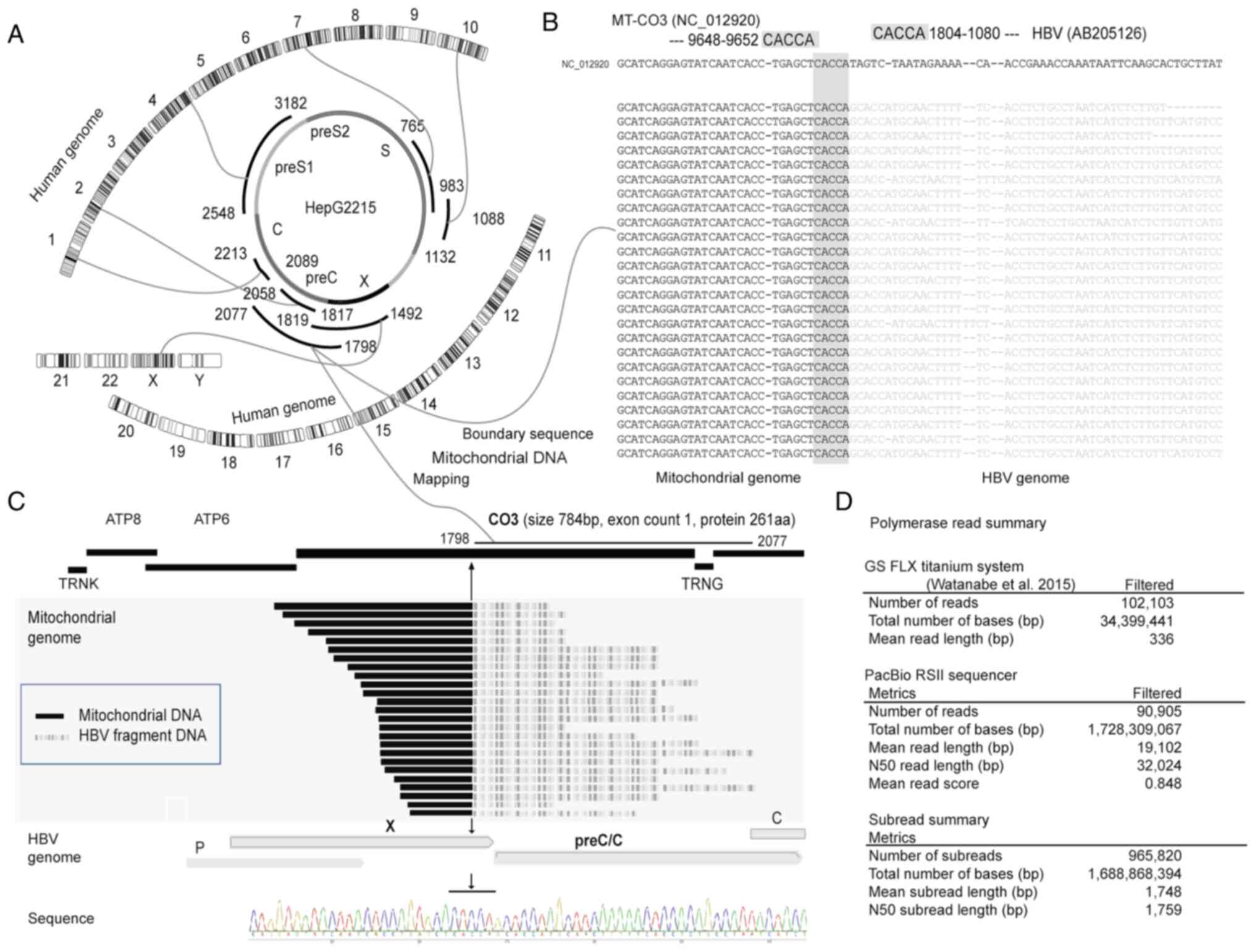
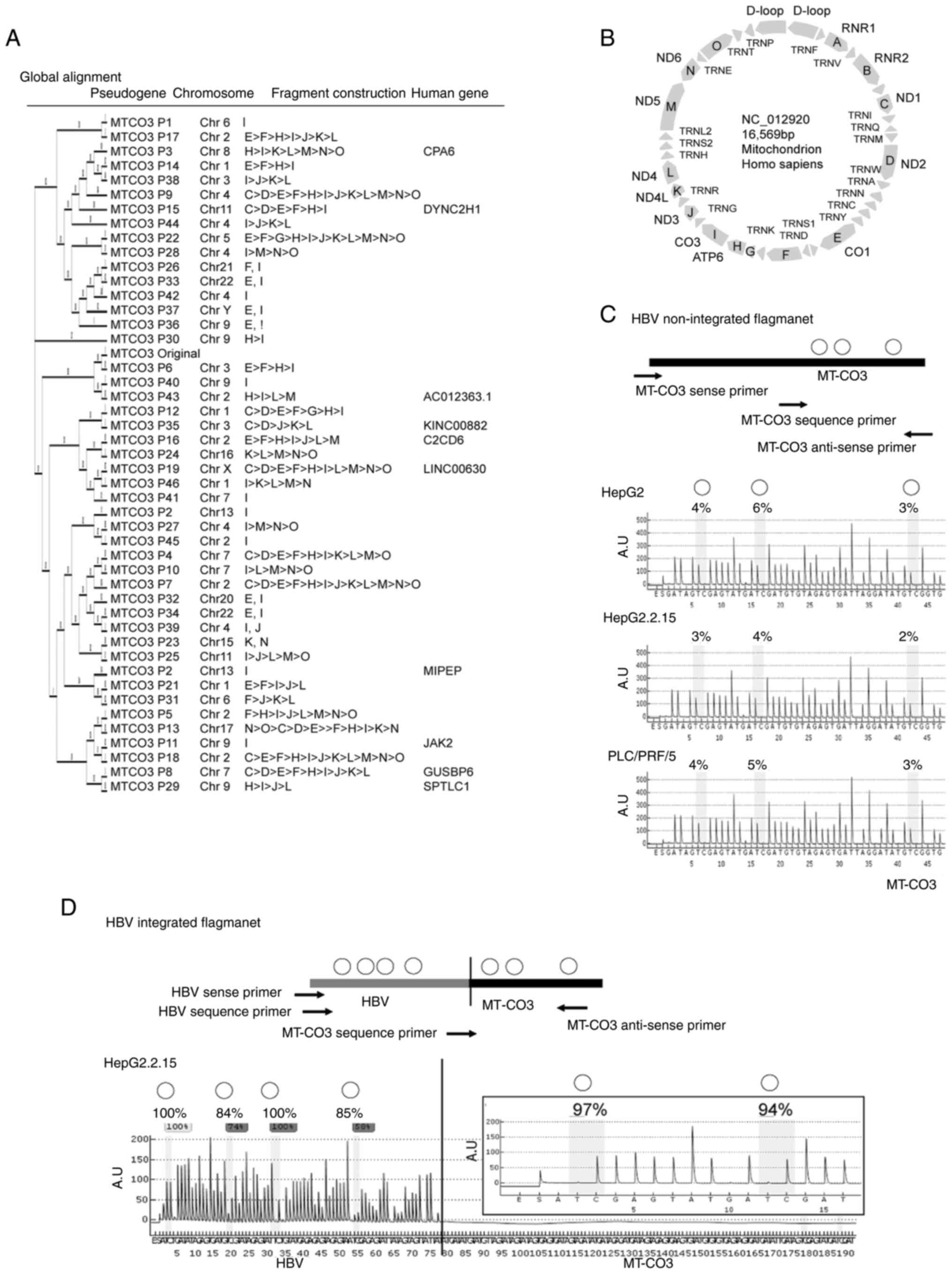
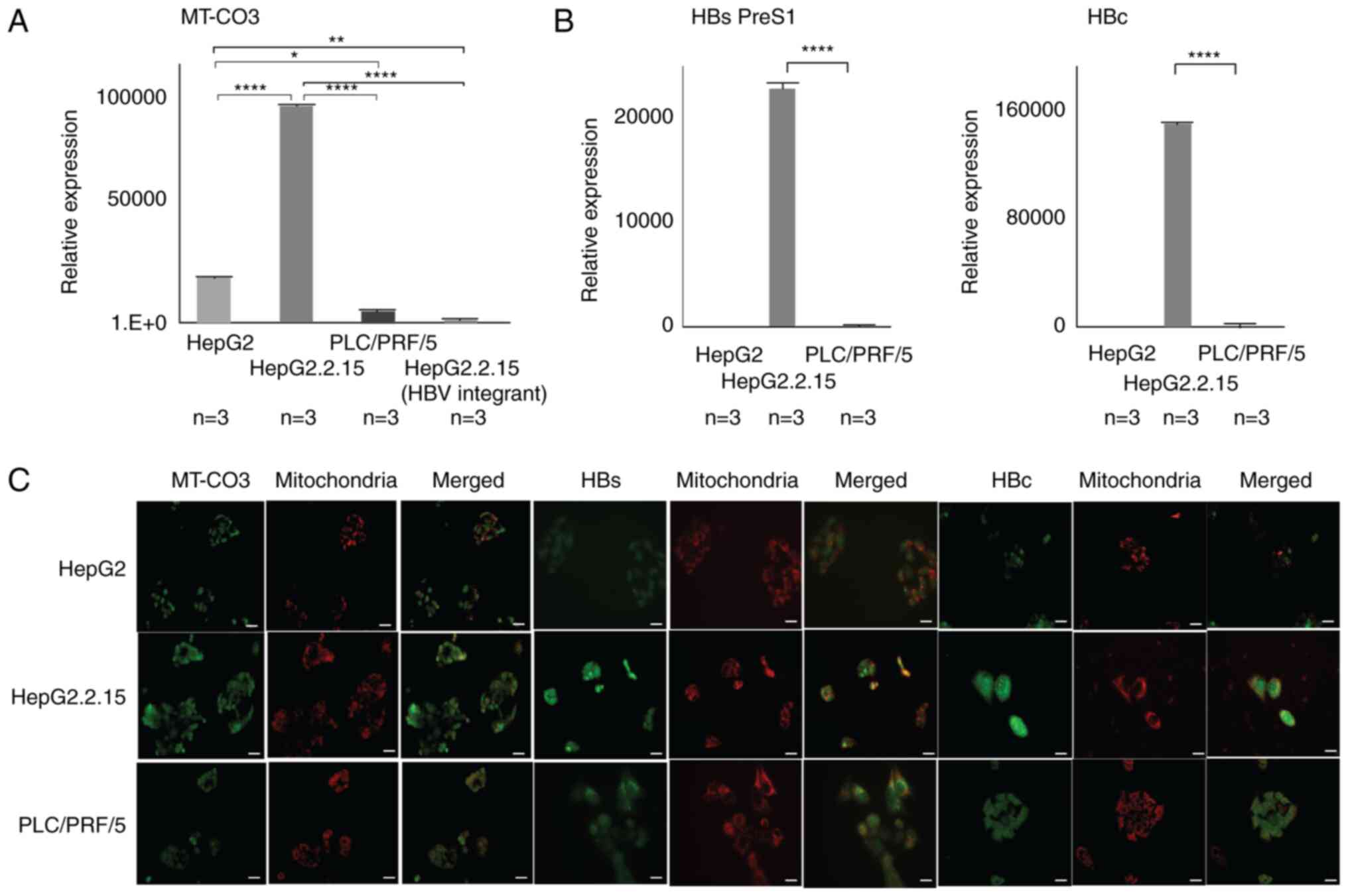
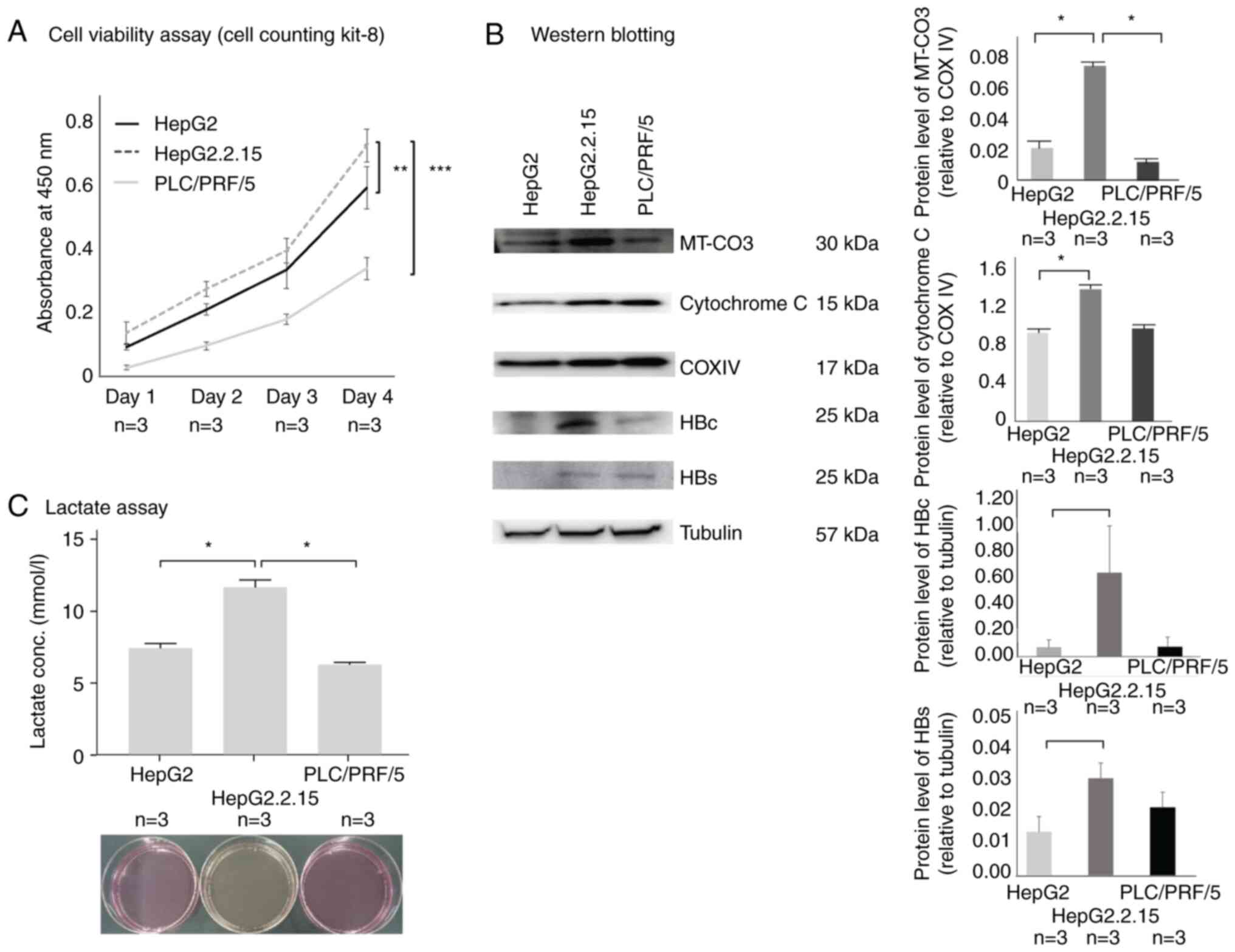
|
1
|
Fattovich G, Bortolotti F and Donato F:
Natural history of chronic hepatitis B: Special emphasis on disease
progression and prognostic factors. J Hepatol. 48:335–352. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ganem D and Prince AM: Hepatitis B virus
infection-natural history and clinical consequences. N Engl J Med.
350:1118–1129. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
McMahon BJ: Natural history of chronic
hepatitis B. Clin Liver Dis. 14:381–396. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shibata T and Aburatani H: Exploration of
liver cancer genomes. Nat Rev Gastroenterol Hepatol. 11:340–349.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lau CC, Sun T, Ching AK, He M, Li JW, Wong
AM, Co NN, Chan AW, Li PS, Lung RWT, et al: Viral-human chimeric
transcript predisposes risk to liver cancer development and
progression. Cancer Cell. 25:335–349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yamamoto H, Watanabe Y, Maehata T, Morita
R, Yoshida Y, Oikawa R, Ishigooka S, Ozawa S, Matsuo Y, Hosoya K,
et al: An updated review of gastric cancer in the next-generation
sequencing era: Insights from bench to bedside and vice versa.
World J Gastroenterol. 20:3927–3937. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Watanabe Y, Yamamoto H, Oikawa R, Toyota
M, Yamamoto M, Kokudo N, Tanaka S, Arii S, Yotsuyanagi H, Koike K
and Itoh F: DNA methylation at hepatitis B viral integrants is
associated with methylation at flanking human genomic sequences.
Genome Res. 25:328–337. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ou JH and Rutter WJ: Hybrid hepatitis B
virus-host transcripts in a human hepatoma cell. Proc Natl Acad Sci
USA. 82:83–87. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sells MA, Zelent AZ, Shvartsman M and Acs
G: Replicative intermediates of hepatitis B virus in HepG2 cells
that produce infectious virions. J Virol. 62:2836–2844. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sells MA, Chen ML and Acs G: Production of
hepatitis B virus particles in Hep G2 cells transfected with cloned
hepatitis B virus DNA. Proc Natl Acad Sci USA. 84:1005–1009. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fotakis G and Timbrell JA: In vitro
cytotoxicity assays: Comparison of LDH, neutral red, MTT and
protein assay in hepatoma cell lines following exposure to cadmium
chloride. Toxicol Lett. 160:171–177. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ishiyama M, Miyazono Y, Sasamoto K, Ohkura
Y and Ueno K: A highly water-soluble disulfonated tetrazolium salt
as a chromogenic indicator for NADH as well as cell viability.
Talanta. 44:1299–1305. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Koike K: Hepatitis B virus X gene is
implicated in liver carcinogenesis. Cancer Lett. 286:60–68. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Al Shahrani M, Heales S, Hargreaves I and
Orford M: Oxidative stress: Mechanistic insights into inherited
mitochondrial disorders and Parkinson's disease. J Clin Med.
6:1002017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Davalli P, Mitic T, Caporali A, Lauriola A
and D'Arca D: ROS, cell senescence, and novel molecular mechanisms
in aging and age-related diseases. Oxid Med Cell Longev.
2016:35651272016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bonilla Guerrero R and Roberts LR: The
role of hepatitis B virus integrations in the pathogenesis of human
hepatocellular carcinoma. J Hepatol. 42:760–777. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kachirskaia I, Shi X, Yamaguchi H, Tanoue
K, Wen H, Wang EW, Appella E and Gozani O: Role for 53BP1 tudor
domain recognition of p53 dimethylated at lysine 382 in DNA damage
signaling. J Biol Chem. 283:34660–34666. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim S, Park SY, Yong H, Famulski JK, Chae
S, Lee JH, Kang CM, Saya H, Chan GK and Cho H: HBV X protein
targets hBubR1, which induces dysregulation of the mitotic
checkpoint. Oncogene. 27:3457–3464. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Martin-Lluesma S, Schaeffer C, Robert EI,
van Breugel PC, Leupin O, Hantz O and Strubin M: Hepatitis B virus
X protein affects S phase progression leading to chromosome
segregation defects by binding to damaged DNA binding protein 1.
Hepatology. 48:1467–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Minami M, Poussin K, Brechot C and
Paterlini P: A novel PCR technique using Alu-specific primers to
identify unknown flanking sequences from the human genome.
Genomics. 29:403–408. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sung WK, Zheng H, Li S, Chen R, Liu X, Li
Y, Lee NP, Lee WH, Ariyaratne PN, Tennakoon C, et al: Genome-wide
survey of recurrent HBV integration in hepatocellular carcinoma.
Nat Genet. 44:765–769. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kawai-Kitahata F, Asahina Y, Tanaka S,
Kakinuma S, Murakawa M, Nitta S, Watanabe T, Otani S, Taniguchi M,
Goto F, et al: Comprehensive analyses of mutations and hepatitis B
virus integration in hepatocellular carcinoma with
clinicopathological features. J Gastroenterol. 51:473–486. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Furuta M, Tanaka H, Shiraishi Y, Uchida T,
Imamura M, Fujimoto A, Fujita M, Sasaki-Oku A, Maejima K, Nakano K,
et al: Correction: Characterization of HBV integration patterns and
timing in liver cancer and HBV-infected livers. Oncotarget.
9:317892018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Machida K, Cheng KTH, Sung VMH, Lee KJ,
Levine AM and Lai MM: Hepatitis C virus infection activates the
immunologic (type II) isoform of nitric oxide synthase and thereby
enhances DNA damage and mutations of cellular genes. J Virol.
78:8835–8843. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yamamoto H, Watanabe Y, Oikawa R, Morita
R, Yoshida Y, Maehata T, Yasuda H and Itoh F: BARHL2 methylation
using gastric wash DNA or gastric juice exosomal dna is a useful
marker for early detection of gastric cancer in an H.
pylori-Independent Manner. Clin Transl Gastroenterol. 7:e1842016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Watanabe Y, Toyota M, Kondo Y, Suzuki H,
Imai T, Ohe-Toyota M, Maruyama R, Nojima M, Sasaki Y, Sekido Y, et
al: PRDM5 identified as a target of epigenetic silencing in
colorectal and gastric cancer. Clin Cancer Res. 13:4786–4794. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Watanabe Y, Kim HS, Castoro RJ, Chung W,
Estecio MR, Kondo K, Guo Y, Ahmed SS, Toyota M, Itoh F, et al:
Sensitive and specific detection of early gastric cancer with DNA
methylation analysis of gastric washes. Gastroenterology.
136:2149–2158. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Orend G, Kuhlmann I and Doerfler W:
Spreading of DNA methylation across integrated foreign (adenovirus
type 12) genomes in mammalian cells. J Virol. 65:4301–4308. 1991.
View Article : Google Scholar : PubMed/NCBI
|