Introduction
Renal cell carcinoma (RCC) accounts for 2–3% of all
malignant tumors in adults, as well as 90% of all malignant kidney
tumors (1,2). The incidence and mortality rates of
kidney cancer have been increasing worldwide. According to the 2021
GLOBOCAN data, 431,288 cases of kidney cancer were diagnosed in
2020 worldwide, accounting for 2.2% of all new cancer cases and
1.8% (179,368) of all new cancer-related deaths (3). Clear cell RCC (ccRCC), which is the
most common subtype of renal cancer (4), is characterized by increased
malignancy and has the highest mortality rate among genitourinary
system cancers (5). Radiation
therapy and chemotherapy are ineffective against ccRCC. Currently,
surgical intervention is the primary treatment for ccRCC. Although
the incidence of kidney cancer is increasing annually, the
development of early detection techniques has markedly decreased
kidney cancer-related mortality rates in recent decades (6). The majority of patients with ccRCC are
asymptomatic and are diagnosed during imaging examinations, such as
computed tomography scan or ultrasound (7). The lack of effective diagnostic
methods prevents early diagnosis and treatment of patients with
ccRCC, contributing to poor prognosis and poor survival rates. The
survival rate of patients with kidney cancer who have distant
metastases is only 12%, while that of patients with localized
cancer is 67% (8). Surgical
resection of locally advanced RCC is the only curative treatment
(5). Approximately 20–30% of
patients with kidney cancer exhibit relapse after nephrectomy
(9,10). Recently, several treatment
strategies, such as immunotherapy, radiotherapy, and molecular
targeted drugs, have been developed, which have markedly improved
the clinical outcomes of advanced diseases (11,12).
However, the clinical outcomes of kidney cancer are poor owing to
the low objective response rates, local recurrences, or distant
metastases. Therefore, there is an urgent need to identify novel
molecular prognostic markers for ccRCC.
Evaluation of the prognostic value of OVO-like
proteins (OVOLs) in patients with ccRCC can potentially improve the
prediction of clinical outcomes and aid in the development of
effective treatments. In mammals, OVOLs encode C2H2 zinc finger
transcription factors (13). OVOLs,
which are members of the zinc finger protein family, function as
transcription factors to regulate gene expression during
differentiation (14). The three
members of the OVOL family are OVOL1, OVOL2, and OVOL3. Molecular
profiling of human tumors revealed that OVOL deregulation is
associated with adverse outcomes in various carcinomas and is
directly related to metastasis (15–17).
The activity of OVOL can stabilize a hybrid phenotype between
epithelial and mesenchymal states, resulting in several benefits
for both tumors and healthy stem cells (18,19).
Epithelial-to-mesenchymal transition (EMT) plays a key role in the
stromal invasion of tumor cells (20). OVOL1 and OVOL2 are reported to be
key regulators of EMT and its mirror process.
mesenchymal-to-epithelial transition (MET) (16). Previous studies have reported that
OVOLs are associated with the clinical stage, EMT, and tumor
metastasis and that they can modulate cancer cell stemness.
Additionally, OVOLs are potential prognostic prediction factors
(17,21–23).
The distinct expression/mutation pattern and prognostic
significance of OVOLs have not been evaluated in ccRCC.
This study evaluated the potential of OVOLs as
predictors of the prognosis of ccRCC using experimental and
bioinformatics approaches.
Materials and methods
Ethics statement
The research protocol used in the present study was
approved by the Ethics Committee of the First Affiliated Hospital
of Nanchang University (approval no. 12-110). The datasets were
retrieved from public databases. All data were collected after
obtaining written consent.
Patient and tumor samples
In total, 20 pairs of kidney renal clear cell
carcinoma (KIRC) and adjacent non-tumorous tissues obtained via
radical nephrectomy were collected from patients who had been
pathologically confirmed to have cancer by two independent
pathologists. The samples collected at the First Affiliated
Hospital of Nanchang between January 2021 and December 2022 were
immediately stored in liquid nitrogen.
Cell lines and cell culture
The KIRC, HK-2, 786-O, Caki-1, and ACHN cell lines
were purchased from the American Type Cell Collection. The cell
lines were maintained as monolayers in minimal essential medium,
RMPI-1640 medium, and Dulbecco's modified essential medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% (v/v) fetal
bovine serum at 37°C in a humidified incubator supplied with 5%
CO2.
RNA isolation and reverse
transcription-quantitative (RT-q)PCR
Total RNA was isolated from ccRCC tissues and
adjacent non-tumorous tissues, as well as from ccRCC cell lines,
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The extracted RNA was reverse-transcribed to
cDNA using the First-Strand cDNA synthesis kit (Qiagen GmbH)
according to the manufacturer's protocol. qPCR was performed using
the SYBR Real-Time PCR kit (Qiagen GmbH). The PCR thermocycling
conditions were as follows: 95°C for 2 min, followed by 40 cycles
of 95°C for 5 sec and 60°C for 10 sec. The relative gene expression
levels were calculated using the 2−ΔΔCq method (24). Each analysis was performed in
triplicate. ACTB served as an internal reference gene. The
sequences of the primers used for qPCR were: GAPDH forward,
5′-GCCACATCGCTCAGACACCAT-3′ and reverse,
5′-CCCATACGACTGCAAAGACCC-3′; human OVOL1 forward,
5′-AGACACGTCCGAACTCACAC-3′ and reverse, 5′-TGCTGCACACCATGGATCTT-3′;
human OVOL2 forward, 5′-CAACGACACCTTCGACCTGA-3′ and reverse,
5′-TCAGGTGGGACTCCAGAGAG-3′; human OVOL3 forward,
5′-TTCGATCTCAAGCGCCACAT-3′ and reverse,
5′-GCTGTCCATGCACCTTAGCA-3′.
Gene Expression Profiling Interactive
Analysis (GEPIA) dataset
GEPIA (http://gepia.cancer-pku.cn/) was used to comparatively
examine the tumor and non-tumor tissue datasets from The Cancer
Genome Atlas (TCGA; http://portal.gdc.com) and the non-tumor datasets from
the Genotype-Tissue Expression database (25). The differential expression levels of
OVOL1, OVOL2, and OVOL3 between ccRCC tissues and
adjacent non-tumor tissues are represented using box plots. The
correlation between gene expression and cancer stage was examined
using the GEPIA software package.
cBioPortal analysis
cBioPortal is a free, open-access online resource
integrating data from large-scale genomic projects, including but
not limited to TCGA and the International Cancer Genome Consortium
(26). In the present study, 512
ccRCC samples (TCGA, provisional) with known mutations, putative
copy-number alterations (identified using the GISTIC module)
(27), and z-scores (RNA Seq V2
RSEM) for mRNA expression were analyzed. Additionally, the
correlation between genetic mutations in OVOL-encoding genes with
the overall survival (OS) of patients with ccRCC was examined. A
log-rank test was used to compare the difference in survival
between the altered and unaltered groups.
STRING analysis
STRING (http://string-db.org) was used to construct
protein-protein interaction networks, including both physical
binding and functional associations, between immune checkpoints and
tumor immune microenvironment-related factors (28). The networks were visualized using R
software (version 4.2.1; http://www.r.project.org).
GSCALite
GSCALite (http://bioinfo.life.hust.edu.cn/web/GSCALite/)
provides a platform for the analysis of gene sets in cancer
(29). In the present study, the
correlation between microRNA (miRNA) and corresponding OVOLs was
analyzed using GSCALite. Drug sensitivity data and gene expression
profiles of cancer cell lines were retrieved from the Genomics of
Drug Sensitivity in Cancer (GDSC) database and the Therapeutics
Response Portal (CTRP).
Statistical analysis
All statistical analyses were performed using R
software (version 3.6.2). The differential expression levels of
OVOLs in ccRCC were analyzed using the ‘limma’ R package and a
Wilcox test. The prognostic significance of OVOLs was evaluated
using Kaplan-Meier survival analysis and Cox proportional hazards
regression analysis. The effect of clinicopathological parameters
and mRNA levels of OVOLs on the survival of patients with ccRCC was
determined using univariate Cox regression analysis. Further
analyses were performed using a P<0.1 threshold. All statistical
analyses were performed using two-sided tests. P<0.05 was
considered to indicate a statistically significant difference. The
RNA-sequencing expression (level 3) profiles and corresponding
clinical information for ccRCC were downloaded from TCGA. The R
software ggstatsplot
packagehttps://CRAN.R-project.org/package=ggstatsplot) was used to
plot the correlation between gene expression and immune score,
while the pheatmap package (https://CRAN.R-project.org/package=pheatmap) was used
to plot multi-gene correlation. Gene Ontology (GO) (30,31)
and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
(32) enrichment analyses were
performed using R. To predict the functional roles of target host
genes, GO enrichment analysis was performed based on biological
process (BP), cellular component (CC) and molecular function (MF)
using the R package ggplot2 v3.3.2 (33).
Results
Differential mRNA levels of OVOLs in
patients with ccRCC
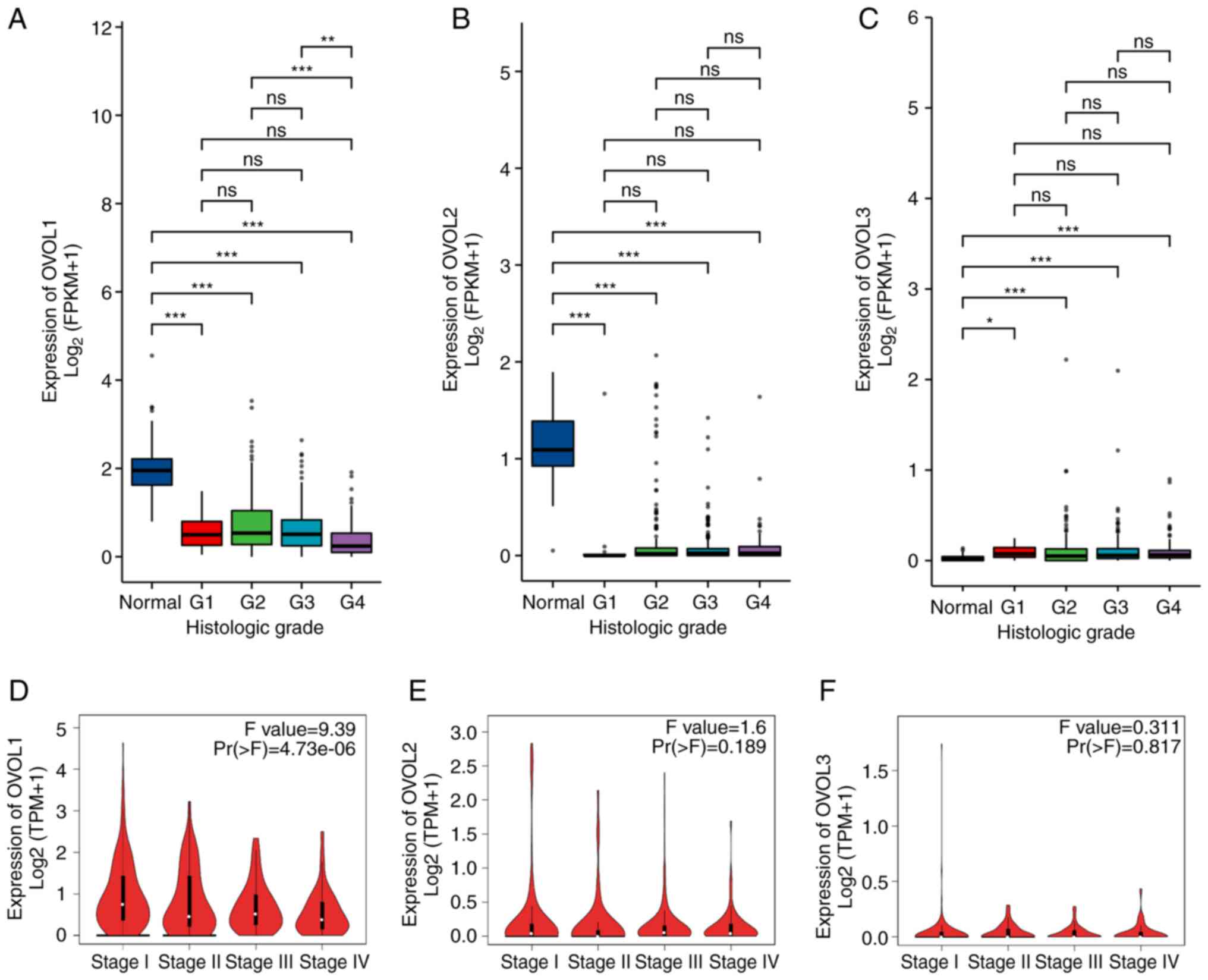
A TCGA-KIRC dataset was analyzed to comparatively
evaluate the expression levels of OVOL family members between ccRCC
samples and 72 paired non-tumor tissue samples. As shown in
Fig. 1A-C, the OVOL1 and
OVOL2 mRNA levels in non-tumor samples were significantly
higher than those in ccRCC tissues. In contrast, the OVOL3
mRNA levels in tumor tissues were upregulated when compared with
those in the non-tumor tissues. Next, the OVOL1, OVOL2, and
OVOL3 expression levels in ccRCC datasets obtained from the
GEPIA database were analyzed. As shown in Fig. 1D-F, the OVOL1 and
OVOL2 mRNA levels in non-tumor tissues were significantly
upregulated when compared with those in ccRCC tissues. However, the
mRNA levels of OVOL3 were not significantly different
between ccRCC and non-tumor tissues. To further validate this
conclusion, 20 pairs of cancerous and adjacent non-cancerous
tissues were subjected to RT-qPCR analysis. As shown in Fig. 1G-I, the OVOL3 mRNA levels in
tumor tissues were higher than those in adjacent non-tumor tissues.
Meanwhile, the OVOL1 and OVOL2 mRNA levels in tumor
tissues were downregulated when compared with those in adjacent
non-tumor tissues. These results indicate that the mRNA levels of
OVOL1 and OVOL2 in paired non-tumor tissues were
significantly lower than those in ccRCC tissues, while those of
OVOL3 exhibited contrasting expression patterns. Healthy
kidney and kidney cancer cell lines were subjected to RT-qPCR
analysis to determine the mRNA levels of OVOL1, OVOL2, and
OVOL3 (Fig. S1). The mRNA
levels of OVOL1 and OVOL2 in kidney cancer cell lines
were lower than those in healthy cell lines. In contrast, the
OVOL3 mRNA levels in kidney cancer cell lines were
upregulated when compared with those in healthy cell lines.
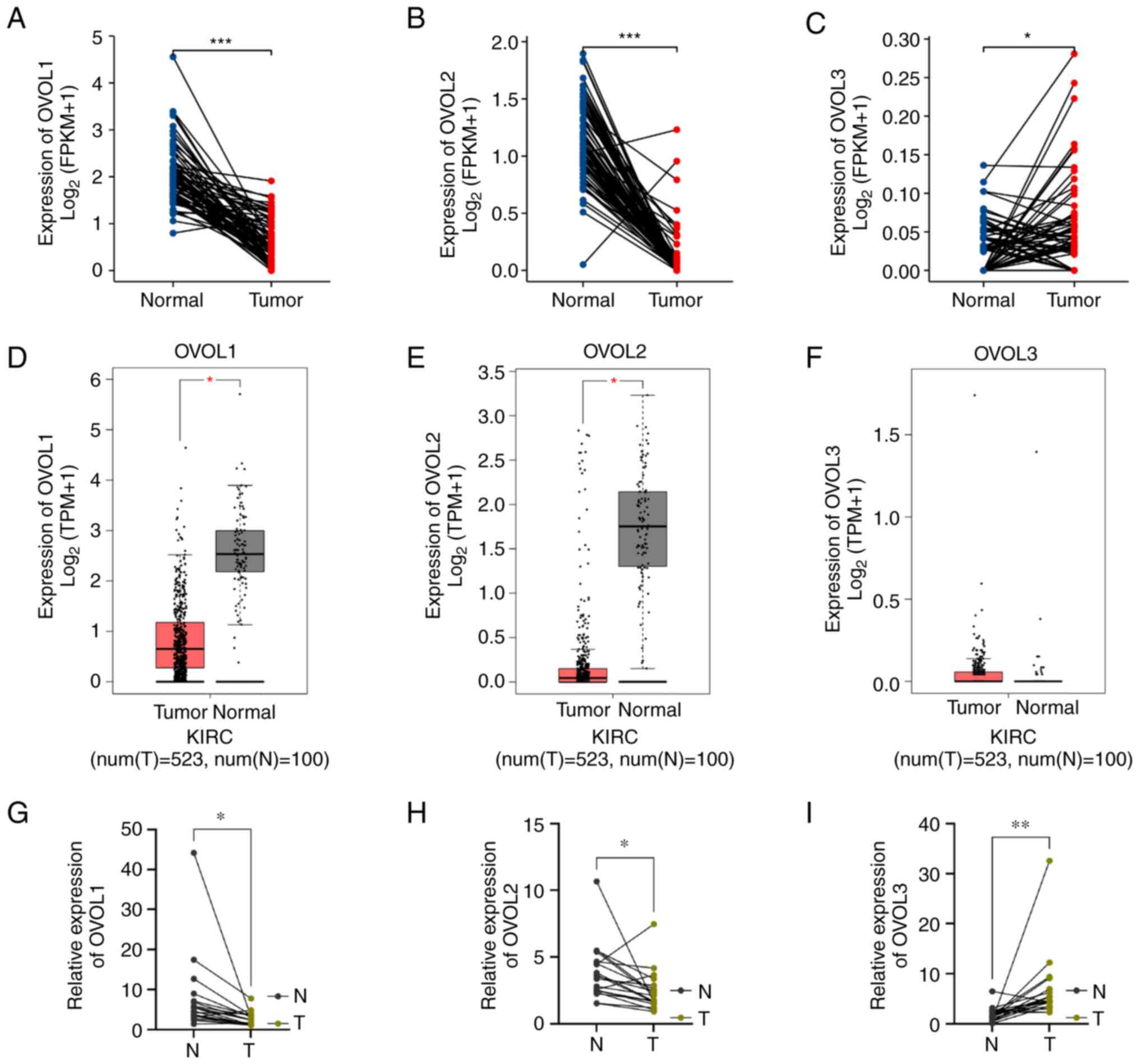 | Figure 1.The expression of distinct OVOL1,
OVOL2 and OVOL3 in KIRC tissues and adjacent normal kidney tissues.
(A-C) The differential mRNA expression levels of OVOL1,
OVOL2, and OVOL3 between 72 pairs of kidney cancer
tissues and adjacent non-cancerous kidney tissues, which were
obtained from The Cancer Genome Atlas and Genotype-Tissue
Expression databases. (D-F) The expression levels of OVOL1,
OVOL2, and OVOL3 in clear cell renal cell carcinoma and
healthy kidney tissues were analyzed using the Gene Expression
Profiling Analysis datasets. (G-I) The mRNA levels of OVOL1,
OVOL2, and OVOL3 in ccRCC tissues and paired adjacent
non-tumor kidney tissues were analyzed using reverse
transcription-quantitative PCR. *P<0.05, **P<0.01,
***P<0.001. ns, not significant; OVOL, OVO-like protein; KIRC,
kidney renal clear cell carcinoma; FPKM, fragments per kilobase
million; TPM, transcripts per million. |
Correlation between OVOLs and the
clinicopathological parameters of patients with ccRCC
Next, the correlation between the OVOL1,
OVOL2, and OVOL3 mRNA levels and clinicopathological
parameters (including histological grade and pathological stage
obtained from TCGA and GEPIA datasets) was evaluated. As shown in
Fig. 2A-C, the OVOL1 mRNA
expression levels were positively correlated with tumor
histological grade. However, the OVOL2 and OVOL3 mRNA
levels were not correlated with the histological grade. As shown in
Fig. 2D-F, the OVOL1 mRNA
levels were correlated with the clinical stage of ccRCC, whereas
the OVOL2 and OVOL3 mRNA levels were not correlated.
Patients at advanced pathological stages exhibited downregulated
OVOL1 levels. Thus, the mRNA expression levels of the OVOL
family members were significantly correlated with several clinical
and pathological parameters in patients with ccRCC.
Prognostic value of OVOLs mRNA levels
in patients with ccRCC
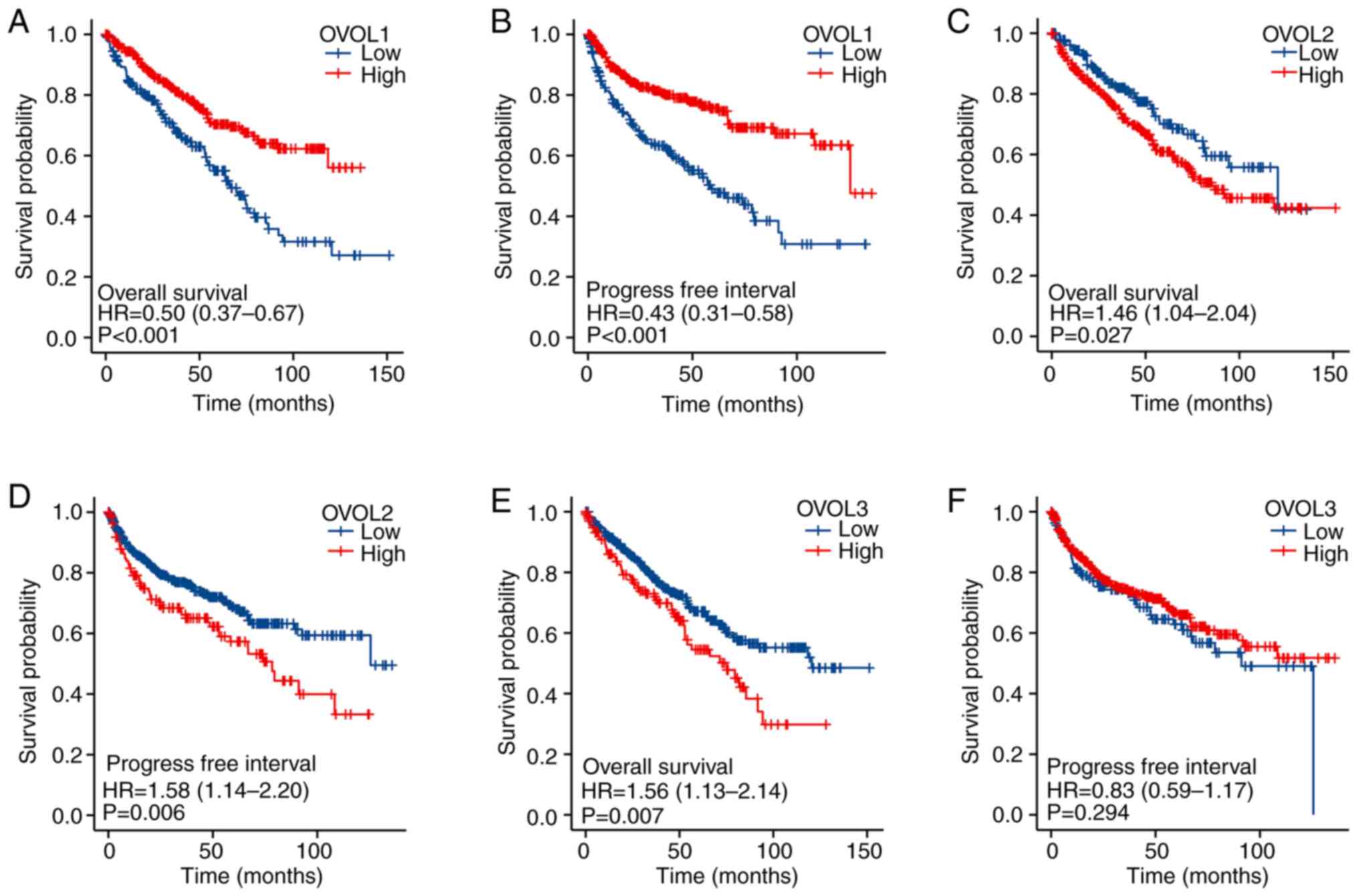
The prognostic value of the expression of
OVOL-encoding mRNAs in patients with ccRCC was analyzed using
Kaplan-Meier survival curves using TCGA datasets. As shown in
Fig. 3, the mRNA levels of OVOL
family members were significantly associated with the prognosis of
patients with ccRCC. The downregulated OVOL1 mRNA levels
[(hazard ratio (HR)=0.50, 95% confidence interval (CI)=0.37-0.67,
and P<0.001] were associated with a poorer OS in patients with
ccRCC. Meanwhile, the upregulated OVOL2 (HR=1.46, 95%
CI=1.04-2.04, P=0.027) and OVOL3 (HR=1.56, 95% CI=1.13-2.14,
P=0.007) mRNA levels were associated with a poorer OS in
patients with ccRCC. The upregulated mRNA levels of OVOL1
(HR=0.43, 95% CI=0.31-0.58, P<0.001) were associated with a
favorable progression-free interval (PFI). However, the
OVOL3 mRNA levels were not associated with the PFI of
patients with ccRCC. The OVOL2 mRNA levels (HR=1.58, 95%
CI=1.14-2.20, P=0.006) were negatively correlated with PFI. Thus,
the mRNA expression levels of OVOLs were significantly correlated
with the prognosis of patients with ccRCC. Additionally, the mRNA
levels may be used as indicators for predicting clinical outcomes,
including the OS, of patients with ccRCC.
Independent prognostic value of OVOL
mRNA levels for predicting OS in patients with ccRCC
The mRNA levels of OVOLs were significantly
associated with the OS of patients with ccRCC. Further, the
independent prognostic value of the mRNA expression levels of OVOL
family members in patients with ccRCC was examined using a TCGA
dataset and Cox survival regression analysis (34). Univariate Cox regression analysis
revealed that the upregulated expression of OVOL1 (HR=0.492,
95% CI=0.347-0.700, P<0.001) was significantly associated with a
favorable OS. Multivariate analysis revealed that the upregulated
mRNA levels of OVOL1 (HR=0.645, 95% CI=0.446-0.933, P=0.020)
were independently correlated with a favorable OS in patients with
ccRCC. Thus, the mRNA levels of OVOL1 are an independent
prognostic factor in patients with ccRCC (Table SI).
Genetic mutations in the OVOL family
and their association with the OS of patients with ccRCC
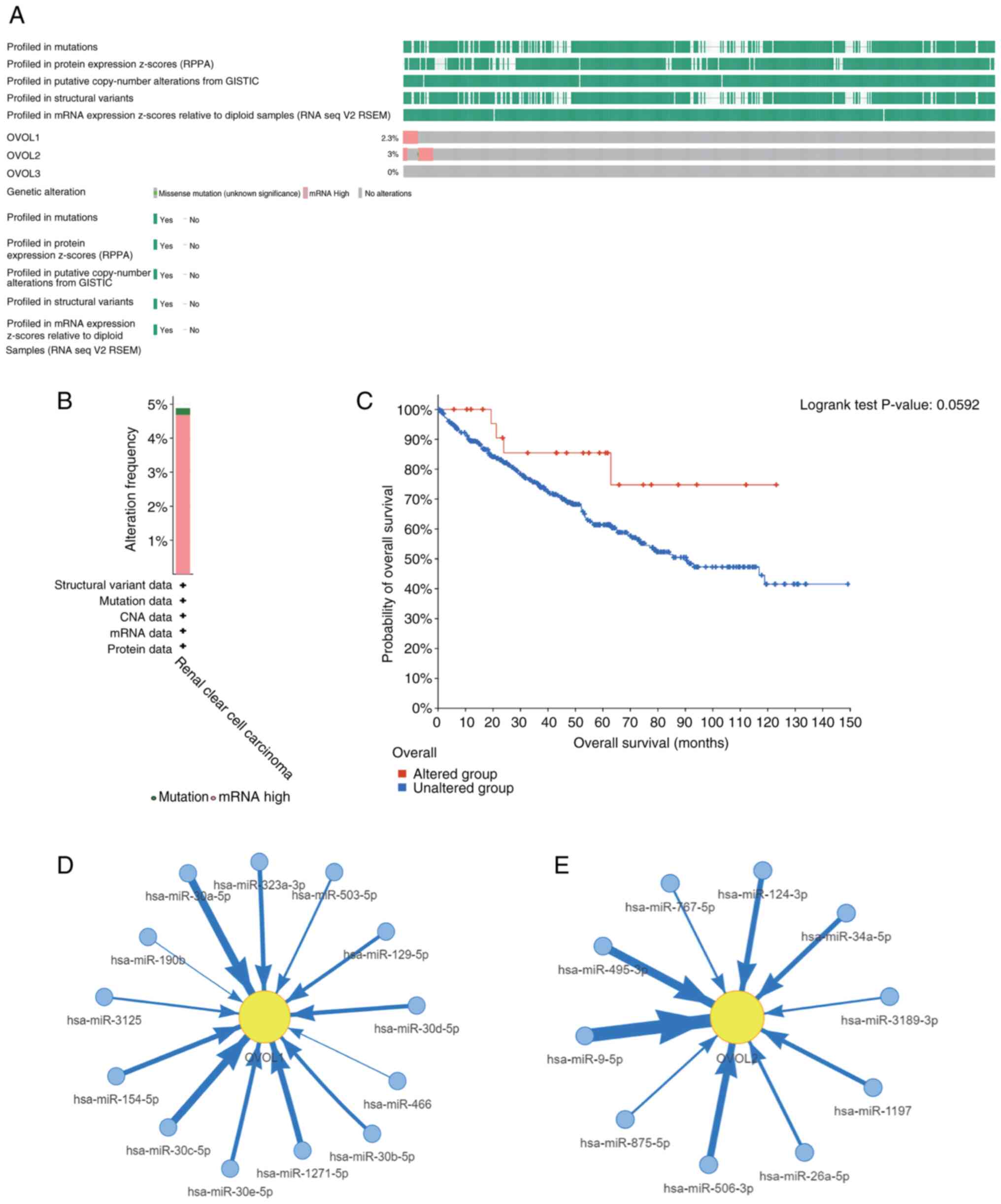
To further evaluate the expression patterns of the
OVOL family members, genetic alterations in OVOL-encoding genes and
their association with the OS of patients with ccRCC were examined
using the cBioPortal online tool. Genetic alterations in
OVOL-encoding genes are shown in Fig.
4A. The frequency of genetic alterations according to the
cBioPortal database is shown in Fig.
4B. The ccRCC dataset analysis indicated that the percentages
of DNA alterations in OVOL1, OVOL2, and OVOL3 were
2.3, 3, and 0%, respectively. Next, the association between
OVOL-encoding gene alterations and survival outcomes was examined.
Mutations in the OVOL-encoding gene family were not associated with
the OS (Fig. 4C). These results
indicated that DNA alterations were not the primary cause for the
dysregulation of OVOL family members. Multiple non-coding RNAs,
including hsa-miR-9-5p and hsa-miR-30a-5p, regulated OVOL family
mRNAs, which suggested that non-coding RNA-mediated regulation may
play a key role in OVOL alterations (Fig. 4D-E).
Predicted functions and pathways of
altered OVOLs and the 100 most frequently altered neighboring genes
in patients with ccRCC
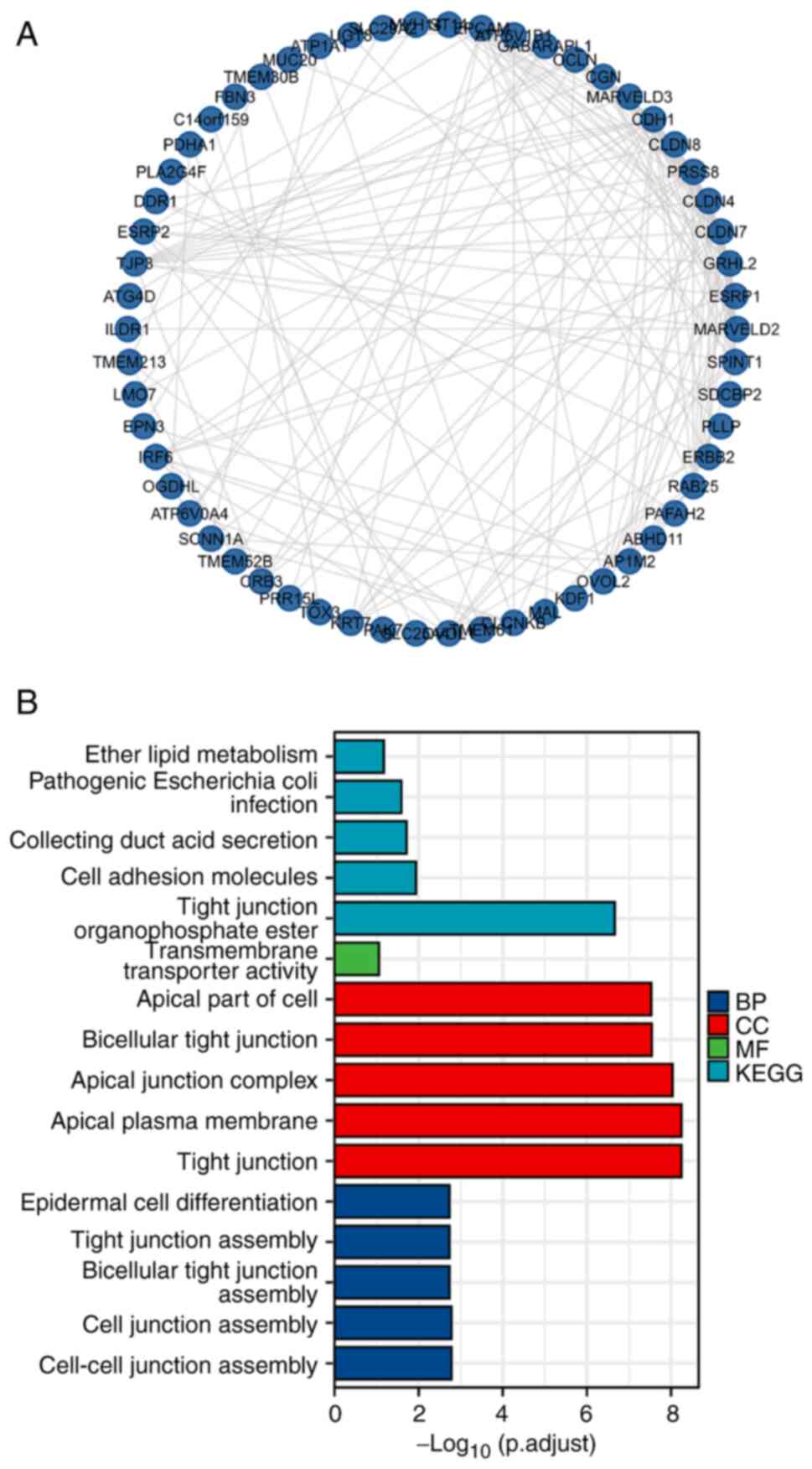
An integrated network was constructed by analyzing
100 genes related to the OVOL mutants. The top 100 genes that were
co-expressed and associated with OVOLs were retrieved from the
cBioPortal database. A protein-protein interaction network was
constructed using R. As shown in Fig.
5A, the cell-cell junction assembly-related genes, including
CDH1, UGT8, GRHL2, MARVELD3, CRB3, MARVELD2, and
OCLN, were significantly associated with OVOL mutations.
BPs, such as GO:0007043 (cell-cell junction assembly), GO:0034329
(cell junction assembly), GO:0070830 (bicellular tight junction
assembly), GO:0120192 (tight junction assembly), and GO:0009913
(epidermal cell differentiation) were significantly associated with
OVOL alterations in ccRCC. CCs, including GO:0070160 (tight
junction), GO:0016324 (apical plasma membrane), GO:0043296 (apical
junction complex), GO:0045177 (apical part of the cell), and
GO:0005923 (bicellular tight junction), were significantly
associated with OVOL alterations. MFs, such as GO:0015605
(organophosphate ester transmembrane transporter activity) were
significantly associated with OVOL alterations. KEGG analysis
revealed that OVOL mutations were enriched in the following five
pathways in ccRCC: has04530 (Tight junction), has04514 (Cell
adhesion molecules), has04966 (Collecting duct acid secretion),
has05130 (Pathogenic Escherichia coli infection), and
ha00565 (Ether lipid metabolism) (Fig.
5B and Table SII).
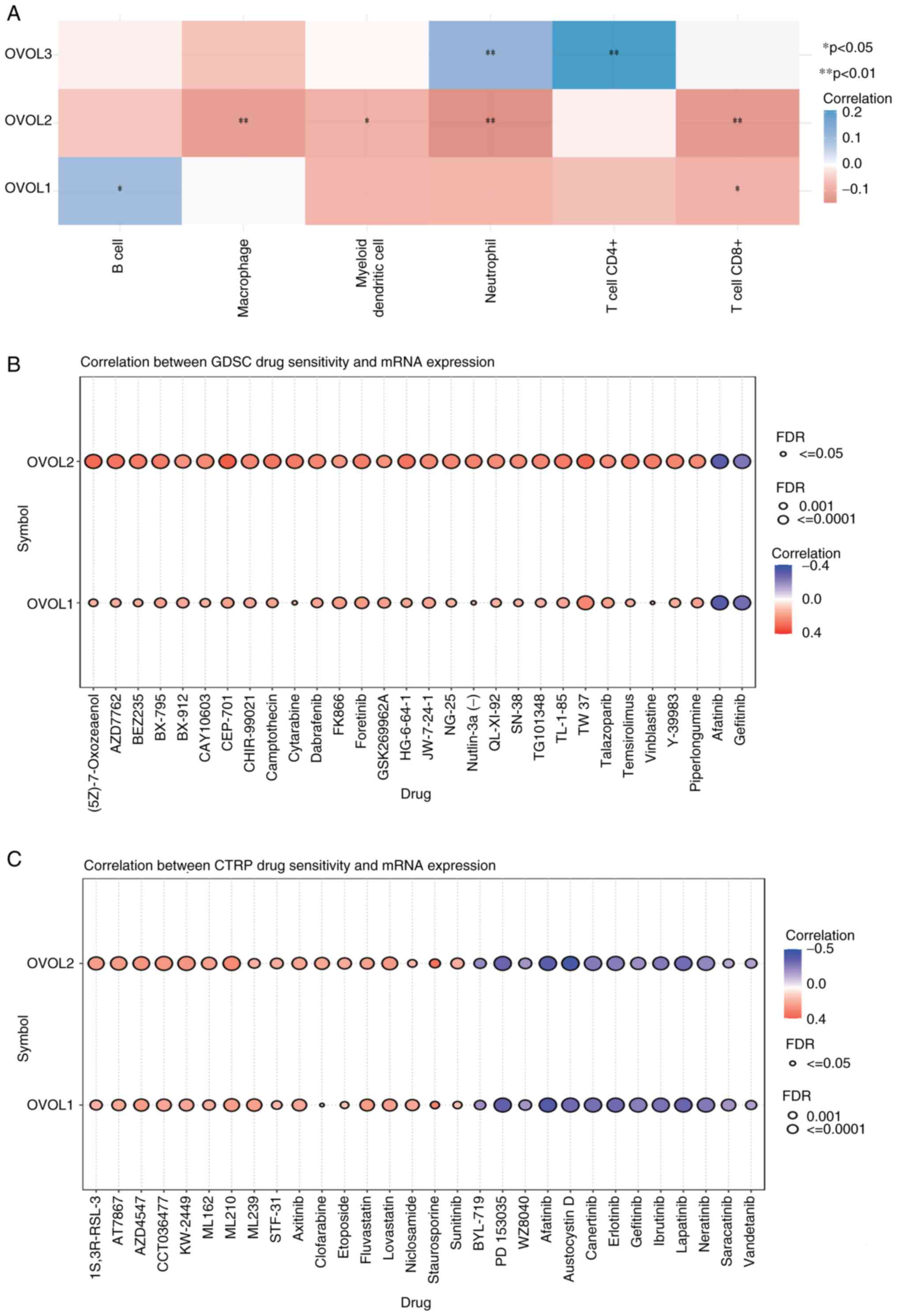
Association between OVOLs and immune
infiltration in ccRCC
The correlation between genes and immune
infiltration was evaluated using the R software pheatmap package.
The abundance of CD8+ T cells was negatively associated with the
expression levels of OVOL1 and OVOL2. Additionally, the abundance
of CD4+ T cells was negatively associated with the expression of
OVOL3. The abundance of neutrophil cells was positively associated
with the expression of OVOL3 but negatively associated with the
expression of OVOL2. Furthermore, the abundance of macrophages and
dendritic cells was negatively associated with the expression of
OVOL2. The abundance of B cells was positively associated with
OVOL1 expression (Fig. 6A). These
data suggested that immune infiltration was closely associated with
the OVOL family members in patients with ccRCC.
Verification of the role of OVOLs in
drug sensitivity
Analysis of the GDSC revealed that the expression
levels of OVOL1 and OVOL2 were positively associated
with certain drugs (Fig. 6B).
Additionally, analysis of CTRP revealed that the mRNA levels of
OVOL1 and OVOL2 were negatively associated with drugs
or small molecules and positively associated with small molecules
(Fig. 6C). Thus, OVOL1 and OVOL2
expression levels were associated with drug resistance, which
indicated that they may be used to determine drug sensitivity.
Discussion
Several studies have reported that OVOLs function as
transcription factors to regulate gene expression during various
differentiation processes (14) and
induce MET in several types of cancer (16). The dysregulation of OVOLs has been
reported in several types of cancer (22,23,35).
In the present study, bioinformatics and experimental studies were
used to investigate the correlation between OVOLs and the prognosis
of ccRCC. A comprehensive analysis of OVOLs in ccRCC has not been
previously performed to the best of our knowledge. The mRNA levels,
protein interactions, and functional enrichment of OVOLs and their
correlation with immune infiltration and prognosis were
investigated in the present study. The results of this study
indicated that OVOLs are potential therapeutic targets and
prognostic markers for ccRCC.
OVOL1 is a key regulator of epithelial lineage
determination and MET. Studies have demonstrated that OVOL1
inhibits breast cancer cell invasion by promoting the degradation
of TGF-β type I receptor (36).
OVOL1 overexpression promoted oral squamous cell carcinoma (OSCC)
progression by inhibiting ZEB1. Thus, OVOL1 is a potential
prognostic marker for OSCC (22).
Additionally, OVOL1 is significantly downregulated in cutaneous
squamous cell carcinoma. Mechanistically, OVOL1 functions as an
upstream suppressor of c-Myc and OVOL2. The OVOL1-OVOL2 axis and a
modulator of c-Myc regulate the invasiveness of cutaneous squamous
cell carcinoma. OVOL1 expression was upregulated in eccrine poroma
and hidradenoma and may play an important role in human skin
morphogenesis and tumorigenesis (37). In the present study, the expression
of OVOL1 in ccRCC tissues was downregulated when compared with that
in adjacent non-tumor tissues. The miRNA network revealed the mRNA
expression of OVOL1 was regulated by multiple miRNAs with
the coordination between several miRNAs regulating the expression
of a single mRNA (38). The miRNAs
bind to their target mRNAs to form RNA-induced silencing complexes
that degrade or inhibit the translation of mRNA (39). The upregulated mRNA expression of
OVOL1 was significantly correlated with prolonged OS and PFI
in ccRCC, suggesting that OVOL1 can function as a tumor suppressor.
Additionally, multivariate Cox regression analyses showed that the
downregulated OVOL1 expression independently predicted poor
outcomes in ccRCC. OVOL1 was also correlated with the infiltration
of immune cells in ccRCC, which suggested that OVOL1 may regulate
cancer immunity. These findings suggest that OVOL1 is a promising
prognostic and therapeutic target for patients with ccRCC.
OVOL2, which functions as a transcription factor to
regulate gene expression by directly binding to the promoter
regions, plays an important role in tumor development and
metastasis. A previous study reported that OVOL2 is closely
associated with EMT during tumor invasion. The expression of OVOL2
is downregulated in non-small cell lung cancer (NSCLC).
Consistently, OVOL2 overexpression inhibited the survival of NSCLC
cells (40). Additionally, a
previous study reported that OVOL2 inhibited EMT in breast cancer
by suppressing the direct transcription of ZEB1. Patients
with nasopharyngeal carcinoma (NPC) exhibiting downregulated OVOL2
levels were associated with poor OS. Thus, OVOL2 is a potential
prognostic indicator for NPC (23).
OVOL2 also inhibits EMT and metastasis in colorectal cancer by
suppressing Wnt signaling (41).
Similarly, OVOL2 inhibits EMT in liver cancer by indirectly
promoting the expression of miR-200 (35). In the present study, the expression
of OVOL2 in non-tumor tissues was higher than that in kidney tumor
tissues. However, the upregulated OVOL2 expression was
significantly correlated with a poorer OS and PFI, suggesting an
oncogenic role of OVOL2. miRNA network analysis revealed that the
oncogenic role of OVOL2 can be attributed to the non-coding
RNA-mediated regulation of OVOL2 mRNA. As the gene
expression levels varied in each cancer cell, tumor heterogeneity
may significantly contribute to differential mRNA expression. It
has been demonstrated that OVOL2 overexpression in macrophages
significantly inhibited M2 polarization and consequently inhibited
breast cancer metastasis by regulating IL10 transcription
and modulating the tumor microenvironment (42). In the present study, OVOL2
expression was negatively correlated with the infiltration of
immune cells. Thus, these results indicate that OVOL2 is a
potential prognostic biomarker and a therapeutic target for ccRCC
and that OVOL2 is an oncogene.
OVOL3 has not been previously studied as it is
expressed only in early embryos. Thus, the correlation between
OVOL3 and cancer is unclear. In the present study, the mRNA
expression levels of OVOL3 in ccRCC tissues were
significantly higher than those in healthy kidney tissues. OVOL3
expression was significantly correlated with poor OS but not with
PFI. The expression of OVOL3 was positively correlated with the
infiltration of immune cells, including neutrophils and CD4+ T
cells. However, there are no studies examining the role of OVOL3 in
different subtypes of ccRCC to the best of our knowledge.
The present study investigated the expression levels
and prognostic value of OVOLs in ccRCC. The findings of this study
improved our understanding of the molecular heterogeneity and
complexity of ccRCC. Additionally, experimental evidence was
generated for the expression of OVOLs in ccRCC tissues. However,
this study has some limitations. Although the mRNA levels of
OVOL1 were demonstrated to be an independent prognostic
factor associated with a short OS in patients with ccRCC, further
studies are needed to validate the findings of this study and
explore the clinical application of OVOLs in ccRCC. Second, the
mechanisms of the different types of OVOLs were not elucidated.
Future studies should explore the potential mechanisms of OVOLs in
ccRCC. Finally, this study was based on retrospective data.
In conclusion, the present study evaluated the
expression levels and prognostic value of OVOLs in ccRCC. The
upregulated mRNA expression levels of OVOL2 and OVOL3
were significantly correlated with the OS in patients with ccRCC.
Additionally, the upregulated mRNA expression of OVOL1 was
associated with a favorable OS. The OVOL1 mRNA levels were
significantly associated with clinical cancer stage and
histological grade in patients with ccRCC. Multivariate analysis
revealed that OVOL1 mRNA expression was independently
associated with a short OS in patients with ccRCC. The mRNA
expression levels of OVOLs were closely associated with immune
infiltration in patients with ccRCC and drug sensitivity.
Therefore, these results indicate that OVOL1 and OVOL2 are
potential therapeutic targets for ccRCC and that OVOL1 is a novel
prognostic factor for ccRCC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank the two
pathologists, Dr Fu Wenda and Dr Wu Haiyan (Department of
Pathology, The First Hospital of Putian City, Putian, China), who
independently confirmed the pathological diagnosis of tumor
tissues.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PZ designed and supervised the project. RC and MJ
performed the bioinformatics analysis and conducted the
experiments. JL and GC were responsible for data interpretation,
literature search, critical revision of the manuscript for
scientific and factual content and confirmed the authenticity of
all the raw data. All the authors have seen and confirmed the
authenticity of the raw data generated during the study. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
All subjects provided written informed consent. The
study was conducted according to the ethical principles of the
Declaration of Helsinki and was approved by the Institutional
Ethics Committee of First Affiliated Hospital of Nanchang
University. (approval no. 202012-110).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moch H, Cubilla AL, Humphrey PA, Reuter VE
and Ulbright TM: The 2016 WHO classification of tumours of the
urinary system and male genital organs-part A: Renal, penile, and
testicular tumours. Eur Urol. 70:93–105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ljungberg B, Albiges L, Abu-Ghanem Y,
Bensalah K, Dabestani S, Fernández-Pello S, Giles RH, Hofmann F,
Hora M, Kuczyk MA, et al: European association of urology
guidelines on renal cell carcinoma: The 2019 update. Eur Urol.
75:799–810. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wong MCS, Goggins WB, Yip BHK, Fung FDH,
Leung C, Fang Y, Wong SYS and Ng CF: Incidence and mortality of
kidney cancer: Temporal patterns and global trends in 39 countries.
Sci Rep. 7:156982017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Padala SA and Barsouk A, Thandra KC,
Saginala K, Mohammed A, Vakiti A, Rawla P and Barsouk A:
Epidemiology of renal cell carcinoma. World J Oncol. 11:79–87.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Williamson TJ, Pearson JR, Ischia J,
Bolton DM and Lawrentschuk N: Guideline of guidelines: Follow-up
after nephrectomy for renal cell carcinoma. BJU Int. 117:555–562.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jamil ML, Keeley J, Sood A, Dalela D,
Arora S, Peabody JO, Trinh QD, Menon M, Rogers CG and Abdollah F:
Long-term risk of recurrence in surgically treated renal cell
carcinoma: A post hoc analysis of the eastern cooperative oncology
group-American college of radiology imaging network E2805 trial
cohort. Eur Urol. 77:277–281. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Unverzagt S, Moldenhauer I, Nothacker M,
Roßmeißl D, Hadjinicolaou AV, Peinemann F, Greco F and Seliger B:
Immunotherapy for metastatic renal cell carcinoma. Cochrane
Database Syst Rev. 5:CD0116732017.PubMed/NCBI
|
|
12
|
Meyer E, Pasquier D, Bernadou G, Calais G,
Maroun P, Bossi A, Theodore C, Albiges L, Stefan D, de Crevoisier
R, et al: Stereotactic radiation therapy in the strategy of
treatment of metastatic renal cell carcinoma: A study of the getug
group. Eur J Cancer. 98:38–47. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tsuji G, Ito T, Chiba T, Mitoma C,
Nakahara T, Uchi H and Furue M: The role of the OVOL1-OVOL2 axis in
normal and diseased human skin. J Dermatol Sci. 90:227–231. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kumar A, Bhandari A, Sinha R, Sardar P,
Sushma M, Goyal P, Goswami C and Grapputo A: Molecular phylogeny of
OVOL genes illustrates a conserved C2H2 zinc finger domain coupled
by hypervariable unstructured regions. PLoS One. 7:e393992012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Watanabe K, Villarreal-Ponce A, Sun P,
Salmans ML, Fallahi M, Andersen B and Dai X: Mammary morphogenesis
and regeneration require the inhibition of EMT at terminal end buds
by Ovol2 transcriptional repressor. Dev Cell. 29:59–74. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Roca H, Hernandez J, Weidner S, McEachin
RC, Fuller D, Sud S, Schumann T, Wilkinson JE, Zaslavsky A, Li H,
et al: Transcription factors OVOL1 and OVOL2 induce the mesenchymal
to epithelial transition in human cancer. PLoS One. 8:e767732013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang ZH, Li Z, Hu M, Yang QJ, Yan S, Wu
RS, Li BA and Guo M: Ovol2 gene inhibits the
epithelial-to-mesenchymal transition in lung adenocarcinoma by
transcriptionally repressing Twist1. Gene. 600:1–8. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jia D, Jolly MK, Boareto M, Parsana P,
Mooney SM, Pienta KJ, Levine H and Ben-Jacob E: OVOL guides the
epithelial-hybrid-mesenchymal transition. Oncotarget.
6:15436–15448. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jolly MK, Jia D, Boareto M, Mani SA,
Pienta KJ, Ben-Jacob E and Levine H: Coupling the modules of EMT
and stemness: A tunable ‘stemness window’ model. Oncotarget.
6:25161–25174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nieto MA: The ins and outs of the
epithelial to mesenchymal transition in health and disease. Annu
Rev Cell Dev Biol. 27:347–376. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu J, Wu Q, Wang Y, Wei Y, Wu H, Duan L,
Zhang Q and Wu Y: Ovol2 induces mesenchymal-epithelial transition
via targeting ZEB1 in osteosarcoma. Onco Targets Ther.
11:2963–2973. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu C, Yan T and Yang J: OVOL1 inhibits
oral squamous cell carcinoma growth and metastasis by suppressing
zinc finger E-box binding homeobox 1. Int J Clin Exp Pathol.
12:2801–2808. 2019.PubMed/NCBI
|
|
23
|
Qi XK, Han HQ, Zhang HJ, Xu M, Li L, Chen
L, Xiang T, Feng QS, Kang T, Qian CN, et al: OVOL2 links stemness
and metastasis via fine-tuning epithelial-mesenchymal transition in
nasopharyngeal carcinoma. Theranostics. 8:2202–2216. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47:W556–W560. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mermel CH, Schumacher SE, Hill B, Meyerson
ML, Beroukhim R and Getz G: GISTIC2.0 facilitates sensitive and
confident localization of the targets of focal somatic copy-number
alteration in human cancers. Genome Biol. 12:R412011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu CJ, Hu FF, Xia MX, Han L, Zhang Q and
Guo AY: GSCALite: A web server for gene set cancer analysis.
Bioinformatics. 34:3771–3772. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
The Gene Ontology Consortium, . The gene
ontology resource: 20 years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu J, Lichtenberg T, Hoadley KA, Poisson
LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee
AV, et al: An integrated TCGA pan-cancer clinical data resource to
drive high-quality survival outcome analytics. Cell.
173:400–416.e11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fu H, Qi L, Chen L, He Y, Zhang N and Guo
H: Expression of ovol2 is related to epithelial characteristics and
shows a favorable clinical outcome in hepatocellular carcinoma.
Onco Targets Ther. 9:5963–5973. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fan C, Wang Q, van der Zon G, Ren J,
Agaser C, Slieker RC, Iyengar PV, Mei H and Ten Dijke P: OVOL1
inhibits breast cancer cell invasion by enhancing the degradation
of TGF-beta type I receptor. Signal Transduct Target Ther.
7:1262022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mitoma C, Nakahara T, Uchi H, Ito T,
Inatomi Y, Ide T, Jinnai S, Jinnai N, Iwasaki N, Sakamoto K, et al:
Preferential expression of OVOL1 in inner root sheath of hair,
sebaceous gland, eccrine duct and their neoplasms in human skin.
Fukuoka Igaku Zasshi. 105:166–173. 2014.PubMed/NCBI
|
|
38
|
Li J, Yu T, Cao J, Liu L, Liu Y, Kong HW,
Zhu MX, Lin HC, Chu DD, Yao M and Yan MX: MicroRNA-148a suppresses
invasion and metastasis of human non-small-cell lung cancer. Cell
Physiol Biochem. 37:1847–1856. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang R, Geng GJ, Guo JG, Mi YJ, Zhu XL,
Li N, Liu HM, Lin JF, Wang JW, Zhao G, et al: An NF-κB/OVOL2
circuit regulates glucose import and cell survival in non-small
cell lung cancer. Cell Commun Signal. 20:402022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ye GD, Sun GB, Jiao P, Chen C, Liu QF,
Huang XL, Zhang R, Cai WY, Li SN, Wu JF, et al: OVOL2, an inhibitor
of WNT signaling, reduces invasive activities of human and mouse
cancer cells and is down-regulated in human colorectal tumors.
Gastroenterology. 150:659–671.e16. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu RS, Lin J, Xing YM, Gao WL, Jiang YX,
Chen LX, Zhang XP and Dai ZL: OVOL2 inhibits macrophage M2
polarization by regulating IL-10 transcription, and thus inhibits
the tumor metastasis by modulating the tumor microenvironment.
Immunol Lett. 242:17–26. 2022. View Article : Google Scholar : PubMed/NCBI
|