Germ cell tumors (GCTs) are a heterogeneous group of
neoplasms arising in the gonads, both the ovaries and the testes.
Due to the migration of primordial germ cells along the midline of
the body, GCTs can also arise in extragonadal sites and the brain
(1). Testicular germ cell tumors
(TGCTs) are rare solid tumors that account for 1% of cancers in
men. However, in young males between the ages of 15 and 44 years,
TGCTs represent the most frequent solid malignancy and have the
highest mortality (2). The
incidence rates of TGCTs have been steadily rising in young men
(3). In general, TGCT treatment by
cisplatin-based therapy is highly successful, even when the disease
is highly metastatic (4).
Unfortunately, acquired resistance to chemotherapy is the major
barrier to curing patients with refractory disease and results in
poor outcomes. Approximately 50% of these patients die from
progressive disease (5). The
mechanism of cisplatin resistance in TGCTs remains unknown,
although some mechanisms have been proposed (6–9).
It has been previously shown that polyadenosine
diphosphate-ribose polymerase (PARP) is overexpressed in TGCTs
compared to normal testis (10).
PARPs represent a family of 17 enzymes associated with several
cellular processes, such as DNA repair, genome maintenance and cell
death (11). The most well-studied
member of the PARP family is PARP1, which has a key role in the
detection of single-strand DNA breaks and repair initiation
(12). Moreover, more than 80% of
overall PARP activity is constituted by PARP1, which has also been
identified as a platinum-DNA damage response protein (13). Therefore, there is a strong
rationale to target the enzymatic activity of PARPs and use PARP
inhibitors (PARPi) as a new therapeutic strategy in the treatment
of cancer.
PARPi are a class of anticancer drugs that compete
with nicotinamide for the catalytically active sites of PARP
molecules. PARP inhibition proved to be a successful strategy in
the treatment of homologous recombination repair (HRR)-deficient
tumors, especially tumors with mutations in the essential HR genes
BRCA1 and BRCA2 (14–17).
This synthetic lethal interaction between PARP inhibition and
mutations in BRCA1 or BRCA2 was discovered by two
independent research groups in 2005 (18,19).
Olaparib, rucaparib, niraparib, talazoparib, pamiparib, and
veliparib are PARPi used in the clinic or under investigation in
several trials (20). The initial
clinical development of PARPi was based on potentiating tumor cell
killing by DNA-damaging agents such as platinum-based
chemotherapeutics. A synergistic effect of cisplatin and PARP
inhibition was shown in three human esophageal cancer cell lines
(21). Another drug combination
experiment revealed that the PARPi olaparib and veliparib
potentiated the killing of non-small cell lung cancer cells by
cisplatin (22). The combination
of veliparib with cisplatin or carboplatin increased the
recurrence-free and overall survival of a genetically engineered
mouse model for BRCA1-associated breast cancer (23). Another study showed synergistic
cytotoxicity of olaparib and cisplatin against BRCA2-deficient
mammary tumor cells (24). Many
clinical trials have been confirmed that the effect of PARP
inhibition observed preclinically could be recapitulated in
patients (25–32).
In this study, we hypothesized that the PARPi
veliparib would synergistically increase the cytotoxicity of
platinum-based drugs and reverse cisplatin resistance in refractory
GCTs. We used a series of cisplatin-resistant variants (referred to
as CisR throughout the manuscript) cell line models and analyzed
the expression of PARP in these cell lines and derived xenografts
as well as their sensitivity to veliparib, and we performed
combined treatment with cisplatin and carboplatin.
Chemicals were purchased from Sigma-Aldrich if not
stated otherwise.
Cell lysates were prepared using RIPA buffer (cat.
no. 9806; Cell Signaling Technology, Inc.) supplemented with
PhosSTOP phosphatase inhibitor (Roche) and cOmplete protease
inhibitor (Roche) and centrifuged for 10 min at 14,000 × g at 4°C,
and protein concentrations were determined by a PierceTM
BCA Protein Assay kit (Thermo Fisher Scientific, Inc.). Protein
extracts were resolved to a final concentration of 30 µg protein
per sample. Protein electrophoretic separation was performed on a
7.5% dodecyl sulfate polyacrylamide gel (SDS-PAGE) and transferred
onto a nitrocellulose membrane (Thermo Fisher Scientific, Inc.).
Subsequently, the membrane was blocked overnight at 4°C in 5%
nonfat dry milk in Tris-buffered saline (TBS). The next day, the
membrane was incubated for 1 h at room temperature with primary
anti-PARP1 antibody (ab137653; Abcam; dilution 1:1,000, 113 kDa).
The membrane was washed in Tris buffer saline with Tween-20 (TBS-T)
and reacted for 1 h at room temperature with horse peroxidase
secondary anti-rabbit antibody (cat. no. 7074S; Cell Signaling
Technology, Inc.; dilution 1:1,000). The bands were viewed by Super
Signal™ West Dura Extended Duration chemiluminescence
detection substrate (Thermo Fisher Scientific, Inc.). Finally,
signals were visualized by a Li-Cor scanner (Image Studio™ Lite
Software). The same membrane was washed in TBS-T and incubated for
1 h at room temperature with β-actin primary antibody (A1978;
Sigma-Aldrich; dilution 1:4,000, 42 kDa). Subsequently, the
membrane was washed again and reacted for 1 h at room temperature
with horse peroxidase secondary anti-mouse antibody (cat. no.
7076S; Cell Signaling Technology, Inc.; dilution 1:2,000). The
signals were visualized in the same way. Western blotting analysis
was repeated three times. The ImageJ 1.46r program (NIH) was used
to measure PARP1 and β-actin densities for densitometric analysis,
and the obtained data were statistically processed.
Six- to 8-week-old SCID beige mice (CD17 Cg-Prkdscid
Lystbg/Crl, Charles River) or NSG mice (The Jackson Laboratory)
were used in accordance with institutional guidelines under
approved protocols. Project was performed in the Animal Facility
for Immunodeficient Mice of the Biomedical Research Center SAS
Bratislava (license no. SK UCH 02017). Project was approved by the
Institutional Ethic Committee of the Biomedical Research Center SAS
Bratislava and by State Veterinary and Food Administration of the
Slovak Republic as the national competence authority under the
registration no. Ro 1030/18-221 in compliance with the Directive
2010/63/EU and the Regulation 377/2012 on the protection of animals
used for scientific purposes. Sacrifice of the animals at the
experiment endpoint was done by induction of anaesthesia by
3.0-3.5% isoflurane for 7–10 min. Signs of muscle relaxation and
loss of consciousness were observed. The deep aneasthesia was
followed by cervical dislocation and the death of animals was
determined as the absence of a corneal reflex, failure to detect
respiration, and the absence of a heartbeat for a period of more
than 5 min to confirm death. The largest long diameter of the
xenograft in this study reached 14.6 mm and the largest short
diameter reached 10.4 mm (for details see the respective Results
subsection). Total number of animals used for this study was
n=22.
To produce GCT cell line xenografts for PARP1
immunohistochemical analysis, a suspension of 2×106 of
NTERA-2, NTERA-2 CisR, NCCIT, NCCIT CisR, TCam-2, TCam-2 CisR,
JEG-3, JEG-3 CisR, NOY-1 and NOY-1 CisR cells in 100 µl of
extracellular matrix (ECM) mixture 1:1 (50 µl serum-free DMEM or
RPMI medium, 50 µl ECM) was injected bilaterally s.c. into each of
the flank of NSG mice (n=2 animals per cell line pair, total n=10).
Xenografts were measured by caliper and animals were sacrificed at
the point when the tumors exceeded 1 cm in diameter. One
representative tumor xenograft was used for IHC staining, details
are listed in the results section.
Slides were deparaffinized, rehydrated and immersed
in phosphate buffered saline solution (10 mM, pH 7.2). No epitope
retrieval was applied to the slides. The slides were incubated for
2 h with primary anti-PARP1 monoclonal antibody (H-250, dilution
1:500; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and
immunostained using anti-mouse/anti-rabbit secondary antibody
(Nichirei Biosciences, Japan) for 30 min according to the
manufacturer's instructions. The reaction was visualized by
diaminobenzidine substrate-chromogen solution (DAB, Dako) for 5
min. Ultimately, the slides were counterstained with hematoxylin.
We used normal tissue as a positive control, with lymphatic tissue
as a positive control for nuclear PARP (40). As a negative control, lymphatic
tissue was used, omitting the primary antibody from the staining
protocol. Representative images were captured with an Olympus BX40
microscope (Olympus Corporation) and a Canon EOS 1000D (Canon
Inc.).
PARP1 expression was scored by the multiplicative
quickscore (QS) method, evaluating both the percentage of positive
cells and the staining intensity of the nuclei. Briefly, the
proportion of cells with nuclear staining was multiplied by the
intensity of staining to provide a final quickscore. The average
intensity of the positively stained cells was given a score from 1
to 3 (1=weak, 2=intermediate, and 3=strong staining). QS was
calculated as follows: QS=(1 × percentage weakly stained) + (2 ×
percentage moderately stained) + (3 × percentage strongly stained)
(10).
For the statistical analysis of studies involving
comparisons between the two groups, the assumption of normality was
tested using the Shapiro-Wilk test, and differences were assessed
by Student's t-test or the Mann-Whitney U test depending on the
normality of the data. For comparisons involving more than two
groups, when the effect of the tested drugs in vivo was analyzed,
multivariate analysis one-way ANOVA Tukey's Honest Significant
Difference test was used. Multiple comparisons were not performed
in cases where the overall test did not indicate significant
differences across samples. GraphPad Prism® software
(GraphPad Inc.) was used. P-values <0.05 were considered
statistically significant.
Five GCT cell lines and their cisplatin-resistant
variants, which have been previously characterized (33–35),
were used in this study. NTERA-2 and NCCIT represent pluripotent
embryonal carcinoma (EC) cell lines, JEG-3 is a choriocarcinoma
(ChC) cell line, and NOY-1 was derived from ovarian yolk sac tumor
(YST). The only available seminoma (SE) model is the TCam-2 cell
line, which was also included in our analyses.
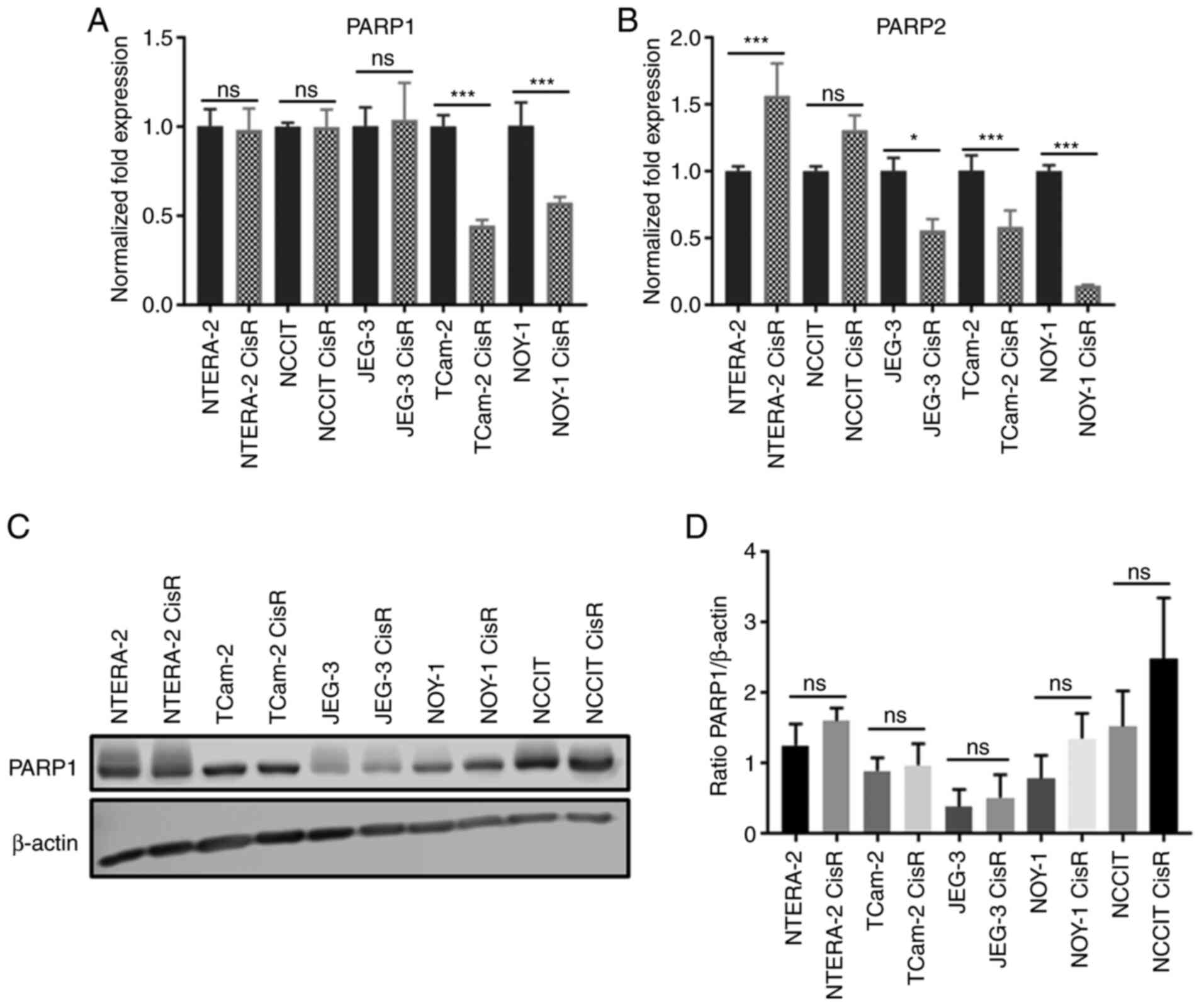
PARP1 and PARP2 gene expression was analyzed in all
five GCT cell lines and their cisplatin-resistant variants using
quantitative RT-PCR. We observed significant downregulation of
PARP1 in TCam-2 CisR and NOY-1 CisR cells compared to parental cell
lines (Fig. 1A). Similarly, a
significant decrease in PARP2 expression was detected in these two
cell lines. PARP2 downregulation was also observed in the JEG-3
CisR cell line. Only cisplatin-resistant NTERA-2 CisR cells showed
significant PARP2 overexpression compared to sensitive cells
(Fig. 1B). Next, we investigated
PARP1 protein levels using Western blot and densitometric analyses.
Western blot analysis revealed the highest levels of PARP1 in the
EC cell lines NTERA-2 and NCCIT and their resistant variants. Only
low levels of PARP1 protein were observed in the ChC cell lines
JEG-3 and JEG-3 CisR (Fig. 1C).
However, no significant changes between parental and resistant cell
lines were detected by densitometric analysis (Fig. 1D).
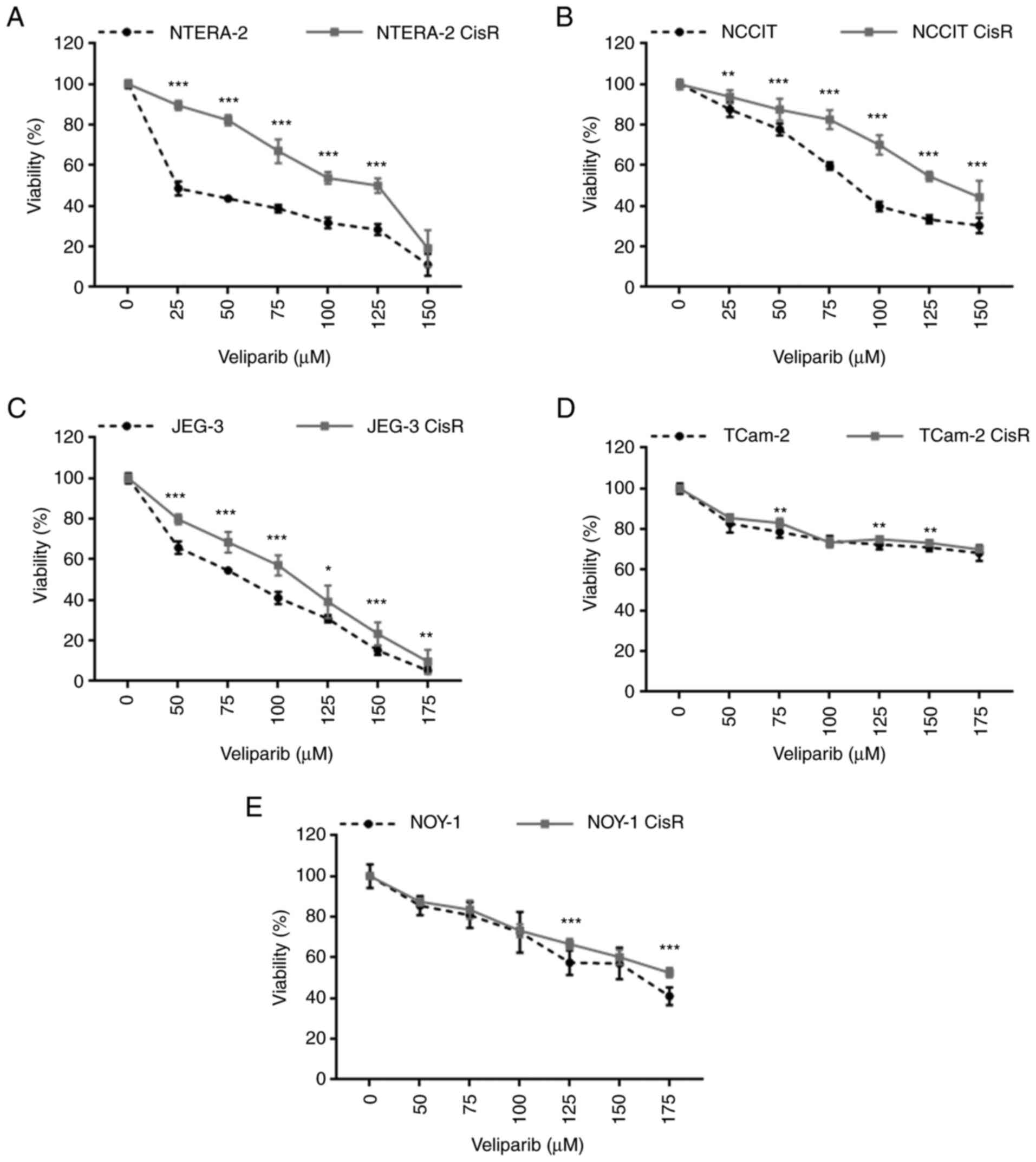
To evaluate the therapeutic potential of PARP
inhibition, we tested the sensitivity of parental and
chemoresistant GCT cell lines to veliparib, an oral PARP1/2
inhibitor. Dose-dependent cytotoxic effects of veliparib were
observed in all cell lines, but they were not prominent in the
TCam-2 pair. The cisplatin-resistant EC cell lines NTERA-2 CisR and
NCCIT CisR were significantly more resistant to veliparib treatment
than the parental cells (Fig. 3A and
B). Similarly, the ChC cell line JEG-3 CisR was significantly
more resistant to this treatment (Fig.
3C). The sensitivity of TCam-2 and NOY-1 cell line pairs to
veliparib was comparable. Resistance to this inhibitor was observed
only at some concentrations (Fig. 3D
and E).
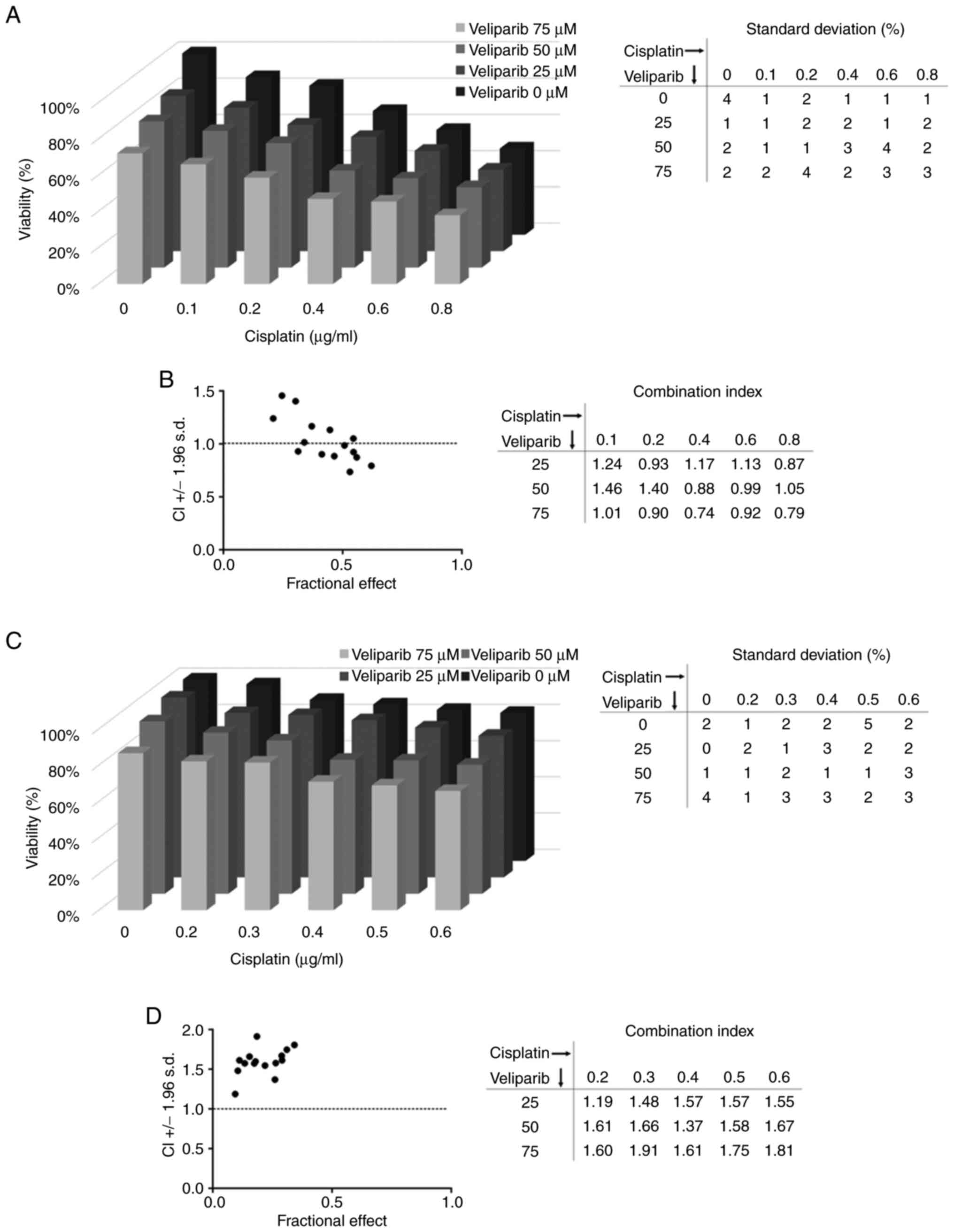
To test whether the PARP inhibitor veliparib could
sensitize chemoresistant cells to cisplatin treatment and yield
synergy, we treated NTERA-2 CisR and NCCIT CisR cells with this
combination. We observed some synergistic effects in NTERA-2 CisR
cells, and the most significant changes were detected when the
highest tested concentration of veliparib was used. The viability
of NTERA-2 CisR cells was decreased by 13% upon treatment with 0.1
µg/ml cisplatin alone, but the combination of 0.1 µg/ml cisplatin
and 75 µM veliparib achieved a 34% reduction in cell viability
(Fig. 4A). The combination index
(CI) was also below 1, indicating a synergistic effect of veliparib
and cisplatin (Fig. 4B). However,
veliparib did not sensitize the NCCIT CisR cell line to cisplatin
treatment (Fig. 4C), and CI above
1 confirmed antagonism of the veliparib and cisplatin combination
in these cells (Fig. 4D).
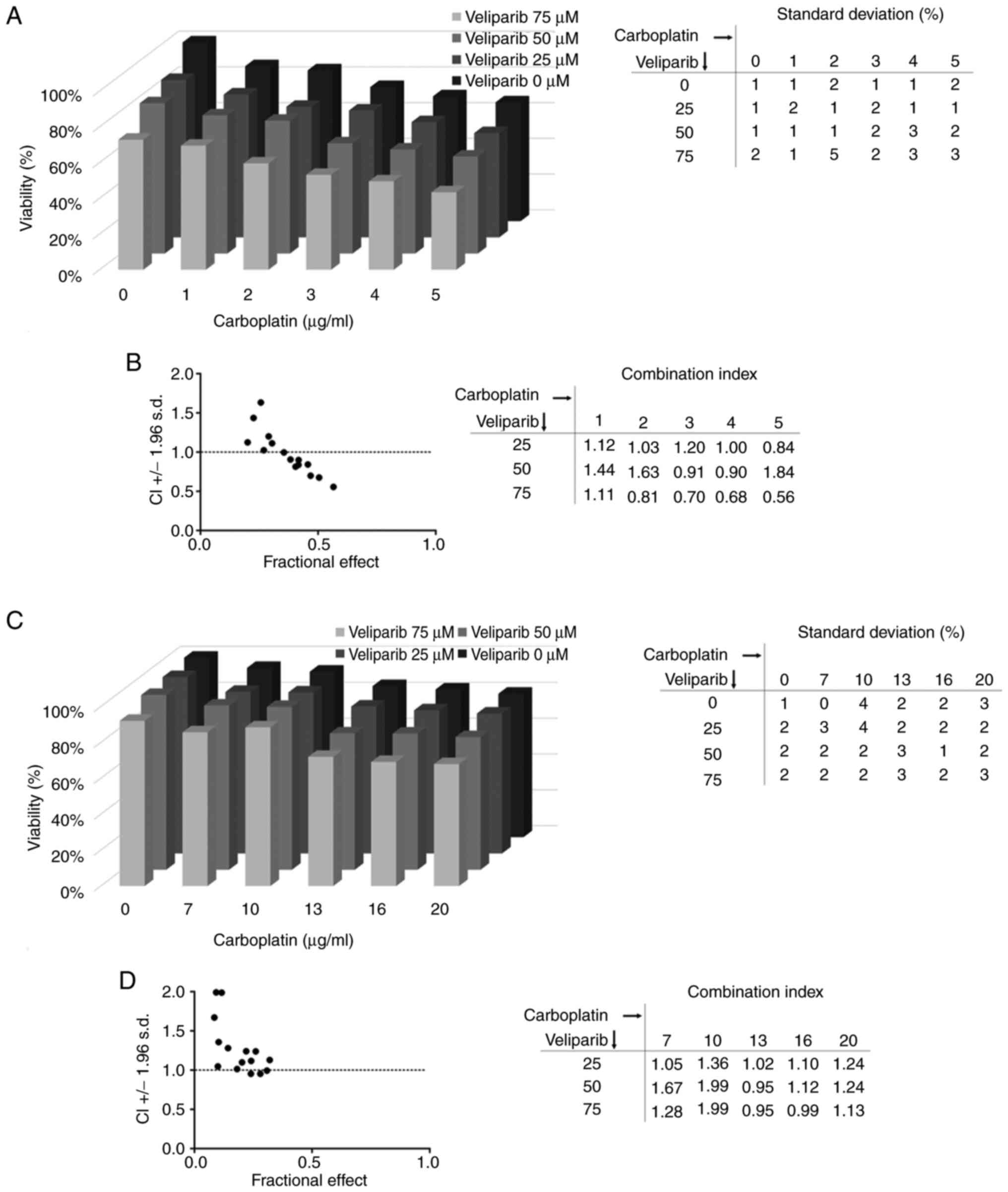
We also tested another platinum-based chemotherapy
drug, carboplatin, and similar results were obtained for both
tested cell lines. Carboplatin alone (1 µg/ml) decreased the
viability of NTERA-2 CisR cells by 13%, but the combination with 75
µM veliparib induced a 30% reduction in cell viability (Fig. 5A). The combination index was again
below 1, indicating synergy of veliparib and carboplatin
combination (Fig. 5B). An
antagonistic effect of this combination was observed in the NCCIT
CisR cell line (Fig. 5C and
D).
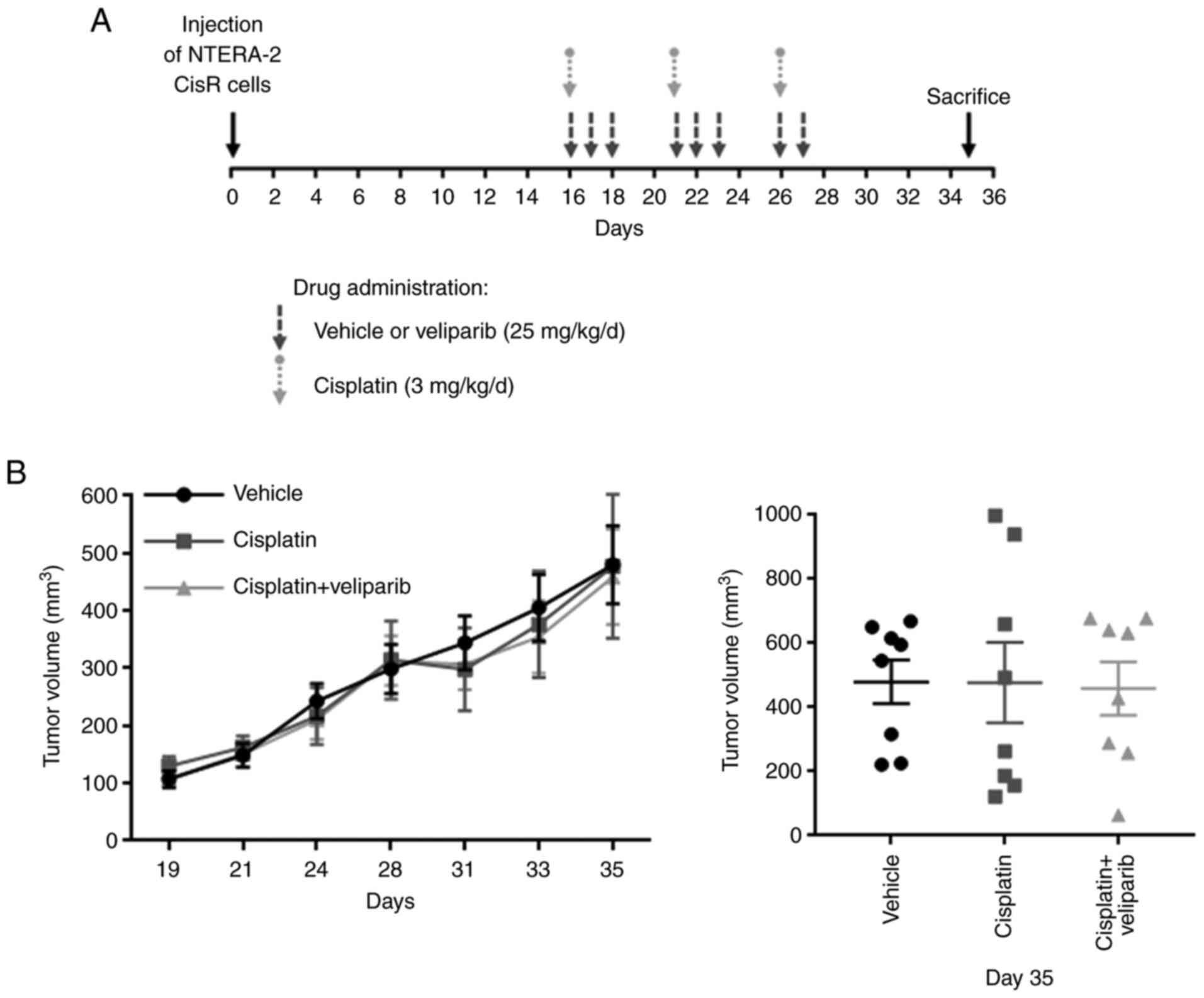
To evaluate the effects of PARP inhibition by
veliparib on NTERA-2 CisR tumor growth, we performed an in
vivo experiment using immunodeficient SCID beige mice. NTERA-2
CisR cells were injected s.c. into mouse flanks to produce tumor
xenografts and the animals were divided into 3 treatment groups: i)
untreated control/vehicle (n=4); ii) cisplatin-3 mg/kg/d (n=4); and
iii) cisplatin + veliparib-25 mg/kg/d (n=4). The treatment started
on Day 16 when all xenografts were palpable (Fig. 6A). Tumor growth was not affected by
cisplatin or the combination treatment, and statistical analysis
did not reveal any significant differences between these three
groups (Fig. 6B). Raw measurements
for the tumors are given in Table
SIII.
The combination of PARPi and polymerase theta (POLθ)
inhibitors was recently shown to overcome PARPi resistance and
increase the cytotoxic effects of PARPi (41,42).
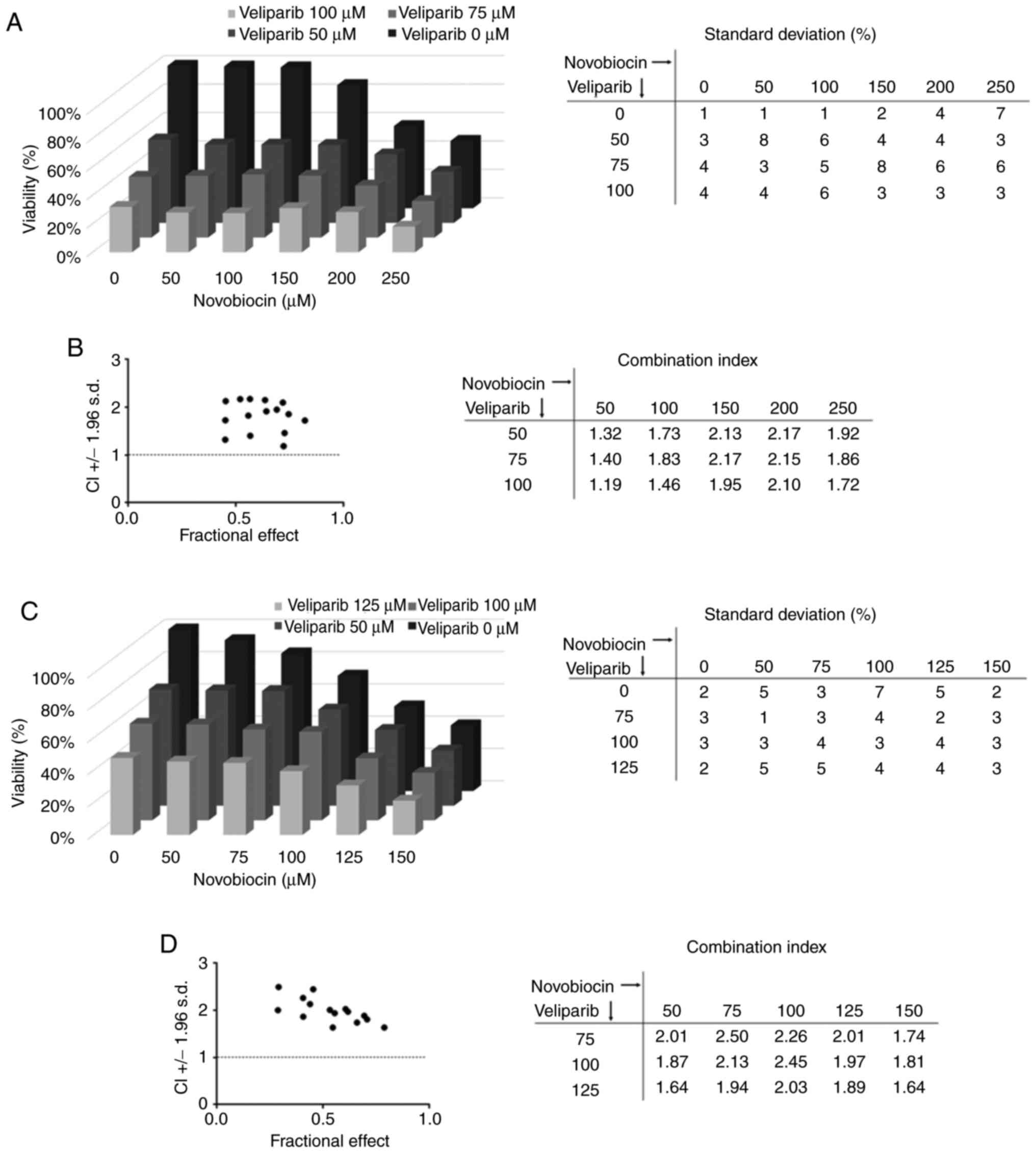
To test whether we could achieve a synergistic effect of this type
of combination we used the antibiotic novobiocin, a specific POLθ
inhibitor (41), combined with the
PARPi veliparib. We treated NTERA-2 CisR cells with this
combination and did not observe any synergy (Fig. 7A). The combination index was above
1, indicating an antagonistic effect of veliparib and novobiocin
(Fig. 7B). Similar results were
observed for other resistant EC cell line, where novobiocin did not
sensitize NCCIT CisR cells to veliparib treatment (Fig. 7C and D).
The enzymes PARP1 and PARP2 have overlapping
functions in the DNA damage response pathway (43,44),
but they differ in their substrate preference (45). PARP1- and PARP2-deficient mice
display postreplicative genomic instability, whereas doubleknockout
mice exhibit lethal genomic instability (46). Our group previously showed that
PARP is overexpressed in TGCTs. Increased PARP expression was
present in early precursors of TGCTs-intratubular germ cell
neoplasia unclassified- and in less differentiated histological
subtypes, such as EC and SE. Its expression decreases with
subsequent tumor tissue differentiation toward choriocarcinoma
(10). Maximal PARP activity was
correlated with PARP1 protein expression in EC cell lines (47). The levels of PARP1 and
poly(ADP-ribosyl)ation were heterogeneous among germ cell tumor
cell lines (48). Expression
analysis confirmed the expression of both PARP1 and
PARP2 in all GCT parental cell lines; however, only in EC
NTERA-2 CisR and NCCIT CisR cell lines the expression on mRNA level
remained or increased. Protein levels of PARP1 did not
significantly differ between parental and resistant cells, however
the lowest level was in ChC cell line pair JEG-3/JEG-3 CisR, which
was also confirmed by IHC on the xenografts. The highest PARP1
protein level was observed in EC cell lines NTERA-2 and NCCIT, as
also visible in IHC analysis. We detected substantial amount of
protein PARP1 in YST cell line pair NOY-1/NOY-1 CisR and seminoma
cell line model TCam-2/TCam-2 CisR pair with corresponding positive
staining in xenografts. Poor correlations between the level of mRNA
and the level of protein were often observed; and may be attributed
to many complex post-transcriptional mechanisms involved in protein
synthesis; proteins may also differ in their in vivo
half-lives; and/or there is also technical difference in protein
and mRNA experiments. Altogether, these results are in line with
our previously published data from patient samples of GCT
demonstrating high PARP1 level in GCT in comparison to healthy
tissue (10).
Clinically approved PARPi vary in their
effectiveness in trapping PARP onto DNA (from the most to the least
potent): talazoparib >> niraparib > olaparib=rucaparib
>> veliparib (49). They are
effective in a synthetically lethal interaction against
HRR-deficient tumors, such as BRCA1/2-mutated tumors (50). PARPi have been approved for the
treatment of breast and metastatic pancreatic cancer and metastatic
castration-resistant prostate cancer (51–55).
Veliparib was tested as a single agent or in combination with
standard chemotherapeutic drugs and markedly improved the
therapeutic efficiency in breast, ovarian and lung cancer (56). A phase I study of veliparib with
cisplatin and vinorelbine showed increased response rates in
advanced triple-negative breast cancer and/or BRCA-mutated breast
cancer (57). In a phase III,
randomized, placebo-controlled BROCADE3 trial, the addition of
veliparib to carboplatin and paclitaxel improved progression-free
survival (PFS) in patients with advanced HER2-negative germline
BRCA1/2-mutated breast cancer (58). The combination of veliparib plus
carboplatin and etoposide demonstrated improved PFS as first-line
treatment in patients with extensive-stage small cell lung cancer
(59). Promising antitumor
activity was also observed in patients with metastatic or advanced
non-small cell lung cancer receiving quadruple therapy with
veliparib, nivolumab, carboplatin and paclitaxel (60). Moreover, combination therapy with
veliparib plus carboplatin and gemcitabine demonstrated promising
PFS and response rates in ovarian cancer patients with germline
BRCA mutations (30).
In GCT model tumor cell lines, olaparib reduced cell
viability in the EC cell lines NCCIT, NTera-2 and 2102Ep, while the
SE cell line TCam-2 was the least sensitive. A clonogenic assay
further confirmed the differential effect of olaparib in TCam-2
cells compared to that in the tested EC cell lines. Moreover, the
least responsive cell lines (NCCIT and TCam-2) exhibited the lowest
BRCA1 methylation levels, and high RAD51C and BRCA1 methylation was
observed in the two most sensitive cell lines (NTera-2 and 2102Ep).
Methylation levels correlated with the expression levels of both of
these targets. Altogether, these findings support the evidence that
promoter methylation of genes involved in HRR could serve as a
predictor of the therapeutic response to PARPi in TGCT patients
(61). Similarly, in the present
study, we observed dose-dependent cytotoxic effects of the PARPi
veliparib in all tested parental and cisplatin-resistant GCT cell
lines, but in the TCam-2 cell line pair, these dose-dependent
effects were not as profound. This cell line pair was also the
least sensitive to veliparib treatment.
Olaparib was able to enhance the toxicity of
cisplatin in combination in EC cells, and sensitivity correlated
with the levels of PARP activity (47). Combination therapy with olaparib
and cisplatin in two cisplatin-resistant EC cell lines, GCT27cis-r
and 2102Epcis-r, was efficient (62). Importantly, a phase II clinical
trial by De Giorgi et al (63) showed that olaparib as a single
agent had only marginal activity in heavily cisplatin-pretreated
and refractory GCT patients. However, an anecdotic 4-month stable
disease was observed in the only patient with a BRCA mutation. The
authors also suggested that future studies with olaparib should be
conducted in combination or following salvage chemotherapy in less
pretreated and more selected GCT patients (63).
At the time of the phase II GCT-SK-004 clinical
trial initiation and based on the data available, using veliparib
in the combination with carboplatin and gemcitabine in multiple
relapsed/refractory germ cell tumors seemed to be promising
strategy (32). At the time of
study initiation, other PARPi, such as olaparib or talazoparib,
were not available, therefore, we analyzed the effect of veliparib
also in our cell line models in vitro and in vivo to
corroborate the results to those from clinical trial. To analyze
the effect of combinatorial treatment with veliparib and cisplatin
or carboplatin, we selected two cisplatin-resistant EC cell lines,
NTERA-2 CisR and NCCIT CisR, as they exhibited high levels of PARP1
protein and were sensitive to veliparib treatment in a
dose-dependent manner. Synergistic effects of veliparib and
cisplatin or carboplatin were observed only in NTERA-2 CisR cells.
However, this combination failed to enhance the cytotoxic effect of
cisplatin in vivo, which is in line with the final results
of a phase II trial determining the efficacy and toxicity of
gemcitabine, carboplatin and veliparib, showing no additive
treatment value of veliparib for refractory GCTs (32). Nevertheless, veliparib still
remains valuable agent due to its different mechanism of action in
comparison to olaparib and talazoparib, and potential synergistic
effect with other treatments. Veliparib is a selective PARP1/2
inhibitor with relatively weak affinity, while olaparib and
talazoparib have relatively strong affinity. Veliparib mainly
selectively inhibits the activity of PARP without holding the PARP
protein to DNA damage repair intermediates (64). Meta-data analysis published
recently suggest activity of veliparib in combination with platinum
agent and chemotherapy in some breast cancer patients with germ
line BRCA mutations (65).
The majority of patients develop PARPi resistance
despite a good initial response; thus, the identification of
potential strategies to overcome these mechanisms could improve the
therapeutic outcome of refractory patients (66,67).
The most common cause is the restoration of HRR in HRR-deficient
tumors, mostly via reversion mutations (68) or epigenetic modifications (69) that induce the re-expression of the
BRCA1/2 protein. Another mechanism is stabilization of the
replication fork by nucleases followed by inhibition of DNA
replication fork degradation in BRCA1/2-deficient cells (70,71).
Several other mechanisms, including the upregulation of the drug
efflux transporter ABCB1 (P-glycoprotein) (72), inhibition of PARP trapping activity
(73) or overexpression of cell
cycle regulators (74), have been
proposed.
Several clinical trials are currently evaluating
possible therapeutic strategies that enhance PARPi sensitivity and
overcome or delay PARPi resistance; however, none of them are
targeting TGCTs. In solid tumors, PARPi were combined with ionizing
radiation (75,76), atezolizumab (77), inhibitors of the G2 checkpoint
kinase WEE1 adavosertib (78) and
AZD1775 (79), HSP90 inhibitor
(80), ATR/CHK1 inhibitors
(81,82) or epigenetic drugs (83,84).
Importantly, the effects of PARPi on the tumor microenvironment
could also pave the way for rational drug combination strategies.
PARPi upregulated expression of PD-L1 in breast cancer cell lines
and animal models. Consequently, anti-PD-L1 therapy resensitized
PARPi-treated cells to T-cell killing, and this combination showed
better therapeutic outcomes than either monotherapy in an in
vivo model (85).
Olaparib induced the differentiation, maturation and
antitumor activation of macrophages with subsequent activation of
the immune-suppressive signaling pathway. However, the combination
of PARPi and macrophage-targeting therapy induced a durable
reprogramming of the tumor microenvironment in triple-negative
breast cancer (86). Recently,
inhibitors of DNA polymerase theta (POLθ) were shown to have
synergistic effects with PARPi in the treatment of HRR-deficient
tumors. ART558, a selective inhibitor of POLθ, induced DNA damage
and synthetic lethality in BRCA1/2-mutated cancer cells and
enhanced the effects of olaparib (42). The specific POLθ inhibitor
novobiocin killed HRR-deficient breast and ovarian cancer cells
in vivo and in patient-derived xenografts. Moreover,
HRR-deficient tumor cells with acquired PARPi resistance were
sensitive to novobiocin in vitro and in vivo
(41). However, in our
experiments, novobiocin failed to exert a synergistic effect with
veliparib in NTERA-2 CisR and NCCIT CisR cells, suggesting that the
synergy will be missing in the subset of tumor cells with increased
proficiency in HRR (62).
In summary, we detected the presence of PARP1
protein in all analyzed GCT cell lines, but the levels were low in
ChC cell lines, which is in line with our previous observations in
clinical samples. GCT cell lines were sensitive to the PARPi
veliparib in a dose-dependent manner; only in the TCam-2 cell line
pair was this effect not as prominent. Moreover, the
cisplatin-resistant EC cell lines NTERA-2 CisR and NCCIT CisR and
the ChC JEG-3 CisR cell line were also more resistant to veliparib
treatment than the parental cells. We observed that veliparib
synergized with cisplatin or carboplatin in NTERA-2 CisR cells, but
this synergy was not confirmed in vivo. Neither combination
with the POLθ inhibitor novobiocin showed synergy. The limitations
of this study can be identified in the lack of direct comparison of
other PARPi such as olaparib or talazoparib, however these were not
available at the time of study initiation. The lack of
high-throughput sequencing of HRR genes in GCT cell lines and
absence PARP2 protein detection also represent study limitation.
Nevertheless, there is still a rationale to use PARPi in more
advanced models including other components of the tumor
microenvironment, as GCTs (cell lines, xenografts and also patient
tumor tissue (10) also showed
PARP1 positivity. Other therapeutic approaches, including
combination with anti-PD-L1 therapy or the use of other PARPi, need
to be tested to determine the therapeutic efficacy of PARPi
combinatorial therapy in GCTs.
The authors would like to thank Ms. Veronika Repaska
(Cancer Research Institute, Biomedical Research Center, Slovak
Academy of Sciences, Bratislava, Slovakia) for taking care of the
mice and Ms. Maria Dubrovcakova (Cancer Research Institute,
Biomedical Research Center, Slovak Academy of Sciences, Bratislava,
Slovakia) for her technical assistance.
The experimental work was supported by the Slovak Research and
Development Agency under contract no. APVV-20-0158 and the
Scientific Grant Agency of The Ministry of Education, Science,
Research and Sport of the Slovak Republic VEGA 2/0124/21 and VEGA
1/0043/18. The experiments mentioned in the studies were enabled
with the kind help and the financial support from the Cancer
Research Foundation (NVR UEO 1993-2021) and the League Against
Cancer (LPR UEO 1990-2021).
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
SS, NU, ZC, SH, KK, MC, LR, MV, LK and MM
contributed to the study conception and design. In vitro
analyses were performed by SS, MC and MV. Expression and western
blot analyses were performed by NU and KK. The preparation of
xenograft models and in vivo experiments were performed by
SS, LR and LK. ZC and SH performed immunohistochemical analysis of
PARP1 in xenograft models. Material preparation and collection of
raw data from measurements were performed by SS and LR, and data
analysis and evaluation were performed by LK and MM. The first
draft of the manuscript was written by SS, LK and MM, and all
authors commented on previous versions of the manuscript. LK and MM
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
This article does not contain any studies with human
participants performed by any of the authors. All procedures
performed in studies involving animals were in accordance with the
ethical standards of the institution or practice at which the
studies were conducted. Studies involving mice were approved by the
Ethic Committee of the Biomedical Research Center, Slovak Academy
of Sciences (Dubravska cesta 9, Bratislava, Slovakia) and by the
national competence authority (State Veterinary and Food
Administration of the Slovak Republic), registration No. Ro
1976/17-221, in compliance with Directive 2010/63/EU of the
European Parliament and the European Council and Regulation
377/2012 for the protection of animals used for scientific
purposes.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Oosterhuis JW and Looijenga LHJ: Human
germ cell tumours from a developmental perspective. Nat Rev Cancer.
19:522–537. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cai Q, Chen Y, Zhang D, Pan J, Xie Z, Xu
C, Li S, Zhang X, Gao Y, Hou J, et al: Estimates of over-time
trends in incidence and mortality of testicular cancer from 1990 to
2030. Transl Androl Urol. 9:182–195. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alsdorf W, Seidel C, Bokemeyer C and Oing
C: Current pharmacotherapy for testicular germ cell cancer. Expert
Opin Pharmacother. 20:837–850. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Feldman DR, Patil S, Trinos MJ, Carousso
M, Ginsberg MS, Sheinfeld J, Bajorin DF, Bosl GJ and Motzer RJ:
Progression-free and overall survival in patients with
relapsed/refractory germ cell tumors treated with single-agent
chemotherapy: Endpoints for clinical trial design. Cancer.
118:981–986. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schmidtova S, Kalavska K and Kucerova L:
Molecular mechanisms of cisplatin chemoresistance and its
circumventing in testicular germ cell tumors. Curr Oncol Rep.
20:882018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lobo J, Jeronimo C and Henrique R:
Cisplatin resistance in testicular germ cell tumors: Current
challenges from various perspectives. Cancers (Basel). 12:16012020.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Singh R, Fazal Z, Freemantle SJ and
Spinella MJ: Mechanisms of cisplatin sensitivity and resistance in
testicular germ cell tumors. Cancer Drug Resist. 2:580–594.
2019.PubMed/NCBI
|
|
9
|
Al-Obaidy KI, Chovanec M and Cheng L:
Molecular characteristics of testicular germ cell tumors:
Pathogenesis and mechanisms of therapy resistance. Expert Rev
Anticancer Ther. 20:75–79. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mego M, Cierna Z, Svetlovska D, Macak D,
Machalekova K, Miskovska V, Chovanec M, Usakova V, Obertova J,
Babal P and Mardiak J: PARP expression in germ cell tumours. J Clin
Pathol. 66:607–612. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
van Beek L, McClay É, Patel S, Schimpl M,
Spagnolo L and Maia de Oliveira T: PARP power: A structural
perspective on PARP1, PARP2, and PARP3 in DNA damage repair and
nucleosome remodelling. Int J Mol Sci. 22:51122021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hottiger MO, Boothby M, Koch-Nolte F,
Lüscher B, Martin NM, Plummer R, Wang ZQ and Ziegler M: Progress in
the function and regulation of ADP-ribosylation. Sci Signal.
4:mr52011.PubMed/NCBI
|
|
13
|
Zhu G and Lippard SJ: Photoaffinity
labeling reveals nuclear proteins that uniquely recognize
cisplatin-DNA interstrand cross-links. Biochemistry. 48:4916–4925.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fong PC, Boss DS, Yap TA, Tutt A, Wu P,
Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et
al: Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA
mutation carriers. N Engl J Med. 361:123–134. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fong PC, Yap TA, Boss DS, Carden CP,
Mergui-Roelvink M, Gourley C, De Greve J, Lubinski J, Shanley S,
Messiou C, et al: Poly(ADP)-ribose polymerase inhibition: Frequent
durable responses in BRCA carrier ovarian cancer correlating with
platinum-free interval. J Clin Oncol. 28:2512–2519. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dockery LE, Gunderson CC and Moore KN:
Rucaparib: The past, present, and future of a newly approved PARP
inhibitor for ovarian cancer. Onco Targets Ther. 10:3029–3037.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Coleman RL, Fleming GF, Brady MF, Swisher
EM, Steffensen KD, Friedlander M, Okamoto A, Moore KN, Efrat
Ben-Baruch N, Werner TL, et al: Veliparib with first-line
chemotherapy and as maintenance therapy in ovarian cancer. N Engl J
Med. 381:2403–2415. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Farmer H, McCabe N, Lord CJ, Tutt AN,
Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I,
Knights C, et al: Targeting the DNA repair defect in BRCA mutant
cells as a therapeutic strategy. Nature. 434:917–921. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bryant HE, Schultz N, Thomas HD, Parker
KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ and Helleday T:
Specific killing of BRCA2-deficient tumours with inhibitors of
poly(ADP-ribose) polymerase. Nature. 434:913–917. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rose M, Burgess JT, O'Byrne K, Richard DJ
and Bolderson E: PARP inhibitors: Clinical relevance, mechanisms of
action and tumor resistance. Front Cell Dev Biol. 8:5646012020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sakogawa K, Aoki Y, Misumi K, Hamai Y, Emi
M, Hihara J, Shi L, Kono K, Horikoshi Y, Sun J, et al: Involvement
of homologous recombination in the synergism between cisplatin and
poly (ADP-ribose) polymerase inhibition. Cancer Sci. 104:1593–1599.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng H, Zhang Z, Borczuk A, Powell CA,
Balajee AS, Lieberman HB and Halmos B: PARP inhibition selectively
increases sensitivity to cisplatin in ERCC1-low non-small cell lung
cancer cells. Carcinogenesis. 34:739–749. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rottenberg S, Jaspers JE, Kersbergen A,
van der Burg E, Nygren AO, Zander SA, Derksen PW, de Bruin M,
Zevenhoven J, Lau A, et al: High sensitivity of BRCA1-deficient
mammary tumors to the PARP inhibitor AZD2281 alone and in
combination with platinum drugs. Proc Natl Acad Sci USa.
105:17079–17084. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Evers B, Drost R, Schut E, de Bruin M, van
der Burg E, Derksen PW, Holstege H, Liu X, van Drunen E, Beverloo
HB, et al: Selective inhibition of BRCA2-deficient mammary tumor
cell growth by AZD2281 and cisplatin. Clin Cancer Res.
14:3916–3925. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Plummer R, Jones C, Middleton M, Wilson R,
Evans J, Olsen A, Curtin N, Boddy A, McHugh P, Newell D, et al:
Phase I study of the poly(ADP-ribose) polymerase inhibitor,
AG014699, in combination with temozolomide in patients with
advanced solid tumors. Clin Cancer Res. 14:7917–7923. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Plummer R, Lorigan P, Steven N, Scott L,
Middleton MR, Wilson RH, Mulligan E, Curtin N, Wang D, Dewji R, et
al: A phase II study of the potent PARP inhibitor, rucaparib
(PF-01367338, AG014699), with temozolomide in patients with
metastatic melanoma demonstrating evidence of chemopotentiation.
Cancer Chemother Pharmacol. 71:1191–1199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bell-McGuinn KM, Brady WE, Schilder RJ,
Fracasso PM, Moore KN, Walker JL, Duska LR, Mathews CA, Chen A,
Shepherd SP, et al: A phase I study of continuous veliparib in
combination with IV carboplatin/paclitaxel or IV/IP
paclitaxel/cisplatin and bevacizumab in newly diagnosed patients
with previously untreated epithelial ovarian, fallopian tube, or
primary peritoneal cancer: An NRG oncology/gynecologic oncology
group study. J Clin Oncol. 33 (Suppl 15):S55072015. View Article : Google Scholar
|
|
28
|
Landrum LM, Brady WE, Armstrong DK, Moore
KN, DiSilvestro PA, O'Malley DM, Tenney ME, Rose PG and Fracasso
PM: A phase I trial of pegylated liposomal doxorubicin (PLD),
carboplatin, bevacizumab and veliparib in recurrent,
platinum-sensitive ovarian, primary peritoneal, and fallopian tube
cancer: An NRG oncology/gynecologic oncology group study. Gynecol
Oncol. 140:204–209. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nishio S, Takekuma M, Takeuchi S, Kawano
K, Tsuda N, Tasaki K, Takahashi N, Abe M, Tanaka A, Nagasawa T, et
al: Phase 1 study of veliparib with carboplatin and weekly
paclitaxel in Japanese patients with newly diagnosed ovarian
cancer. Cancer Sci. 108:2213–2220. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gray HJ, Bell-McGuinn K, Fleming GF,
Cristea M, Xiong H, Sullivan D, Luo Y, McKee MD, Munasinghe W and
Martin LP: Phase I combination study of the PARP inhibitor
veliparib plus carboplatin and gemcitabine in patients with
advanced ovarian cancer and other solid malignancies. Gynecol
Oncol. 148:507–514. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wilson RH, Evans TJ, Middleton MR, Molife
LR, Spicer J, Dieras V, Roxburgh P, Giordano H, Jaw-Tsai S, Goble S
and Plummer R: A phase I study of intravenous and oral rucaparib in
combination with chemotherapy in patients with advanced solid
tumours. Br J Cancer. 116:884–892. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mego M, Svetlovska D, Reckova M, Angelis
D, Kalavska K, Obertova J, Palacka P, Rejlekova K, Sycova-Mila Z,
Chovanec M and Mardiak J: Gemcitabine, carboplatin and veliparib in
multiple relapsed/refractory germ cell tumours: The GCT-SK-004
phase II trial. Invest New Drugs. 39:1664–1670. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schmidtova S, Kalavska K, Gercakova K,
Cierna Z, Miklikova S, Smolkova B, Buocikova V, Miskovska V,
Durinikova E, Burikova M, et al: Disulfiram overcomes cisplatin
resistance in human embryonal carcinoma cells. Cancers (Basel).
11:12242019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schmidtova S, Dorssers LCJ, Kalavska K,
Gillis AJM, Oosterhuis JW, Stoop H, Miklikova S, Kozovska Z,
Burikova M, Gercakova K, et al: Napabucasin overcomes cisplatin
resistance in ovarian germ cell tumor-derived cell line by
inhibiting cancer stemness. Cancer Cell Int. 20:3642020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Schmidtova S, Kalavska K, Liskova V, Plava
J, Miklikova S, Kucerova L, Matuskova M, Rojikova L, Cierna Z,
Rogozea A, et al: Targeting of deregulated Wnt/β-catenin signaling
by PRI-724 and LGK974 inhibitors in germ cell tumor cell lines. Int
J Mol Sci. 22:42632021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pfaffl MW, Horgan GW and Dempfle L:
Relative expression software tool (REST) for group-wise comparison
and statistical analysis of relative expression results in
real-time PCR. Nucleic Acids Res. 30:e362002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chou TC: Theoretical basis, experimental
design, and computerized simulation of synergism and antagonism in
drug combination studies. Pharmacol Rev. 58:621–681. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Stehlik P, Paulikova H and Hunakova L:
Synthetic isothiocyanate indole-3-ethyl isothiocyanate (homoITC)
enhances sensitivity of human ovarian carcinoma cell lines A2780
and A2780/CP to cisplatin. Neoplasma. 57:473–481. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Klauschen F, von Winterfeld M, Stenzinger
A, Sinn BV, Budczies J, Kamphues C, Bahra M, Wittschieber D,
Weichert W, Striefler J, et al: High nuclear
poly-(ADP-ribose)-polymerase expression is prognostic of improved
survival in pancreatic cancer. Histopathology. 61:409–416. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou J, Gelot C, Pantelidou C, Li A, Yücel
H, Davis RE, Färkkilä A, Kochupurakkal B, Syed A, Shapiro GI, et
al: A first-in-class polymerase theta inhibitor selectively targets
homologous-recombination-deficient tumors. Nat Cancer. 2:598–610.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zatreanu D, Robinson HMR, Alkhatib O,
Boursier M, Finch H, Geo L, Grande D, Grinkevich V, Heald RA,
Langdon S, et al: Poltheta inhibitors elicit BRCA-gene synthetic
lethality and target PARP inhibitor resistance. Nat Commun.
12:36362021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ray Chaudhuri A and Nussenzweig A: The
multifaceted roles of PARP1 in DNA repair and chromatin
remodelling. Nat Rev Mol Cell Biol. 18:610–621. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang M, Wu W, Wu W, Rosidi B, Zhang L,
Wang H and Iliakis G: PARP-1 and Ku compete for repair of DNA
double strand breaks by distinct NHEJ pathways. Nucleic Acids Res.
34:6170–6182. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Langelier MF, Riccio AA and Pascal JM:
PARP-2 and PARP-3 are selectively activated by 5′ phosphorylated
DNA breaks through an allosteric regulatory mechanism shared with
PARP-1. Nucleic Acids Res. 42:7762–7775. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ménissier de Murcia J, Ricoul M, Tartier
L, Niedergang C, Huber A, Dantzer F, Schreiber V, Amé JC, Dierich
A, LeMeur M, et al: Functional interaction between PARP-1 and
PARP-2 in chromosome stability and embryonic development in mouse.
EMBO J. 22:2255–2263. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cavallo F, Graziani G, Antinozzi C,
Feldman DR, Houldsworth J, Bosl GJ, Chaganti RS, Moynahan ME, Jasin
M and Barchi M: Reduced proficiency in homologous recombination
underlies the high sensitivity of embryonal carcinoma testicular
germ cell tumors to cisplatin and poly (adp-ribose) polymerase
inhibition. PLoS One. 7:e515632012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ogino H, Nakayama R, Sakamoto H, Yoshida
T, Sugimura T and Masutani M: Analysis of poly(ADP-ribose)
polymerase-1 (PARP1) gene alteration in human germ cell tumor cell
lines. Cancer Genet Cytogenet. 197:8–15. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Murai J, Huang SY, Das BB, Renaud A, Zhang
Y, Doroshow JH, Ji J, Takeda S and Pommier Y: Trapping of PARP1 and
PARP2 by CLinical PARP inhibitors. Cancer Res. 72:5588–5599. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dedes KJ, Wilkerson PM, Wetterskog D,
Weigelt B, Ashworth A and Reis-Filho JS: Synthetic lethality of
PARP inhibition in cancers lacking BRCA1 and BRCA2 mutations. Cell
Cycle. 10:1192–1199. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Moore K, Colombo N, Scambia G, Kim BG,
Oaknin A, Friedlander M, Lisyanskaya A, Floquet A, Leary A, Sonke
GS, et al: Maintenance olaparib in patients with newly diagnosed
advanced ovarian cancer. N Engl J Med. 379:2495–2505. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Golan T, Hammel P, Reni M, Van Cutsem E,
Macarulla T, Hall MJ, Park JO, Hochhauser D, Arnold D, Oh DY, et
al: Maintenance olaparib for germline BRCA-mutated metastatic
pancreatic cancer. N Engl J Med. 381:317–327. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Robson ME, Tung N, Conte P, Im SA, Senkus
E, Xu B, Masuda N, Delaloge S, Li W, Armstrong A, et al: OlympiAD
final overall survival and tolerability results: Olaparib versus
chemotherapy treatment of physician's choice in patients with a
germline BRCA mutation and HER2-negative metastatic breast cancer.
Ann Oncol. 30:558–566. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
de Bono J, Mateo J, Fizazi K, Saad F,
Shore N, Sandhu S, Chi KN, Sartor O, Agarwal N, Olmos D, et al:
Olaparib for metastatic castration-resistant prostate cancer. N
Engl J Med. 382:2091–2102. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Abida W, Campbell D, Patnaik A, Shapiro
JD, Sautois B, Vogelzang NJ, Voog EG, Bryce AH, McDermott R, Ricci
F, et al: Non-BRCA DNA damage repair gene alterations and response
to the PARP inhibitor rucaparib in metastatic castration-resistant
prostate cancer: Analysis from the phase II TRITON2 study. Clin
Cancer Res. 26:2487–2496. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
George RR, Thomas R, Davice A and Mathew
MS: Veliparib for the treatment of solid malignancies. J Oncol
Pharm Pract. 28:924–934. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Rodler ET, Kurland BF, Griffin M, Gralow
JR, Porter P, Yeh RF, Gadi VK, Guenthoer J, Beumer JH, Korde L, et
al: Phase I study of veliparib (ABT-888) combined with cisplatin
and vinorelbine in advanced triple-negative breast cancer and/or
BRCA mutation-associated breast cancer. Clin Cancer Res.
22:2855–2864. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Diéras V, Han HS, Kaufman B, Wildiers H,
Friedlander M, Ayoub JP, Puhalla SL, Bondarenko I, Campone M,
Jakobsen EH, et al: Veliparib with carboplatin and paclitaxel in
BRCA-mutated advanced breast cancer (BROCADE3): A randomised,
double-blind, placebo-controlled, phase 3 trial. Lancet Oncol.
21:1269–1282. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Byers LA, Bentsion D, Gans S, Penkov K,
Son C, Sibille A, Owonikoko TK, Groen HJM, Gay CM, Fujimoto J, et
al: Veliparib in combination with carboplatin and etoposide in
patients with treatment-Naïve extensive-stage small cell lung
cancer: A phase 2 randomized study. Clin Cancer Res. 27:3884–3895.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Clarke JM, Patel JD, Robert F, Kio EA,
Thara E, Ross Camidge D, Dunbar M, Nuthalapati S, Dinh MH and Bach
BA: Veliparib and nivolumab in combination with platinum doublet
chemotherapy in patients with metastatic or advanced non-small cell
lung cancer: A phase 1 dose escalation study. Lung Cancer.
161:180–188. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lobo J, Constancio V, Guimaraes-Teixeira
C, Leite-Silva P, Miranda-Gonçalves V, Sequeira JP, Pistoni L,
Guimarães R, Cantante M, Braga I, et al: Promoter methylation of
DNA homologous recombination genes is predictive of the
responsiveness to PARP inhibitor treatment in testicular germ cell
tumors. Mol Oncol. 15:846–865. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Caggiano C, Cavallo F, Giannattasio T,
Cappelletti G, Rossi P, Grimaldi P, Feldman DR, Jasin M and Barchi
M: Testicular germ cell tumors acquire cisplatin resistance by
rebalancing the usage of DNA repair pathways. Cancers (Basel).
13:7872021. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
De Giorgi U, Schepisi G, Gurioli G, Pisano
C, Basso U, Lolli C, Petracci E, Casadei C, Cecere SC, Attademo L,
et al: Olaparib as salvage treatment for advanced germ cell tumors
after chemotherapy failure: Results of the open-label, single-arm,
IGG-02 phase II trial. J Clin Oncol. 38 (Suppl 15):S50582020.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Shen Y, Rehman FL, Feng Y, Boshuizen J,
Bajrami I, Elliott R, Wang B, Lord CJ, Post LE and Ashworth A: BMN
673, a novel and highly potent PARP1/2 inhibitor for the treatment
of human cancers with DNA repair deficiency. Clin Cancer Resh.
19:5003–5015. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Jiang Y, Meng XY, Deng NN, Meng C, Li LH,
He ZK, Wang XY, Song ZY and Cui RJ: Effect and safety of
therapeutic regimens for patients with germline BRCA
mutation-associated breast cancer: A network meta-analysis. Front
Oncol. 11:7187612021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Kim DS, Camacho CV and Kraus WL: Alternate
therapeutic pathways for PARP inhibitors and potential mechanisms
of resistance. Exp Mol Med. 53:42–51. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Dias MP, Moser SC, Ganesan S and Jonkers
J: Understanding and overcoming resistance to PARP inhibitors in
cancer therapy. Nat Rev Clin Oncol. 18:773–791. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Edwards SL, Brough R, Lord CJ, Natrajan R,
Vatcheva R, Levine DA, Boyd J, Reis-Filho JS and Ashworth A:
Resistance to therapy caused by intragenic deletion in BRCA2.
Nature. 451:1111–1115. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ter Brugge P, Kristel P, van der Burg E,
Boon U, de Maaker M, Lips E, Mulder L, de Ruiter J, Moutinho C,
Gevensleben H, et al: Mechanisms of therapy resistance in
patient-derived xenograft models of BRCA1-deficient breast cancer.
J Natl Cancer Inst. 108:2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Taglialatela A, Alvarez S, Leuzzi G,
Sannino V, Ranjha L, Huang JW, Madubata C, Anand R, Levy B, Rabadan
R, et al: Restoration of replication fork stability in BRCA1- and
BRCA2-deficient cells by inactivation of SNF2-family fork
remodelers. Mol Cell. 68:414–430.e8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Rondinelli B, Gogola E, Yücel H, Duarte
AA, van de Ven M, van der Sluijs R, Konstantinopoulos PA, Jonkers
J, Ceccaldi R, Rottenberg S and D'Andrea AD: EZH2 promotes
degradation of stalled replication forks by recruiting MUS81
through histone H3 trimethylation. Nat Cell Biol. 19:1371–1378.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Jaspers JE, Sol W, Kersbergen A, Schlicker
A, Guyader C, Xu G, Wessels L, Borst P, Jonkers J and Rottenberg S:
BRCA2-deficient sarcomatoid mammary tumors exhibit multidrug
resistance. Cancer Res. 75:732–741. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Gogola E, Duarte AA, de Ruiter JR, Wiegant
WW, Schmid JA, de Bruijn R, James DI, Guerrero Llobet S, Vis DJ,
Annunziato S, et al: Selective loss of PARG restores PARylation and
counteracts PARP inhibitor-mediated synthetic lethality. Cancer
Cell. 35:950–952. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Bajrami I, Frankum JR, Konde A, Miller RE,
Rehman FL, Brough R, Campbell J, Sims D, Rafiq R, Hooper S, et al:
Genome-wide profiling of genetic synthetic lethality identifies
CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity.
Cancer Res. 74:287–297. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Jiang J, Yang ES, Jiang G, Nowsheen S,
Wang H, Wang T, Wang Y, Billheimer D, Chakravarthy AB, Brown M, et
al: p53-dependent BRCA1 nuclear export controls cellular
susceptibility to DNA damage. Cancer Res. 71:5546–5557. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yang ES, Nowsheen S, Rahman MA, Cook RS
and Xia F: Targeting BRCA1 localization to augment breast tumor
sensitivity to poly(ADP-Ribose) polymerase inhibition. Cancer Res.
72:5547–5555. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
LoRusso P, Pilat MJP, Santa-Maria CA,
Connolly, Roesch EE, Afghahi A, Han HS, Nanda R, Wulf GM, Assad H,
et al: Trial in progress: A phase II open-label, randomized study
of PARP inhibition (olaparib) either alone or in combination with
anti-PD-L1 therapy (atezolizumab) in homologous DNA repair (HDR)
deficient, locally advanced or metastatic non-HER2-positive breast
cancer. J Clin Oncol. 38 (Suppl 15):TPS11022020. View Article : Google Scholar
|
|
78
|
Moore KN, Chambers SK, Hamilton EP, Chen
LM, Oza AM, Ghamande SA, Konecny GE, Plaxe SC, Spitz DL, Geenen
JJJ, et al: Adavosertib with chemotherapy in patients with primary
platinum-resistant ovarian, fallopian tube, or peritoneal cancer:
An open-label, four-arm, phase II study. Clin Cancer Res. 28:36–44.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Tutt A, Stephens C, Frewer P, Pierce A,
Rhee J, Edgington S, Ottesen L, Ah-See ML, Hollingsworth SJ and
Dean E: VIOLETTE: A randomized phase II study to assess the DNA
damage response inhibitors AZD6738 or AZD1775 in combination with
olaparib (Ola) versus Ola monotherapy in patients (pts) with
metastatic, triple-negative breast cancer (TNBC). J Clin Oncol. 37
(Suppl 15):TPS11122019. View Article : Google Scholar
|
|
80
|
Choi YE, Battelli C, Watson J, Liu J,
Curtis J, Morse AN, Matulonis UA, Chowdhury D and Konstantinopoulos
PA: Sublethal concentrations of 17-AAG suppress homologous
recombination DNA repair and enhance sensitivity to carboplatin and
olaparib in HR proficient ovarian cancer cells. Oncotarget.
5:2678–2687. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Kim H, George E, Ragland R, Rafail S,
Zhang R, Krepler C, Morgan M, Herlyn M, Brown E and Simpkins F:
Targeting the ATR/CHK1 axis with PARP inhibition results in tumor
regression in BRCA-mutant ovarian cancer models. Clin Cancer Res.
23:3097–3108. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Murai J, Feng Y, Yu GK, Ru Y, Tang SW,
Shen Y and Pommier Y: Resistance to PARP inhibitors by SLFN11
inactivation can be overcome by ATR inhibition. Oncotarget.
7:76534–76550. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Muvarak NE, Chowdhury K, Xia L, Robert C,
Choi EY, Cai Y, Bellani M, Zou Y, Singh ZN, Duong VH, et al:
Enhancing the cytotoxic effects of PARP inhibitors with DNA
demethylating agents-a potential therapy for cancer. Cancer Cell.
30:637–650. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Pulliam N, Fang F, Ozes AR, Tang J,
Adewuyi A, Keer H, Lyons J, Baylin SB, Matei D, Nakshatri H, et al:
An effective epigenetic-PARP inhibitor combination therapy for
breast and ovarian cancers independent of BRCA mutations. Clin
Cancer Res. 24:3163–3175. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Jiao S, Xia W, Yamaguchi H, Wei Y, Chen
MK, Hsu JM, Hsu JL, Yu WH, Du Y, Lee HH, et al: PARP inhibitor
upregulates PD-L1 expression and enhances cancer-associated
immunosuppression. Clin Cancer Res. 23:3711–3720. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Mehta AK, Cheney EM, Hartl CA, Pantelidou
C, Oliwa M, Castrillon JA, Lin JR, Hurst KE, de Oliveira Taveira M,
Johnson NT, et al: Targeting immunosuppressive macrophages
overcomes PARP inhibitor resistance in BRCA1-associated
triple-negative breast cancer. Nat Cancer. 2:66–82. 2021.
View Article : Google Scholar : PubMed/NCBI
|