Introduction
Breast cancer, a phenomenon of uncontrolled
proliferation of breast epithelial cells caused by carcinogenic
factors, is the most common malignancy and ranks second among the
causes of tumor-associated death in females; it has become a public
health issue and endangers women's health worldwide (1). According to the data released by the
International Agency for Research on Cancer of the World Health
Organization, breast cancer surpassed lung cancer in 2020 and
became the world's most significant cancer type with the most
newly-diagnosed cases and deaths among females (2). Women with early breast cancer or
locoregional relapse are usually able to be cured by
multidisciplinary remedies, such as surgery, radiotherapy,
chemotherapy and pharmacotherapy. However, for those with
metastatic breast cancer, the clinical treatment outcomes are still
far from satisfactory and only palliative care (e.g., focusing on
the prolongation of survival and alleviation of symptoms) may be
possible, largely due to the metastatic heterogeneity and the
elusive pathogenesis of breast cancer. For instance, as reviewed by
Li et al (3),
triple-negative breast cancer (TNBC) may have worse cause-specific
survival and overall survival than the non-TNBC counterpart in all
stages and substages, regardless of influencing factors from
univariate and multivariate analyses (e.g., tumor grade, age,
ethnicity, surgery and radiation treatments).
Runt-related transcription factor 2 (RUNX2) belongs
to the RUNX family (including RUNX1-3) and has been recognized as a
key modulator and master transcription factor for osteogenesis, as
well as prostate and skeletal development (4–7). To
date, RUNX2 has been involved in diverse physiological processes,
including osteogenic differentiation of mesenchymal stem/stromal
cells, chondrocyte hypertrophy, immunomodulation, vascular invasion
and endothelial cell migration via modulating a variety of
signaling cascades (e.g., MAPK and NK-κB pathway macrophage
reprogramming) (8–11). Meanwhile, the dysregulation and
alteration of RUNX2 expression or activity may result in
arteriosclerosis, skeletal dysplasia (e.g., cleidocranial
dysplasia) and tumorigenesis (4,12–14).
For instance, RUNX2 is involved in the progression of various tumor
types, such as osteosarcoma, renal cell carcinoma, gastric cancer
and breast cancer (15–20). For instance, a recent study by our
group reported the facilitating effect of RUNX2 during
aggressiveness and chemoresistance of TNBC cells via activating
MMP1, which was significantly associated with poor prognosis
(21). Of note, other studies have
also indicated the involvement of RUNX2 in breast cancer stem cells
(BCSCs) and breast cancer progression (22,23).
For instance, Zhang et al (23,24)
found that RUNX2 was required for the activity of
CD44+/CD24−/low BCSCs during breast cancer
development, while miR-205/RUNX2 axis was further identified with s
negative regulatory effect upon the activity of
CD44+/CD24− BCSCs. Taken together, these
studies indicated the involvement of RUNX2 in BCSCs and its roles
in breast cancer diagnosis and drug resistance, revealing its
promising prospective clinical application and utility as an
antitumor drug target in the future.
The present review article mainly focused on the
roles of RUNX2 and BCSCs in the progression and management of
breast cancer. Furthermore, current advances in therapeutic
approaches for breast cancer, ranging from RUNX2-based remedies to
drug resistance, were summarized. Collectively, the systematic and
detailed research on RUNX2 has constituted a prospective area of
diagnosis and treatment strategy innovation for breast cancer.
Breast cancer and classification
Breast cancers are divided into different molecular
subtypes, including luminal A, luminal B, human epidermal growth
factor receptor 2 (HER2)-positive and basal-like breast cancer.
Among them, luminal A is a class of breast cancer with an
immunohistochemical index showing estrogen receptor (ER)
positivity, progesterone receptor (PR) positivity and HER2
negativity. Of the aforementioned subtypes, luminal A breast cancer
is the most common type of breast cancer, which may be targeted by
hormonal therapy with a favorable prognosis (25,26).
Distinct from luminal A breast cancer, the
HER2+ luminal B subtype features poor differentiation
and a worse prognosis (27).
HER2-positive breast cancer accounts for 12–20% of all invasive
breast cancers, which exhibits multifaceted deteriorative
characteristics, including a higher degree of malignancy, faster
disease progression, greater likelihood of relapse and metastasis,
and poor prognosis (28).
HER2-targeted therapies have been widely used for the treatment of
HER2-positive breast cancer, such as monoclonal antibodies, kinase
inhibitors and antibody-drug conjugates (ADCs). Basal-like breast
cancer is positive for basal cytokeratin and epidermal growth
factor receptors and/or c-Kit expression, and it accounts for ~15%
of breast cancers. It usually features rapid development of local
and distant metastases, and thus, it has relatively high mortality
rates (29). Gene expression
profiling typically classifies triple-negative breast cancer (TNBC)
as a major subtype of basal-like breast cancer without the
expression of ER, PR or HER2 (30),
which is clinically characterized by a strong aggressive nature,
high metastatic potential, proneness to relapse and poor prognosis
(31). Although poly(ADP ribose)
polymerase inhibitors (e.g., programmed cell death-1 monoclonal
antibody, trophoblast cell surface antigen 2 ADC) have been used
for a subset of TNBCs, the prognosis of patients with TNBC is still
far from satisfactory (Table
I).
 | Table I.Molecular typing of breast
cancer. |
Table I.
Molecular typing of breast
cancer.
| Subtype | ER | PR | HER2 |
|---|
| Luminal A | + | + | - |
| Luminal B
(HER2-negative) | + | - | - |
| HER2-positive
(ER-positive) | + | Any | + |
| HER2-positive
(ER-negative) | - | - | + |
| Basal-like (Triple
negative) | - | - | - |
Status of clinical treatment of breast
cancer
Breast cancer is a heterogeneous disease with highly
aggressive and complex biological features, and the clinical
treatment and prognosis of different patients vary greatly
(32). As mentioned above, patients
with early breast cancer may be effectively cured and the focus of
treatment is to avoid overtreatment and undertreatment (33). However, metastatic breast cancer
cannot be radically cured by current clinical remedies and its
management mainly focuses on prolonging survival time and
maintaining quality of life instead (34).
Currently, the major clinical remedies of breast
cancer are endocrine therapy, molecular targeted therapy and
chemotherapy (26). For instance,
patients with stage I and II breast cancers are recommended to
undergo breast-conserving surgery and radiation treatment, whereas
those with node-positive breast cancer are systemically treated
with chemotherapy, endocrine therapy and trastuzumab. As to
patients with stage III breast cancer, chemotherapy is applied to
facilitate breast-conserving surgery (35).
However, as the molecular phenotypes vary across
breast cancers, breast cancer is not sensitive to either
conventional endocrine therapy or molecular targeted therapies
(36). Therefore, chemotherapy
remains the primary strategy for breast cancer management. However,
the efficacy of conventional postoperative adjuvant chemotherapy is
unreliable, and residual metastatic lesions eventually lead to
tumor recurrence, which largely attributes to secondary tumor
recurrence and metastasis caused by acquired chemotherapy
resistance (37,38). Thus, there is an urgent need to
explore the drug resistance mechanisms and to identify novel
therapeutic targets to finally overcome the resistance of breast
cancer to chemotherapy, which is of great clinical significance for
clinical practice in the future. Of note, state-of-the-art updates
have indicated the role of the transcription factor RUNX2 in BCSCs
and drug resistance in breast cancer, which will provide
overwhelming new references for the development of the next
generation of therapeutic targets and innovative drugs for breast
cancer management.
The RUNX family
RUNX is a highly conserved transcription factor
family that has an important role in the regulation of gene
expression involved in embryonic development and cell
differentiation. Recent studies have demonstrated that members of
the RUNX transcription factor family are involved in the
differentiation of a variety of hematopoietic cells. RUNX2 is
required for osteogenesis, whereas RUNX1 and RUNX3 control blood
cell development at different stages of cell lineage
specification.
RUNX1 is also known as acute myeloid leukemia gene
1. Mutations in this gene are common in patients with lymphoblastic
and myeloid leukemia (39). It has
been suggested that RUNX1 deficiency is associated with familial
platelet disorders. Familial platelet disorders predispose to
myeloid leukemia and are associated with thrombocytopenia, as well
as marked reductions in B-lymphoid, T-lymphoid and myeloid lineages
(40).
RUNX2 has a key role in the development of bone and
cartilage tissue, while abnormal expression of RUNX2 is associated
with a variety of cancer types, and in particular, the process of
tumor bone metastasis (41). To
date, RUNX2 has been proven to be involved in melanoma, thyroid
cancer and hepatocellular carcinoma (42–44).
For instance, Guan et al (44) found that circular (circ) RNA_102272
promoted cisplatin resistance of hepatocellular carcinoma via
inhibiting the targeting effect of microRNA (miR)-326 upon RUNX2.
Of note, Matthijssens et al (45) reported that RUNX2 was able to
significantly induce glycolysis and oxidative phosphorylation, and
resulted in increased mitochondrial activity, which collectively
suggested the promoting effect of RUNX2 in accelerating tumor cell
metabolism, and in particular, in benefiting the invasion and
migration of leukemic cells via mediating the interaction between
glycolysis and mitochondrial respiration. RUNX2 also functions in
the development of gastric cancer, including tumor invasion and
metastasis by simultaneously facilitating the proliferation of
gastric cancer cells and increasing the self-renewal potential
(19). Ji et al (46) found that metastasis associated lung
adenocarcinoma transcript 1 was able to modulate the transcription
and translation of RUNX2 to further increase the metastasis of
recurrent colorectal cancer via binding to miR-15 family members,
inhibiting LDL receptor related protein 6 expression and enhancing
β-catenin signaling, or binding to splicing factor proline and
glutamine rich (SFPQ) and dissociating the SFPQ/polypyrimidine
tract binding protein 2 dimer. Collectively, RUNX2 holds the
potential to function as an important indicator for the detection
of multiple cancers in the early stage, which also serves as a
promising therapeutic target for clinical practice.
RUNX3 has a significant role in the occurrence and
development of a variety of human cancers. Zhang et al
(47) indicated that RUNX3 inhibits
colorectal cancer proliferation and metastasis. Liu et al
(48) found that RUNX3 inhibits
glutamine metabolism in gastric cancer. A study identified a
NO•/RUNX3/kynurenine metabolic axis, which enhances disease
aggressiveness in pancreatic cancer (49).
To summarize, the RUNX family not only plays a role
in normal physiological functions, but abnormal expression of any
member of the family may cause a variety of different diseases.
Physiological and pathological roles of
RUNX2
RUNX2, together with RUNX1 and RUNX3, is an
essential transcription factor of the RUNX family (50). To date, studies have indicated the
role of RUNX2 in numerous physiological and pathological processes,
such as osteoblast differentiation, chondrocyte maturation
(51), skeletal and mammary gland
development (22), osteoarthritis,
osteosarcoma, prostatic carcinoma, gastric cancer and even breast
cancer (19,52–55).
Roles of RUNX2 in physiological
processes
RUNX2 has important roles in various types of cells,
such as chondrocytes, osteoblasts and mesenchymal stem/stromal
cells (MSCs) (56). Meanwhile,
RUNX2 has been reported essential for breast development (57), which also exhibits good interaction
with twist family BHLH transcription factor (TWIST)1 in regulating
cranial neural crest-derived cell fate and thus guides craniofacial
muscle development (58).
For decades, RUNX2 has been recognized as a key
transcription factor for bone formation and osteoblast
differentiation, as identified by cell sorting, lineage tracing and
single-cell transcriptome analysis. For instance, Shu et al
(59) developed a dual-recombinase
fate-mapping system for the capture of the spatio-temporal skeletal
progenitor transition during postnatal bone formation. During
intramembranous ossification, RUNX2 promotes the differentiation of
MSCs into anterior osteoblasts and immature osteoblasts (12). Furthermore, Liu and Lee
(60) reviewed the regulatory
network of key transcription factors governing bone formation, such
as Msh homeobox 2, TWIST and promyelocytic leukemia zinc-finger
protein. During endochondral ossification, RUNX2 maintains the
survival of terminal hypertrophic cartilage and promotes its
trans-differentiation into osteoblasts (61). Despite the fast-growing
understanding of the transcriptional mechanisms of osteogenesis,
the detailed mechanisms of RUNX2-related skeletal development and
precise orchestration of osteogenesis remain largely elusive.
RUNX2 has also been reported to have a crucial
function during breast development via directly regulating the
expression pattern of a number of genes related to mammary gland
development (62). Studies have
revealed the expression of RUNX2 in mammary tissues, including
basal cells and luminal cells, which thereby participates in breast
development. For instance, Inman and Shore (63) and Sato et al (64) verified that osteopontin may act as a
target of RUNX2 and exert its function in breast differentiation in
breast epithelial cells during pregnancy and lactation.
RUNX2 is associated with breast
cancer
Besides its critical role in physiological
development, RUNX2 also functions as an oncogene in numerous types
of cancer. RUNX2 has been associated with a wide variety of
processes, including tumor progression and heterogeneity, via
mediating the responses of cells to signaling pathways hyperactive
in tumors (65), such as the
connective tissue growth factor-RUNX2-RANKL axis (66), miR-130a-5p-RUNX2-serine/threonine
kinase 32A network (67), the bone
morphogenetic protein (BMP)/TGF-β-RUNX2 loop (65) and the Zic family member
2-RUNX2-nucleolar and coiled-body phosphoprotein 1 signaling axis
(68). To date, RUNX2 has emerged
as a key mediator in the metastasis of cancers with preinvasive and
promigratory behaviors in osteosarcoma, breast cancer, thyroid
cancer, prostatic carcinoma and melanoma cells (69). Furthermore, numerous investigations
of the molecular mechanisms of RUNX2 have revealed the mode of
action in tumor metastasis and growth via activating the expression
of bone matrix and adhesion proteins, matrix metalloproteinase
(MMP) and angiogenic factors in cancer cells (70,71).
As mentioned above, breast cancer is composed of
distinct subtypes with multiple stages during tumor progression. Of
note, RUNX2 has been reported to have an important role in bone
metastasis, which is the final stage of breast cancer development
(72). For instance,
phosphorylation of RUNX2 mediated by tyrosine kinase ABL is
adequate to promote breast cancer invasion (73), while the interaction with core
binding factor subunit β protein is sufficient to guide breast
cancer cell invasion (53).
During tumor metastasis, cancer cells function via
autophagy to recover nutrients to maintain their own survival and
RUNX2 promotes autophagy by increasing acetylation of α-tubulin
sub-units of microtubules, and thus promotes the metastasis of
breast cancer cells (74). It has
been indicated that the integrin subunit α5 (ITGA5)β3 expressed on
the surface of breast cancer cells was able to anchor cells to
osteoblasts by binding the tripeptide Arg-Gly-Asp motif of bone
matrix proteins (75). During the
aforementioned process, RUNX2 facilitated breast cancer cell
recruitment and colonization in bone in an ITGA5-dependent manner
(76). SET domain containing 7,
histone lysine methyltransferase (SET7)/9 in breast cancer is able
to activate target gene expression through histone methylation or
directly act as a target via non-histone methylation. It has also
been indicated that SET7/9 is able to promote multiple malignant
biological behaviors in the development of breast cancer by
activating RUNX2 (77). TGF-β is
indispensable in normal physiological function (78) and had a complex ‘double-edged sword’
role in the occurrence and development of tumors (79). Furthermore, RUNX2 may promote bone
metastasis in breast cancer cells through the activation of the
TGF-β signaling pathway (80). In
addition, accumulating evidence has indicated that multiple miRNAs
(e.g., miR-30, miR-135, miR-203, miR-205, miR-505-3P and
miR-590-3P) are adequate to inhibit the development and metastasis
of breast cancer via targeting RUNX2 (23,71,81–84).
Taken together, abnormal expression of RUNX2 may be associated with
the occurrence and progression of breast cancer, and the underlying
molecular mechanisms, including target genes and the concomitant
signaling pathways, remain to be fully elucidated.
BCSCs
CSCs and epithelial-mesenchymal transition (EMT) are
the major elements contributing to the metastasis and recurrence of
cancers (85,86). In general, CSCs are unique
subpopulations of solid tumors (e.g., breast cancer, lung cancer
and stomach cancer) with stem cell-related characteristics, such as
self-renewal, differentiation and tumorigenic potential, which have
re-emerged as a hot topic of increased interest in the field
(87,88). In breast cancer, BCSCs promote
angiogenesis in mammary tissues by dedifferentiating to endothelial
cells and secreting proangiogenic or angiogenic factors, which
collectively facilitates metastasis and therapy resistance of
breast cancer (89). Of note,
studies have demonstrated that a series of signaling pathways are
involved in orchestrating the phenotypes of CSCs, including the
Hippo, Wnt, Notch and Hedgehog pathways (90). Meanwhile, drug efflux transporters
and multi-drug resistance genes expressed in BCSCs also confer
resistance against conventional chemotherapeutic drugs (90).
Breast cancer and BCSCs
Despite the inspiring advancements in radiation and
chemotherapies, breast cancer is still an intractable disease owing
to tumor relapse and drug resistance caused by BCSCs (90). BCSCs participate in the occurrence
and development of breast cancer and usually result in cancer
relapse with enhanced aggressiveness (91). Although only a small proportion of
breast cancer cells may lead to tumors xenografts, these
tumor-initiating cells are able to reconstruct tumors with a
similar heterogeneity to that of the primary tumor, which indicates
the distinct stem cell-like plasticity of BCSCs (92). To date, a large number of studies
have indicated the pivotal role of the different pathways involved
in the regulation of BCSCs, such as Wnt, Hedgehog and Notch
signaling. For instance, Katoh (93) summarized Wnt signaling cascades
cross-talk with the fibroblast growth factor, Notch, Hedgehog and
TGF-β/BMP signaling cascades and regulate the expression of
functional CSC markers. Furthermore, Ibrahim et al (94) verified syndecan-1 as a novel
biomarker for inflammatory TNBC and modulating the BCSC phenotype
via orchestrating the IL-6/STAT3-Notch and EGFR signal
cascades.
According to the currently recognized stem cell
markers for breast cancer, BCSCs may be divided into the
CD44+/CD24− and aldehyde dehydrogenase
(ALDH)+ subtypes (95).
The most rapidly proliferating ALDH+ BCSCs have an
epithelial cell morphology, whereas the
CD44+/CD24− mesenchymal subsets display
declined proliferation but high invasion and metastasis (96). In general, based on the expression
levels of CD44, breast cancers of different molecular subtypes show
variations in the proportion of CD44+/CD24−
BCSCs (97). For instance, the
proportion of CD44+/CD24− BCSCs in basal-like
breast cancer is significantly higher than that in luminal A and
luminal B breast cancers (98),
which may be accountable for the worse prognosis of basal-like
breast cancer as compared with that of the luminal A and luminal B
subtypes (Table II). Collectively,
the existence of BCSCs has tremendously increased the metastasis,
angiogenesis and therapy resistance of breast cancer, as well as
the secondary tumor formation in patients. Therefore, devising
therapeutic interventions with multidisciplinary strategies to
target BCSCs would be of great help in boosting patients' survival
rates and in increasing the sensitivity to anti-tumor drugs for
breast cancer.
 | Table II.BCSC markers. |
Table II.
BCSC markers.
| First author,
year | Marker | BCSCs | Location | Function | (Refs.) |
|---|
| Dzobo, 2021 | CD44 | + | Cell membrane | Cell-cell
interactions, cell adhesion and migration | (133) |
| Fillmore, 2007 | CD24 | - | Cell membrane | Cell
differentiation | (134) |
| Tomita, 2016 | ALDH1 | + | Cytoplasm | Oxidize xenobiotic
and intracellular aldehydes | (135) |
RUNX2 and BCSCs
State-of-the-art renewal has indicated the
involvement of RUNX2 in various cancer types. The pro-cancer role
of RUNX2 in breast cancer is known to be related to BCSCs (7). Furthermore, several studies have
indicated a potential link between RUNX2 and CD44. For instance, in
colorectal cancer, RUNX2 interacts with brahma-related gene 1 to
target CD44 for promoting invasion and migration of colorectal
cancer cells (99). In prostate
cancer cells, RUNX2 forms a complex with intracellular domain of
CD44 as a co-transcriptional factor, activates the expression of
metastasis-related genes, and contributes to migration and tumor
formation (54).
As a surface marker of BCSCs, CD44 rather than CD24
is positively correlated with RUNX2 expression, which suggests the
association of RUNX2 with CD44+/CD24− BCSCs.
For instance, Zhang et al (24) found that RUNX2 promoted the
malignant biological behavior of breast cancer cells by regulating
the proportion of BCSCs. As a common event in breast cancer
progression, PR Ser294 phosphorylation is required to maintain
BCSCs fate via cooperation with the growth factor-initiated
signaling pathway and key phosphor-PR targets (e.g., solute carrier
family 37 member 2 and RUNX2). With the aid of the microsphere
formation test, RUNX2 has been proven to act as an important driver
of BCSC formation in vitro (100). In detail, RUNX2 has a critical
influence in both EMT and BCSCs, and the ectopic expression of
RUNX2 may subsequently induce the occurrence of EMT via the
regulation of key pathways (e.g., TGF-β, Wnt) (101). Meanwhile, RUNX2 also has a
critical role in CD44+/CD24− MCF10AT1 cells
with BCSC characteristics (102).
Taken together, RUNX2 has a critical role in
promoting the development and metastasis of breast cancer via
regulating the proportion of BCSCs, which provides new insight for
the further dissection of the pathogenesis and therapeutic remedies
for breast cancer.
Drug resistance in breast cancer
Attributed to the rapid development of technologies,
the diagnosis and treatment of breast cancer have markedly
improved; however, drug resistance in breast cancer remains an
issue, as the mechanisms comprise disorders in the orchestration of
hormones, apoptosis and efflux pump activation, signaling pathways
and oncogenes (103).
Relationship between CSCs and drug
resistance
CSCs are self-renewal cells with high tumorigenic
potential. CSCs are able to adapt to changes in the surrounding
environment and are more resistant to radiotherapy and chemotherapy
than other cells in the tumor. Several studies have indicated that
the dual effects of intrinsic and extrinsic factors contributes to
CSC-mediated therapy resistance.
CSCs have a very slow cell cycle and possess an
anti-apoptotic machinery, DNA repair systems and persistent
stemness, which lead to drug resistance during cancer treatment
(104). Besides, CSC drug
resistance may also be caused by external factors, such as the
tumor microenvironment (TME). When the TME is always in a state of
nutrition, metabolism and oxygen deficiency, it promotes the
adaptation of CSCs to the TME (105). CSC drug resistance is complex.
CSCs promote tumor progression, treatment resistance and disease
recurrence, which may achieved by their sustained proliferation,
invasion into normal tissue, promotion of angiogenesis, evasion of
the immune system and resistance to conventional anticancer
therapies (106,107).
BCSCs and drug resistance in breast
cancer
Cancer metastasis and drug resistance currently
remain major challenges in cancer therapy (106). Increasing evidence suggests that
CSCs favor cancer metastasis and drug resistance, leading to
relapse of cancer and death of patients (108). On the one hand, BCSCs have been
considered the result of overactivation of mutant normal breast
stem cells during self-renewal. On the other hand, BCSCs are
resultant of the dedifferentiation of cancer cells caused by
somatic mutations or microenvironmental components during treatment
(109).
The proportion of BCSCs was found to be
significantly increased in chemoresistant and radioresistant breast
cancer cell lines and human tissues (110). Furthermore, overactivation of the
anti-apoptotic PI3K signaling pathway and the antioxidant nuclear
factor E2-related factor 2 signaling pathway collectively
contribute to cellular resistance and resistance of BCSCs to
radiation-induced ROS attack and apoptosis compared with non-BCSCs
(111,112). Further mechanistic studies
revealed that BCSCs with a high level of free radical scavenger
expression were more tolerant to the hypoxic environment and thus
conferred a radiation-resistant phenotype by reducing the
accumulation of intracellular ROS (113). In addition, certain cell surface
pumps in BCSCs (e.g., ATP binding cassette subfamily G member 2)
are able to impair the intracellular accumulation of anticancer
drugs (114). Collectively, the
variations in BCSCs are responsible for increased drug resistance
of breast cancer.
RUNX2 and drug resistance of breast
cancers
Overexpression of RUNX2 commonly leads to reduced
sensitivity of cancer cells (e.g., osteosarcoma) to chemotherapy,
which may thus be used as a reliable marker for the evaluation of
chemotherapy resistance (115).
Conversely, loss of RUNX2 expression is adequate to increase the
sensitivity of osteosarcoma cells to the chemotherapeutic agent
doxorubicin (116). First,
Sugimoto et al (117) found
that silencing RUNX2 was able to enhance the sensitivity of AsPC-1
cells in P53-deficient human pancreatic cancer to gemcitabine (GEM)
by stimulating Tap63 (isoform of p63 gene)-mediated cell death.
Furthermore, Ozaki et al (118) verified that increased
Tap73-dependent cell death mediated by RUNX2 depletion enhanced the
sensitivity of P53-mutant pancreatic cancer MiaPaCa-2 cells to GEM.
In addition, RUNX2 was found to weaken the proapoptotic activity
and enhance the sensitivity to GEM drugs of P53-mutant Panc-1
pancreatic cancer cells via suppressing Tap63 expression (118). These findings demonstrate that
RUNX2 is associated with drug sensitivity to GEM in pancreatic
cancer cells lacking functional P53.
In breast cancer, Tamoxifen (TAM) resistance is
responsible for a large proportion of breast cancer-associated
mortalities. Jeselsohn et al (119) found that RUNX2 was able to
interact with ERα to directly induce SOX9 transcription and reduce
the sensitivity of breast cancer cells to TAM. Of note, Geter et
al (120) identified RUNX2 as
the only RUNX family member that was transcriptionally or
translationally upregulated in both TAM-resistant LCC9 cells and
patient-derived xenograft derived from tamoxifen-resistant cell
lines, suggesting that RUNX2 has a core role in the drug resistance
of breast cancer. The study's results revealed that RUNX2 promoted
the metastasis of breast cancer, and knockdown of RUNX2 was able to
significantly restore the sensitivity of breast cancer to TAM
(120). Furthermore, Othman et
al (121) found that silencing
of RUNX2 increased the sensitivity of breast cancer cells to
microtubule-targeting agents. Wang et al (122) demonstrated that serum miR-4530 may
sensitize breast cancer to taxane- and anthracycline-based
neoadjuvant chemotherapy by suppressing RUNX2. A recent study
revealed that RUNX2 directly targeted MMP1 and facilitates
aggressiveness and chemoresistance of TNBC cells (21).
However, the specific mechanisms of RUNX2-associated
drug resistance in breast cancer still require confirmation and
further elucidation. Systematic and detailed dissection of the
underlying mechanisms will contribute to conquering drug resistance
and provide new references for the treatment of breast cancer
(Fig. 1).
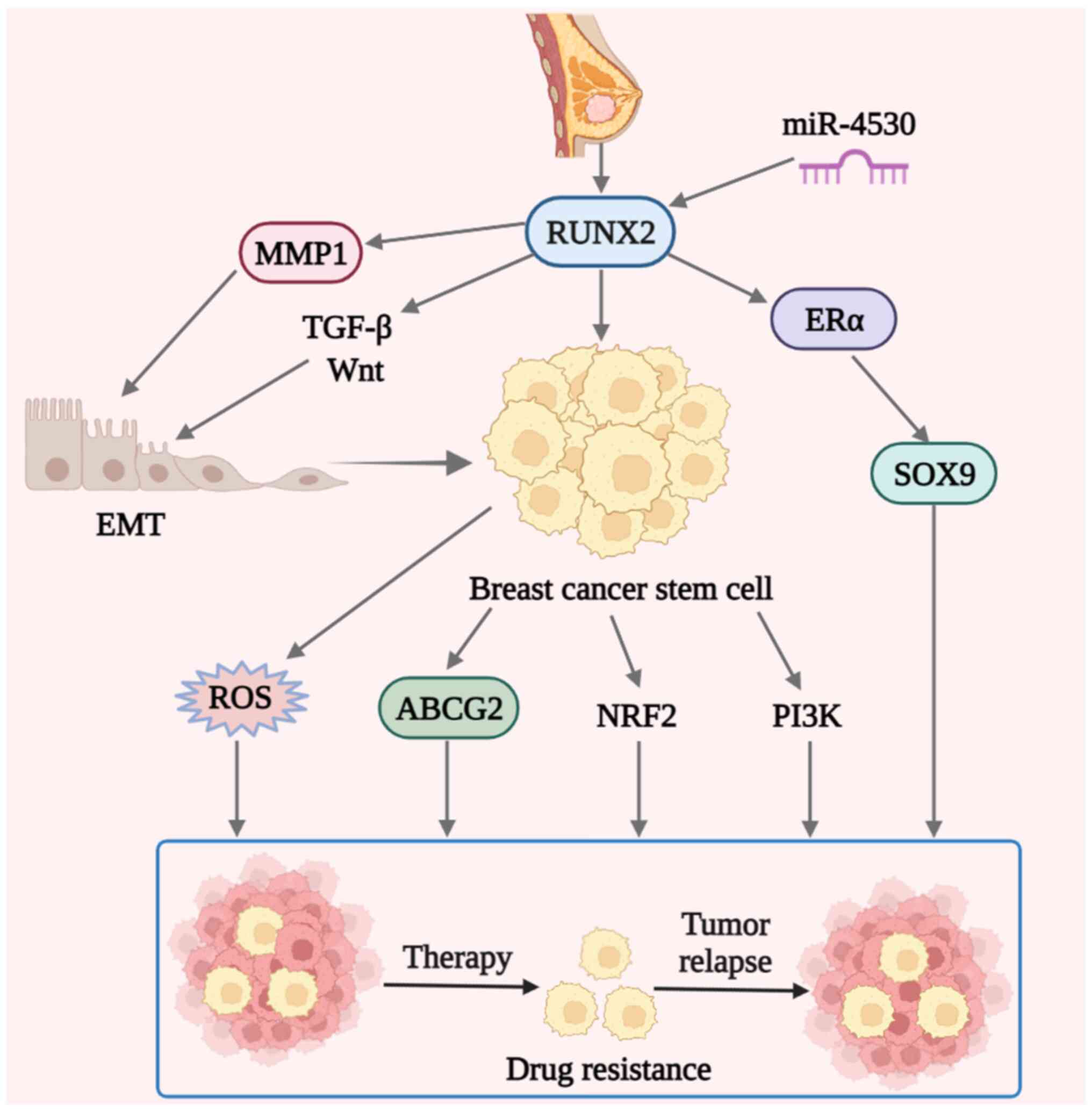 | Figure 1.Schematic diagram of possible drug
resistance mechanisms of RUNX2 in breast cancer. In breast cancer,
aberrant RUNX2 expression contributes to drug resistance. Elevated
serum miR-4530 levels may sensitize breast cancer to drugs by
suppressing RUNX2. The drug resistance of breast cancer may be
initiated by the direct transcription of SOX9 induced by the
interaction of RUNX2 with ERα. In addition, RUNX2 may regulate
BCSCs, which cause drug resistance through protein ABCG2 or the
PI3K and NRF2 signaling pathways. ROS in BCSCs also have a certain
role. Finally, RUNX2 may affect EMT through secreted protein MMP1
or signaling pathways, such as TGF-β and Wnt, and EMT regulates
BCSCs and eventually leads to breast cancer resistance. RUNX2,
Runt-related transcription factor 2; miR, microRNA; ER, estrogen
receptor; NRF2, nuclear factor erythroid 2-related factor 2; BCSC,
breast cancer stem cell; ROS, reactive oxygen species; ABCG2, ATP
binding cassette subfamily G member 2; EMT, epithelial to
mesenchymal transition. |
Discussion
RUNX2 has been recognized as a master transcription
factor for osteogenesis and bone metastases of invasive breast
cancer (72). In highly metastatic
breast cancer, RUNX2 phosphorylation has associations with the
invasive capacity of MDA-MB-231 cells by modulating the activity of
tyrosine kinase ABL, which is involved in the activation of the
classical BMP-SMAD signaling cascades (17). It was further indicated that RUNX2
facilitates the aggressiveness and chemoresistance of TNBC and the
concomitant tumorigenesis and cancer progression via activating
MMP1, which thus provides a basis for developing RUNX2-MMP1
axis-based novel candidates for breast cancer diagnostics (21).
Longitudinal studies have highlighted the potential
utility of RUNX2 as a pivotal contributor to bone-specific
metastasis during breast cancer. For instance, Li et al
(76) verified the facilitating
effect of RUNX2 and the targeted ITGA5 during the adhesion and
attraction of breast cancer cells, as well as osteotropism and bone
colonization. In contrast to the aforementioned studies indicating
the critical role of RUNX2 in bone-specific metastasis or
progression of breast cancer, the present review mainly focused on
the latest updates on the interaction between RUNX2 and BCSCs and
their biofunctions, which contribute to the aggressiveness and drug
resistance of breast cancer. Furthermore, the systematic and
detailed investigation of the RUNX2-BCSCs axis in breast cancer and
knowledge regarding its utility in diagnosis and treatment are
still far from satisfactory and further preclinical and clinical
investigations are required in the future.
Of note, current progress has also highlighted the
potential application of developing computational models based on
the data fusion paradigm for identification of the disease-related
non-coding RNAs (ncRNAs) as a future direction in disease
assessment (123,124). For instance, Huang et al
(123) summarized the advances in
miRNAs and algorithm design for complex diseases by computational
models, which may help overcome the obstacles to disease
prediction. Simultaneously, Chen et al (125) and Wang et al (126) put forward the limitations and
future directions of the disease prediction utility of long ncRNAs
(lncRNAs) or circular RNAs (circRNAs) for complex diseases by
combing experimental technology with computational prediction
algorithms. Similarly, a series of ncRNAs have been identified to
be linked to the disease progression of breast cancer or TNBC, such
as the miR-20a-5p/high mobility group AT-hook 2 axis,
lncRNA-CDC6/miRNA-215 and circRNA_002502/miR-182-5p/forkhead box
O3a axis (127–130). In addition, Yin et al
(131) performed a weighted gene
co-expression network analysis and competing endogenous RNA network
analysis based on the Cancer Genome Atlas database and identified
the prospective association of 3,301 key modules and 453 genes with
breast cancer prognosis. Therewith, it would be of great interest
and importance to perform further studies to predict the
association of RUNX2 and BCSCs with breast cancer development and
drug resistance based on powerful computational model construction,
which may also benefit the development of the framework of oncology
diagnostics and therapeutics from a different perspective in the
future. The present review mainly summarized the research progress
on RUNX2 and BCSC in terms of breast cancer progression and
diagnosis, may benefit the identification of key mechanisms of
breast cancer and supply references for the concomitant anticancer
treatment and drug development in the future.
As a malignant tumor type with high morbidity and
mortality rates, breast cancer persistently endangers the health
and lives of females. While chemotherapy remains the major remedy
for breast cancer, drug resistance severely limits the efficacy and
usually results in tumor metastasis and recurrence.
In recent years, RUNX2 in breast cancer has
attracted considerable attention, as increasing evidence indicated
its pivotal role in tumor development and metastasis, as well as
its association with BCSCs-related drug resistance. Based on the
current literature, it may be presumed that RUNX2 enhances drug
resistance in breast cancer by regulating the proportion and
biofunction of BCSCs, which will vastly enhance the current
understanding of the pathogenesis and provide reliable targets for
clinical treatment and drug development in the future.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science
Foundation of China (grant nos. 82173377 and 81302319 to FL), Major
Research Project of the Education Department of Anhui Province
(grant no. KJ2018ZD018), the National Natural Science Foundation of
China (grant no. 82260031), the Non-profit Central Research
Institute Fund of Chinese Academy of Medical Sciences (grant no.
2019PT320005) and the 2021 Central-Guided Local Science and
Technology Development Fund (grant no. ZYYDDFFZZJ-1).
Availability of data and materials
Not applicable.
Authors' contributions
WS and FL designed the study, performed data
analysis and interpretation, wrote the manuscript and gave final
approval of the manuscript. WS and CK performed the literature
search and wrote the manuscript. FL and LZ reviewed the manuscript.
All authors contributed to the review article, and have read and
approved the final manuscript. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Akram M, Iqbal M, Daniyal M and Khan AU:
Awareness and current knowledge of breast cancer. Biol Res.
50:332017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li X, Yang J, Peng L, Sahin AA, Huo L,
Ward KC, O'Regan R, Torres MA and Meisel JL: Triple-negative breast
cancer has worse overall survival and cause-specific survival than
non-triple-negative breast cancer. Breast Cancer Res Treat.
161:279–287. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gomathi K, Akshaya N, Srinaath N, Moorthi
A and Selvamurugan N: Regulation of Runx2 by post-translational
modifications in osteoblast differentiation. Life Sci.
245:1173892020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu DD, Zhang CY, Liu Y, Li J, Wang YX and
Zheng SG: RUNX2 regulates osteoblast differentiation via the BMP4
signaling pathway. J Dent Res. 101:1227–1237. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Komori T: Runx2, a multifunctional
transcription factor in skeletal development. J Cell Biochem.
87:1–8. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Y, Ge C and Franceschi RT: Role of
Runx2 in prostate development and stem cell function. Prostate.
81:231–241. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Westendorf JJ: Transcriptional
co-repressors of Runx2. J Cell Biochem. 98:54–64. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang L, Wei Y, Chi Y, Liu D, Yang S, Han
Z and Li Z: Two-step generation of mesenchymal stem/stromal cells
from human pluripotent stem cells with reinforced efficacy upon
osteoarthritis rabbits by HA hydrogel. Cell Biosci. 11:62021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang L, Wang H, Liu C, Wu Q, Su P, Wu D,
Guo J, Zhou W, Xu Y, Shi L and Zhou J: MSX2 initiates and
accelerates mesenchymal stem/stromal cell specification of hPSCs by
regulating TWIST1 and PRAME. Stem Cell Reports. 11:497–513. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu J and Zhang H, Pan J, Hu Z, Liu L, Liu
Y, Yu X, Bai X, Cai D and Zhang H: Fargesin ameliorates
osteoarthritis via macrophage reprogramming by downregulating MAPK
and NF-κB pathways. Arthritis Res Ther. 23:1422021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim HJ, Kim WJ and Ryoo HM:
Post-translational regulations of transcriptional activity of
RUNX2. Mol Cells. 43:160–167. 2020.PubMed/NCBI
|
|
13
|
Zhang Y and Duan X: A novel 90-kbp
deletion of RUNX2 associated with cleidocranial dysplasia. Genes
(Basel). 13:11282022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ukkat J, Hoang-Vu C, Trojanowicz B and
Rebelo A: Osteocalcin, osteopontin and RUNX2 expression in
patients' leucocytes with arteriosclerosis. Diseases. 9:192021.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang X, Ren Z, Liu B and Wei S: RUNX2
mediates renal cell carcinoma invasion through calpain2. Biol Pharm
Bull. 45:1653–1659. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wysokinski D, Blasiak J and Pawlowska E:
Role of RUNX2 in breast carcinogenesis. Int J Mol Sci.
16:20969–20993. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He F, Matsumoto Y, Asano Y, Yamamura Y,
Katsuyama T, La Rose J, Tomonobu N, Komalasari NLGY, Sakaguchi M,
Rottapel R and Wada J: RUNX2 phosphorylation by tyrosine kinase ABL
promotes breast cancer invasion. Front Oncol. 11:6652732021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Song X, Liu J, Liu B, Piao C, Kong C and
Li Z: RUNX2 interacts with SCD1 and activates Wnt/β-catenin
signaling pathway to promote the progression of clear cell renal
cell carcinoma. Cancer Med. Oct 6–2022.(Epub ahead of print).
|
|
19
|
Guo Z, Zhou K, Wang Q, Huang Y, Ji J, Peng
Y, Zhang X, Zheng T, Zhang Z, Chong D and Yang Z: The transcription
factor RUNX2 fuels YAP1 signaling and gastric cancer tumorigenesis.
Cancer Sci. 112:3533–3544. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li N, Luo D, Hu X, Luo W, Lei G, Wang Q,
Zhu T, Gu J, Lu Y and Zheng Q: RUNX2 and osteosarcoma. Anticancer
Agents Med Chem. 15:881–887. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Si W, Xu X, Wan L, Lv F, Wei W, Xu X, Li
W, Huang D, Zhang L and Li F: RUNX2 facilitates aggressiveness and
chemoresistance of triple negative breast cancer cells via
activating MMP1. Front Oncol. 12:9960802022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ferrari N, McDonald L, Morris JS, Cameron
ER and Blyth K: RUNX2 in mammary gland development and breast
cancer. J Cell Physiol. 228:1137–1142. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang L, Liu L, Xu X, He X, Wang G, Fan C,
Zheng Q and Li F: miR-205/RunX2 axis negatively regulates
CD44+/CD24− breast cancer stem cell activity.
Am J Cancer Res. 10:1871–1887. 2020.PubMed/NCBI
|
|
24
|
Zhang P, Liu L, Zhang L, He X, Xu X, Lu Y
and Li F: Runx2 is required for activity of
CD44+/CD24−/low breast cancer stem cell in
breast cancer development. Am J Transl Res. 12:2305–2318.
2020.PubMed/NCBI
|
|
25
|
Kudela E, Samec M, Koklesova L, Liskova A,
Kubatka P, Kozubik E, Rokos T, Pribulova T, Gabonova E, Smolar M
and Biringer K: miRNA expression profiles in luminal A breast
cancer-implications in biology, prognosis, and prediction of
response to hormonal treatment. Int J Mol Sci. 21:76912020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Harbeck N and Gnant M: Breast cancer.
Lancet. 389:1134–1150. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ades F, Zardavas D, Bozovic-Spasojevic I,
Pugliano L, Fumagalli D, de Azambuja E, Viale G, Sotiriou C and
Piccart M: Luminal B breast cancer: Molecular characterization,
clinical management, and future perspectives. J Clin Oncol.
32:2794–2803. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Loibl S and Gianni L: HER2-positive breast
cancer. Lancet. 389:2415–2429. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Alexandrou S, George SM, Ormandy CJ, Lim
E, Oakes SR and Caldon CE: The proliferative and apoptotic
landscape of basal-like breast cancer. Int J Mol Sci. 20:6672019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wolff AC, Hammond ME, Hicks DG, Dowsett M,
McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M,
Fitzgibbons P, et al: Recommendations for human epidermal growth
factor receptor 2 testing in breast cancer: American society of
clinical oncology/college of American pathologists clinical
practice guideline update. Arch Pathol Lab Med. 138:241–256. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dagogo-Jack I and Shaw AT: Tumour
heterogeneity and resistance to cancer therapies. Nat Rev Clin
Oncol. 15:81–94. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cardoso F, Kyriakides S, Ohno S,
Penault-Llorca F, Poortmans P, Rubio IT, Zackrisson S and Senkus E;
ESMO Guidelines Committee. Electronic address, : simpleclinicalguidelines@esmo.org:
Early breast cancer: ESMO clinical practice guidelines for
diagnosis, treatment and follow-up†. Ann Oncol. 30:1194–1220. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tosello G, Torloni MR, Mota BS, Neeman T
and Riera R: Breast surgery for metastatic breast cancer. Cochrane
Database Syst Rev. 3:CD0112762018.PubMed/NCBI
|
|
35
|
Maughan KL, Lutterbie MA and Ham PS:
Treatment of breast cancer. Am Fam Physician. 81:1339–1346.
2010.PubMed/NCBI
|
|
36
|
Pondé NF, Zardavas D and Piccart M:
Progress in adjuvant systemic therapy for breast cancer. Nat Rev
Clin Oncol. 16:27–44. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Abderrahman B and Jordan VC: Telling
details of breast-cancer recurrence. Nature. 553:1552018.
View Article : Google Scholar
|
|
39
|
Miyoshi H, Shimizu K, Kozu T, Maseki N,
Kaneko Y and Ohki M: t(8;21) breakpoints on chromosome 21 in acute
myeloid leukemia are clustered within a limited region of a single
gene, AML1. Proc Natl Acad Sci USA. 88:10431–10434. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Schlegelberger B and Heller PG: RUNX1
deficiency (familial platelet disorder with predisposition to
myeloid leukemia, FPDMM). Semin Hematol. 54:75–80. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ito Y, Bae SC and Chuang LS: The RUNX
family: developmental regulators in cancer. Nat Rev Cancer.
15:81–95. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cecconi D, Brandi J, Manfredi M, Serena M,
Dalle Carbonare L, Deiana M, Cheri S, Parolini F, Gandini A,
Marchetto G, et al: Runx2 stimulates neoangiogenesis through the
Runt domain in melanoma. Sci Rep. 9:80522019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vitale E, Sauta E, Gugnoni M, Torricelli
F, Manicardi V and Ciarrocchi A: A multimodal integrative approach
to model transcriptional addiction of thyroid cancer on RUNX2.
Cancer Commun (Lond). 42:892–896. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Guan Y, Zhang Y, Hao L and Nie Z:
CircRNA_102272 promotes cisplatin-resistance in hepatocellular
carcinoma by decreasing MiR-326 targeting of RUNX2. Cancer Manag
Res. 12:12527–12534. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Matthijssens F, Sharma ND, Nysus M, Nickl
CK, Kang H, Perez DR, Lintermans B, Van Loocke W, Roels J, Peirs S,
et al: RUNX2 regulates leukemic cell metabolism and chemotaxis in
high-risk T cell acute lymphoblastic leukemia. J Clin Invest.
131:e1415662021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ji Q, Cai G, Liu X, Zhang Y, Wang Y, Zhou
L, Sui H and Li Q: MALAT1 regulates the transcriptional and
translational levels of proto-oncogene RUNX2 in colorectal cancer
metastasis. Cell Death Dis. 10:3782019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang F, Su T and Xiao M: RUNX3-regulated
circRNA METTL3 inhibits colorectal cancer proliferation and
metastasis via miR-107/PER3 axis. Cell Death Dis. 13:5502022.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu H, Xue Q, Cai H, Jiang X, Cao G, Chen
T, Chen Y and Wang D: RUNX3-mediated circDYRK1A inhibits glutamine
metabolism in gastric cancer by up-regulating
microRNA-889-3p-dependent FBXO4. J Transl Med. 20:1202022.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang L, Tang W, Yang S, He P, Wang J,
Gaedcke J, Ströbel P, Azizian A, Ried T, Gaida MM, et al:
NO•/RUNX3/kynurenine metabolic signaling enhances
disease aggressiveness in pancreatic cancer. Int J Cancer.
146:3160–3169. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Mevel R, Draper JE, Lie-A-Ling M, Kouskoff
V and Lacaud G: RUNX transcription factors: Orchestrators of
development. Development. 146:dev1482962019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Komori T: Regulation of proliferation,
differentiation and functions of osteoblasts by Runx2. Int J Mol
Sci. 20:16942019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Nagata K, Hojo H, Chang SH, Okada H, Yano
F, Chijimatsu R, Omata Y, Mori D, Makii Y, Kawata M, et al: Runx2
and Runx3 differentially regulate articular chondrocytes during
surgically induced osteoarthritis development. Nat Commun.
13:61872022. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Villanueva F, Araya H, Briceño P, Varela
N, Stevenson A, Jerez S, Tempio F, Chnaiderman J, Perez C,
Villarroel M, et al: The cancer-related transcription factor RUNX2
modulates expression and secretion of the matricellular protein
osteopontin in osteosarcoma cells to promote adhesion to
endothelial pulmonary cells and lung metastasis. J Cell Physiol.
234:13659–13679. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Senbanjo LT, AlJohani H, Majumdar S and
Chellaiah MA: Characterization of CD44 intracellular domain
interaction with RUNX2 in PC3 human prostate cancer cells. Cell
Commun Signal. 17:802019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yin X, Teng X, Ma T, Yang T, Zhang J, Huo
M, Liu W, Yang Y, Yuan B, Yu H, et al: RUNX2 recruits the
NuRD(MTA1)/CRL4B complex to promote breast cancer progression and
bone metastasis. Cell Death Differ. 29:2203–2217. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chan WCW, Tan Z, To MKT and Chan D:
Regulation and role of transcription factors in osteogenesis. Int J
Mol Sci. 22:54452021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Coffman JA: Runx transcription factors and
the developmental balance between cell proliferation and
differentiation. Cell Biol Int. 27:315–324. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Han X, Feng J, Guo T, Loh YE, Yuan Y, Ho
TV, Cho CK, Li J, Jing J, Janeckova E, et al: Runx2-Twist1
interaction coordinates cranial neural crest guidance of soft
palate myogenesis. Elife. 10:e623872021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shu HS, Liu YL, Tang XT, Zhang XS, Zhou B,
Zou W and Zhou BO: Tracing the skeletal progenitor transition
during postnatal bone formation. Cell Stem Cell. 28:2122–2136.e3.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Liu TM and Lee EH: Transcriptional
regulatory cascades in Runx2-dependent bone development. Tissue Eng
Part B Rev. 19:254–263. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Qin X, Jiang Q, Nagano K, Moriishi T,
Miyazaki T, Komori H, Ito K, Mark KV, Sakane C, Kaneko H and Komori
T: Runx2 is essential for the transdifferentiation of chondrocytes
into osteoblasts. PLoS Genet. 16:e10091692020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Owens TW, Rogers RL, Best S, Ledger A,
Mooney AM, Ferguson A, Shore P, Swarbrick A, Ormandy CJ, Simpson
PT, et al: Runx2 is a novel regulator of mammary epithelial cell
fate in development and breast cancer. Cancer Res. 74:5277–5286.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Inman CK and Shore P: The osteoblast
transcription factor Runx2 is expressed in mammary epithelial cells
and mediates osteopontin expression. J Biol Chem. 278:48684–48689.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Sato M, Morii E, Komori T, Kawahata H,
Sugimoto M, Terai K, Shimizu H, Yasui T, Ogihara H, Yasui N, et al:
Transcriptional regulation of osteopontin gene in vivo by
PEBP2alphaA/CBFA1 and ETS1 in the skeletal tissues. Oncogene.
17:1517–1525. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Pratap J, Lian JB, Javed A, Barnes GL, van
Wijnen AJ, Stein JL and Stein GS: Regulatory roles of Runx2 in
metastatic tumor and cancer cell interactions with bone. Cancer
Metastasis Rev. 25:589–600. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Kim B, Kim H, Jung S, Moon A, Noh DY, Lee
ZH, Kim HJ and Kim HH: A CTGF-RUNX2-RANKL axis in breast and
prostate cancer cells promotes tumor progression in bone. J Bone
Miner Res. 35:155–166. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ma F, Xie Y, Lei Y, Kuang Z and Liu X: The
microRNA-130a-5p/RUNX2/STK32A network modulates tumor invasive and
metastatic potential in non-small cell lung cancer. BMC Cancer.
20:5802020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wu CY, Li L, Chen SL, Yang X, Zhang CZ and
Cao Y: A Zic2/Runx2/NOLC1 signaling axis mediates tumor growth and
metastasis in clear cell renal cell carcinoma. Cell Death Dis.
12:3192021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Cohen-Solal KA, Boregowda RK and Lasfar A:
RUNX2 and the PI3K/AKT axis reciprocal activation as a driving
force for tumor progression. Mol Cancer. 14:1372015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Tandon M, Gokul K, Ali SA, Chen Z, Lian J,
Stein GS and Pratap J: Runx2 mediates epigenetic silencing of the
bone morphogenetic protein-3B (BMP-3B/GDF10) in lung cancer cells.
Mol Cancer. 11:272012. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Pranavkrishna S, Sanjeev G, Akshaya RL,
Rohini M and Selvamurugan N: A computational approach on studying
the regulation of TGF-β1-stimulated Runx2 expression by MicroRNAs
in human breast cancer cells. Comput Biol Med. 137:1048232021.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Vishal M, Swetha R, Thejaswini G, Arumugam
B and Selvamurugan N: Role of Runx2 in breast cancer-mediated bone
metastasis. Int J Biol Macromol. 99:608–614. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Fang Y, Xue Z, Zhao L, Yang X, Yang Y,
Zhou X, Feng S and Chen K: Calycosin stimulates the osteogenic
differentiation of rat calvarial osteoblasts by activating the
IGF1R/PI3K/Akt signaling pathway. Cell Biol Int. 43:323–332. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Tandon M, Othman AH, Ashok V, Stein GS and
Pratap J: The role of Runx2 in facilitating autophagy in metastatic
breast cancer cells. J Cell Physiol. 233:559–571. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Schneider JG, Amend SR and Weilbaecher KN:
Integrins and bone metastasis: Integrating tumor cell and stromal
cell interactions. Bone. 48:54–65. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Li XQ, Lu JT, Tan CC, Wang QS and Feng YM:
RUNX2 promotes breast cancer bone metastasis by increasing integrin
α5-mediated colonization. Cancer Lett. 380:78–86. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Si W, Zhou J, Zhao Y, Zheng J and Cui L:
SET7/9 promotes multiple malignant processes in breast cancer
development via RUNX2 activation and is negatively regulated by
TRIM21. Cell Death Dis. 11:1512020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Morikawa M, Derynck R and Miyazono K:
TGF-β and the TGF-β family: Context-dependent roles in cell and
tissue physiology. Cold Spring Harb Perspect Biol. 8:a0218732016.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Larson C, Oronsky B, Carter CA, Oronsky A,
Knox SJ, Sher D and Reid TR: TGF-beta: A master immune regulator.
Expert Opin Ther Targets. 24:427–438. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Li XQ, Du X, Li DM, Kong PZ, Sun Y, Liu
PF, Wang QS and Feng YM: ITGBL1 is a Runx2 transcriptional target
and promotes breast cancer bone metastasis by activating the TGFβ
signaling pathway. Cancer Res. 75:3302–3313. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Taipaleenmäki H, Browne G, Akech J, Zustin
J, van Wijnen AJ, Stein JL, Hesse E, Stein GS and Lian JB:
Targeting of Runx2 by miR-135 and miR-203 impairs progression of
breast cancer and metastatic bone disease. Cancer Res.
75:1433–1444. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Croset M, Pantano F, Kan CWS, Bonnelye E,
Descotes F, Alix-Panabières C, Lecellier CH, Bachelier R, Allioli
N, Hong SS, et al: miRNA-30 family members inhibit breast cancer
invasion, osteomimicry, and bone destruction by directly targeting
multiple bone metastasis-associated genes. Cancer Res.
78:5259–5273. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhao P, Guan H, Dai Z, Ma Y, Zhao Y and
Liu D: Long noncoding RNA DLX6-AS1 promotes breast cancer
progression via miR-505-3p/RUNX2 axis. Eur J Pharmacol.
865:1727782019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Rohini M, Gokulnath M, Miranda PJ and
Selvamurugan N: miR-590-3p inhibits proliferation and promotes
apoptosis by targeting activating transcription factor 3 in human
breast cancer cells. Biochimie. 154:10–18. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Batlle E and Clevers H: Cancer stem cells
revisited. Nat Med. 23:1124–1134. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Babaei G, Aziz SGG and Jaghi NZZ: EMT,
cancer stem cells and autophagy; the three main axes of metastasis.
Biomed Pharmacother. 133:1109092021. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Peitzsch C, Tyutyunnykova A, Pantel K and
Dubrovska A: Cancer stem cells: The root of tumor recurrence and
metastases. Semin Cancer Biol. 44:10–24. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Ishiguro T, Ohata H, Sato A, Yamawaki K,
Enomoto T and Okamoto K: Tumor-derived spheroids: Relevance to
cancer stem cells and clinical applications. Cancer Sci.
108:283–289. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Baccelli I, Schneeweiss A, Riethdorf S,
Stenzinger A, Schillert A, Vogel V, Klein C, Saini M, Bäuerle T,
Wallwiener M, et al: Identification of a population of blood
circulating tumor cells from breast cancer patients that initiates
metastasis in a xenograft assay. Nat Biotechnol. 31:539–544. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Butti R, Gunasekaran VP, Kumar TVS,
Banerjee P and Kundu GC: Breast cancer stem cells: Biology and
therapeutic implications. Int J Biochem Cell Biol. 107:38–52. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Dittmer J: Breast cancer stem cells:
Features, key drivers and treatment options. Semin Cancer Biol.
53:59–74. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Katoh M: Canonical and non-canonical WNT
signaling in cancer stem cells and their niches: Cellular
heterogeneity, omics reprogramming, targeted therapy and tumor
plasticity (review). Int J Oncol. 51:1357–1369. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Ibrahim SA, Gadalla R, El-Ghonaimy EA,
Samir O, Mohamed HT, Hassan H, Greve B, El-Shinawi M, Mohamed MM
and Götte M: Syndecan-1 is a novel molecular marker for triple
negative inflammatory breast cancer and modulates the cancer stem
cell phenotype via the IL-6/STAT3, Notch and EGFR signaling
pathways. Mol Cancer. 16:572017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Colacino JA, Azizi E, Brooks MD, Harouaka
R, Fouladdel S, McDermott SP, Lee M, Hill D, Madden J, Boerner J,
et al: Heterogeneity of human breast stem and progenitor cells as
revealed by transcriptional profiling. Stem Cell Reports.
10:1596–1609. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu
Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, et al:
Breast cancer stem cells transition between epithelial and
mesenchymal states reflective of their normal counterparts. Stem
Cell Reports. 2:78–91. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zöller M: CD44: Can a cancer-initiating
cell profit from an abundantly expressed molecule? Nat Rev Cancer.
11:254–267. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Vikram R, Chou WC, Hung SC and Shen CY:
Tumorigenic and metastatic role of
CD44−/low/CD24−/low cells in luminal breast
cancer. Cancers (Basel). 12:12392020. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Yan X, Han D, Chen Z, Han C, Dong W, Han
L, Zou L, Zhang J, Liu Y and Chai J: RUNX2 interacts with BRG1 to
target CD44 for promoting invasion and migration of colorectal
cancer cells. Cancer Cell Int. 20:5052020. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Knutson TP, Truong TH, Ma S, Brady NJ,
Sullivan ME, Raj G, Schwertfeger KL and Lange CA:
Posttranslationally modified progesterone receptors direct
ligand-specific expression of breast cancer stem cell-associated
gene programs. J Hematol Oncol. 10:892017. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Valenti MT, Serafini P, Innamorati G, Gili
A, Cheri S, Bassi C and Dalle Carbonare L: Runx2 expression: A
mesenchymal stem marker for cancer. Oncol Lett. 12:4167–4172. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Fritz AJ, Hong D, Boyd J, Kost J, Finstaad
KH, Fitzgerald MP, Hanna S, Abuarqoub AH, Malik M, Bushweller J, et
al: RUNX1 and RUNX2 transcription factors function in opposing
roles to regulate breast cancer stem cells. J Cell Physiol.
235:7261–7272. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Muley H, Fadó R, Rodriguez-Rodriguez R and
Casals N: Drug uptake-based chemoresistance in breast cancer
treatment. Biochem Pharmacol. 177:1139592020. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Dingwall S, Lee JB, Guezguez B, Fiebig A,
McNicol J, Boreham D, Collins TJ and Bhatia M: Neoplastic human
embryonic stem cells as a model of radiation resistance of human
cancer stem cells. Oncotarget. 6:22258–22269. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Najafi M, Mortezaee K and Majidpoor J:
Cancer stem cell (CSC) resistance drivers. Life Sci.
234:1167812019. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Huang T, Song X, Xu D, Tiek D, Goenka A,
Wu B, Sastry N, Hu B and Cheng SY: Stem cell programs in cancer
initiation, progression, and therapy resistance. Theranostics.
10:8721–8743. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Garcia-Mayea Y, Mir C, Masson F, Paciucci
R and LLeonart ME: Insights into new mechanisms and models of
cancer stem cell multidrug resistance. Semin Cancer Biol.
60:166–180. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Smith AG and Macleod KF: Autophagy, cancer
stem cells and drug resistance. J Pathol. 247:708–718. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Pece S, Tosoni D, Confalonieri S, Mazzarol
G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG and Di Fiore
PP: Biological and molecular heterogeneity of breast cancers
correlates with their cancer stem cell content. Cell. 140:62–73.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Phillips TM, McBride WH and Pajonk F: The
response of CD24(−/low)/CD44+ breast cancer-initiating cells to
radiation. J Natl Cancer Inst. 98:1777–1785. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Ryoo IG, Choi BH and Kwak MK: Activation
of NRF2 by p62 and proteasome reduction in sphere-forming breast
carcinoma cells. Oncotarget. 6:8167–8184. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Bai J, Chen WB, Zhang XY, Kang XN, Jin LJ,
Zhang H and Wang ZY: HIF-2α regulates CD44 to promote cancer stem
cell activation in triple-negative breast cancer via PI3K/AKT/mTOR
signaling. World J Stem Cells. 12:87–99. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Bai X, Ni J, Beretov J, Graham P and Li Y:
Cancer stem cell in breast cancer therapeutic resistance. Cancer
Treat Rev. 69:152–163. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Leccia F, Del Vecchio L, Mariotti E, Di
Noto R, Morel AP, Puisieux A, Salvatore F and Ansieau S: ABCG2, a
novel antigen to sort luminal progenitors of BRCA1- breast cancer
cells. Mol Cancer. 13:2132014. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Sadikovic B, Thorner P, Chilton-Macneill
S, Martin JW, Cervigne NK, Squire J and Zielenska M: Expression
analysis of genes associated with human osteosarcoma tumors shows
correlation of RUNX2 overexpression with poor response to
chemotherapy. BMC Cancer. 10:2022010. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Roos A, Satterfield L, Zhao S, Fuja D,
Shuck R, Hicks MJ, Donehower LA and Yustein JT: Loss of Runx2
sensitises osteosarcoma to chemotherapy-induced apoptosis. Br J
Cancer. 113:1289–1297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Sugimoto H, Nakamura M, Yoda H, Hiraoka K,
Shinohara K, Sang M, Fujiwara K, Shimozato O, Nagase H and Ozaki T:
Silencing of RUNX2 enhances gemcitabine sensitivity of
p53-deficient human pancreatic cancer AsPC-1 cells through the
stimulation of TAp63-mediated cell death. Cell Death Discov.
1:150102015. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Ozaki T, Nakamura M, Ogata T, Sang M, Yoda
H, Hiraoka K, Sang M and Shimozato O: Depletion of pro-oncogenic
RUNX2 enhances gemcitabine (GEM) sensitivity of p53-mutated
pancreatic cancer Panc-1 cells through the induction of
pro-apoptotic TAp63. Oncotarget. 7:71937–71950. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Jeselsohn R, Cornwell M, Pun M, Buchwalter
G, Nguyen M, Bango C, Huang Y, Kuang Y, Paweletz C, Fu X, et al:
Embryonic transcription factor SOX9 drives breast cancer endocrine
resistance. Proc Natl Acad Sci USA. 114:E4482–E4491. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Geter PA, Ernlund AW, Bakogianni S, Alard
A, Arju R, Giashuddin S, Gadi A, Bromberg J and Schneider RJ:
Hyperactive mTOR and MNK1 phosphorylation of eIF4E confer tamoxifen
resistance and estrogen independence through selective mRNA
translation reprogramming. Genes Dev. 31:2235–2249. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Othman A, Winogradzki M, Patel S, Holmes
W, Blank A and Pratap J: The role of Runx2 in microtubule
acetylation in bone metastatic breast cancer cells. Cancers
(Basel). 14:34362022. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Wang XX, Ye FG, Zhang J, Li JJ, Chen QX,
Lin PY and Song CG: Serum miR-4530 sensitizes breast cancer to
neoadjuvant chemotherapy by suppressing RUNX2. Cancer Manag Res.
10:4393–4400. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Huang L, Zhang L and Chen X: Updated
review of advances in microRNAs and complex diseases: Taxonomy,
trends and challenges of computational models. Brief Bioinform.
23:bbac3582022. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Chen X, Xie D, Zhao Q and You ZH:
MicroRNAs and complex diseases: From experimental results to
computational models. Brief Bioinform. 20:515–539. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Chen X, Yan CC, Zhang X and You ZH: Long
non-coding RNAs and complex diseases: From experimental results to
computational models. Brief Bioinform. 18:558–576. 2017.PubMed/NCBI
|
|
126
|
Wang CC, Han CD, Zhao Q and Chen X:
Circular RNAs and complex diseases: From experimental results to
computational models. Brief Bioinform. 22:bbab2862021. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Zhao W, Geng D, Li S, Chen Z and Sun M:
LncRNA HOTAIR influences cell growth, migration, invasion, and
apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer
Med. 7:842–855. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Xu J, Wu KJ, Jia QJ and Ding XF: Roles of
miRNA and lncRNA in triple-negative breast cancer. J Zhejiang Univ
Sci B. 21:673–689. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Kong X, Duan Y, Sang Y, Li Y, Zhang H,
Liang Y, Liu Y, Zhang N and Yang Q: LncRNA-CDC6 promotes breast
cancer progression and function as ceRNA to target CDC6 by sponging
microRNA-215. J Cell Physiol. 234:9105–9117. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Sang Y, Chen B, Song X, Li Y, Liang Y, Han
D, Zhang N, Zhang H, Liu Y, Chen T, et al: circRNA_0025202
regulates tamoxifen sensitivity and tumor progression via
regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther.
27:1638–1652. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Yin X, Wang P, Yang T, Li G, Teng X, Huang
W and Yu H: Identification of key modules and genes associated with
breast cancer prognosis using WGCNA and ceRNA network analysis.
Aging (Albany NY). 13:2519–2538. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Joshi H and Press MF: Molecular oncology
of breast cancer. The Breast. Elsevier; Amsterdam, The Netherlands:
pp. pp282–307.e5. 2018, Available online:. https://www.sciencedirect.com/science/article/pii/B978032335955900022230–May.
2021 View Article : Google Scholar
|
|
133
|
Dzobo K and Sinkala M: Cancer stem cell
marker CD44 plays multiple key roles in human cancers: immune
suppression/evasion, drug resistance, epithelial-mesenchymal
transition, and metastasis. OMICS. 25:313–332. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Fillmore C and Kuperwasser C: Human breast
cancer stem cell markers CD44 and CD24: Enriching for cells with
functional properties in mice or in man? Breast Cancer Res.
9:3032007. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Tomita H, Tanaka K, Tanaka T and Hara A:
Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget.
7:11018–11032. 2016. View Article : Google Scholar : PubMed/NCBI
|















