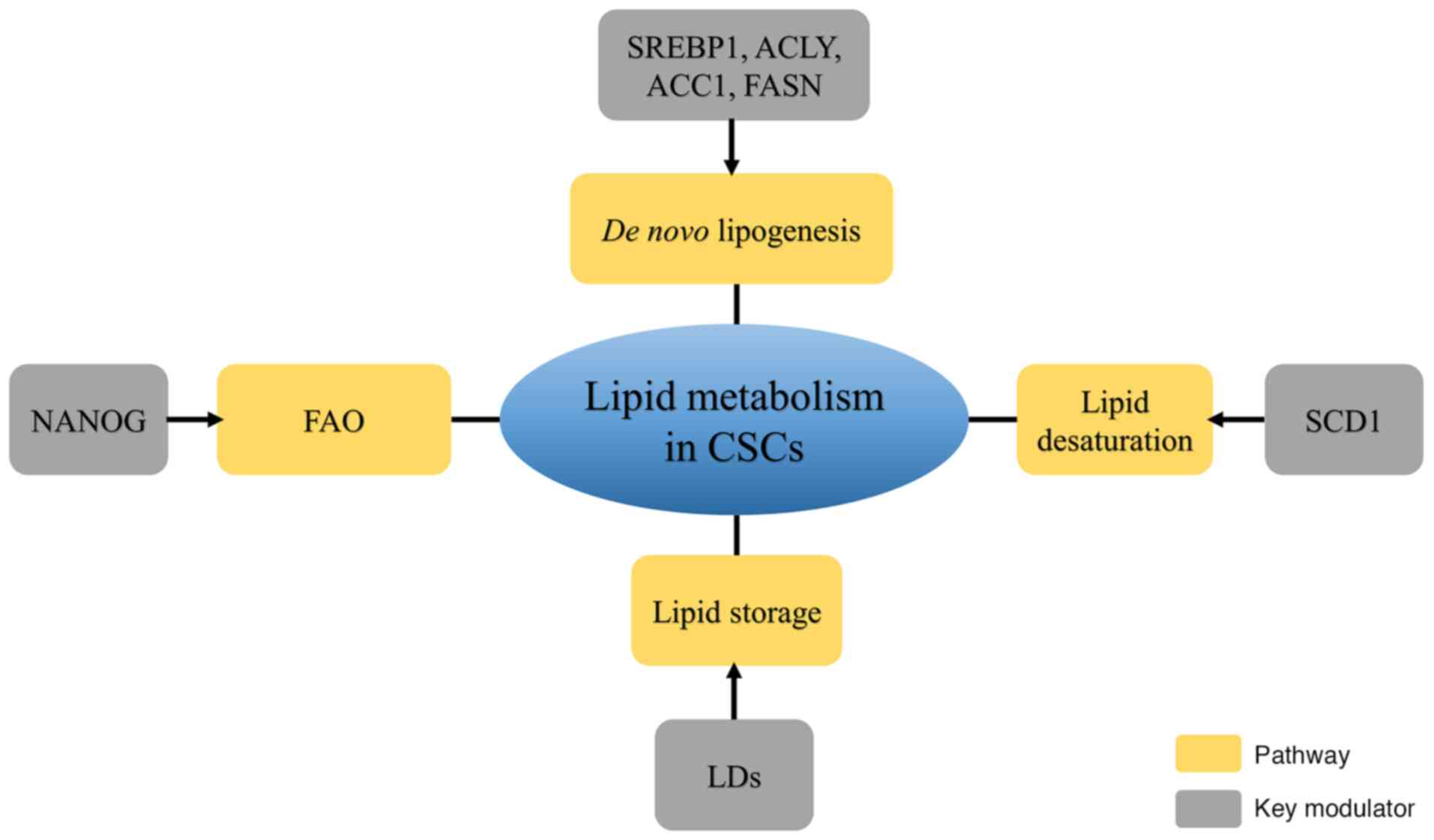
CSCs, with their self-renewal and tumor-initiating
abilities, serve an important role in metastatic dissemination,
radioresistance, chemoresistance and recurrence. A review on
metabolomics demonstrated the contribution of lipid metabolism to
the generation and maintenance of CSCs (76). Lipid metabolism reprogramming,
including de novo lipogenesis and the formation of LDs and
FAO, is involved in CSC generation and stemness maintenance
(Fig. 1).
Thus, understanding the mechanisms underlying CSC
lipid metabolism reprogramming, as well as identifying the
differences in lipid metabolism between CSCs and non-stem cancer
cells will be of significance for improving the current clinical
treatment of cancer. Several therapeutic targets of lipid
metabolism have been developed to enhance antitumor effects
(99). For instance, SCD1
inhibitors, CAY10566 and A939572, targeting FA desaturation process
effectively suppress cancer stemness and tumor progression
(64). As CSCs have been widely
investigated, the development of effective agents targeting lipid
metabolism and ameliorating radioresistance or chemoresistance is
important.
Not applicable.
The present study was supported by grants from the Postgraduate
Innovation Project of Jiangsu Province (grant nos. SJCX20_1435 and
SJCX20_1436).
Data sharing is not applicable.
YL and JG designed the structure of the review. LS
revised the manuscript critically for important intellectual
content. HL and ZZ wrote and reviewed the article. All authors have
read and approved the final manuscript. Data authentication is not
applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang
J, Zhang G, Wang X, Dong Z, Chen F and Cui H: Targeting cancer stem
cell pathways for cancer therapy. Signal Transduct Target Ther.
5:82020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jung Y and Kim WY: Cancer stem cell
targeting: Are we there yet? Arch Pharm Res. 38:414–422. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dando I, Dalla Pozza E, Biondani G,
Cordani M, Palmieri M and Donadelli M: The metabolic landscape of
cancer stem cells. IUBMB Life. 67:687–693. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sancho P, Barneda D and Heeschen C:
Hallmarks of cancer stem cell metabolism. Br J Cancer.
114:1305–1312. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martinez-Outschoorn UE, Peiris-Pages M,
Pestell RG, Sotgia F and Lisanti MP: Cancer metabolism: A
therapeutic perspective. Nat Rev Clin Oncol. 14:11–31. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Capp JP: Cancer stem cells: From
historical roots to a new perspective. J Oncol. 2019:51892322019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang S, Balch C, Chan MW, Lai HC, Matei
D, Schilder JM, Yan PS, Huang TH and Nephew KP: Identification and
characterization of ovarian cancer-initiating cells from primary
human tumors. Cancer Res. 68:4311–4320. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dalerba P, Dylla SJ, Park IK, Liu R, Wang
X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al:
Phenotypic characterization of human colorectal cancer stem cells.
Proc Natl Acad Sci USA. 104:10158–10163. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hermann PC, Huber SL, Herrler T, Aicher A,
Ellwart JW, Guba M, Bruns CJ and Heeschen C: Distinct populations
of cancer stem cells determine tumor growth and metastatic activity
in human pancreatic cancer. Cell Stem Cell. 1:313–323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai
P, Chu PW, Lam CT, Poon RT and Fan ST: Significance of
CD90+ cancer stem cells in human liver cancer. Cancer
Cell. 13:153–166. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meacham CE and Morrison SJ: Tumour
heterogeneity and cancer cell plasticity. Nature. 501:328–337.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen K, Huang YH and Chen JL:
Understanding and targeting cancer stem cells: Therapeutic
implications and challenges. Acta Pharmacol Sin. 34:732–740. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dean M, Fojo T and Bates S: Tumour stem
cells and drug resistance. Nat Rev Cancer. 5:275–284. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dalla Pozza E, Dando I, Biondani G, Brandi
J, Costanzo C, Zoratti E, Fassan M, Boschi F, Melisi D, Cecconi D,
et al: Pancreatic ductal adenocarcinoma cell lines display a
plastic ability to bidirectionally convert into cancer stem cells.
Int J Oncol. 46:1099–1108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Toh TB, Lim JJ and Chow EK: Epigenetics in
cancer stem cells. Mol Cancer. 16:292017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bröske AM, Vockentanz L, Kharazi S, Huska
MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, et
al: DNA methylation protects hematopoietic stem cell multipotency
from myeloerythroid restriction. Nat Genet. 41:1207–1215. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Morita R, Hirohashi Y, Suzuki H, Takahashi
A, Tamura Y, Kanaseki T, Asanuma H, Inoda S, Kondo T, Hashino S, et
al: DNA methyltransferase 1 is essential for initiation of the
colon cancers. Exp Mol Pathol. 94:322–329. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wongtrakoongate P: Epigenetic therapy of
cancer stem and progenitor cells by targeting DNA methylation
machineries. World J Stem Cells. 7:137–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Esteller M: Epigenetic gene silencing in
cancer: The DNA hypermethylome. Hum Mol Genet. 16:Spec No: 1.
R50–R59. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hoffmeyer K, Raggioli A, Rudloff S, Anton
R, Hierholzer A, Del Valle I, Hein K, Vogt R and Kemler R:
Wnt/β-catenin signaling regulates telomerase in stem cells and
cancer cells. Science. 336:1549–1554. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Myant KB, Cammareri P, McGhee EJ, Ridgway
RA, Huels DJ, Cordero JB, Schwitalla S, Kalna G, Ogg EL, Athineos
D, et al: ROS production and NF-κB activation triggered by RAC1
facilitate WNT-driven intestinal stem cell proliferation and
colorectal cancer initiation. Cell Stem Cell. 12:761–773. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Beachy PA, Karhadkar SS and Berman DM:
Tissue repair and stem cell renewal in carcinogenesis. Nature.
432:324–331. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Andersson ER, Sandberg R and Lendahl U:
Notch signaling: Simplicity in design, versatility in function.
Development. 138:3593–3612. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Eyler CE and Rich JN: Survival of the
fittest: Cancer stem cells in therapeutic resistance and
angiogenesis. J Clin Oncol. 26:2839–2845. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou HM, Zhang JG, Zhang X and Li Q:
Targeting cancer stem cells for reversing therapy resistance:
Mechanism, signaling, and prospective agents. Signal Transduct
Target Ther. 6:622021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dick JE: Stem cell concepts renew cancer
research. Blood. 112:4793–4807. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gerlinger M, Rowan AJ, Horswell S, Math M,
Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N,
Stewart A, et al: Intratumor heterogeneity and branched evolution
revealed by multiregion sequencing. N Engl J Med. 366:883–892.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Robey RW, Pluchino KM, Hall MD, Fojo AT,
Bates SE and Gottesman MM: Revisiting the role of ABC transporters
in multidrug-resistant cancer. Nat Rev Cancer. 18:452–464. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Arnold CR, Mangesius J, Skvortsova II and
Ganswindt U: The role of cancer stem cells in radiation resistance.
Front Oncol. 10:1642020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
van Jaarsveld MTM, Deng D, Ordoñez-Rueda
D, Paulsen M, Wiemer EAC and Zi Z: Cell-type-specific role of CHK2
in mediating DNA damage-induced G2 cell cycle arrest. Oncogenesis.
9:352020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Patil M, Pabla N and Dong Z: Checkpoint
kinase 1 in DNA damage response and cell cycle regulation. Cell Mol
Life Sci. 70:4009–4021. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hambardzumyan D, Squatrito M and Holland
EC: Radiation resistance and stem-like cells in brain tumors.
Cancer Cell. 10:454–456. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
LaCasse EC, Mahoney DJ, Cheung HH,
Plenchette S, Baird S and Korneluk RG: IAP-targeted therapies for
cancer. Oncogene. 27:6252–6275. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Morrison R, Schleicher SM, Sun Y, Niermann
KJ, Kim S, Spratt DE, Chung CH and Lu B: Targeting the mechanisms
of resistance to chemotherapy and radiotherapy with the cancer stem
cell hypothesis. J Oncol. 2011:9418762011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Luo M and Wicha MS: Targeting cancer stem
cell redox metabolism to enhance therapy responses. Semin Radiat
Oncol. 29:42–54. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gealy R, Zhang L, Siegfried JM, Luketich
JD and Keohavong P: Comparison of mutations in the p53 and K-ras
genes in lung carcinomas from smoking and nonsmoking women. Cancer
Epidemiol Biomarkers Prev. 8:297–302. 1999.PubMed/NCBI
|
|
39
|
Liberti MV and Locasale JW: The Warburg
effect: How does it benefit cancer cells? Trends Biochem Sci.
41:211–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Guppy M, Greiner E and Brand K: The role
of the Crabtree effect and an endogenous fuel in the energy
metabolism of resting and proliferating thymocytes. Eur J Biochem.
212:95–99. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ciavardelli D, Rossi C, Barcaroli D, Volpe
S, Consalvo A, Zucchelli M, De Cola A, Scavo E, Carollo R,
D'Agostino D, et al: Breast cancer stem cells rely on fermentative
glycolysis and are sensitive to 2-deoxyglucose treatment. Cell
Death Dis. 5:e13362014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pasto A, Bellio C, Pilotto G, Ciminale V,
Silic-Benussi M, Guzzo G, Rasola A, Frasson C, Nardo G, Zulato E,
et al: Cancer stem cells from epithelial ovarian cancer patients
privilege oxidative phosphorylation, and resist glucose
deprivation. Oncotarget. 5:4305–4319. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shen YA, Wang CY, Hsieh YT, Chen YJ and
Wei YH: Metabolic reprogramming orchestrates cancer stem cell
properties in nasopharyngeal carcinoma. Cell Cycle. 14:86–98. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liao J, Qian F, Tchabo N,
Mhawech-Fauceglia P, Beck A, Qian Z, Wang X, Huss WJ, Lele SB,
Morrison CD and Odunsi K: Ovarian cancer spheroid cells with stem
cell-like properties contribute to tumor generation, metastasis and
chemotherapy resistance through hypoxia-resistant metabolism. PLoS
One. 9:e849412014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Palorini R, Votta G, Balestrieri C,
Monestiroli A, Olivieri S, Vento R and Chiaradonna F: Energy
metabolism characterization of a novel cancer stem cell-like line
3AB-OS. J Cell Biochem. 115:368–379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhou Y, Zhou Y, Shingu T, Feng L, Chen Z,
Ogasawara M, Keating MJ, Kondo S and Huang P: Metabolic alterations
in highly tumorigenic glioblastoma cells: Preference for hypoxia
and high dependency on glycolysis. J Biol Chem. 286:32843–32853.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Song IS, Jeong YJ and Han J: Mitochondrial
metabolism in cancer stem cells: A therapeutic target for colon
cancer. BMB Rep. 48:539–540. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Folmes CD, Nelson TJ, Martinez-Fernandez
A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C and
Terzic A: Somatic oxidative bioenergetics transitions into
pluripotency-dependent glycolysis to facilitate nuclear
reprogramming. Cell Metab. 14:264–271. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Snaebjornsson MT, Janaki-Raman S and
Schulze A: Greasing the wheels of the cancer machine: The Role of
lipid metabolism in cancer. Cell Metab. 31:62–76. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Luo X, Cheng C, Tan Z, Li N, Tang M, Yang
L and Cao Y: Emerging roles of lipid metabolism in cancer
metastasis. Mol Cancer. 16:762017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Santos CR and Schulze A: Lipid metabolism
in cancer. FEBS J. 279:2610–2623. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Rohrig F and Schulze A: The multifaceted
roles of fatty acid synthesis in cancer. Nat Rev Cancer.
16:732–749. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Geng F, Cheng X, Wu X, Yoo JY, Cheng C,
Guo JY, Mo X, Ru P, Hurwitz B and Kim SH: Inhibition of SOAT1
suppresses glioblastoma growth via blocking SREBP-1-Mediated
lipogenesis. Clin Cancer Res. 22:5337–5348. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gopal K, Grossi E, Paoletti P and Usardi
M: Lipid composition of human intracranial tumors: A biochemical
study. Acta Neurochir (Wien). 11:333–347. 1963. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Currie E, Schulze A, Zechner R, Walther TC
and Farese RV Jr: Cellular fatty acid metabolism and cancer. Cell
Metab. 18:153–161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shimano H and Sato R: SREBP-regulated
lipid metabolism: Convergent physiology-divergent pathophysiology.
Nat Rev Endocrinol. 13:710–730. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Schlosser HA, Drebber U, Urbanski A, Haase
S, Baltin C, Berlth F, Neiss S, von Bergwelt-Baildon M, Fetzner UK,
Warnecke-Eberz U, et al: Glucose transporters 1, 3, 6, and 10 are
expressed in gastric cancer and glucose transporter 3 is associated
with UICC stage and survival. Gastric Cancer. 20:83–91. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sharen G, Peng Y, Cheng H, Liu Y, Shi Y
and Zhao J: Prognostic value of GLUT-1 expression in pancreatic
cancer: Results from 538 patients. Oncotarget. 8:19760–19767. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sun HW, Yu XJ, Wu WC, Chen J, Shi M, Zheng
L and Xu J: GLUT1 and ASCT2 as predictors for prognosis of
hepatocellular carcinoma. PLoS One. 11:e01689072016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Williams NC and O'Neill LAJ: A Role for
the krebs cycle intermediate citrate in metabolic reprogramming in
innate immunity and inflammation. Front Immunol. 9:1412018.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Mancini R, Noto A, Pisanu ME, De Vitis C,
Maugeri-Saccà M and Ciliberto G: Metabolic features of cancer stem
cells: The emerging role of lipid metabolism. Oncogene.
37:2367–2378. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang T, Fahrmann JF, Lee H, Li YJ,
Tripathi SC, Yue C, Zhang C, Lifshitz V, Song J, Yuan Y, et al:
JAK/STAT3-Regulated fatty Acid β-oxidation is critical for breast
cancer stem cell self-renewal and chemoresistance. Cell Metab.
27:136–150.e5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Li J, Condello S, Thomes-Pepin J, Ma X,
Xia Y, Hurley TD, Matei D and Cheng JX: Lipid Desaturation is a
metabolic marker and therapeutic target of ovarian cancer stem
cells. Cell Stem Cell. 20:303–314.e5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Brandi J, Dando I, Pozza ED, Biondani G,
Jenkins R, Elliott V, Park K, Fanelli G, Zolla L, Costello E, et
al: Proteomic analysis of pancreatic cancer stem cells: Functional
role of fatty acid synthesis and mevalonate pathways. J Proteomics.
150:310–322. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chen CL, Uthaya Kumar DB, Punj V, Xu J,
Sher L, Tahara SM, Hess S and Machida K: NANOG metabolically
reprograms tumor-initiating Stem-like cells through tumorigenic
changes in oxidative phosphorylation and fatty acid metabolism.
Cell Metab. 23:206–219. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ito K, Carracedo A, Weiss D, Arai F, Ala
U, Avigan DE, Schafer ZT, Evans RM, Suda T, Lee CH and Pandolfi PP:
A PML-PPAR-δ pathway for fatty acid oxidation regulates
hematopoietic stem cell maintenance. Nat Med. 18:1350–1358. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Beloribi-Djefaflia S, Vasseur S and
Guillaumond F: Lipid metabolic reprogramming in cancer cells.
Oncogenesis. 5:e1892016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Tirinato L, Liberale C, Di Franco S,
Candeloro P, Benfante A, La Rocca R, Potze L, Marotta R, Ruffilli
R, Rajamanickam VP, et al: Lipid droplets: A new player in
colorectal cancer stem cells unveiled by spectroscopic imaging.
Stem Cells. 33:35–44. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
de Gonzalo-Calvo D, López-Vilaró L,
Nasarre L, Perez-Olabarria M, Vázquez T, Escuin D, Badimon L,
Barnadas A, Lerma E and Llorente-Cortés V: Intratumor cholesteryl
ester accumulation is associated with human breast cancer
proliferation and aggressive potential: A molecular and
clinicopathological study. BMC Cancer. 15:4602015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yue S, Li J, Lee SY, Lee HJ, Shao T, Song
B, Cheng L, Masterson TA, Liu X, Ratliff TL and Cheng JX:
Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT
activation underlies human prostate cancer aggressiveness. Cell
Metab. 19:393–406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yasumoto Y, Miyazaki H, Vaidyan LK, Kagawa
Y, Ebrahimi M, Yamamoto Y, Ogata M, Katsuyama Y, Sadahiro H, Suzuki
M and Owada Y: Inhibition of fatty acid synthase decreases
expression of stemness markers in glioma stem cells. PLoS One.
11:e01477172016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Li H, Feng Z and He ML: Lipid metabolism
alteration contributes to and maintains the properties of cancer
stem cells. Theranostics. 10:7053–7069. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Clémot M, Sênos Demarco R and Jones DL:
Lipid mediated regulation of adult stem cell behavior. Front Cell
Dev Biol. 8:1152020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Castro LF, Wilson JM, Goncalves O,
Galante-Oliveira S, Rocha E and Cunha I: The evolutionary history
of the stearoyl-CoA desaturase gene family in vertebrates. BMC Evol
Biol. 11:1322011. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yi M, Li J, Chen S, Cai J, Ban Y, Peng Q,
Zhou Y, Zeng Z, Peng S, Li X, et al: Emerging role of lipid
metabolism alterations in Cancer stem cells. J Exp Clin Cancer Res.
37:1182018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Colacino JA, McDermott SP, Sartor MA,
Wicha MS and Rozek LS: Transcriptomic profiling of curcumin-treated
human breast stem cells identifies a role for stearoyl-coa
desaturase in breast cancer prevention. Breast Cancer Res Treat.
158:29–41. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Lai KKY, Kweon SM, Chi F, Hwang E, Kabe Y,
Higashiyama R, Qin L, Yan R, Wu RP, Lai K, et al: Stearoyl-CoA
desaturase promotes liver fibrosis and tumor development in mice
via a wnt positive-signaling loop by stabilization of low-density
lipoprotein-receptor-related proteins 5 and 6. Gastroenterology.
152:1477–1491. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Bansal S, Berk M, Alkhouri N, Partrick DA,
Fung JJ and Feldstein A: Stearoyl-CoA desaturase plays an important
role in proliferation and chemoresistance in human hepatocellular
carcinoma. J Surg Res. 186:29–38. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Mason P, Liang B, Li L, Fremgen T, Murphy
E, Quinn A, Madden SL, Biemann HP, Wang B, Cohen A, et al: SCD1
inhibition causes cancer cell death by depleting mono-unsaturated
fatty acids. PLoS One. 7:e338232012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Pandey PR, Xing F, Sharma S, Watabe M, Pai
SK, Iiizumi-Gairani M, Fukuda K, Hirota S, Mo YY and Watabe K:
Elevated lipogenesis in epithelial stem-like cell confers survival
advantage in ductal carcinoma in situ of breast cancer. Oncogene.
32:5111–5122. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Sun Y, He W, Luo M, Zhou Y, Chang G, Ren
W, Wu K, Li X, Shen J, Zhao X and Hu Y: SREBP1 regulates
tumorigenesis and prognosis of pancreatic cancer through targeting
lipid metabolism. Tumour Biol. 36:4133–4141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Raulien N, Friedrich K, Strobel S, Rubner
S, Baumann S, von Bergen M, Körner A, Krueger M, Rossol M and
Wagner U: Fatty acid oxidation compensates for
lipopolysaccharide-induced warburg effect in glucose-deprived
monocytes. Front Immunol. 8:6092017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Carracedo A, Cantley LC and Pandolfi PP:
Cancer metabolism: Fatty acid oxidation in the limelight. Nat Rev
Cancer. 13:227–232. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Mullen PJ, Yu R, Longo J, Archer MC and
Penn LZ: The interplay between cell signalling and the mevalonate
pathway in cancer. Nat Rev Cancer. 16:718–731. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Ginestier C, Monville F, Wicinski J,
Cabaud O, Cervera N, Josselin E, Finetti P, Guille A, Larderet G,
Viens P, et al: Mevalonate metabolism regulates Basal breast cancer
stem cells and is a potential therapeutic target. Stem Cells.
30:1327–1337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Mak D and Kramvis A: Epidemiology and
aetiology of hepatocellular carcinoma in Sub-Saharan Africa.
Hepatoma Res. 7:392021.
|
|
88
|
Afify SM, Sanchez Calle A, Hassan G, Kumon
K, Nawara HM, Zahra MH, Mansour HM, Khayrani AC, Alam MJ, Du J, et
al: A novel model of liver cancer stem cells developed from induced
pluripotent stem cells. Br J Cancer. 122:1378–1390. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Song K, Kwon H, Han C, Zhang J, Dash S,
Lim K and Wu T: Active glycolytic metabolism in CD133(+)
hepatocellular cancer stem cells: Regulation by MIR-122.
Oncotarget. 6:40822–40835. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Rawla P, Sunkara T and Barsouk A:
Epidemiology of colorectal cancer: Incidence, mortality, survival,
and risk factors. Prz Gastroenterol. 14:89–103. 2019.PubMed/NCBI
|
|
91
|
Vázquez-Iglesias L, Barcia-Castro L,
Rodríguez-Quiroga M, Páez de la Cadena M, Rodríguez-Berrocal J and
Cordero OJ: Surface expression marker profile in colon cancer cell
lines and sphere-derived cells suggests complexity in CD26+ cancer
stem cells subsets. Biology Open. 8:bio0416732019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Chen KY, Liu X, Bu P, Lin CS, Rakhilin N,
Locasale JW and Shen X: A metabolic signature of colon cancer
initiating cells. Annu Int Conf IEEE Eng Med Biol Soc.
2014:4759–4762. 2014.PubMed/NCBI
|
|
93
|
Huang B, Li X, Li Y, Zhang J, Zong Z and
Zhang H: Current immunotherapies for glioblastoma multiforme. Front
Immunol. 11:6039112021. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Menard JA, Christianson HC, Kucharzewska
P, Bourseau-Guilmain E, Svensson KJ, Lindqvist E, Indira Chandran
V, Kjellén L, Welinder C, Bengzon J, et al: Metastasis stimulation
by hypoxia and acidosis-induced extracellular lipid uptake is
mediated by proteoglycan-dependent endocytosis. Cancer Res.
76:4828–4840. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Rawla P, Sunkara T and Gaduputi V:
Epidemiology of pancreatic cancer: Global trends, etiology and risk
factors. World J Oncol. 10:10–27. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Li C, Heidt DG, Dalerba P, Burant CF,
Zhang L, Adsay V, Wicha M, Clarke MF and Simeone DM: Identification
of pancreatic cancer stem cells. Cancer Res. 67:1030–1037. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Arasanz H, Hernández C, Bocanegra A,
Chocarro L, Zuazo M, Gato M, Ausin K, Santamaría E,
Fernández-Irigoyen J, Fernandez G, et al: Profound reprogramming
towards stemness in pancreatic cancer cells as adaptation to AKT
inhibition. Cancers. 12:21812020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Bian Y, Yu Y, Wang S and Li L:
Up-regulation of fatty acid synthase induced by EGFR/ERK activation
promotes tumor growth in pancreatic cancer. Biochem Biophys Res
Commun. 463:612–617. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Visweswaran M, Arfuso F, Warrier S and
Dharmarajan A: Aberrant lipid metabolism as an emerging therapeutic
strategy to target cancer stem cells. Stem Cells. 38:6–14. 2020.
View Article : Google Scholar : PubMed/NCBI
|