Introduction
Colorectal cancer (CRC) is the third most common
type of cancer worldwide in terms of the number of patients
affected and the second most common in terms of the number of
deaths, and its prevalence is increasing (1). Although a number of CRC cases can be
cured by surgery, it is reported worldwide that recurrence occurs
in ~30% of cases, even after curative resection (2), and prevention and early detection of
recurrence are still major issues.
Local recurrence (LR) after resection for CRC is a
serious issue. LR is defined as a recurrent lesion in or around the
primary tumor site, including the pericolic tissue, the adjacent
mesentery, lymph nodes or the suture line of anastomosis.
Anastomotic recurrence (AR) has often been considered a type of LR,
and its incidence has been reported to be 1–2% worldwide (3). AR has been shown to have a poorer
prognosis and can progress to more advanced pathological stages
than primary tumors (3). AR is
thought to be caused by the implantation of exfoliated tumor cells
into the anastomotic line, and the risk of AR is high in rectal
cancer (3,4); however, to the best of our knowledge,
the relationship with other clinicopathological factors has not
been established. At present, there have been two reports on the
genetic analysis of AR cases, but none of them have presented with
any specific genetic features of AR (5,6). Our
previous study reported on two cases of repeated AR following
curative resection of CRC and both were revealed to have the
KRAS G13D mutation (7).
Although only two cases were reported, it may be hypothesized that
the KRAS G13D mutation could contribute to the development
of AR, as well as other aspects of recurrence, such as resistance
and dormancy. The present study aimed to clarify the relationship
between the KRAS G13D mutation and AR in CRC.
Patients and methods
Patients
The present study assessed 21 patients who underwent
curative resection for CRC at the Department of Surgical Oncology,
Faculty of Medicine, The University of Tokyo (Tokyo, Japan) between
January 2005 and December 2019, and were diagnosed with AR. A total
of 67 patients who were diagnosed with non-anastomotic LR (NALR)
after curative resection were also included. Patients with
hereditary CRC and colitis-associated cancer were excluded from the
study. In the present study, AR was defined as ‘recurrence on the
anastomotic line’; the recurrence site must be located on the
anastomotic line and pathologically proven with a resection
specimen or endoscopic biopsy. Recurrent lesions that were in
contact with the anastomotic line, but mainly located outside of
the bowel wall were not considered as AR and were classified as
NALR. NALR did not include pelvic peritoneal dissemination or
lateral lymph node recurrence in the present study. NALR was
observed in 67 cases, and 21 cases of NALR matched to 21 cases of
AR were used as a control group to compare the prevalence of the
KRAS G13D mutation. In addition to the prevalence of the
KRAS G13D mutation, the following clinicopathological
findings were retrospectively evaluated: Sex, age, gross appearance
type (classification of gross appearance), tumor size,
histopathological type, tumor depth, lymph node metastasis, venous
invasion, lymphatic invasion, preoperative carcinoembryonic antigen
and carbohydrate antigen 19-9 levels and the association of the
KRAS G13D mutation with prognosis (Table I). The clinicopathological findings
were described according to the American Joint Committee on
Cancer/International Union Against Cancer TNM classification, 8th
edition (8).
 | Table I.Clinicopathological characteristics
of patients with AR and NALR after matching. |
Table I.
Clinicopathological characteristics
of patients with AR and NALR after matching.
| Variable | AR (n=21) | NALR (n=21) | P-value |
|---|
| Sex |
|
| 1.00 |
| Male
(%) | 13 (61.9) | 13 (61.9) |
|
| Age |
|
| 0.76 |
| ≥65
years (%) | 10 (47.6) | 11 (52.4) |
|
| Tumor location |
|
| 1.00 |
| Rectum
(%) | 8 (38.1) | 8 (38.1) |
|
| Pre-operative
CEA |
|
| 0.53 |
| ≥5
ng/ml (%) | 11 (52.4) | 14 (66.7) |
|
| Pre-operative CA
19-9 |
|
| 0.70 |
| ≥37
ng/ml (%) | 3 (14.3) | 5 (23.8) |
|
| T stage |
|
| 0.61 |
| T3-4
(%) | 20 (95.2) | 18 (85.7) |
|
| N stage |
|
| 1.00 |
| N1-2
(%) | 15 (71.4) | 15 (71.4) |
|
| Tumor diameter |
|
| 0.76 |
| ≥50 mm
(%) | 11 (52.4) | 12 (57.1) |
|
| Histological
type |
|
| 1.00 |
| Tub
(%) | 20 (95.2) | 20 (95.2) |
|
| Lymphatic
invasion |
|
| 0.53 |
|
Positive (%) | 10 (47.6) | 8 (38.1) |
|
| Venous
invasion |
|
| 0.41 |
|
Positive (%) | 16 (76.2) | 19 (90.5) |
|
| Pathological
stage |
|
| 1.00 |
| I–II
(%) | 6 (28.6) | 6 (28.6) |
|
| III
(%) | 11 (52.4) | 12 (57.1) |
|
| IV
(%) | 4 (19.0) | 3 (14.3) |
|
| KRAS G13D
mutation |
|
| 0.05 |
|
Positive (%) | 7 (33.3) | 1 (4.8) |
|
The present study was conducted according to The
Declaration of Helsinki and the study protocol was approved by the
ethics committee of The University of Tokyo [approval no.
3252-(13)]. Informed consent was
obtained in the form of an opt-out option on the website
(http://all-1su.umin.jp/research/files/04_2.pdf).
DNA extraction from formalin-fixed,
paraffin-embedded (FFPE) specimens
DNA was extracted from 10-µm FFPE specimens obtained
from the University of Tokyo. A Maxwell® RSC DNA FFPE
Kit (Promega Corporation) was used for DNA extraction. All samples
were extracted after an overnight proteinase K digestion step at
70°C, and all extractions were performed according to the
manufacturer's protocol.
KRAS G13D mutation detection by
droplet digital polymerase chain reaction (ddPCR)
Mutation of the KRAS gene was examined by
ddPCR using the QX200™ Droplet Digital™ PCR system (Bio-Rad
Laboratories, Inc.). Each DNA sample was diluted to 3,000 ng/ml, as
measured by a Qubit 3.0 Fluorometer (Thermo Fisher Scientific,
Inc.). PCR reaction mixtures contained 12 µl ddPCR Supermix for
Probes (Bio-Rad Laboratories, Inc.), 1.2 µl PrimePCR for ddPCR, 1.2
µl Uracil-DNA Glycosylase (UDG; New England BioLabs, Inc.) and 9.6
µl diluted DNA sample; 20 µl of the 24-µl reaction mixture was
loaded in a DG8™ Cartridges for QX200™/QX100™ Droplet Generator
(Bio-Rad Laboratories, Inc.) and droplets were generated. The
entire droplet emulsion volume was further loaded in ddPCR™ 96-Well
Plates (Bio-Rad Laboratories, Inc.). The loaded 96-well PCR plate
was then heat-sealed with pierceable foil in the PX1™ PCR Plate
Sealer and placed in a T100™ Thermal Cycler (both from Bio-Rad
Laboratories, Inc.). Amplification was conducted as follows: 95°C
for 1 min, followed by 40 cycles at 55°C for 10 min, 94°C for 30
sec and a final extension step at 98°C for 10 min. Commercial
primers (PrimePCR for ddPCR KRAS G13D, assay ID:
dHsaMDV2510598; Bio-Rad Laboratories, Inc.) were used. UDG was used
to limit the chances of artifacts due to formalin-fixation
(9).
KRAS mutation detection in clinical
practice
RAS mutation status (KRAS exon 2, 3,
or 4 mutation, NRAS exon 2, 3, or 4 mutation) was evaluated
using the PCR-reverse sequence-specific oligonucleotide method
(BML, Inc.).
Statistical analysis
All statistical analyses were carried out using EZR
version 4.1.2 (Saitama Medical Center, Jichi Medical University,
Saitama, Japan), which is a graphical user interface for R version
3.0.2 (The R Foundation) (10).
Comparisons were performed using χ2 test or Fisher's
exact test for categorical variables. In univariate analysis using
Fisher's exact test, odds ratio and their 95% confidence intervals
were estimated. Survival curves were drawn using the Kaplan-Meier
method and compared using the log-rank test. P<0.05 was
considered to indicate a statistically significant difference.
To reduce potential confounding effects and
treatment selection bias, case matching was conducted in the
comparative analysis of AR and NALR. The following five factors
that could affect KRAS status were selected: Age, sex,
cancer location, histological type and cancer stage. R version
3.0.2 package ‘optmatch (https://cran.r-project.org/web/packages/optmatch)’
was used for the matching.
Results
The study population comprised 21 AR cases and 21
matched NALR cases. The clinicopathological characteristics of
patients with AR and NALR before matching are shown in Table SI. The median follow-up period was
52.3 months. The clinicopathological characteristics of patients
with AR or NALR after matching are shown in Table I.
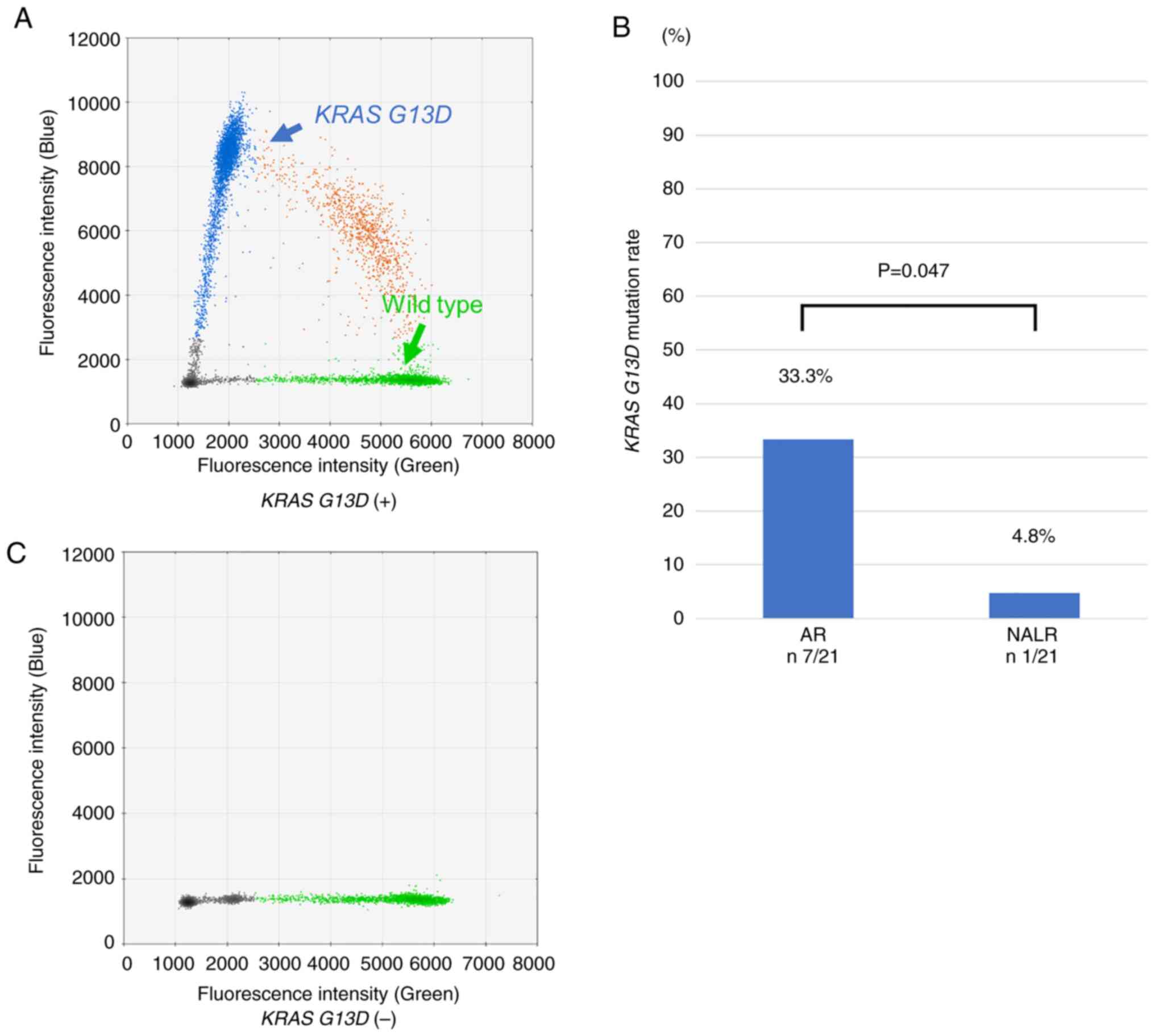
There was no significant difference in the
clinicopathological characteristics between the matched AR and the
NALR groups, whereas the KRAS G13D mutation rate was
significantly higher in the AR group; AR 33.3% (7/21) vs. NALR 4.8%
(1/21) (P=0.047; Fig. 1).
The pathological findings on the recurrent lesions
and details of treatment and prognosis in the AR group are shown in
Table II. Of the 14 patients who
tested negative for the KRAS G13D mutation in the present
study, the KRAS status of 8 cases was evaluated for clinical
purposes; 2 cases were positive for the KRAS G12D mutation,
and the remaining 6 cases presented with wild-type KRAS. All
of the 7 patients who tested positive for the KRAS G13D
mutation were evaluated for KRAS status in the clinical
setting, and there were no cases with double KRAS mutations.
The RAS status of the matched NALR group is shown in
Table SII. Of the 20 patients who
tested negative for the KRAS G13D mutation, the KRAS
status of 10 cases was evaluated for clinical purposes; three cases
were positive for the KRAS G12D mutation, two cases were
positive for the KRAS G12V mutation, one case was positive
for the KRAS G12A mutation and the remaining four cases presented
with wild-type KRAS.
 | Table II.Information on tumor recurrence and
therapy for patients with AR. |
Table II.
Information on tumor recurrence and
therapy for patients with AR.
| No. | KRAS
G13D | RAS
status | Age, years | Sex | Tumor location | NAC | Surgery | Depth | Histology | pN | ly | v | AC | Anti-EGFR |
|---|
| 1 | + | G13D | 78 | F | Ascending | - | T/C | MP | Tub | 1 | 2 | 2 | - | - |
| 2 | + | G13D | 61 | M | Sigmoid | - | LAR | SE | Tub | 1 | 0 | 3 | + | - |
| 3 | + | G13D | 33 | F | Rectum | - | APR | SS | Tub | 0 | 1 | 1 | + | - |
| 4 | + | G13D | 42 | F | Sigmoid | - | LAR | SS | Tub | 0 | 0 | 2 | + | - |
| 5 | + | G13D | 39 | M | Rectum | + | LAR | MP | Tub | 0 | 0 | 0 | + | - |
| 6 | + | G13D | 68 | F | Ascending | - | - | - | Tub | - | - | - | - | - |
| 7 | + | G13D | 59 | M | Rectum | - | - | - | Por | - | - | - | - | - |
| 8 | - | G12D | 66 | M | Cecum | - | RHC | SE | Tub | 0 | 0 | 0 | - | - |
| 9 | - | G12D | 63 | M | Sigmoid | - | LAR | A | Tub | 0 | 1 | - | - | - |
| 10 | - | WT | 55 | F | Rectum | + | LAR | A | Tub | 0 | 0 | 1 | + | + |
| 11 | - | WT | 77 | M | Sigmoid | - | S/C | SI | Tub | 1 | 1 | 0 | + | - |
| 12 | - | WT | 70 | M | Rectum | - | LAR | A | Tub | 1 | 0 | 1 | + | + |
| 13 | - | WT | 66 | M | Descending | - | LHC | SS | Tub | 1 | 2 | 2 | - | - |
| 14 | - | WT | 79 | M | Sigmoid | + | LAR | A | Tub | 0 | 0 | 1 | - | - |
| 15 | - | WT | 55 | M | Rectum | - | - | - | Tub | - | - | - | - | - |
| 16 | - | Not tested | 60 | F | Rectum | - | Hartmann | A | Tub | 0 | 0 | 0 | + | - |
| 17 | - | Not tested | 75 | F | Sigmoid | - | S/C | MP | Tub | 0 | 0 | 0 | - | - |
| 18 | - | Not tested | 60 | M | Sigmoid | - | LHC | SS | Tub | 0 | 1 | 2 | + | - |
| 19 | - | Not tested | 67 | M | Sigmoid | + | APR | A | Tub | 0 | 0 | 1 | + | - |
| 20 | - | Not tested | 69 | F | Transverse | - | RHC | SE | Tub | 1 | 0 | 3 | - | - |
| 21 | - | Not tested | 60 | M | Rectum | - | - | - | Tub | - | - | - | - | - |
Comparing the KRAS G13D mutation-positive
(KRAS G13D+) and KRAS G13D
mutation-negative (KRAS G13D−) patients in the AR
group, there was no significant difference in the
clinicopathological background (Table
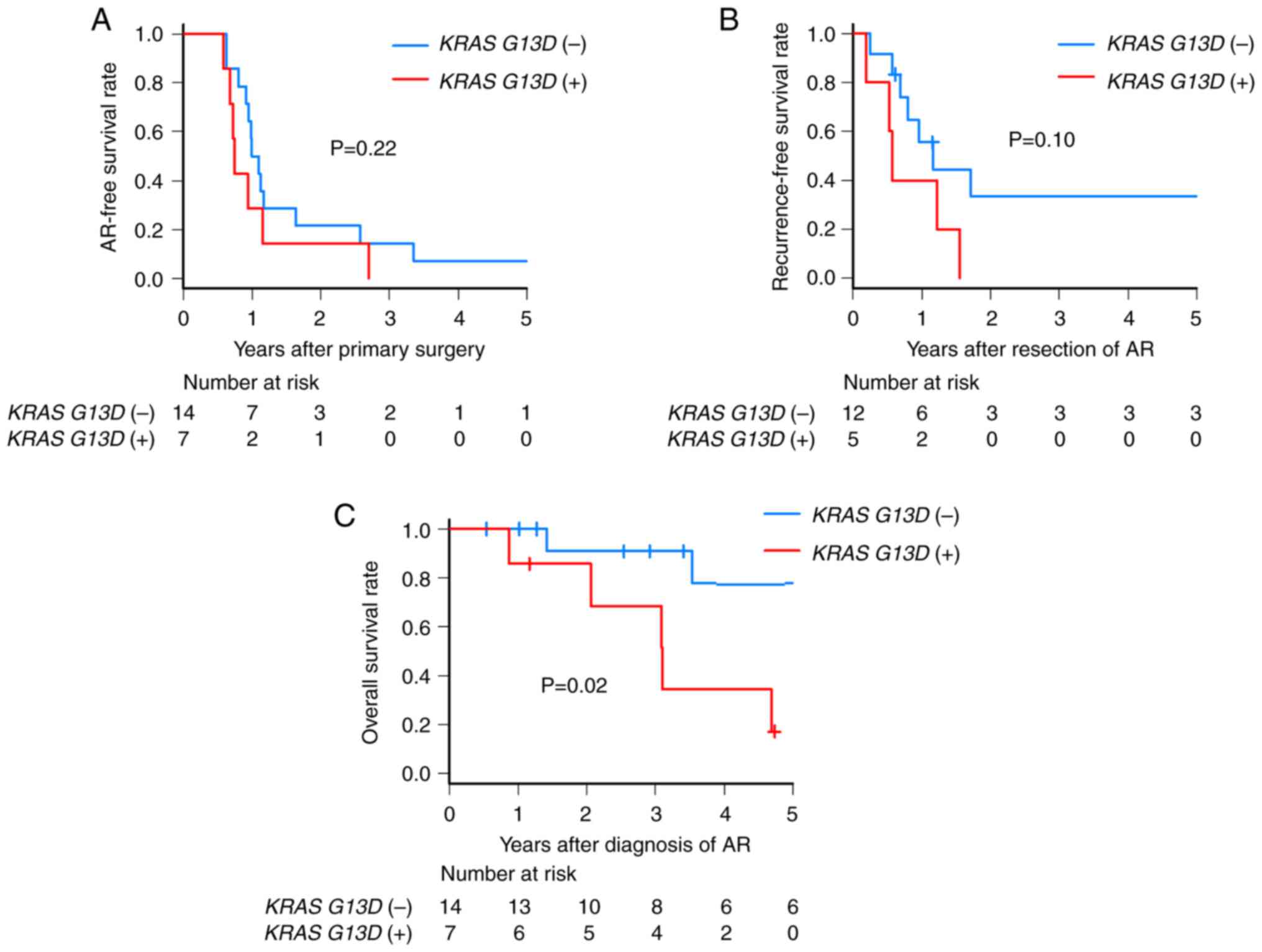
III), interval from initial surgery to AR (Fig. 2A) and recurrence resection rate
(Table III). On analyzing the 17
cases who underwent surgical resection of AR, even though there was
no significant difference in recurrence-free survival (RFS) after
resection (2-year RFS after resection: KRAS G13D+
0% vs. KRAS G13D− 33.3%; P=0.10), all KRAS
G13D+ patients experienced subsequent recurrence
within 2 years (Fig. 2B). Notably,
KRAS G13D+ patients had a significantly poorer
overall survival (OS) (3-year OS: KRAS G13D+
68.6% vs. KRAS G13D− 90.9%; P=0.02) (Fig. 2C). The rate of synchronous
recurrence at the diagnosis of AR was 28.6% (2/7) in KRAS
G13D+ patients and 42.9% (6/14) in KRAS
G13D− patients. There was no significant difference
in the synchronous recurrence patterns between patients with and
without the KRAS G13D mutation (Table IV). By contrast, KRAS
G13D+ patients tended to experience more instances
of metachronous locoregional recurrences (P=0.10), and repetitive
AR was observed in KRAS G13D+ patients alone
(P=0.07) (Table V); 80.0% (4/5) of
the KRAS G13D+ patients experienced subsequent
distant metastatic recurrences.
 | Table III.Clinicopathological characteristics
of patients with AR. |
Table III.
Clinicopathological characteristics
of patients with AR.
| Variable | KRAS G13D
(+) (n=7) | KRAS G13D
(−) (n=14) | P-value |
|---|
| Sex |
|
| 0.346 |
| Male
(%) | 3 (42.9) | 10 (71.4) |
|
| Age |
|
| 0.183 |
| ≥65
years (%) | 2 (28.6) | 9 (64.3) |
|
| Tumor location |
|
| 1.000 |
| Rectum
(%) | 3 (42.9) | 5 (35.7) |
|
| Pre-operative
CEA |
|
| 0.362 |
| ≥5
ng/ml (%) | 5 (71.4) | 6 (42.9) |
|
| Pre-operative CA
19-9 |
|
| 0.527 |
| ≥37
ng/ml (%) | 2 (28.6) | 1 (7.14) |
|
| T stage |
|
| 0.333 |
|
T3-4 | 7 (100) | 13 (92.9) |
|
| N stage |
|
| 0.613 |
|
N1-2 | 6 (85.7) | 9 (64.3) |
|
| Tumor diameter |
|
| 0.362 |
| ≥50 mm
(%) | 5 (71.4) | 6 (42.9) |
|
| Histological
type |
|
| 0.333 |
| Tub
(%) | 6 (85.7) | 14 (100) |
|
| Lymphatic
invasion |
|
| 1.000 |
|
Positive (%) | 3 (42.9) | 7 (50.0) |
|
| Venous
invasion |
|
| 1.000 |
|
Positive | 5 (71.4) | 11 (78.6) |
|
| Pathological
stage |
|
| 0.589 |
|
I–II | 1 | 5 |
|
|
III | 4 | 7 |
|
| IV | 2 | 2 |
|
| Simultaneous other
metastases |
|
| 1.000 |
|
Yes | 3 (42.9) | 6 (42.9) |
|
| Resection for
AR |
|
| 0.574 |
|
Yes | 5 (71.4) | 12 (85.7) |
|
| Use of anti-EGFR
antibody |
|
| 0.533 |
|
Yes | 0 (0.0) | 2 (14.3) |
|
 | Table IV.Distribution of synchronous recurrent
sites of patients with AR. |
Table IV.
Distribution of synchronous recurrent
sites of patients with AR.
| Recurrent
sites | KRAS G13D
(+) (n=7) | KRAS G13D
(−) (n=14) | OR (95% CI) | P-value |
|---|
| Locoregional
(%) | 0 (0.0) | 0 (0.0) | - | - |
| Distant metastasis
(%) | 2 (28.6) | 6 (42.9) | 0.549
(0.039-5.020) | 0.66 |
|
Liver | 1 (14.3) | 5 (35.7) | 0.316
(0.005-4.004) | 0.61 |
|
Lung | 1 (14.3) | 1 (7.1) | 2.082
(0.023-182.6) | 1.00 |
|
Dissemination | 1 (14.3) | 2 (14.3) | 1.000
(0.015-23.10) | 1.00 |
|
Extra-regional LN | 0 (0.0) | 0 (0.0) | - | - |
| Total (%) | 2 (28.6) | 6 (42.9) | 0.549
(0.039-5.020) | 0.66 |
 | Table V.Distribution of subsequent recurrent
sites in patients with AR. |
Table V.
Distribution of subsequent recurrent
sites in patients with AR.
| Recurrent
sites | KRAS G13D
(+) (n=5) | KRAS G13D
(−) (n=12) | OR (95% CI) | P-value |
|---|
| Locoregional
(%) | 4 (80.0) | 3 (25.0) | 10.05
(0.668–651.0) | 0.10 |
|
Anastomotic (repetitive) | 2 (40.0) | 0 (0.0) | Inf
(0.495-Inf) | 0.07 |
|
Non-anastomotic | 2 (40.0) | 3 (25.0) | 1.915
(0.110–28.29) | 0.60 |
| Distant metastasis
(%) | 4 (80.0) | 5 (41.7) | 5.059
(0.352–313.6) | 0.29 |
|
Liver | 0 (0.0) | 2 (16.7) | 0 (0–13.32) | 1.00 |
|
Lung | 2 (40.0) | 1 (8.3) | 6.321
(0.251–468.8) | 0.19 |
|
Dissemination | 1 (20.0) | 2 (16.7) | 1.233
(0.017–30.77) | 1.00 |
|
Extra-regional LN | 1 (20.0) | 1 (8.3) | 2.569
(0.028–234.6) | 0.52 |
| Total (%) | 5 (100.0) | 7 (58.3) | Inf
(0.410-Inf) | 0.25 |
Discussion
The present study observed a high KRAS G13D
mutation rate in patients with AR (AR 33.3% vs. matched NALR 4.8%;
P=0.047). All KRAS G13D+ patients who underwent
curative resection for AR had subsequent recurrence within 2 years,
and the OS was poorer than that of KRAS G13D−
patients (3-year OS: 68.6% vs. 90.9%; P=0.02).
KRAS is one of the key driver genes in CRC
and is detected early in the carcinogenesis of CRC. The KRAS
mutation rate in CRC is reported to be 30–40% worldwide, and
KRAS codon 13 mutations, including KRAS G13D, are
reported to occur in 6–8% of cases worldwide (11–13).
In a large-scale study in Japan, the total KRAS mutation
rate was 37.6%, and the KRAS codon 13 mutation rate was
7.7%, which was comparable to the results reported in Western
countries (14).
The 33.3% KRAS G13D mutation rate among
patients with AR in the present study was higher than that reported
in previous studies, suggesting that there may be a certain
association between the KRAS G13D mutation and AR. In
addition, there was a significant difference in the KRAS
G13D mutation rate between AR and NALR, suggesting that this is
a specific characteristic of AR rather than of LR. Andreyev et
al (11) reported that G to A
mutations in the KRAS gene, which include the KRAS
G13D mutation, were more frequent (58.3%) among patients with
AR than among patients with other types of recurrence (~22%)
(P=0.02), and the results of this previous study are consistent
with the current findings.
Although several reports have stated that CRC with
KRAS mutations has a poor prognosis (14–16),
there are few reports on the association between prognosis and each
KRAS subtype, and consistent results have not been obtained
(17–20). Notably, the association between
KRAS subtypes and the clinical significance of CRC has been
reported in few studies. Kodaz et al (21) reported that the KRAS G13D
mutation was more frequent in the left colon and in patients <70
years old, whereas the KRAS G12D mutation was more frequent
in the right colon and in patients >50 years old. Bazan et
al (18) reported that codon 12
KRAS mutations were associated with mucinous histology,
whereas codon 13 KRAS mutations were associated with lymph
node metastasis and advanced Dukes' stage.
In contrast to previous reports, the present study
focused on AR cases, which may be responsible for the difference in
prognosis between the cases with KRAS G13D mutation and
those without the mutation. Several in vitro analyses have
shown the characteristics of KRAS subtypes using colon
cancer cell lines. Organ et al (22) reported that DLD1, a colon cancer
cell line that is positive for the KRAS G13D mutation,
showed a higher adhesion ability to the extracellular matrix and
migration, in contrast to DKO4, a cell line in which KRAS
G13D was knocked out of DLD1 cells. It was hypothesized that
the KRAS G13D mutant may have an enhanced adhesion ability
to the extracellular matrix and migration compared with wild-type
KRAS, and these characteristics may contribute to the
implantation of tumor cells into the anastomotic line, which is the
pathogenic mechanism of AR. Stolze et al (23) reported that KRAS G13D, unlike
other subtypes of KRAS mutation, showed a high expression of
epidermal growth factor receptors (EGFRs) and high activation of
proliferative signaling in the presence of EGF. In the tissue
repair process at the anastomotic site, the role of growth factors
is important; therefore, these characteristics of the KRAS
G13D mutation may be responsible for AR.
In the present study, KRAS G13D+
patients with AR had a poor prognosis, probably because all KRAS
G13D+ patients with AR experienced a subsequent
recurrence after undergoing resection for AR. Margonis et al
(24) reported that the KRAS
codon 13 mutation was a risk factor for extrahepatic and pulmonary
recurrence after curative resection of liver metastasis of CRC, and
this was not observed for all KRAS mutations. Owing to the
small number of AR cases in the present study, statistically
significant difference was not be observed; however, it was
observed that 80% (4/5) of the KRAS G13D+
patients experienced subsequent distant metastatic recurrence after
curative resection for AR. As aforementioned, it has been reported
that the KRAS G13D mutation enhances the adhesion and
migratory ability, and is associated with increased proliferative
signaling. These characteristics may be responsible for the
pathogenesis of AR, in addition to the subsequent recurrences and
poor prognoses.
The poor prognosis of KRAS G13D+
patients could also be attributed to the absence of an indication
for a regimen including anti-EGFR antibodies (21). However, only 2 of the 14 patients
with AR diagnosed as KRAS G13D− received
chemotherapy including anti-EGFR antibodies in the present study;
therefore, the effect of anti-EGFR antibodies may be limited. Few
reports have suggested that the KRAS G13D mutation differs
from other KRAS subtypes and may benefit from treatment with
the anti-EGFR antibody cetuximab (25,26),
but this has been doubted by some reports (27) and no conclusion has been reached.
The therapeutic efficacy of anti-EGFR antibody therapy in KRAS
G13D+ requires further study.
There were several limitations to the present study.
First, it was a single-center, retrospective study with a small
number of patients. Second, the study only analyzed the KRAS
G13D mutation and did not consider other KRAS subtypes
or BRAF mutations. Although 8 KRAS G13D−
patients were tested for the RAS status in the clinical
setting, 6 KRAS G13D− patients were not tested.
Therefore, the KRAS G13D− group may have included
patients with other KRAS subtypes or BRAF mutations.
Additionally, 2 cases belonging to the KRAS G13D−
group possessed the KRAS G12D mutation. Third, there was no
analysis of the effect of AR on patient survival compared with the
patients with NALR.
In conclusion, the KRAS G13D mutation rate
was significantly higher in patients with AR, and patients with AR
and the KRAS G13D mutation had a poorer prognosis than
KRAS G13D− patients with AR. Although the role of
the KRAS G13D mutation in the development of AR requires
further investigation, postoperative surveillance and treatment
strategies should be considered with attention to the possibility
of AR and subsequent recurrence in KRAS G13D
mutation-positive patients. Although a definitive conclusion could
not be reached due to the small sample size and the fact that it
was not considered that mutations other than KRAS G13D may
affect the outcome, the present results may be worth confirming in
future studies containing a larger number of patients.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Shinya Abe and Dr
Yuzo Nagai (Department of Surgical Oncology, The University of
Tokyo, Tokyo, Japan) for their advisory assistance.
Funding
The present study was supported by Grants-in-Aid for Scientific
Research (grant nos. 21H02778, 18K07194, 19K09114, 19K09115 and
20K09051) and Challenging Research (Exploratory; grant no.
20K21626) from the Japan Society for the Promotion of Science and
by the Project for Cancer Research and Therapeutic Evolution (grant
no. JP 19cm0106502) from the Japan Agency for Medical Research and
Development.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are not publicly available due to a license agreement
with the University of Tokyo, but are available from the
corresponding author on reasonable request.
Authors' contributions
KeM, KS and KH substantially contributed to the
study conception, design and analysis of data. KeM, KU, SK and TY
substantially contributed to the acquisition of laboratory data.
HN, KK, KoM, SE, YY, HS substantially contributed to the
acquisition of clinical data. HY and SI substantially contributed
to the interpretation of data. KeM and KS confirm the authenticity
of all the raw data. HY and SI gave final approval of the version
to be published. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the ethics
committee of The University of Tokyo [approval no. 3252-(13); Tokyo, Japan]. This study was
conducted in accordance with The Declaration of Helsinki.
Patient consent for publication
Informed consent was obtained in the form of an
opt-out option on the website for the participation in the research
(http://all-1su.umin.jp/custom8.html).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Böhm B, Schwenk W, Hucke HP and Stock W:
Does methodic long-term follow-up affect survival after curative
resection of colorectal carcinoma? Dis Colon Rectum. 36:280–286.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jung WB, Yu CS, Lim SB, Park IJ, Yoon YS
and Kim JC: Anastomotic recurrence after curative resection for
colorectal cancer. World J Surg. 41:285–294. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
McGregor JR, Galloway DJ, McCulloch P and
George WD: Anastomotic suture materials and implantation
metastasis: An experimental study. Br J Surg. 76:331–334. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Costi R, Santi C, Bottarelli L, Azzoni C,
Le Bian AZ, Ricco M, Sarli L, Silini EM and Violi V: Anastomotic
recurrence of colon cancer: Genetic analysis challenges the widely
held theories of cancerous cells' intraluminal implantation and
metachronous carcinogenesis. J Surg Oncol. 114:228–236. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vakiani E, Shah RH, Berger MF,
Makohon-Moore AP, Reiter JG, Ostrovnaya I, Attiyeh MA, Cercek A,
Shia J, Lacobuzio-Donahue CA, et al: Local recurrences at the
anastomotic area are clonally related to the primary tumor in
sporadic colorectal carcinoma. Oncotarget. 8:42487–42494. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Okada S, Hata K, Kawai K, Yamomoto Y,
Tanaka T, Nishikawa T, Sasaki K, Kaneko M, Emoto S, Murono K and
Nozawa H: Association between KRAS G13D mutations and anastomotic
recurrence in colorectal cancer: Two case reports. Medicine
(Baltimore). 98:e147812019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brierley JD, Gospodarowicz MK and
Wittekind C: UICC TNM classification of malignant tumours. 8th
edition. Wiley-Blackwell; Oxford: 2017
|
|
9
|
Do H and Dobrovic A: Dramatic reduction of
sequence artefacts from DNA isolated from formalin-fixed cancer
biopsies by treatment with uracil- DNA glycosylase. Oncotarget.
3:546–558. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kanda Y: Investigation of the freely
available easy-to-use software ‘EZR’ for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Andreyev HJ, Norman AR, Cunningham D,
Oates JR and Clarke PA: Kirsten ras mutations in patients with
colorectal cancer: The multicenter ‘RASCAL’ study. J Natl Cancer
Inst. 90:675–684. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Roth AD, Tejpar S, Delorenzi M, Yah P,
Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C,
et al: Prognostic role of KRAS and BRAF in stage II and III
resected colon cancer: Results of the translational study on the
PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 28:466–474.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hinoi T: Cancer genomic profiling in
colorectal cancer: Current challenges in subtyping colorectal
cancers based on somatic and germline variants. J Anus Rectum
Colon. 5:213–228. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Watanabe T, Yoshino T, Uetake H, Yamazaki
K, Ishiguro M, Kurokawa T, Saijo N, Ohashi Y and Sugihara K: KRAS
mutational status in Japanese patients with colorectal cancer:
Results from a nationwide, multicenter, cross-sectional study. Jpn
J Clin Oncol. 43:706–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kadowaki S, Kakuta M, Takahashi S,
Takahashi A, Arai Y, Nishimura Y, Yatsuoka T, Ooki A, Yamaguchi K,
Matsuo K, et al: Prognostic value of KRAS and BRAF mutations in
curatively resected colorectal cancer. World J Gastroenterol.
21:1275–1283. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Passiglia F, Bronte G, Bazan V, Galvano A,
Vincenzi B and Russo A: Can KRAS and BRAF mutations limit the
benefit of liver resection in metastatic colorectal cancer
patients? A systematic review and meta-analysis. Crit Rev Oncol
Hematol. 99:150–157. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Inoue Y, Saigusa S, Iwata T, Okugawa Y,
Toiyama Y, Tanaka K, Uchida K, Mohri Y and Kusunoki M: The
prognostic value of KRAS mutations in patients with colorectal
cancer. Oncol Rep. 28:1579–1584. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bazan V, Migliavacca M, Zanna I, Tubiolo
C, Grassi N, Latteri MA, La Farina M, Albanese I, Dardanoni G,
Salerno S, et al: Specific codon 13 K-ras mutations are predictive
of clinical outcome in colorectal cancer patients, whereas codon 12
K-ras mutations are associated with mucinous histotype. Ann Oncol.
13:1438–1446. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hayama T, Hashiguchi Y, Okamoto K, Okada
Y, Ono K, Shimada R, Ozawa T, Toyoda T, Tsuchiya T, Iinuma H, et
al: G12V and G12C mutations in the gene KRAS are associated with a
poorer prognosis in primary colorectal cancer. Int J Colorectal
Dis. 34:1491–1496. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Imamura Y, Morikawa T, Liao X, Lochhead P,
Kuchida A, Yamauchi M, Qian ZE, Nishihara R, Meyerhardt JA, Haigis
KM, et al: Specific mutations in KRAS codons 12 and 13, and patient
prognosis in 1075 BRAF wild-type colorectal cancers. Clin Cancer
Res. 18:4753–4763. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kodaz H, Hacibekiroglu I, Erdogan B,
Turkmen E, Tozkir H, Albayrak D, Uzunoglu S and Cicin I:
Association between specific KRAS mutations and the
clinicopathological characteristics of colorectal tumors. Mol Clin
Oncol. 3:179–184. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Organ SL, Hai J, Radulovich N, Marshall
CB, Leung L, Sasazuki T, Shirasawa S, Zhu CQ, Navab R, Ikura M and
Tsao MS: p120RasGAP is a mediator of rho pathway activation and
tumorigenicity in the DLD1 colorectal cancer cell line. PLoS One.
9:e861032014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stolze B, Reinhart S, Bulllinger L,
Frohling S and Scholl C: Comparative analysis of KRAS codon 12, 13,
18, 61, and 117 mutations using human MCF10A isogenic cell lines.
Sci Rep. 5:85352015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Margonis GA, Kim Y, Sasaki K, Samaha M,
Amini N and Pawlik TM: Codon 13 KRAS mutation predicts patterns of
recurrence in patients undergoing hepatectomy for colorectal liver
metastases. Cancer. 122:2698–2707. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
De Roock W, Jonker DJ, Di Nicolantonio F,
Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M,
Piessevaux H, et al: Association of KRAS p.G13D mutation with
outcome in patients with chemotherapy-refractory metastatic
colorectal cancer treated with cetuximab. JAMA. 304:1812–1820.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tejpar S, Celik I, Schlichting M,
Sartorius U, Bokemeyer C and Van Cutsem E: Association of KRAS G13D
tumor mutations with outcome in patients with metastatic colorectal
cancer treated with first-line chemotherapy with or without
cetuximab. J Clin Oncol. 30:3570–3577. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rowland A, Dias MM, Wiese MD, Kichenadasse
G, McKinnon RA, Karapetis CS and Sorich MJ: Meta-analysis comparing
the efficacy of anti-EGFR monoclonal antibody therapy between KRAS
G13D and other KRAS mutant metastatic colorectal cancer tumours.
Eur J Cancer. 55:122–130. 2016. View Article : Google Scholar : PubMed/NCBI
|