Introduction
The farnesyl diphosphate synthase (FDPS) gene
encodes an enzyme involved in the mevalonate pathway that catalyzes
the sequential condensation of dimethylallyl pyrophosphate with two
molecules of isopentenyl pyrophosphate to form farnesyl
pyrophosphate (1). FDPS contributes
to the biosynthesis of cholesterol and steroid hormones, dolichols,
heme A and ubiquinone, which are important intermediary metabolites
that participate in numerous biological processes, including
response to environmental changes. Disordered FDPS expression also
leads to multiple types of cancer that threaten human health
(2). For example, to investigate
whether detectable FDPS activity was present in human colorectal
cancer (CRC), Notarnicola et al (3) conducted a radiochemical assay using
the tissues of 50 patients and compared the FDPS activity level in
CRC tissues with that in normal surrounding mucosa. The results of
the assay demonstrated that FDPS activity and its mRNA level were
increased in the cancer samples compared with in normal mucosa. In
addition, higher FDPS activity inhibited cellular apoptosis in CRC.
Inhibiting farnesyl biosynthesis may lead to blocking of Ras
signaling and the lowering of MAPK activity, thus inhibiting
proliferation of glioma cells (4).
FDPS also serves an important role in the apoptosis of cancer cells
by blocking the JNK signaling cascade and activating mevalonate
metabolism in paclitaxel-treated glioblastoma cells (5).
FDPS is also a crucial enzyme implicated in other
diseases. The association between FDPS polymorphisms and
osteoporosis has been extensively investigated (6–8). Due
to the important functions of FDPS in the mevalonate pathway, an
increasing number of studies have evaluated its potential as a drug
target (9–11). Since FDPS has an affinity for bone
minerals and inhibitory effects on osteoclasts, it was previously
identified as a main biochemical target of the bisphonate (BP)
drugs widely used to treat osteoporosis (12–15).
FDPS generates isoprenoid lipids involved in the post-translational
modification of small GTP-binding proteins essential for osteoclast
function (16). Therefore,
inhibiting FDPS results in the antiresorptive action of BPs and
prevents the biosynthesis of isoprenoid lipids, ultimately inducing
cellular dysfunction and osteoclast death (17,18).
It has been demonstrated that FDPS could maintain
the resorption activity of the osteoclasts (7). FDPS may promote cancer progression in
PTEN-deficient prostate cancer through the GTPase/AKT axis
(19). However, current studies
have mainly focused on its role as a synthetase in the mevalonate
pathway. A previous review study demonstrated that numerous
metabolic enzymes influence RNAs or their own functions through
their RNA binding activity (20).
Using an RNA interactome method, researchers found that FDPS
interacted with RNAs in HeLa cells (21), implying that FDPS may serve as an
RNA-binding protein (RBP). By interacting with their targeted RNAs
through a series of canonical RNA binding domains, RBPs serve key
roles in post-transcriptional events, including alternative
splicing (AS) (22), alternative
polyadenylation (23,24), gene translational regulation
(25) and RNA modification
(26). Therefore, any disruption to
RBPs that regulate crucial cellular functions may cause disease,
particularly cancer cachexia (27–30).
Furthermore, dysregulated AS is implicated in multiple types of
human cancer, including lung and liver cancer (31), and can aberrantly activate oncogenes
and cancer pathways (32). However,
as an emerging RBP, how FDPS regulates AS is poorly understood and
its genome-wide RNA targets have not been fully investigated.
In the present study, to identify the
transcriptome-wide targets of FDPS, the FDPS gene was overexpressed
in HeLa cells to investigate behavioral changes. In addition, the
impact of FDPS on gene expression levels and AS was analyzed by
exploring the transcriptome of the overexpression cells and control
cells. To verify the findings in HeLa cells, the targeted genes
were validated in human osteosarcoma (HOS) cells, which have been
previously studied to investigate the molecular mechanisms of
osteoporosis (33). The results
revealed that FDPS extensively regulated the RNA levels and AS
patterns of numerous genes that are involved in cell proliferation
or are related to the cell cycle, which broadens the understanding
of FDPS-mediated essential biological processes in diseases.
Materials and methods
Cell culture and transfection
The human HeLa and HOS cell lines (cat. nos. CL-0350
and CL-0360, respectively; Procell Life Science & Technology
Co., Ltd.) were cultured at 37°C with 5% CO2 in Minimum
Essential Medium (cat. no. PM150410; Procell Life Science &
Technology Co., Ltd.) supplemented with 10% FBS (cat. no. 10091148;
Gibco; Thermo Fisher Scientific, Inc.), 100 µg/ml streptomycin and
100 U/ml penicillin (cat. no. SV30010; HyClone; Cytiva). HeLa and
HOS cells (5×104) were cultured in 24-well plates with
500 µl cell growth medium. The vector was transfected into HeLa
cells using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) after cells reached 70% confluency
according to the manufacturer's protocol. The cells were then
incubated at 37°C for 48 h. To construct FDPS overexpression
(FDPS-OE) and empty vector control samples, HeLa and HOS cells were
transfected with a FDPS-OE plasmid or empty vector (500 ng/well)
using Lipofectamine® 2000 (cat. no. 11668019;
Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. For FDPS-OE, the coding sequence of
FDPS was cloned into the pIRES-hrGFP-1a vector (cat. no. 240031;
Agilent Technologies, Inc.). The plasmid was constructed according
to a previously published method (34). The primer sequences for FDPS-OE
construction were as follows: Forward primer,
5′-AGCCCGGGCGGATCCGAATTCATGGATTCATCCCTTACCCGC-3′ and reverse
primer, 5′-GTCATCCTTGTAGTCCTCGAGCTTTCTCCGCTTGTAGATTTTGC-3′. The
transfection mixture was prepared at 37°C for ~30 min, and then
added to the cells for incubation for 6 h. The transfection mixture
was then replaced with fresh medium and the cells were cultured
until 48 h. After 48 h of transfection, the supernatant was removed
from the well plate, and the cells were rinsed with PBS. The cells
were then lysed with TRIzol® (cat. no. 15596-018;
Ambion; Thermo Fisher Scientific, Inc.) for RNA extraction and
lysed with RIPA buffer (cat. no. PR20001; Proteintech Group, Inc.)
for the subsequent experiments.
Western blotting
To prepare total cell lysates, the cells were lysed
on ice for 30 min using RIPA buffer (cat. no. PR20001; Proteintech
Group, Inc.) containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1%
deoxycholate, 1% Triton X-100, 1 mM EDTA and 0.1% SDS. The samples
were centrifuged (12,000 × g for 5 min at 4°C) and 20 µl
supernatant was analyzed on a 10% SDS-PAGE gel after boiling
(100°C) for 10 min. Protein concentration was determined using the
BCA method, and 20 µg protein was loaded per lane. Following this,
the proteins in the gel were transferred onto 0.45-mm PVDF
membranes (MilliporeSigma). The membranes were blocked with 5%
skimmed milk (in buffer containing 10 mM Tris pH 8.0, 150 mM NaCl
and 0.05% Tween 20) for 1 h at room temperature. The membranes were
incubated overnight with primary antibody at 4°C and then incubated
with HRP-conjugated secondary antibody (anti-rabbit, 1:5,000; cat.
no. SA00001-2; Proteintech Group, Inc.) for 1 h at room
temperature. The protein bands were visualized by chemiluminescence
instrument (cat. no. 5200; Tanon Science and Technology Co., Ltd.).
FDPS was detected using a monoclonal Flag antibody (1:2,000; cat.
no. F7425; Sigma-Aldrich; Merck KGaA) diluted in TBS with 0.1%
Tween 20. Actin (1:2,000; cat. no. AC026; ABclonal Biotech Co.,
Ltd.) was used as the loading control.
MTT assay
Cell proliferation or cytotoxicity was evaluated
using an MTT assay. Subsequently, 25 µl MTT solution (5 mg/ml) was
added to each well and the cells were incubated (37°C) for a
further 4 h. The supernatant was removed from each well after
centrifugation with 2,504 × g at room temperature for 15 min. DMSO
was used to dissolve the colored formazan crystals produced from
the MTT added to each well (0.15 ml/well), and the optical density
(OD) values were measured at 490 nm.
RNA extraction and sequencing
RNA was extracted from transfected cells using
TRIzol reagent (cat. no. 15596-018; Ambion; Thermo Fisher
Scientific, Inc.) and was purified twice with phenol-chloroform.
RNA quality was determined by examining A260/A280 with a Nanodrop™
OneCspectrophotometer (Thermo Fisher Scientific, Inc.). RNA
Integrity was confirmed by 1.5% agarose gel electrophoresis.
Qualified RNAs were finally quantified by Qubit3.0 with a Qubit™
RNA Broad Range Assay kit (cat. no. Q10210; Thermo Fisher
Scientific, Inc.). In total, two biological replicates were
prepared for both FDPS-OE and empty vector control samples. For
each sample, 1 µg total RNA was used for RNA sequencing (RNA-seq)
library preparation with a VAHTS Stranded mRNA-seq Library Prep Kit
(cat. no. NR605-02; Vazyme Biotech Co., Ltd.). The libraries were
prepared according to the manufacturer's instructions and applied
to Hiseq X Ten Kit and HiSeq X/HD Reagent Kit v2.5 (300/Cycles)
(Illumina, Inc.) for library construction. An Illumina HiSeq X Ten
system was used for 150 nucleotide paired-end sequencing. The
loading concentration was 3 ng/µl, and the concentration was
finally quantified by Qubit3.0 (Thermo Fisher Scientific, Inc.).
For each RNA-seq sample, the FASTX-Toolkit (version 0.0.13;
http://hannonlab.cshl.edu/fastx_toolkit/) was used to
remove adaptors and low-quality reads. The filtered reads were
aligned onto the human genome (GRCh38 assembly) using TopHat2
v2.1.1 (35).
Differentially expressed gene (DEG)
and AS analysis
The gene expression levels were calculated as
fragments per kilobase of transcript per million fragments mapped
(FPKM) values. The statistical power of this experimental design,
calculated using RNASeqPower v1.38.0 (36), was 0.99 with two biological
replicates. The Bioconductor package edgeR v3.32.1 (37) was used to screen out the DEGs
between FDPS-OE and control cells. A false discovery rate (FDR)
<0.05 and fold change >2 or <0.5 were set as the cut-off
criteria for DEGs. A final power value of 0.90 was used to detect a
2-fold change in expression. Furthermore, Pearson's correlation
analysis was performed to assess sample distance.
The AS events (ASEs) and regulated ASEs (RASEs)
between the samples were defined and quantified using the ABLas
pipeline as previously described (38). A total of 10 types of ASEs were
detected based on the splice junction reads, including exon
skipping (ES), alternative 5′ splice site (A5SS), alternative 3′
splice site (A3SS), intron retention, mutually exclusive exons
(MXE), mutually exclusive 5′ untranslated regions, mutually
exclusive 3′ untranslated regions, cassette exon (CE), A3SS + ES
and A5SS + ES.
Fisher's exact test was used to calculate the
P-value. P<0.05 was considered to indicate a statistically
significant difference. The RASE ratio was calculated as the
changed ratio of alternatively spliced reads and constitutively
spliced reads between FDPS-OE and control samples. A RASE ratio
>0.2 and P<0.05 were set as the threshold for RASE
detection.
Reverse transcription-quantitative
(RT-q) PCR validation of DEGs and AS events in HOS cells
To validate the RNA-seq data and assess gene
overexpression, RT-qPCR was performed using FDPS-OE HOS cells for
selected DEGs. The primers for RT-qPCR analysis are listed in
Table I. A total of three
biological replicates for FDPS-OE and control samples were used for
RT-qPCR. cDNA synthesis was performed using a reverse transcription
kit (cat. no. R323-01; Vazyme Biotech Co., Ltd.) at 42°C for 5 min,
37°C for 15 min and 85°C for 5 sec, performed on a T100
thermocycler (Bio-Rad Laboratories, Inc.). qPCR was performed on
the ABI QuantStudio 5 (Thermo Fisher Scientific, Inc.) with the
following thermocycling conditions: Denaturing at 95°C for 10 min,
followed by 40 cycles of denaturing at 95°C for 15 sec and
annealing and extension at 60°C for 1 min. PCR amplifications were
performed in triplicate for each sample. qPCR was performed on a
Bio-Rad S1000 (Bio-Rad Laboratories, Inc.) with reverse
transcription kit (R323-01, Vazyme, China). The PCR conditions
consisted of denaturation at 95°C for 10 min, and 40 cycles of
denaturation at 95°C for 15 sec, and annealing and extension at
60°C for 1 min. For each sample, PCR amplifications were performed
in triplicate. By normalizing the cycle threshold of the control
housekeeping gene GAPDH, the expression levels of selected genes
were calculated using the 2−ΔΔCq formula (39).
 | Table I.Reverse transcription-quantitative
PCR primers used for gene expression and alternative splicing
quantification. |
Table I.
Reverse transcription-quantitative
PCR primers used for gene expression and alternative splicing
quantification.
| Gene name | Primer | Sequence
(5′-3′) |
|---|
| GAPDH | Forward |
CGGAGTCAACGGATTTGGTCGTAT |
|
| Reverse |
AGCCTTCTCCATGGTGGTGAAGAC |
| FDPS | Forward |
AGGGCAATGTGGATCTTGTC |
|
| Reverse |
GAAAGAACTCCCCCATCTCC |
| BMP1 | M/AS-Forward |
ATGGCAAGTTCTGTGGTTC |
|
| AS-Reverse |
GGCCTCTTTTCTGAGAAGAAG |
|
| M-Reverse |
TCGTCCTTGTCTGAGAAGAAG |
| SEMA4D | M/AS-Forward |
TCCACATTTCCCAGTTCTCC |
|
| AS-Reverse |
TAAGATACAGCATTTCTTCTG |
|
| M-Reverse |
CTGATGGTTTGCATTTCTTCTG |
| ANXA2 | M/AS-Forward |
CAGGTGCCTTTTGTATCC |
|
| AS-Reverse |
GCTTTCAAAAAGGGTGAAAATG |
|
| M-Reverse |
CACGGCCCAGGGTGAAAATG |
| SIRT2 | M-Forward |
AATCTGAGTCGGTCTGGCTC |
|
| AS-Forward |
AGGGTGAGAGGGGTCTGGCTC |
|
| M/AS-Reverse |
GTAGTTCTGTGCCCTATCACG |
In addition, RT-qPCR was also performed as
aforementioned to analyze ASEs. The primers for detection of ASEs
are shown in Table I. A
boundary-spanning primer was used for the sequence encompassing the
junction of the constitutive exon and alternative exon, and an
opposing primer encompassing the constitutive exon was used for
detection of alternative isoforms. The boundary-spanning primer of
the alternative exon was designed according to the ‘model exon’ to
detect model splicing or according to the ‘altered exon’ to detect
altered splicing.
Functional annotation
The KOBAS 2.0 server (http://kobas.cbi.pku.edu.cn./) was used to carry out
Gene Ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes
(KEGG) enrichment analyses to examine the primary functions of DEGs
and differentially alternatively spliced genes (40). A hypergeometric test was used to
identify the enrichment of each pathway, while the FDR (<0.05)
was used to define the threshold of significance.
Other methods and statistical
analysis
Ggplot2 v3.3.5 (https://github.com/tidyverse/ggplot2) was used to
generate a volcano plot, heat maps and hierarchical clustering.
After normalizing the reads of each gene, an in-house script
(sogen) was used to visualize next-generation sequence data and
genomic annotations. An unpaired two-tailed Student's t-test was
conducted for comparisons between two groups using Excel (Microsoft
Corporation). The data are presented as the mean ± SD of least
three biological replicates, except for the RNA-seq data (two
biological replicates). P<0.05 was considered to indicate a
statistically significant difference. The Cancer Genome Atlas
(TCGA) database was used to verify the FDPS expression levels and
the prognostic effect of FDPS in patients with cervical cancer and
other cancers via the Gene Expression Profiling Interactive
Analysis 2 (GEPIA2) web server (http://gepia2.cancer-pku.cn/#index) (41). For overall survival analysis,
cervical squamous cell carcinoma (CESC) patients (n=3,934) were
divided into two groups by auto select best cutoff of FDPS
expression level using the Kaplan-Meier Plotter web site (42).
Results
FDPS may promote proliferation in HeLa
cells and is associated with multiple types of cancer
A previous RNA interactome study revealed the RNA
binding ability of FDPS in HeLa cells (21), while the regulatory functions of
FDPS binding to RNA remained unknown. Thus, HeLa cells were used to
investigate FDPS functions on the transcriptome, and HOS cells were
used to validate its potential targets. An FDPS-OE plasmid was
constructed and transfected into HeLa and HOS cells. The effect of
FDPS-OE in both HeLa (Fig. 1A) and
HOS cells (Fig. 1B) was assessed by
RT-qPCR. Western blot analysis was also conducted to confirm
FDPS-OE in HeLa cells (Fig. 1C).
Using an MTT assay as the detection method, it was observed that
FDPS-OE resulted in an increase in OD value (Fig. 1D), suggesting that HeLa cells may
proliferate more quickly in the FDPS-OE group (P<0.01) than in
the control group. Using the GEPIA2 web server, the RNA levels of
FDPS were determined to be higher in CESC tumor samples than in
control samples (Fig. 1E), and
patients with a higher FDPS expression level had a worse prognosis
(Fig. 1F). According to GEPIA2
analysis result of TCGA data, FDPS expression was also dysregulated
in other types of cancer, including colon adenocarcinoma, lymphoid
neoplasm diffuse large B-cell lymphoma, liver hepatocellular
carcinoma, pancreatic adenocarcinoma, rectum adenocarcinoma,
thymoma, kidney chromophobe and acute myeloid leukemia (Fig. S1). These results suggest that FDPS
serves important regulatory functions in multiple types of
cancer.
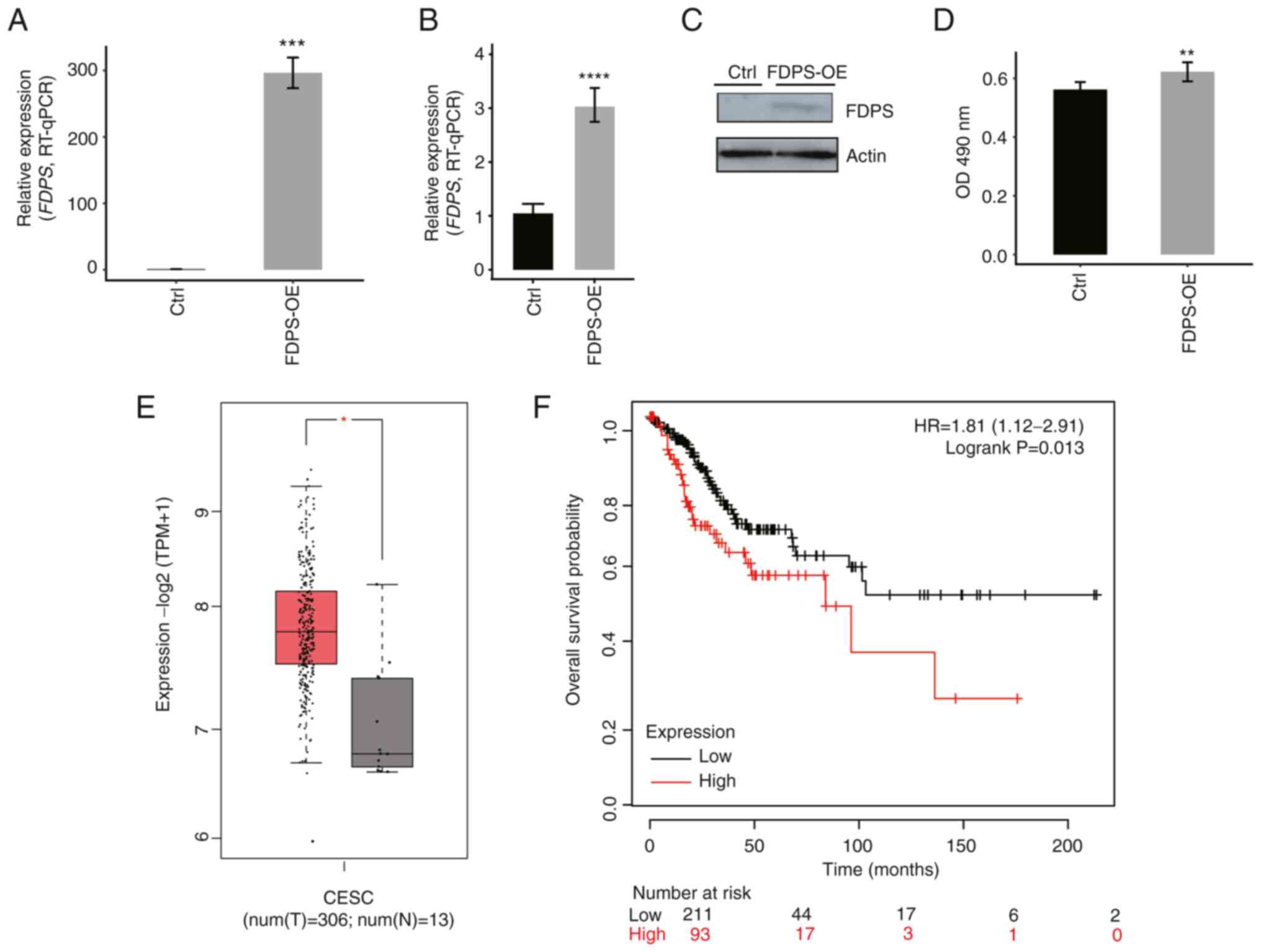 | Figure 1.FDPS-OE promotes the proliferation
rate of HeLa cells and influences patients with CESC. (A) FDPS-OE
in HeLa cells was quantified by RT-qPCR. (B) FDPS-OE in HOS cells
was quantified by RT-qPCR. (C) FDPS-OE in HeLa cells was examined
by western blotting. (D) An MTT assay demonstrated that FDPS-OE
promoted the proliferation of HeLa cells. (E) According to The
Cancer Genome Atlas data, higher FDPS expression was observed in
the tumor tissues of patients with CESC. (F) When analyzing the
overall survival time, patients with CESC with higher FDPS
expression were found to have a worse prognosis than patients with
lower FDPS expression. Data are presented as the mean ± SD.
*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Ctrl, control; CESC, cervical squamous cell carcinoma; FDPS,
farnesyl diphosphate synthase; HR, hazard ratio; num(N), number of
normal tissues; num(T), number of tumor tissues; OD, optical
density; OE, overexpression; RT-qPCR, reverse
transcription-quantitative PCR; TPM, transcripts per million. |
FDPS-OE alters the global expression
profiles in HeLa cells
To investigate the RNA regulatory functions of FDPS
in HeLa cells, whole transcriptome sequencing (RNA-seq) experiments
were performed to identify its potential targets. The
polyadenylated RNAs from HeLa cells were captured after 48 h of
transfection with control or FDPS-OE plasmids. For each RNA-seq
sample, 70±5 million high-quality reads were obtained.
Subsequently, the filtered reads were aligned onto the human genome
(GRCh38 assembly) with a total aligned ratio of 91.18-92.63% and a
uniquely aligned ratio of 96.73-97.61%. The uniquely aligned reads
were then used for further analysis.
To compare the gene expression patterns between
FDPS-OE and control samples, the expression value of each gene was
calculated as FPKM, with 28,558 expressed genes being identified
from the RNA-seq analysis. The overexpression of FDPS was then
further validated in a parallel RNA-seq analysis using FPKM values
(Fig. 2A). All expressed genes were
used to calculate a correlation matrix based on Pearson's
correlation coefficient (PCC) among the samples. The PCC between
the FDPS-OE and control samples are presented in the diagonal of
the heat map in Fig. 2B, where the
two biological replicates were highly correlated.
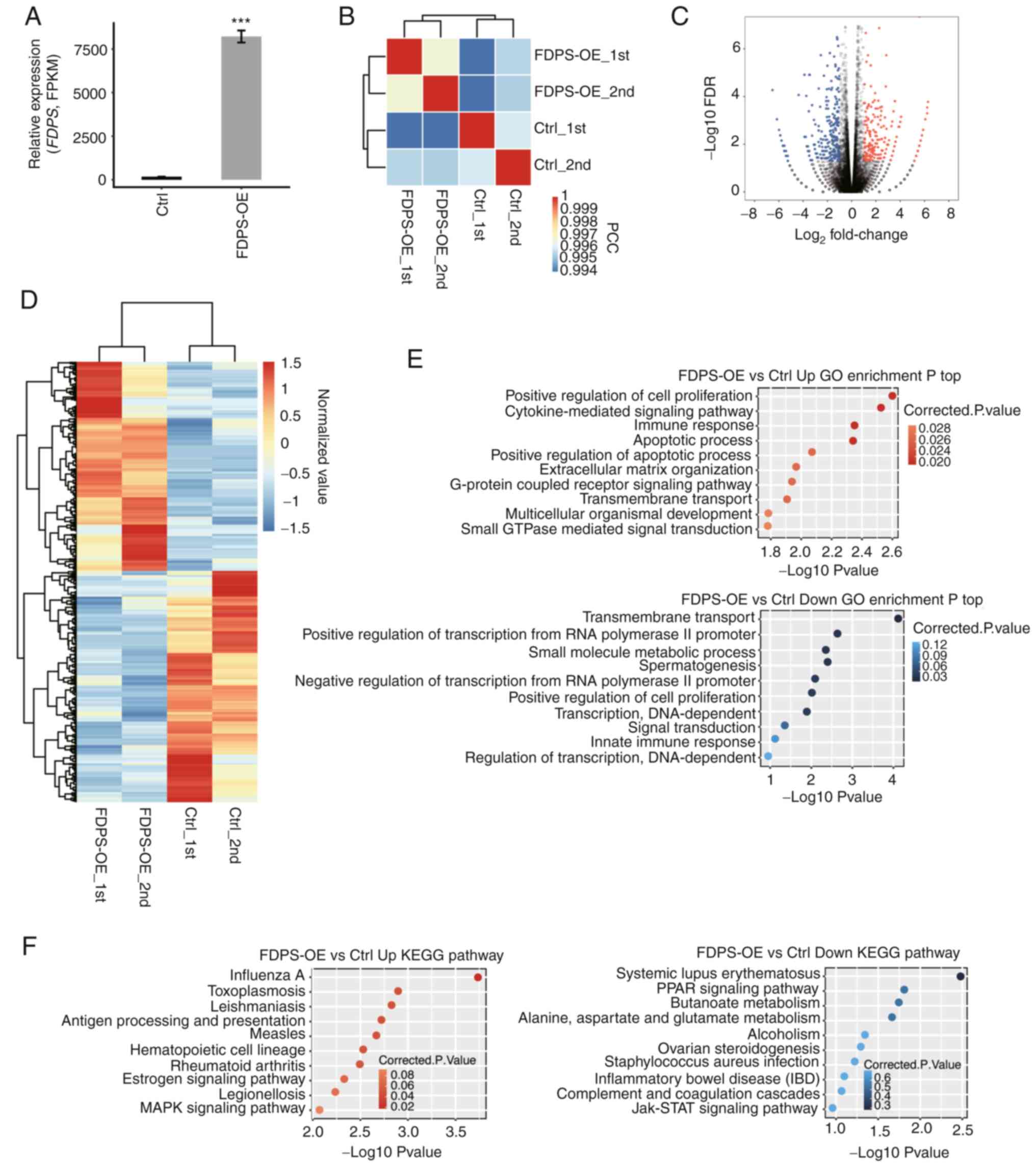 | Figure 2.RNA-seq analysis of FDPS-regulated
transcriptome profiles. (A) FDPS expression was quantified by
RNA-seq. Data are presented as the mean ± SD. (B) Heat map showing
the hierarchically clustered Pearson correlation matrix by
comparing the transcript expression values of Ctrl and FDPS-OE
samples. (C) Volcano plot showing the identified DEGs of FDPS-OE
compared with Ctrl samples. Upregulated genes are shown in red and
downregulated genes are shown in blue. (D) Hierarchical clustering
heatmap showing all DEGs in the Ctrl and FDPS-OE samples. FPKM
values of each gene were log2-transformed and median-centered. (E)
Top 10 representative GO biological processes of upregulated and
downregulated genes. (F) Top 10 representative KEGG pathways of
upregulated and downregulated genes. ***P<0.001. Ctrl, control;
DEGs, differentially expressed genes; FDPS, farnesyl diphosphate
synthase; FDR, false discovery rate; FPKM, fragments per kilobase
of transcript per million fragments mapped; GO, Gene Ontology; IBD,
inflammatory bowel disease; KEGG, Kyoto Encyclopedia of Genes and
Genomes; OE, overexpression; PPAR, peroxisome
proliferator-activated receptor; RNA-seq, RNA sequencing. |
To further investigate genes dysregulated by FDPS-OE
at the transcriptional level, edgeR was used to identify DEGs
between FDPS-OE and control samples. When the cut-off was set at
fold change >2 or <0.5 with a 5% FDR, the number of
upregulated and downregulated genes was 290 and 321, respectively,
indicating that FDPS had a binary effect on transcriptional
regulation (Fig. 2C; Table SI). In addition, according to the
heat map analysis of the expression patterns of all DEGs, there was
a clear separation between FDPS-OE and control samples and a high
consistency in both datasets (Fig.
2D). In short, the aforementioned results suggest that FDPS-OE
extensively regulates gene expression in HeLa cells.
To reveal the DEG-enriched functional pathways, GO
and KEEG enrichment analyses were performed to annotate all 611
DEGs. The upregulated and downregulated genes were enriched in 16
and 10 GO terms, respectively. Based on the biological process
terms of the GO analysis, the upregulated DEGs were mainly
associated with ‘positive regulation of cell proliferation’,
‘cytokine-mediated signaling pathway’, ‘immune response’,
‘apoptotic process’ and ‘extracellular matrix organization’, which
were important biological processes. By contrast, the downregulated
genes were mainly enriched in ‘transmembrane transport’, ‘positive
regulation of transcription from RNA polymerase II promoter’,
‘small molecule metabolic process’ and ‘positive regulation of cell
proliferation’(Fig. 2E). The
results of the KEGG enrichment analysis are shown in Fig. 2F, the results of which were similar
with that of the GO analysis.
Validation of DEGs associated with
osteoporosis in HOS cells
Previous studies have demonstrated that cell
proliferation (19,27), apoptosis (43,44)
and migration (28) are associated
with osteoporosis. Thus, several DEGs enriched in the ‘positive
regulation of cell proliferation’, ‘apoptotic process’ and
‘extracellular matrix organization’ pathways, including
IL24, interferon-induced proteins with tetratricopeptide
repeats 2 (IFIT2), colony stimulating factor 2, MMP19
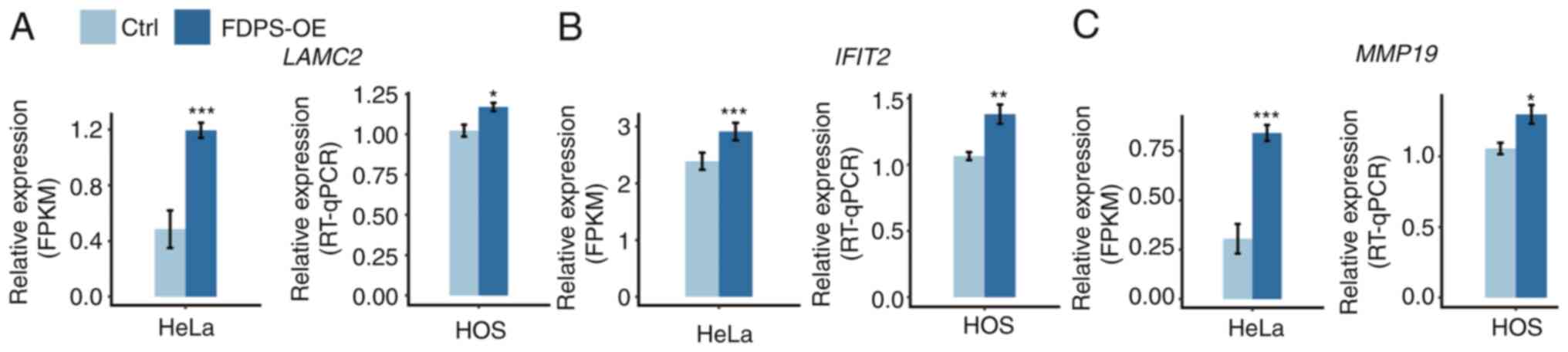
and laminin subunit γ2 (LAMC2), were screened in HeLa cells.
To determine whether these genes were also regulated by FDPS in
osteoporosis cell lines, HOS cells were transfected with FDPS-OE
plasmid and RT-qPCR experiments were performed. The results of
these experiments demonstrated that three out of the five selected
genes (LAMC2, IFIT2, and MMP19) exhibited similar
trends after FDPS-OE, consistent with the result of the RNA-seq
analysis in HeLa cells (Fig.
3).
FDPS regulates ASEs in HeLa cells
To comprehensively investigate the role of FDPS in
AS regulation, RNA-seq in HeLa cells was also used to explore
FDPS-meditated ASEs. The mean values of 65.3±4.0 million uniquely
mapped reads were obtained from FDPS-OE and control cells, of which
40.52-41.97% were junction reads. In total, ~68.40% annotated exons
(251,254 out of 367,321) were detected, together with 166,644 known
splice junctions and 233,935 novel splice junctions. Using an ABLas
program tool (38), 83,415 known
ASEs and 61,886 novel ASEs were found.
A stringent cut-off of P<0.05 and changed ratio
>0.2 was applied to identify RASEs with a high confidence
(Tables SII and SIII). The most prevalent FDPS RASEs were
A5SS (136 events), A3SS (95 events), CE (79 events) and ES (76
events) (Fig. 4A). These results
suggested that FDPS extensively regulated ASEs in HeLa cells. By
analyzing the intersection between DEGs and AS genes, it was found
that four genes were shared between DEGs and regulated AS genes
(RASGs). These four genes were dominated by long non-coding RNAs,
including RP1-179N16.6, RP5-884G6.2 and RP11-115D19.1
(Fig. 4B). Notably, these results
demonstrated that FDPS was also an RASG (Fig. 4B), suggesting that FDPS regulates
its own AS pattern.
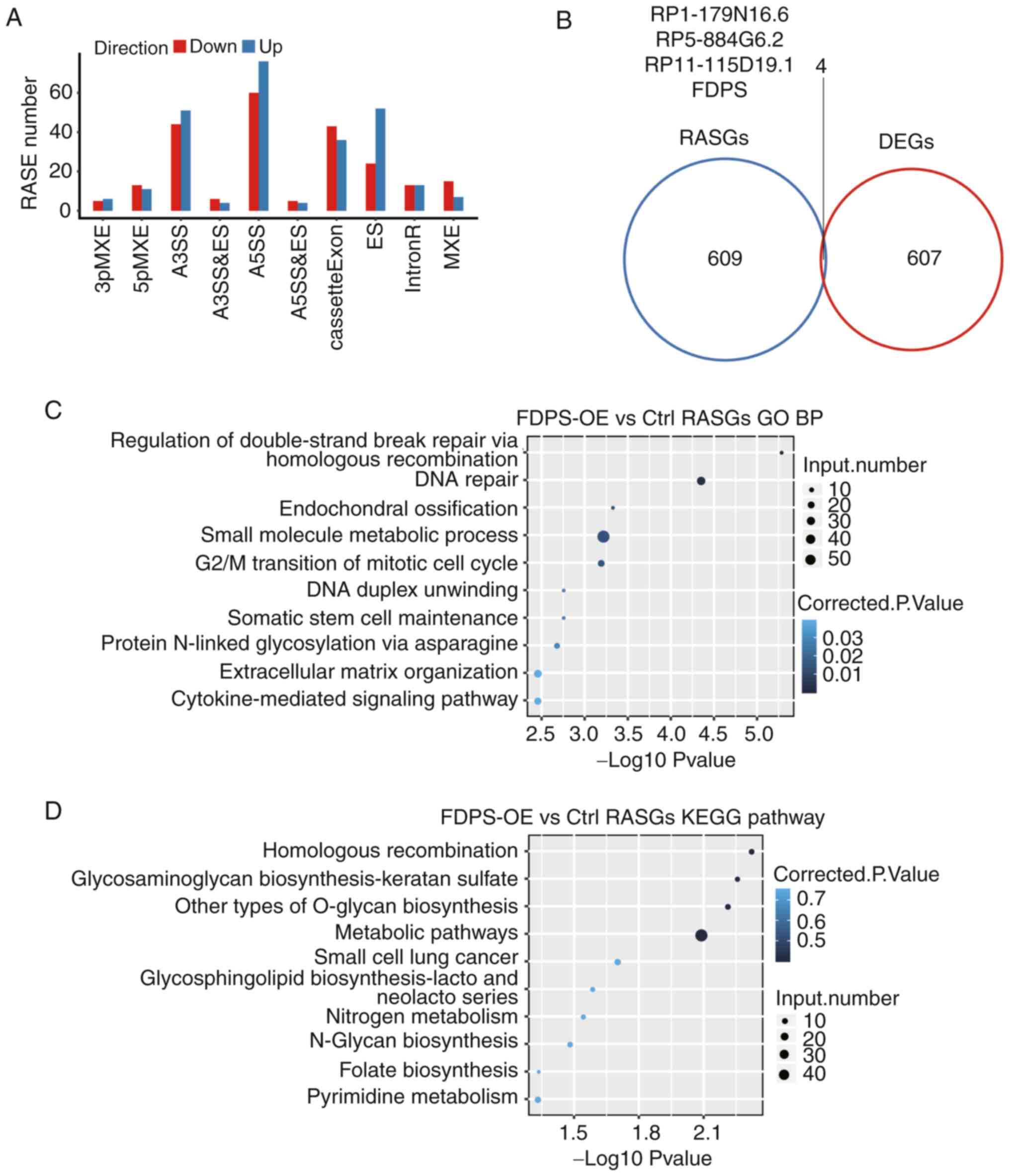 | Figure 4.Identification and functional
analysis of FDPS-regulated genes and ASEs. (A) Bar plot showing the
classification of FDPS-regulated ASEs. (B) Venn diagram showing the
overlap of FDPS-regulated DEGs and RASGs. (C) Top 10 enriched GO
biological processes of FDPS-regulated alternatively spliced genes.
(D) Top 10 enriched KEGG pathways of FDPS-regulated alternatively
spliced genes. 3pMXE, MXE 3′ untranslated regions; 5pMXE, MXE 5′
untranslated regions; A3SS, alternative 3′ splice site; A5SS,
alternative 5′ splice site; ASEs, alternative splicing events;
Ctrl, control; DEGs, differentially expressed genes; ES, exon
skipping; FDPS, farnesyl diphosphate synthase; GO, Gene Ontology;
IntronR, intron retention; KEGG, Kyoto Encyclopedia of Genes and
Genomes; MXE, mutually exclusive; RASE, regulated ASE; RASGs,
regulated alternative splicing genes. |
GO functional clustering analysis demonstrated that
the AS genes were enriched in the ‘regulation of double-strand
break repair via homologous recombination’, ‘DNA repair’ (not via
homologous recombination), ‘endochondral ossification’, ‘G2/M
transition of mitotic cell cycle’ and ‘DNA duplex unwinding’
(Fig. 4C). Enriched KEGG pathways
(P<0.05) included pathways involved in ‘homologous
recombination’, ‘glycosaminoglycan biosynthesis-keratan sulfate’,
‘other types of O-glycan biosynthesis’, ‘glycosphingolipid
biosynthesis-lacto and neolacto series’ and ‘N-Glycan
biosynthesis’, which were important pathways (Fig. 4D).
As shown in the GO functional clustering analysis,
FDPS-regulated AS genes were enriched in the ‘endochondral
ossification’ pathway, which is associated with altering bone mass
in osteoporosis (29). Therefore,
three FDPS-regulated splicing events located in the alkaline
phosphatase biomineralization associated, fibroblast growth factor
receptor 3 and NGF1-A binding protein 1 genes were validated by
RT-qPCR (data not shown). Several alternatively spliced genes were
also validated by RT-qPCR in HOS cells, including bone morphogenic
protein 1 (BMP1), semaphorin 4D (SEMA4D), annexin A2
(ANXA2) and sirtuin 2 (SIRT2). Although these genes
were not enriched in the top 10 pathways, they are associated with
osteoporosis. The RT-qPCR results of four of the seven RASGs
(BMP1, SEMA4D, ANXA2 and SIRT2) were consistent with
the results of the transcriptome analysis in HeLa cells (Fig. 5).
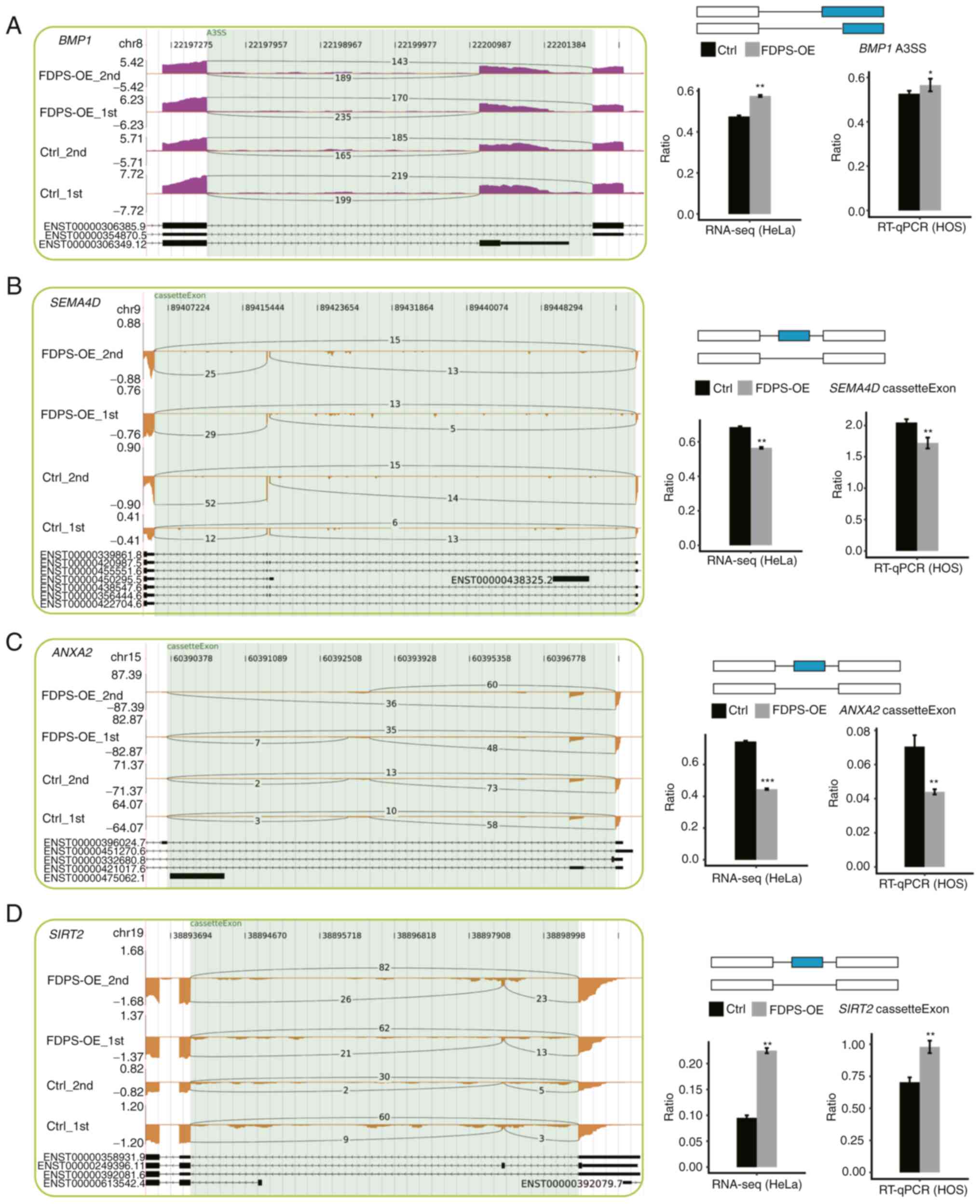 | Figure 5.FDPS regulates the AS of genes
involved in osteoporosis. Integrative genomics viewer (IGV)-sashimi
plots showing different ASEs in four genes, including (A)
BMP1, (B) SEMA4D, (C) ANXA2 and (D)
SIRT2. The reads distribution of each RASE is plotted in the
left panel with the transcript ID and structure of each gene shown
below. The schematic diagrams depict the structures of ASEs, AS1
(top) and AS2 (bottom) of top right panel. The constitutive exons
are denoted by white boxes, intron sequences by a horizontal line
(right; top panel) and alternative exons by blue boxes. RNA-seq
quantification in HeLa cells and RT-qPCR validation of ASEs in HOS
cells are shown in the bottom right panel. The data are presented
as the mean ± SD. *P<0.05, **P<0.01, ***P<0.001. A3SS,
alternative 3′ splice site; ANXA2, annexin A2; AS, alternative
splicing; ASE, AS event; BMP1, bone morphogenic protein 1; Ctrl,
control; ENST, prefix of transcript ID ensembl transcript; FDPS,
farnesyl diphosphate synthase; OE, overexpression; RNA-seq, RNA
sequencing; RT-qPCR, reverse transcription-quantitative PCR;
SEMA4D, semaphorin 4D; SIRT2, sirtuin 2. |
Discussion
As an essential enzyme in the isoprenoid
biosynthetic pathway, FDPS supplies precursors to synthesize
important isoprenoids, such as sterols, ubiquinones, carotenoids
and dolichols (45). Aberrant FDPS
expression is associated with disease, particularly cancer. For
example, elevated expression of FDPS has been found in a number of
human malignant tumors, including glioblastoma (23) and prostate cancer (19). FDPS has been revealed to be a
potential RBP, while the genome-wide target genes of FDPS remain to
be determined (3). We hypothesize
that FDPS may globally regulate gene expression and AS, eventually
participating in osteoporosis through binding to RNAs.
In the present study, RNA-seq was used to perform
the transcriptome analysis of FDPS-OE in HeLa cells. To the best of
our knowledge, this was the first study to investigate the role of
FDPS as a transcriptome regulator from a genome-wide perspective.
Furthermore, FDPS-regulated DEGs and RASGs were validated in
FDPS-OE HOS cells, indicating that FDPS may modulate transcriptome
profiles in bone cells. FDPS has been demonstrated to promote bone
resorption, and thus, is indispensable in osteoporosis development
(7). In the present study, it was
demonstrated that FDPS regulated the mRNA levels and the AS of
genes involved in the ‘positive regulation of cell proliferation’,
‘apoptotic process’, ‘extracellular matrix organization’ and
‘endochondral ossification’ pathways, which may provide a novel
perspective in understanding FDPS biology and regulatory
mechanisms.
The level of cell proliferation was demonstrated to
be increased significantly in FDPS-OE cells, which was consistent
with the previous finding that FDPS had an important role in
promoting cell proliferation (23,46).
Additionally, due to FDPS-OE, several genes associated with
‘positive regulation of cell proliferation’, ‘immune response’ and
‘extracellular matrix organization’ pathways were upregulated,
including IFIT2, MMP19 and LAMC2, which may benefit
the survival, proliferation and migration of cancer cells. IFIT2,
an IFN-stimulated gene, is a tumor suppressor that inhibits
proliferation and migration, while promoting the apoptosis of
cancer cells in a number of tumor types (47–50).
In addition to cancer, IFIT2 dysregulation has also been reported
to be associated with osteoporosis. Gao et al (51) demonstrated that IFIT2 serves
crucial roles in the fracture repair process in osteoporosis,
although the exact mechanisms remain elusive. In the present study,
IFIT2 upregulation was induced by FDPS-OE, implying novel
regulating mechanisms of FDPS in osteoporosis development.
In the present study, it was demonstrated that FDPS
regulated the expression of genes involved in ‘extracellular matrix
organization’. MMP1, MMP2, MMP9 and MMP13 are expressed in bone
tissues and serve key roles in the digestion of bone matrix by
osteoblasts. MMP13 is also positively associated with bone mineral
density (52,53). By contrast, MMP2 and MMP9 are
negatively associated with bone mineral density (22). MMP19 is also involved in the
breakdown of extracellular matrix in normal physiological
processes, such as embryonic development, reproduction and tissue
remodeling (54). However, to the
best of our knowledge, the function of MMP19 in osteoporosis
remains unknown. Upregulation of MMP19 induced by FDPS-OE in the
present study indicated that MMP19 may decrease bone mineral
density in a manner analogous to MMP2 and MMP9. However, further
investigation is required. LAMC2, a member of the extracellular
matrix glycoprotein family, is the major component of basement
membranes. LAMC2 is involved in various biological
processes, including cell adhesion, differentiation, migration,
invasion, traction force and metastasis (55). LAMC2 expression was
upregulated by FDPS-OE in the present study. However, to the best
of our knowledge, the role of LAMC2 in osteoporosis has not
yet been reported. In summary, as an RBP, FDPS may participate in
osteoporosis by regulating the expression levels of genes involved
in bone mineral density or osteoclastogenesis.
AS removes pre-mRNA introns to generate mature mRNAs
and is associated with the development of numerous diseases,
including cancer (53),
cardiovascular diseases (54) and
neurological diseases (56,57). Systematic analysis has demonstrated
that AS changes in tumors may represent independent oncogenic
processes that greatly affect cancer transformations (58). Additionally, AS also serves
important roles in skeletal diseases (59–61).
The cytokine, TGF-β, which controls bone density, exists as three
isoforms, with only upregulation of TGF-β3 being associated with
osteoporosis in patients (62).
Similarly, specific isoforms of the human calcitonin receptor and
tartrate-resistant acid phosphatase are associated with
osteoporosis (63,64). Taken together, these results suggest
that the AS of genes is important for osteoporosis formation.
However, there are relatively few studies on AS in osteoporosis
(59). In the present study, large
numbers of ASEs were detected in HeLa cells after FDPS-OE,
including A3SS of BMP1 and CE of SRIT2, SEMA4D and
ANXA2. Furthermore, these RASEs were validated in FDPS-OE
HOS cells, which is a popular cell line for the study of bone
formation (65,66) or osteoporosis (67). Jing et al (68) observed an increase in bone mass
density and bone volume fraction in SIRT2 knockout rats.
Their study suggested that SIRT2 serves a role in age-related bone
loss, likely via regulation of osteoclastogenesis. SEMA4D,
previously regarded as an axon guidance protein, has been
demonstrated to inhibit bone formation (69) and promote bone resorption (70). Previously, a molecular and cellular
basis of BMP1-dependent osteoporosis has been defined both in
humans and in zebrafish (71,72).
These findings indicate that BMP1 is essential for bone formation
and stability. The BMP1 gene encodes several isoforms,
including BMP1 and mammalian tolloid, which proteolytically remove
the C-terminal propetide from procollagen (72). In addition, BMP1-3, an isoform of
the BMP1 gene, is elevated in patients with acute bone
fracture, and may be involved in bone repair (73). Whether these two isoforms of
BMP1 have an impact on osteoporosis formation needs further
validation through knockdown studies or through overexpressing
different isoforms. Another finding of the present study was that
FDPS may regulate its own AS, which may be a feedback mechanism for
FDPS upregulation, and thus, FDPS may regulate its own functions in
HeLa cells. However, how FDPS regulates AS and gene expression has
not been investigated in the present study. Experiments, such as
pull-down or gel shift assays, should be performed in future
studies.
In conclusion, in the present study, RNA-seq
technology was applied to demonstrate how FDPS may regulate gene
expression and AS in HeLa cells. It was demonstrated that genes
critical in cell proliferation were upregulated by FDPS-OE. It was
also demonstrated that the AS of genes implicated in the cell cycle
was also regulated by FDPS. These results suggest that, as an RBP,
FDPS may serve an important role in HeLa and HOS cells by
modulating mRNA expression at transcriptional and
post-transcriptional levels through binding to precursor mRNAs.
Further studies are required to identify the molecular mechanisms
in which FDPS regulates gene expression and AS, such as the
regulatory mechanism of FDPS, to make up the flaws in the present
study. In summary, the present study contributes to the
understanding of FDPS-targeted therapies.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by The Natural Science Foundation of
Jilin Province (grant no. 2018010113JC).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Gene Expression Omnibus
repository, with accession number GSE151605 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151605).
Authors' contributions
LW and HC contributed to the study conception and
design. Material preparation, data collection and analyses were
performed by BK, DC, ZC and YH. The first draft of the manuscript
was written by LW, ZC and HC. LW, ZC, DC, BK, YH and HC confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liang PH, Ko TP and Wang AHJ: Structure,
mechanism and function of prenyltransferases. Eur J Biochem.
269:3339–3354. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Waller DD, Park J and Tsantrizos YS:
Inhibition of farnesyl pyrophosphate (FPP) and/or geranylgeranyl
pyrophosphate (GGPP) biosynthesis and its implication in the
treatment of cancers. Crit Rev Biochem Mol Biol. 54:41–60. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Notarnicola M, Messa C, Cavallini A,
Bifulco M, Tecce MF, Eletto D, Di Leo A, Montemurro S, Laezza C and
Caruso MG: Higher farnesyl diphosphate synthase activity in human
colorectal cancer inhibition of cellular apoptosis. Oncology.
67:351–358. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bouterfa HL, Sattelmeyer V, Czub S,
Vordermark D, Roosen K and Tonn JC: Inhibition of Ras farnesylation
by lovastatin leads to downregulation of proliferation and
migration in primary cultured human glioblastoma cells. Anticancer
Res. 20:2761–2771. 2000.PubMed/NCBI
|
|
5
|
Woo IS, Eun SY, Kim HJ, Kang ES, Kim HJ,
Lee JH, Chang KC, Kim JH, Hong SC and Seo HG: Farnesyl diphosphate
synthase attenuates paclitaxel-induced apoptotic cell death in
human glioblastoma U87MG cells. Neurosci Lett. 474:115–120. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marini F, Falchetti A, Silvestri S, Bagger
Y, Luzi E, Tanini A, Christiansen C and Brandi ML: Modulatory
effect of farnesyl pyrophosphate synthase (FDPS) rs2297480
polymorphism on the response to long-term amino-bisphosphonate
treatment in postmenopausal osteoporosis. Curr Med Res Opin.
24:2609–2615. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Olmos JM, Zarrabeitia MT, Hernández JL,
Sañudo C, González-Macías J and Riancho JA: Common allelic variants
of the farnesyl diphosphate synthase gene influence the response of
osteoporotic women to bisphosphonates. Pharmacogenomics J.
12:227–232. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ciubean AD, Ungur RA, Irsay L, Ciortea VM,
Borda IM, Dogaru GB, Trifa AP, Vesa SC and Buzoianu AD:
Polymorphisms of FDPS, LRP5, SOST and VKORC1 genes and their
relation with osteoporosis in postmenopausal Romanian women. PLoS
One. 14:e02257762019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wiemer A, Hohl R and Wiemer D: The
intermediate enzymes of isoprenoid metabolism as anticancer
targets. Anticancer Agents Med Chem. 9:526–542. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dunford JE, Kwaasi AA, Rogers MJ, Barnett
BL, Ebetino FH, Russell RG, Oppermann U and Kavanagh KL:
Structure-activity relationships among the nitrogen containing
bisphosphonates in clinical use and other analogues: Time-dependent
inhibition of human farnesyl pyrophosphate synthase. J Med Chem.
51:2187–2195. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang J, Zhu HH, Chen GP, Ye Y, Zhao CZ,
Mou Y and Hu SJ: Inhibition of farnesyl pyrophosphate synthase
attenuates angiotensin II-induced cardiac hypertrophy and fibrosis
in vivo. Int J Biochem Cell Biol. 45:657–666. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fontalis A and Eastell R: The challenge of
long-term adherence: The role of bone turnover markers in
monitoring bisphosphonate treatment of osteoporosis. Bone.
136:1153362020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kamimura M, Ikegami S, Mukaiyama K, Koiwai
H, Nakamura Y, Taguchi A and Kato H: Additive effects of
eldecalcitol in poorly responding long-term bisphosphonate
treatment for osteoporosis. Osteoporos Sarcopenia. 5:57–61. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kavanagh KL, Guo K, Dunford JE, Wu X,
Knapp S, Ebetino FH, Rogers MJ, Russell RG and Oppermann U: The
molecular mechanism of nitrogen-containing bisphosphonates as
antiosteoporosis drugs. Proc Natl Acad Sci USA. 103:7829–7834.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
LaFleur J, DuVall SL, Willson T, Ginter T,
Patterson O, Cheng Y, Knippenberg K, Haroldsen C, Adler RA, Curtis
JR, et al: Analysis of osteoporosis treatment patterns with
bisphosphonates and outcomes among postmenopausal veterans. Bone.
78:174–185. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Coxon FP and Rogers MJ: The role of
prenylated small GTP-binding proteins in the regulation of
osteoclast function. Calcif Tissue Int. 72:80–84. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Russell RG, Watts NB, Ebetino FH and
Rogers MJ: Mechanisms of action of bisphosphonates: Similarities
and differences and their potential influence on clinical efficacy.
Osteoporos Int. 19:733–759. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jobke B, Milovanovic P, Amling M and Busse
B: Bisphosphonate-osteoclasts: Changes in osteoclast morphology and
function induced by antiresorptive nitrogen-containing
bisphosphonate treatment in osteoporosis patients. Bone. 59:37–43.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Seshacharyulu P, Rachagani S, Muniyan S,
Siddiqui JA, Cruz E, Sharma S, Krishnan R, Killips BJ, Sheinin Y,
Lele SM, et al: FDPS cooperates with PTEN loss to promote prostate
cancer progression through modulation of small GTPases/AKT axis.
Oncogene. 38:5265–5280. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Castello A, Hentze MW and Preiss T:
Metabolic enzymes enjoying new partnerships as RNA-binding
proteins. Trends Endocrinol Metab. 26:746–757. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Castello A, Fischer B, Eichelbaum K, Horos
R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T,
Steinmetz LM, et al: Insights into RNA biology from an atlas of
mammalian mRNA-binding proteins. Cell. 149:1393–1406. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li H, Xie S, Liu X, Wu H, Lin X, Gu J,
Wang H and Duan Y: Matrine alters microRNA expression profiles in
SGC-7901 human gastric cancer cells. Oncol Rep. 32:2118–2126. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Abate M, Laezza C, Pisanti S, Torelli G,
Seneca V, Catapano G, Montella F, Ranieri R, Notarnicola M,
Gazzerro P, et al: Deregulated expression and activity of farnesyl
diphosphate synthase (FDPS) in glioblastoma. Sci Rep. 7:141232017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chatrikhi R, Mallory MJ, Gazzara MR,
Agosto LM, Zhu WS, Litterman AJ, Ansel KM and Lynch KW: RNA binding
protein CELF2 regulates signal-induced alternative polyadenylation
by competing with enhancers of the polyadenylation machinery. Cell
Rep. 28:2795–2806.e3. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fuller K, Owens JM, Jagger CJ, Wilson A,
Moss R and Chambers TJ: Macrophage colony-stimulating factor
stimulates survival and chemotactic behavior in isolated
osteoclasts. J Exp Med. 178:1733–1744. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shinoda K, Suda A, Otonari K, Futaki S and
Imanishi M: Programmable RNA methylation and demethylation using
PUF RNA binding proteins. Chem Commun (Camb). 56:1365–1368. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fagoonee S, Picco G, Orso F, Arrigoni A,
Longo DL, Forni M, Scarfò I, Cassenti A, Piva R, Cassoni P, et al:
The RNA-binding protein ESRP1 promotes human colorectal cancer
progression. Oncotarget. 8:10007–10024. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rauwel B, Degboé Y, Diallo K, Sayegh S,
Baron M, Boyer JF, Constantin A, Cantagrel A and Davignon JL:
Inhibition of osteoclastogenesis by the RNA-binding protein QKI5: A
novel approach to protect from bone resorption. J Bone Miner Res.
35:753–765. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kang MH, Jeong KJ, Kim WY, Lee HJ, Gong G,
Suh N, Győrffy B, Kim S, Jeong SY, Mills GB and Park YY: Musashi
RNA-binding protein 2 regulates estrogen receptor 1 function in
breast cancer. Oncogene. 36:1745–1752. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sherman EJ, Mitchell DC and Garner AL: The
RNA-binding protein SART3 promotes miR-34a biogenesis and G1 cell
cycle arrest in lung cancer cells. J Biol Chem. 294:17188–17196.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang E and Aifantis I: RNA splicing and
cancer. Trends Cancer. 6:631–644. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen J and Weiss WA: Alternative splicing
in cancer: Implications for biology and therapy. Oncogene. 34:1–14.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sugiyama M, Kodama T, Konishi K, Abe K,
Asami S and Oikawa S: Compactin and simvastatin, but not
pravastatin, induce bone morphogenetic protein-2 in human
osteosarcoma cells. Biochem Biophys Res Commun. 271:688–692. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tu Y, Wu X, Yu F, Dang J, Wang J, Wei Y,
Cai Z, Zhou Z, Liao W, Li L and Zhang Y: Tristetraprolin
specifically regulates the expression and alternative splicing of
immune response genes in HeLa cells. BMC Immunol. 20:132019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim D, Pertea G, Trapnell C, Pimentel H,
Kelley R and Salzberg SL: TopHat2: Accurate alignment of
transcriptomes in the presence of insertions, deletions and gene
fusions. Genome Biol. 14:R362013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Therneau T, Hart S and Kocher J:
Calculating samplesSize estimates for RNA Seq studies. R package
version 1.36.0. 2022.
|
|
37
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xia H, Chen D, Wu Q, Wu G, Zhou Y, Zhang Y
and Zhang L: CELF1 preferentially binds to exon-intron boundary and
regulates alternative splicing in HeLa cells. Biochim Biophys Acta
Gene Regul Mech. 1860:911–921. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39((Web Server Issue)): W316–W322. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47((W1)): W556–W560.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nagy Á, Munkácsy G and Győrffy B:
Pancancer survival analysis of cancer hallmark genes. Sci Rep.
11:60472021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fujiwara T, Zhou J, Ye S and Zhao H:
RNA-binding protein Musashi2 induced by RANKL is critical for
osteoclast survival. Cell Death Dis. 7:e23002016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li X, Ominsky MS, Villasenor KS, Niu QT,
Asuncion FJ, Xia X, Grisanti M, Wronski TJ, Simonet WS and Ke HZ:
Sclerostin antibody reverses bone loss by increasing bone formation
and decreasing bone resorption in a rat model of male osteoporosis.
Endocrinology. 159:260–271. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dhar MK, Koul A and Kaul S: Farnesyl
pyrophosphate synthase: A key enzyme in isoprenoid biosynthetic
pathway and potential molecular target for drug development. N
Biotechnol. 30:114–123. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ishimoto K, Tachibana K, Hanano I,
Yamasaki D, Nakamura H, Kawai M, Urano Y, Tanaka T, Hamakubo T,
Sakai J, et al: Sterol-regulatory-element-binding protein 2 and
nuclear factor Y control human farnesyl diphosphate synthase
expression and affect cell proliferation in hepatoblastoma cells.
Biochem J. 429:347–357. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shen H, Zhan M, Zhang Y, Huang S, Xu S,
Huang X, He M, Yao Y, Man M and Wang J: PLZF inhibits proliferation
and metastasis of gallbladder cancer by regulating IFIT2. Cell
Death Dis. 9:712018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen L, Zhai W, Zheng X, Xie Q, Zhou Q,
Tao M, Zhu Y, Wu C and Jiang J: Decreased IFIT2 expression promotes
gastric cancer progression and predicts poor prognosis of the
patients. Cell Physiol Biochem. 45:15–25. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ohsugi T, Yamaguchi K, Zhu C, Ikenoue T
and Furukawa Y: Decreased expression of interferon-induced protein
2 (IFIT2) by Wnt/β-catenin signaling confers anti-apoptotic
properties to colorectal cancer cells. Oncotarget. 8:100176–100186.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Su W, Xiao W, Chen L, Zhou Q, Zheng X, Ju
J, Jiang J and Wang Z: Decreased IFIT2 expression in human
non-small-cell lung cancer tissues is associated with cancer
progression and poor survival of the patients. Onco Targets Ther.
12:8139–8149. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gao F, Xu F, Wu D, Cheng J and Xia P:
Identification of novel genes associated with fracture healing in
osteoporosis induced by Krm2 overexpression or Lrp5 deficiency. Mol
Med Rep. 15:3969–3976. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Paiva KB and Granjeiro JM: Bone tissue
remodeling and development: Focus on matrix metalloproteinase
functions. Arch Biochem Biophys. 561:74–87. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Mazur CM, Woo JJ, Yee CS, Fields AJ,
Acevedo C, Bailey KN, Kaya S, Fowler TW, Lotz JC, Dang A, et al:
Osteocyte dysfunction promotes osteoarthritis through
MMP13-dependent suppression of subchondral bone homeostasis. Bone
Res. 7:342019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jara P, Calyeca J, Romero Y, Plácido L, Yu
G, Kaminski N, Maldonado V, Cisneros J, Selman M and Pardo A:
Matrix metalloproteinase (MMP)-19-deficient fibroblasts display a
profibrotic phenotype. Am J Physiol Lung Cell Mol Physiol.
308:L511–L522. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Garg M, Braunstein G and Koeffler HP:
LAMC2 as a therapeutic target for cancers. Taylor & Francis;
pp. 979–982. 2014, PubMed/NCBI
|
|
56
|
Vuong CK, Black DL and Zheng S: The
neurogenetics of alternative splicing. Nat Rev Neurosci.
17:265–281. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Raj T, Li YI, Wong G, Humphrey J, Wang M,
Ramdhani S, Wang YC, Ng B, Gupta I, Haroutunian V, et al:
Integrative transcriptome analyses of the aging brain implicate
altered splicing in Alzheimer's disease susceptibility. Nat Genet.
50:1584–1592. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Climente-Gonzalez H, Porta-Pardo E, Godzik
A and Eyras E: The functional impact of alternative splicing in
cancer. Cell Rep. 20:2215–2226. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fan X and Tang L: Aberrant and alternative
splicing in skeletal system disease. Gene. 528:21–26. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Han Y, Wang D, Guo J, Xiong Q, Li P, Zhou
YA and Zhao B: A novel splicing pathogenic variant in COL1A1
causing osteogenesis imperfecta (OI) type I in a Chinese family.
Mol Genet Genomic Med. 8:e13662020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kimura T, Lueck JD, Harvey PJ, Pace SM,
Ikemoto N, Casarotto MG, Dirksen RT and Dulhunty AF: Alternative
splicing of RyR1 alters the efficacy of skeletal EC coupling. Cell
Calcium. 45:264–274. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Grainger DJ, Percival J, Chiano M and
Spector TD: The role of serum TGF-beta isoforms as potential
markers of osteoporosis. Osteoporos Int. 9:398–404. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Beaudreuil J, Taboulet J, Orcel P, Graulet
AM, Denne MA, Baudoin C, Jullienne A and De Vernejoul MC:
Calcitonin receptor mRNA in mononuclear leucocytes from
postmenopausal women: Decrease during osteoporosis and link to bone
markers with specific isoform involvement. Bone. 27:161–168. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Janckila AJ, Takahashi K, Sun SZ and Yam
LT: Tartrate-resistant acid phosphatase isoform 5b as serum marker
for osteoclastic activity. Clin Chem. 47:74–80. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang L, Park P, La Marca F, Than K, Rahman
S and Lin CY: Bone formation induced by BMP-2 in human osteosarcoma
cells. Int J Oncol. 43:1095–1102. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Anderson PH, Atkins GJ, Findlay DM,
Oloughlin PD, Welldon K, Vincent C and Morris HA: RNAi-mediated
silencing of CYP27B1 abolishes 1,25(OH)2D3 synthesis and reduces
osteocalcin and CYP24 mRNA expression in human osteosarcoma (HOS)
cells. J Steroid Biochem Mol Biol. 103:601–605. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Trost Z, Trebse R, Prezelj J, Komadina R,
Logar DB and Marc J: A microarray based identification of
osteoporosis-related genes in primary culture of human osteoblasts.
Bone. 46:72–80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Jing Y, Zhou Y, Zhou F, Wang X, Tao B, Sun
L, Liu J and Zhao H: SIRT2 deficiency prevents age-related bone
loss in rats by inhibiting osteoclastogenesis. Cell Mol Biol
(Noisy-le-grand). 65:66–71. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Negishi-Koga T, Shinohara M, Komatsu N,
Bito H, Kodama T, Friedel RH and Takayanagi H: Suppression of bone
formation by osteoclastic expression of semaphorin 4D. Nat Med.
17:1473–1480. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Terpos E, Ntanasis-Stathopoulos I,
Christoulas D, Bagratuni T, Bakogeorgos M, Gavriatopoulou M,
Eleutherakis-Papaiakovou E, Kanellias N, Kastritis E and Dimopoulos
MA: Semaphorin 4D correlates with increased bone resorption,
hypercalcemia, and disease stage in newly diagnosed patients with
multiple myeloma. Blood Cancer J. 8:422018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Asharani P, Keupp K, Semler O, Wang W, Li
Y, Thiele H, Yigit G, Pohl E, Becker J, Frommolt P, et al:
Attenuated BMP1 function compromises osteogenesis, leading to bone
fragility in humans and zebrafish. Am J Hum Genet. 90:661–674.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Martínez-Glez V, Valencia M,
Caparrós-Martín JA, Aglan M, Temtamy S, Tenorio J, Pulido V,
Lindert U, Rohrbach M, Eyre D, et al: Identification of a mutation
causing deficient BMP1/mTLD proteolytic activity in autosomal
recessive osteogenesis imperfecta. Hum Mutat. 33:343–350. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Grgurevic L, Macek B, Mercep M, Jelic M,
Smoljanovic T, Erjavec I, Dumic-Cule I, Prgomet S, Durdevic D, Vnuk
D, et al: Bone morphogenetic protein (BMP)1-3 enhances bone repair.
Biochem Biophys Res Commun. 408:25–31. 2011. View Article : Google Scholar : PubMed/NCBI
|