Introduction
Lung cancer is one of the most common malignancies
(1). For more than a decade, lung
cancer has remained the leading cause of cancer death worldwide
(1). Lung cancer is divided into
small-cell lung cancer (SCLC) and non-small-cell lung cancer
(NSCLC), of which NSCLC accounts for about 85–90% and mainly
includes lung adenocarcinoma (LUAD) and lung squamous cell
carcinoma (LUSC) (2). Although the
overall treatment effect of patients has been improved with the
continuous progress of diagnosis and treatment technology in recent
years, the prognosis of lung cancer is still poor (2,3).
Therefore, exploring the molecular mechanism of lung cancer
pathogenesis and progression is necessary for improving the
survival rate of patients with lung cancer.
Tumor invasion and metastasis are important reasons
for the poor prognosis of patients with NSCLC (4). A critical intrinsic factor for
epithelial tumors to infiltrate and metastasize is the
epithelial-mesenchymal transition (EMT) of tumor cells, which can
be abnormally activated at various stages of cancer progression and
plays critical roles in cell growth, invasion, metastasis, and
resistance to the therapy of NSCLC (5–7).
Consequently, it is of great significance to find the key factors
that activate EMT for the treatment of NSCLC.
Heterogeneous nuclear ribonucleoproteins (hnRNPs)
are a kind of RNA binding proteins (RBPs) closely related to the
function and metabolism of RNA, such as transcription, alternative
splicing, nucleocytoplasmic transport, stability, and translation
(8). Studies have shown that the
dysregulation of hnRNPs expression is closely related to the EMT of
various malignant tumor cells (9).
HnRNPAB is a member of the hnRNPs family. Zhou et al
(10) and Yang et al
(11) reported that hnRNPAB was
overexpressed in hepatocellular carcinoma (HCC) and promoted the
EMT of tumor cells by regulating the transcriptions of snail family
transcriptional repressor 1 (SNAI1, SNAIL) and long-noncoding RNA
(lncRNA) ELF209. In addition, the expression of hnRNPAB is
increased in breast cancer, colon cancer, and gastric cancer, in
which hnRNPAB could serve as a prognostic biomarker and therapeutic
target (12–14). However, the function of hnRNPAB in
NSCLC has not yet been reported.
In this study, the expression and localization of
hnRNPAB in NSCLC tissues and its correlation with
clinicopathological parameters of patients with NSCLC were analyzed
through public databases. Then the role of hnRNPAB silencing in the
malignant transformation of NSCLC cells was investigated in
vitro, and finally the molecular mechanism of hnRNPAB promoting
the proliferation and metastasis of NSCLC cells was investigated by
bioinformatics and RT-qPCR. This study may contribute to
discovering a new prognostic biomarker and therapeutic target for
NSCLC patients.
Materials and methods
HnRNPAB expression analysis
The expression and localization of hnRNPAB in lung
cancer and normal tissues were analyzed by the human protein atlas
database (http://www.proteinatlas.org/) and UALCAN database
(http://ualcan.path.uab.edu/). The data
of 585 LUAD and 550 LUSC cases were downloaded from the TCGA
database (https://tcga-data.nci.nih.gov/tcga/), including mRNA
expression profile, sex, age, tumor-node-metastases (TNM)
classification, and overall survival. The median expression level
of hnRNPAB mRNA in 585 LUAD or 550 LUSC cases was used as the
cutoff. Then the survival curve was drawn by the Kaplan-Meier
method and compared by the log-rank test. The relationships between
hnRNPAB expression and clinicopathological parameters were analyzed
by the Chi-square (χ2) test. Univariate and multivariate
analyses of potential prognostic factors for overall survival were
preformed using the Cox proportional hazard model. Genes associated
with hnRNPAB expression in NSCLC were analyzed using the Linked
Omics database (http://linkedomics.org/) based on the Pearson
correlation coefficient.
Cell Culture
The human NSCLC cell line NCI-H292 and the human
renal epithelial cell line 293T were purchased from Kunming Cell
Bank of Chinese Academy of sciences. The human NSCLC cell line PC-9
was obtained from FuHeng Biology Company (Shanghai, China).
NCI-H292 and PC-9 were maintained in RPMI-1640 medium (C11875500BT,
Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented
with 10% fetal bovine serum (10270-106, FBS, Gibco). 293T was
cultured in Dulbecco's Modified Eagle Medium (DMEM, C11995500BT,
Gibco) with 10% FBS. All cell lines were cultured at 37°C in a 5%
CO2 humidified atmosphere.
Plasmid construction
To construct hnRNPAB knockdown recombinants, hnRNPAB
and negative control (NC) shRNA sequences (sense and antisense)
were designed (shown in Table I)
and synthesized, which were dissolved in Tris-EDTA buffer to a
final concentration of 100 µM. For each shRNA, the sense and
antisense strands were mixed with a 1:1 ratio and then annealed
through a PCR program (95°C for 2 min, 72°C for 2 min, 37°C for 2
min, 25°C for 2 min). The paired shRNA was cloned into the vector
psi-LVRU6GP (GeneCopoeia, Germantown, MD, USA) digested with
BamH I and EcoR I (1010S and 1040S, Takara, Beijing,
China). All constructs were confirmed by DNA sequencing (Sangon
Biotech, Shanghai, China).
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Gene | Direction | Primer sequences
(5′-3′) |
|---|
| hnRNPAB (NM_004499)
shRNA-1 | Sense |
GATCCGTGGAAGCAAGTGTGAGATCAATTCAA |
|
|
|
GAGATTGATCTCACACTTGCTTCCATTTTTTG |
|
| Antisense |
AATTCAAAAAATGGAAGCAAGTGTGAGATCAA |
|
|
|
TCTCTTGAATTGATCTCACACTTGCTTCCACG |
| hnRNPAB (NM_004499)
shRNA-2 | Sense |
GATCCGGTTTGGGTTTATCCTGTTCATTCAAGA |
|
|
|
GATGAACAGGATAAACCCAAACCTTTTTTG |
|
| Antisense |
AATTCAAAAAAGGTTTGGGTTTATCCTGTTCA |
|
|
|
TCTCTTGAATGAACAGGATAAACCCAAACCG |
| NC shRNA | Sense |
GATCCGCTTCGCGCCGTAGTCTTATTCAAG |
|
|
|
AGATAAGACTACGGCGCGAAGCTTTTTTG |
| | Antisense |
AATTCAAAAAAGCTTCGCGCCGTAGTCTTA |
|
|
|
TCTCTTGAATAAGACTACGGCGCGAAGCG |
| hnRNPAB (NM_004499)
qPCR | Sense |
TTTGGCGAGTTTGGGGAGATT |
|
| Antisense |
GCCATACTGCTGCTGATAGAC |
| NOP16
(NM_001256539) qPCR | Sense |
AAAGGAAATACTCTGTCTCGGG |
| | Antisense |
CTTCTGCAAAGAATCGAGGAAG |
| DDX41
(NM_001321732) qPCR | Sense |
CCACTACCTTCATCAACAAAGC |
| | Antisense |
ACCTTCTGCTTGGCTTCTAG |
| NPM1 (NM_001355007)
qPCR | Sense |
AGTATATCTGGAAAGCGGTCTG |
| | Antisense |
CATTTTTGGCTGGAGTATCTCG |
| TCOF1
(NM_001135243) qPCR | Sense |
TTTGTCGACCCTAATCGTAGTC |
| | Antisense |
CAGGAGTCAAGCACTGTGTAG |
| TMED9 (NM_017510)
qPCR | Sense |
GAGACCATGGTCATAGGAAACT |
| | Antisense |
TATGGGAAGTGAAAGTGAACCT |
| CENPH (NM_022909)
qPCR | Sense |
TTCCAGAACCTTATTTTGGGGA |
| | Antisense |
CTTCTCAAGCTGCAGAACAATT |
| CCNB1
(NM_001354844) qPCR | Sense |
CTTGCAGTAAATGATGTGGATG |
| | Antisense |
GTGACTTCCCGACCCAGTAG |
| PPM1G (NM_177983)
qPCR | Sense |
AACTCCTTCACAAGAAAATGGC |
| | Antisense |
TGTCCTCAAAGAACTTGGACTT |
| SFXN1
(NM_001322977) qPCR | Sense |
TGGGATCAAAGCACTTTCATTG |
| | Antisense |
TTTCTGTAAGACCAGGAGGAAC |
| HSPA9 (NM_004134)
qPCR | Sense |
GTCAGATTGGAGCATTTGTGTT |
| | Antisense |
CGAGTCATTGAAATAAGCTGGG |
| GAPDH
(NM_001357943) qPCR | Sense |
GGAGCGAGATCCCTCCAAAAT |
| | Antisense |
GGCTGTTGTCATACTTCTCATGG |
| ZEB1 (NM_001128128)
qPCR | Sense |
TTACACCTTTGCATACAGAACCC |
| | Antisense |
TTTACGATTACACCCAGACTGC |
| E-cadherin
(NM_001317185) qPCR | Sense |
CGAGAGCTACACGTTCACGG |
| | Antisense |
GGGTGTCGAGGGAAAAATAGG |
| N-cadherin
(NM_001308176) qPCR | Sense |
AGCCAACCTTAACTGAGGAGT |
| | Antisense |
GGCAAGTTGATTGGAGGGATG |
| Vimentin
(NM_003380) qPCR | Sense |
TGCCGTTGAAGCTGCTAACTA |
| | Antisense |
CCAGAGGGAGTGAATCCAGATTA |
| SNAI1 (NM_005985)
qPCR | Sense |
ACTGCAACAAGGAATACCTCAG |
| | Antisense |
GCACTGGTACTTCTTGACATCTG |
HnRNPAB knockdown
293T cells were plated in a 6-well plate and the
cell confluence reached 90% after 24 h. The hnRNPAB shRNA or NC
shRNA recombinants, lentivirus packaging plasmid psPASX2 and PMD2G
were mixed in a 4:3:1 ratio and then transfected into 293T cells
using an Entranster™-R4000 reagent (4000-4, Engreen Biosystem,
Beijing, China) following the manufacturer's instructions. At 48 h
after transfection, the viral supernatant was collected and
filtered with 0.22 µm filters. The virus solution was mixed with
lentivirus infection reagent (30001-2-0.5, Engreen Biosystem)
according to the manufacturer's instructions and then added into
the NCI-H292 or PC-9 cells with a 40–60% cell confluence. After 8 h
of lentivirus infection, the infection solution was replaced by
fresh RPMI-1640 medium (10% FBS, no antibiotics). After 72 h of
infection, puromycin (final concentration 2.0 µg/ml) was added to
screen the stable cell lines.
Western blotting
Cells were lysed in RIPA buffer (P0013B, Beyotime,
Shanghai, China) for 20 min on ice and then centrifuged at 12,000 ×
g for 10 min at 4°C. The proteins in harvested supernatants were
separated on 10% SDS-PAGE gels and then transferred onto
polyvinylidene difluoride membranes (IPVH00010, Millipore, Merck,
Billerica, MA, USA), which were successively probed with indicated
antibodies and detected with an Enhanced Chemiluminescence Reagent
(WBKLS0100, Millipore). Rabbit polyclonal antibodies against
hnRNPAB (A17497, 1:2,000, ABclonal, Wuhan, China), Glyceraldehyde
3-phosphate dehydrogenase (GAPDH, 60004-1-Ig, 1:5,000, Proteintech,
Wuhan, China), ZEB1 (21544-1-AP, 1:1,000, Proteintech), N-cadherin
(22018-1-AP, 1:5,000, Proteintech), E-cadherin (20874-1-AP,
1:5,000, Proteintech), Vimentin (10366-1-AP, 1:10,000, Proteintech)
and SNAI1 (13099-1-AP, 1:2,000, Proteintech), and a horseradish
peroxidase-conjugated secondary antibody (SA00001-2, 1:5,000,
Proteintech) were used.
Real-time quantitative PCR (RT-qPCR)
analysis
Total RNA was extracted using a TRIzol reagent
(15596-026, Invitrogen, Thermo Fisher Scientific) and reverse
transcribed into cDNA by a PrimeScript™ RT reagent Kit with gDNA
Eraser (RR047A, Takara). Fluorescence quantitative PCR was
performed using a 2× SYBR Green quantitative PCR Master Mix
(B21203, Bimake, Shanghai, China) according to the manufacturer's
protocol in an Applied Biosystems StepOne Plus Real-Time PCR System
(Applied Biosystems, Foster, CA, USA). The primer sequences of each
gene were shown in Table I. GAPDH
was used as a reference gene. The relative mRNA expression was
calculated by the 2−ΔΔCt method. All reactions were
performed in triplicate.
CCK8 assay
Cells were seeded into 96-well plates at a
concentration of 2×103 cells per well and then cultured
for 24, 48, and 72 h, respectively. At the indicated time, 10 µl
CCK-8 solution was added to each well and mixed. The cells were
incubated for 1–4 h at 37°C and the absorbance at 450 nm was
measured with a Biotek ELx800 (Biotek Winooski, Vermont, USA).
Adherent colony formation
Cells were seeded in 6-well plates at a
concentration of 1×103 cells per well and cultured for 2
weeks. The cells were washed twice with 1× phosphate buffered
solution (PBS), fixed with 4% paraformaldehyde for 30 min, and
stained with 0.01% crystal violet for 15 min. The number of
colonies was counted using ImageJ.
Wound healing assay
Cells were plated in 6-well plates and reached
confluence after 24 h. A straight wound was scratched on the cell
layer with a 200 µl pipette. Then the cells were washed twice with
1× PBS and cultured in the medium with 4% FBS. The photographs were
taken at 0 and 24 h for the same visual fields. Cell migration was
expressed as the percentage of scratch healing, which was
calculated by [(scratch area at 0 h-scratch area at 24 h)/scratch
area at 0 h] ×100%.
Invasion assay
The upper Transwell chamber (CLS3464, Corning
Costar, Tewksbury, MA, USA) was coated with 100 µl 10% BD Matrigel
matrix (356234, BD Biosciences, San Jose, CA, USA) diluted with the
serum-free medium, and then incubated in a cell incubator for 2 h.
2×104 cells per well suspended in the serum-free medium
were plated into the upper chamber. The bottom chamber was filled
with 700 µl medium containing 10% FBS. After incubation for 48 h,
cells in the upper membrane of chamber were removed and cells
adhering to the lower membrane were fixed with 4% paraformaldehyde
for 30 min and stained with crystal violet for 15 min. Five random
areas for each chamber were selected to count cell numbers.
Cell cycle analysis
Cells were collected, washed twice with precooled 1×
PBS, and fixed overnight at 4°C with 75% alcohol (diluted with 1×
PBS). Then the cells were harvested again, washed twice with
precooled PBS, and incubated with 500 µl staining solution (50
µg/ml propidium iodide, 100 µg/ml RNase A, 0.2% Triton X-100, 1×
PBS) for 30 min. Cell-cycle was detected on a BD Accuri C6 flow
cytometry (BD Biosciences, San Jose, CA, USA) and the results were
analyzed with the software Modifit (Verity Software House, Topsham,
ME, USA).
Cell apoptosis analysis
Cells were harvested and washed twice with precooled
1× PBS. After staining with Annexin V-APC and 7-AAD (AP105,
MULTISCIENCES, Hangzhou, China), cells were analyzed on a BD Accuri
C6 flow cytometry (BD Biosciences). All samples were assayed in
triplicate.
Statistical analysis
Software SPSS 21.0 (IBM, Armonk, NY, USA) was used
for the statistical analysis. Differences between two groups were
determined by Student's t-test, and analysis of one-way ANOVA
followed by the post hoc Dunnett's test was used for comparisons of
more than two. Survival curves were analyzed by the Kaplan-Meier
log-rank test. Genes expression correlation was analyzed using the
Pearson correlation coefficient. Experiments were performed in
triplicate. All experimental data are presented as the mean ± SD.
P<0.05 indicates a statistically significant difference.
Results
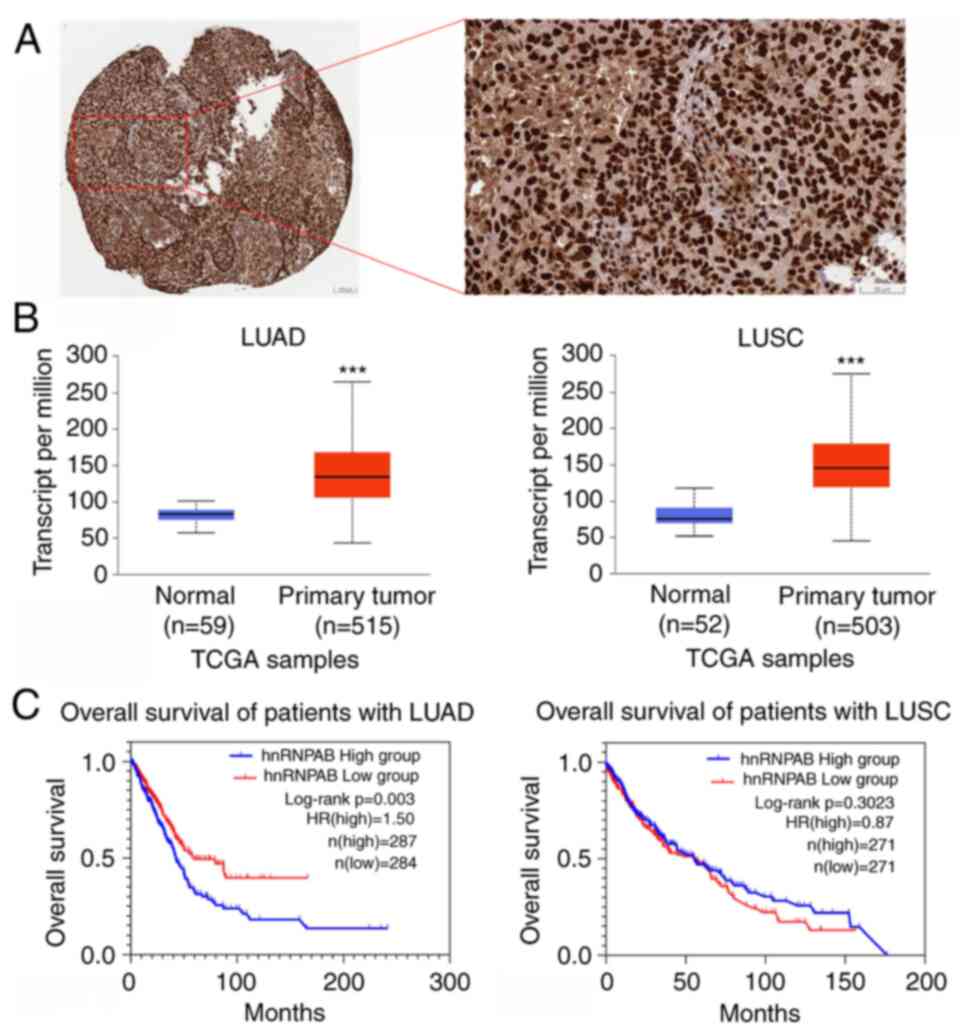
HnRNPAB expression is up-regulated in
NSCLC tissues and associated with the poor prognosis of
patients
Firstly, the expression and role of hnRNPAB in NSCLC
tissues were examined. Immunohistochemical staining (IHC) analysis
from the human protein atlas database indicated that hnRNPAB
protein was mainly expressed in the nucleus of NSCLC cells
(Fig. 1A). Compared with normal
tissues, hnRNPAB mRNA was highly expressed in both LUAD and LUSC
tissues (Fig. 1B). Overall survival
analysis showed that high hnRNPAB mRNA expression was significantly
associated with a poor prognosis in the patients with LUAD
(Fig. 1C). Moreover, hnRNPAB mRNA
expression was strongly associated with the sex and TNM
classification of patients with LUAD (Table II). Univariate and multivariate
analyses indicated that hnNRPAB expression was an independent
prognostic factor for patients with LUAD (Table III). Therefore, the analysis
suggested that hnRNPAB may be a useful biomarker in the prognosis
of patients with LUAD.
 | Table II.Correlations between hnRNPAB mRNA
expression level and clinicopathological parameters in NSCLC. |
Table II.
Correlations between hnRNPAB mRNA
expression level and clinicopathological parameters in NSCLC.
|
| hnRNPAB expression
in LUAD | hnRNPAB expression
in LUSC |
|---|
|
|
|
|
|---|
| Variable | Low (n=292) | High (n=293) | χ2 | P-value | Low (n=275) | High (n=275) | χ2 | P-value |
|---|
| Sex |
|
| 6.419 | 0.011a |
|
| 1.133 | 0.287 |
|
Female | 119 | 150 |
|
| 208 | 197 |
|
|
|
Male | 173 | 143 |
|
| 67 | 78 |
|
|
| Age |
|
| 1.410 | 0.235 |
|
| 1.029 | 0.310 |
|
≤60 | 94 | 108 |
|
| 58 | 68 |
|
|
|
>60 | 198 | 185 |
|
| 217 | 207 |
|
|
| T
classification |
|
| 10.487 | 0.033a |
|
| 4.077 | 0.253 |
| T1 | 112 | 79 |
|
| 55 | 71 |
|
|
| T2 | 150 | 171 |
|
| 172 | 151 |
|
|
| T3 | 21 | 29 |
|
| 38 | 39 |
|
|
| T4 | 7 | 13 |
|
| 10 | 14 |
|
|
|
TX | 2 | 1 |
|
| 0 | 0 |
|
|
| N
classification |
|
| 13.298 | 0.010a |
|
| 2.258 | 0.688 |
| N0 | 194 | 178 |
|
| 181 | 170 |
|
|
| N1 | 46 | 61 |
|
| 65 | 79 |
|
|
| N2 | 38 | 49 |
|
| 23 | 20 |
|
|
| N3 | 0 | 2 |
|
| 2 | 3 |
|
|
|
NX | 14 | 3 |
|
| 4 | 3 |
|
|
| M
classification |
|
| 6.050 | 0.049a |
|
| 3.144 | 0.208 |
| M0 | 188 | 212 |
|
| 227 | 221 |
|
|
| M1 | 12 | 15 |
|
| 6 | 2 |
|
|
|
MX | 92 | 66 |
|
| 42 | 52 |
|
|
 | Table III.Univariate and multivariate analyses
of various prognostic parameters in patients with LUAD. |
Table III.
Univariate and multivariate analyses
of various prognostic parameters in patients with LUAD.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
| Variable | Number of
patients |
|
|
|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Sex |
| 0.905 | 0.690-1.186 | 0.468 | 0.935 | 0.706-1.239 | 0.640 |
|
Female | 262 |
|
|
|
|
|
|
|
Male | 309 |
|
|
|
|
|
|
| Age |
| 1.236 | 0.925-1.653 | 0.151 | 1.191 | 0.889-1.1594 | 0.242 |
|
≤60 | 192 |
|
|
|
|
|
|
|
>60 | 379 |
|
|
|
|
|
|
| T
classification |
| 1.511 | 1.285-1.776 | 0.000a | 1.340 | 1.136-1.581 | 0.001b |
| T1 | 189 |
|
|
|
|
|
|
| T2 | 312 |
|
|
|
|
|
|
| T3 | 48 |
|
|
|
|
|
|
| T4 | 19 |
|
|
|
|
|
|
|
TX | 3 |
|
|
|
|
|
|
| N
classification |
| 1.338 | 1.192-1.503 | 0.000a | 1.298 | 1.142-1.474 | 0.000a |
| N0 | 365 |
|
|
|
|
|
|
| N1 | 106 |
|
|
|
|
|
|
| N2 | 82 |
|
|
|
|
|
|
| N3 | 2 |
|
|
|
|
|
|
|
NX | 16 |
|
|
|
|
|
|
| M
classification |
| 0.943 | 0.803-1.107 | 0.471 | 1.0121 | 0.857-1.194 | 0.890 |
| M0 | 387 |
|
|
|
|
|
|
| M1 | 26 |
|
|
|
|
|
|
|
MX | 158 |
|
|
|
|
|
|
| hnRNPAB |
| 1.509 | 1.147-1.984 | 0.003b | 1.505 | 1.138-1.989 | 0.004b |
|
Low | 287 |
|
|
|
|
|
|
|
High | 284 |
|
|
|
|
|
|
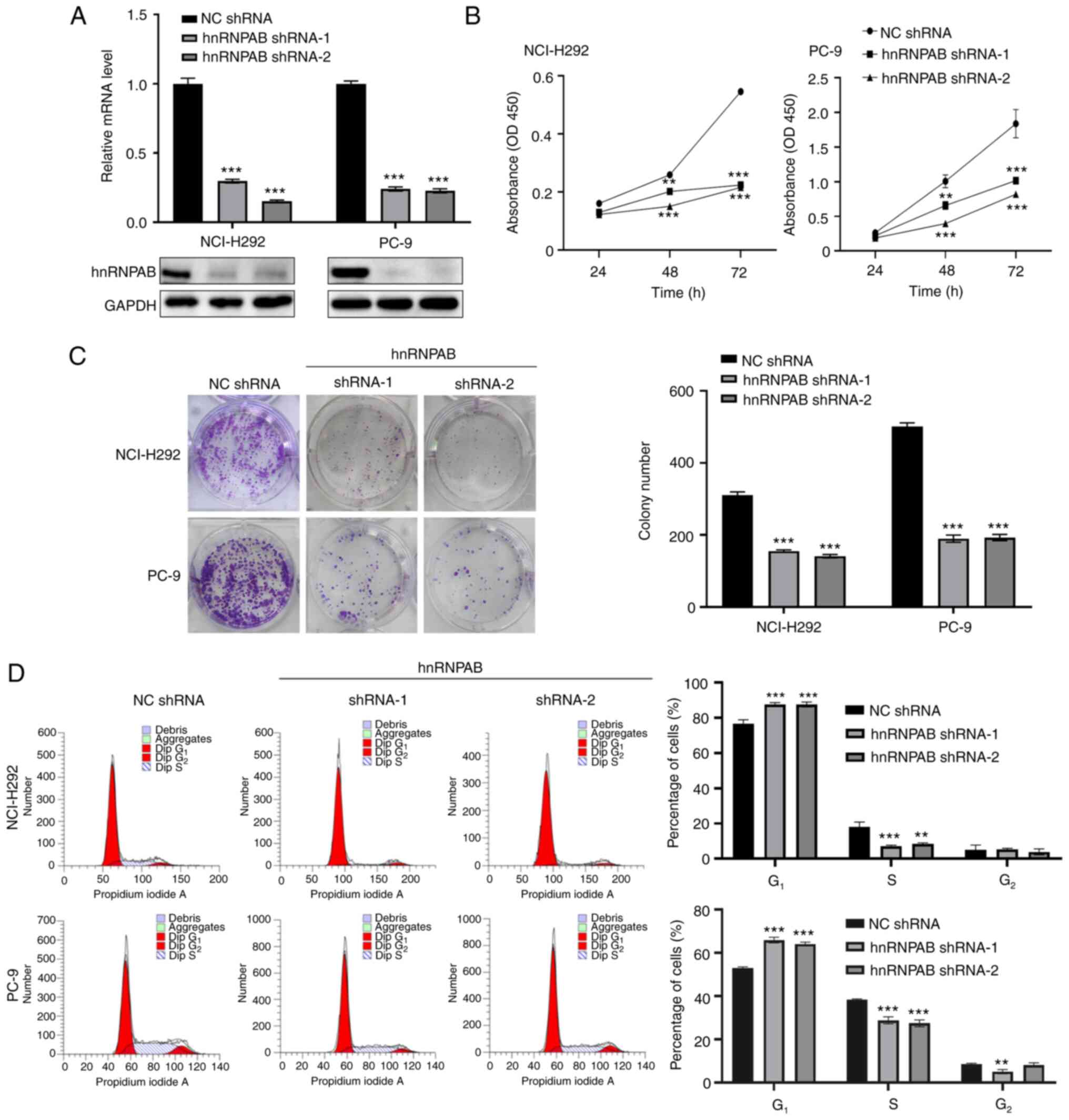
HnRNPAB silencing inhibits the
proliferation of NSCLC cells
The role of hnRNPAB expression dysregulation in
NSCLC cells was further explored by constructing two stable hnRNPAB
knockdown cell lines. Compared with the control group (NC shRNA),
hnRNPAB protein and mRNA expression levels in NCI-H292 and PC-9
cells with hnRNPAB knockdown (hnRNPAB shRNA-1 and shRNA-2) were
significantly reduced (Fig. 2A).
CCK-8 and adherent colony formation assays indicated that hnRNPAB
knockdown decreased cell viability and proliferation (Fig. 2B and C). In addition, the flow
cytometry showed that silencing of hnRNPAB arrested the cell cycle
at G0/G1 phase but did not promote cell
apoptosis, which indicated that hnRNPAB silence suppressed cell
proliferation by arresting the cell cycle at
G0/G1 phase (Figs. 2D and S1).
HnRNPAB knockdown inhibits cell
migration, invasion, and EMT of NSCLC
Next, the effects of hnRNPAB knockdown on cell
migration, invasion, and EMT were detected. The scratch test showed
that the wound healing degree of hnRNPAB shRNA group was
significantly lower than that of NC shRNA group (Fig. 3A). The invasion assay showed that
the cell number passing through the Matrigel matrix in hnRNPAB
shRNA group was obviously less than that in NC shRNA group
(Fig. 3B). Western blotting and
RT-qPCR showed that the protein and mRNA expression levels of
EMT-related markers ZEB1, SNAI1, N-cadherin and Vimentin in hnRNPAB
shRNA group were decreased, while the protein and mRNA expression
levels of E-cadherin were increased (Fig. 3C). Collectively, the results
suggested that hnRNPAB knockdown significantly inhibited the
migration, invasion, and EMT of NSCLC cells.
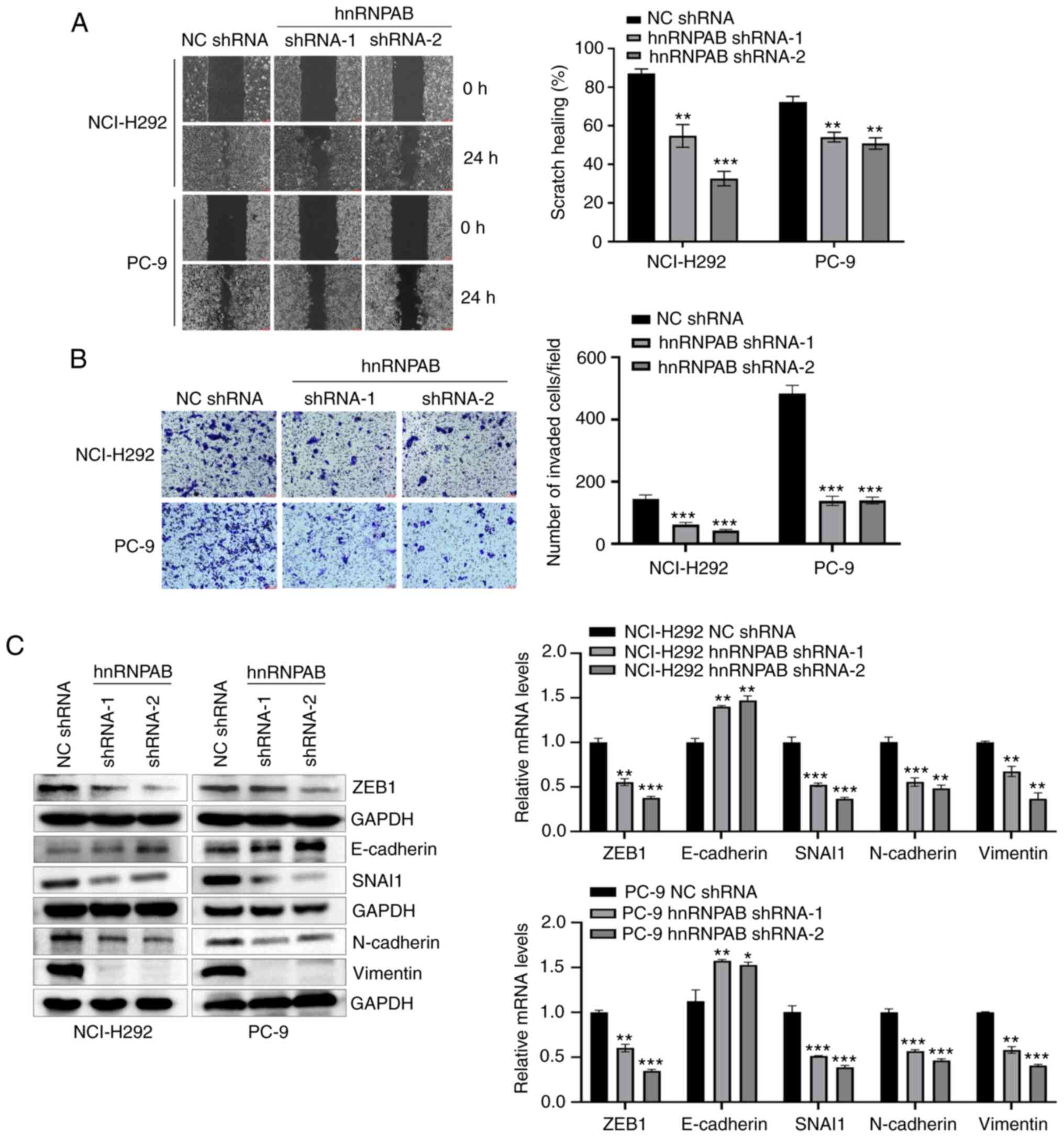 | Figure 3.HnRNPAB-silencing inhibits the
migration, invasion and EMT of NSCLC cells. (A) Cell migration was
evaluated using wound healing assay (magnification, ×100). (B) Cell
invasion was evaluated using Matrigel matrix-coated Transwell assay
(magnification, ×100). (C) Protein and mRNA expression levels of
EMT-related markers ZEB1, E-cadherin, SNAI1, N-cadherin and
Vimentin were determined using western blotting and reverse
transcription-quantitative-PCR. GAPDH was used as a loading
control. For western blotting, multiple membranes from the same
sample at the same time were used to expose different target
proteins and so >1 GAPDH were provided. Results are
representative of three independent experiments. *P<0.05,
**P<0.01 and ***P<0.001 compared with respective NC shRNA
group. HnRNPAB, heterogeneous nuclear ribonucleoprotein A/B; NC,
negative control; shRNA, short hairpin RNA; EMT,
epithelial-mesenchymal transition; ZEB1, zinc finger E-box binding
homeobox 1. |
Analysis of genes related to hnRNPAB
expression in NSCLC
To explore the molecular mechanism of hnRNPAB
promoting the malignant transformation of NSCLC cells, the genes
related to hnRNPAB expression in NSCLC were analyzed using the
LinkedOmics database. As shown in Fig.
4A and B, the expressions of 9764 genes were positively
correlated with the expression of hnRNPAB, and 10214 genes were
negatively correlated with hnRNPAB. Several hnRNPAB-positive
correlated genes with high Pearson Correlations that have been
reported to be associated with tumorigenesis were screened to
perform the co-expression analysis with hnRNPAB using the mRNA
expression data of 585 LUAD and 550 LUSC samples obtained from the
TCGA database, including NOP16 nucleolar protein (NOP16),
nucleophosmin 1 (NPM1), treacle ribosome biogenesis factor 1
(TCOF1), centromere protein H (CENPH), cyclin B1 (CCNB1), heat
shock protein family A (Hsp70) member 9 (HSPA9), sideroflexin 1
(SFXN1), protein phosphatase, Mg2+/Mn2+
dependent 1G (PPM1G), transmembrane p24 trafficking protein 9
(TMED9), and DEAD-box helicase 41 (DDX41) (15–26).
As shown in Fig. 4C, the screened
10 genes were positively co-expressed with hnRNPAB in both LUAD and
LUSC samples. Moreover, the expressions of these genes were
verified in hnRNPAB knockdown cells using RT-qPCR technology and
the results showed that 5 genes except for HSPA9, SFXN1, PPM1G,
TMED9, and DDX41 were down-regulated (Figs. 5 and S2), suggesting that hnRNPAB may promote
the proliferation, migration, invasion, and EMT of NSCLC cells
partially by up-regulating the expressions of these 5 genes.
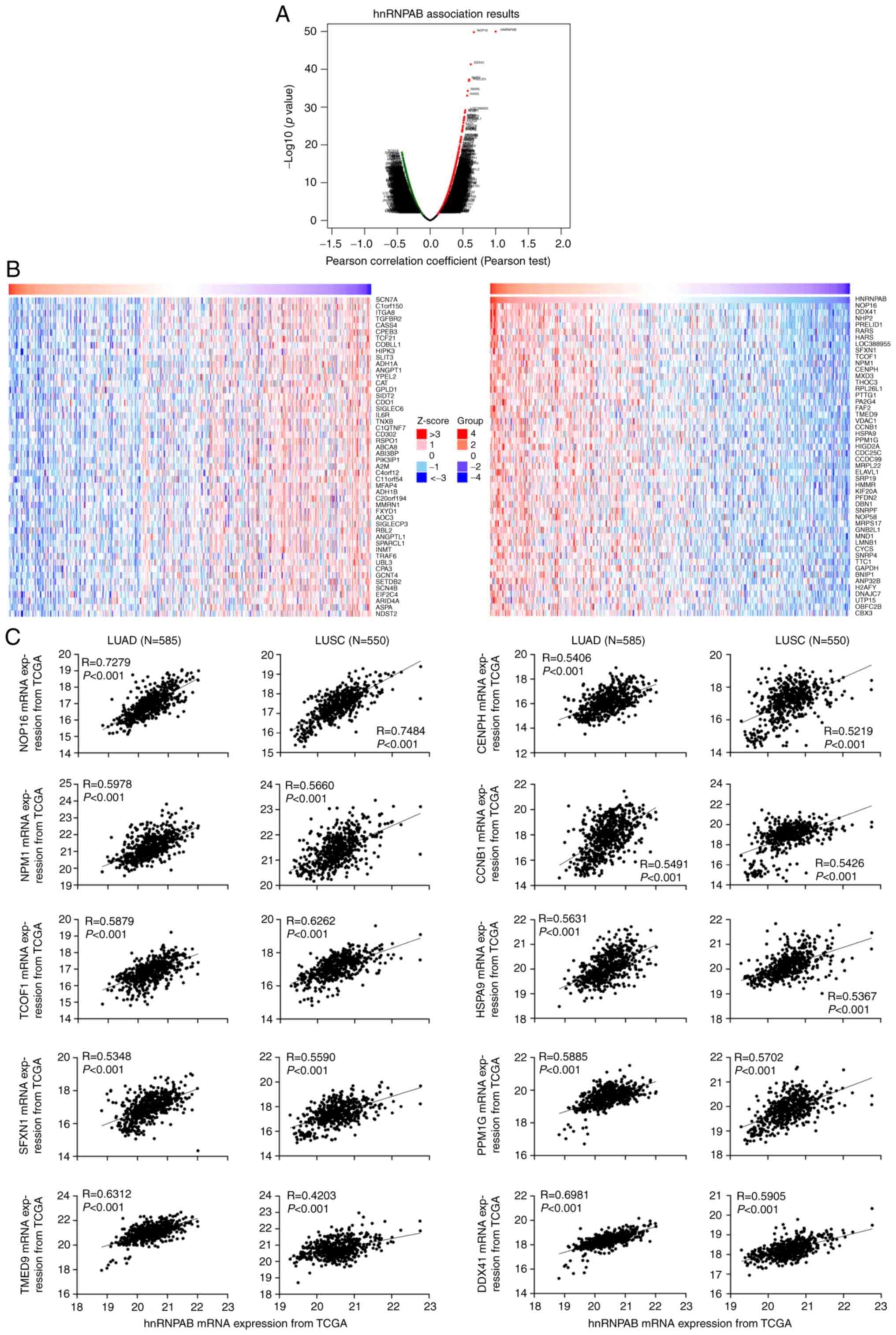 | Figure 4.Analysis of genes related to hnRNPAB
expression in NSCLC. (A) Analysis of hnRNPAB-related genes using
LinkedOmics database based on Pearson correlation coefficient. (B)
Analysis of positive and negative correlated genes with hnRNPAB.
(C) Co-expression analysis between hnRNPAB and the screened genes
using the mRNA expression data of NSCLC samples from The Cancer
Genome Atlas database. HnRNPAB, heterogeneous nuclear
ribonucleoprotein A/B; NSCLC, non-small cell lung cancer; LUAD,
lung adenocarcinoma; LUSC, lung squamous cell carcinoma; NOP16,
NOP16 nucleolar protein; NPM1, nucleophosmin 1; TCOF1, treacle
ribosome biogenesis factor 1; CENPH, centromere protein H; CCNB1,
cyclin B1; HSP, heat shock protein family; SFXN1, sideroflexin 1;
PPM1G, protein phosphatase, Mg2+/Mn2+
dependent 1G; TMED9, transmembrane p24 trafficking protein 9;
DDX41, DEAD-box helicase 41. |
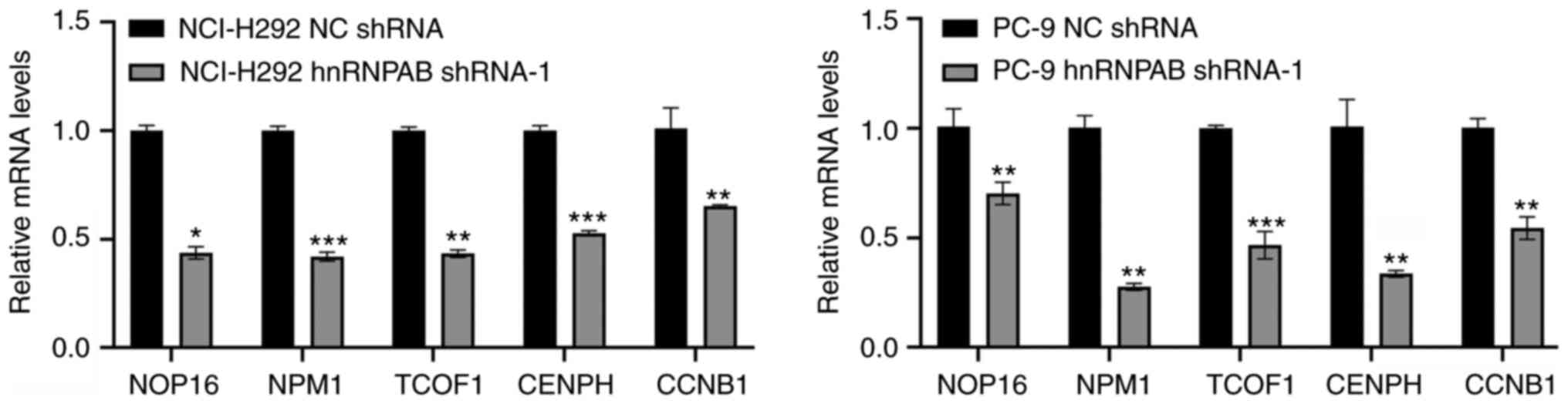 | Figure 5.Expression levels of the screened
genes in stable hnRNPAB-silenced cells were verified using RT-qPCR.
The total RNA was extracted and the mRNA expression levels of
NOP16, NPM1, TCOF1, CENPH and CCNB1 in stable non-small cell lung
cancer cell lines were determined using RT-qPCR. GAPDH was used as
an internal control. Results are representative of three
independent experiments. *P<0.05, **P<0.01 and ***P<0.001
compared with respective NC shRNA group. HnRNPAB, heterogeneous
nuclear ribonucleoprotein A/B; RT-qPCR, reverse
transcription-quantitative PCR; NOP16, NOP16 nucleolar protein;
NPM1, nucleophosmin 1; TCOF1, treacle ribosome biogenesis factor 1;
CENPH, centromere protein H; CCNB1, cyclin B1; NC, negative
control; shRNA, short hairpin RNA. |
Discussion
NSCLC has seriously threatened human health. Once
NSCLC was diagnosed, most patients usually were in the late stage
or their tumors have metastasized, thus losing the opportunity for
surgery (27). Therefore, finding
new diagnostic and therapeutic targets is of great importance to
improve the survival rate of NSCLC patients.
HnRNPs are a kind of RNA binding proteins. So far,
more than 20 hnRNP members have been identified, including hnRNPA ~
U (28). With the deepening of the
research, it is found that hnRNP members have been involved in the
development and progression of tumors by regulating cell
proliferation, metastasis, self-renewal of cancer stem cells and so
on (9). For example, hnRNPL
promotes the progression of prostate cancer by regulating
circCSPP1/microRNA-520h/early growth response factor 1 (EGR1) axis
(29). HnRNPA1, A2/B1, and K
binding to the promoter of the tumor suppressor Annexin A7 (ANXA7)
alter the splicing pattern of ANXA7, leading to the progression of
prostate cancer (30).
HnRNPAB is a member of the hnRNP family. Previous
studies have shown that hnRNPAB is dysregulated in multiple tumors,
which is associated with various malignant phenotypes of tumors
(10–12,31).
However, the expression and role of hnRNPAB in NSCLC remain
unclear. According to our results, hnRNPAB exhibited a relatively
high expression in NSCLC samples, showing significant correlation
with the overall survival, sex and TNM classification of patients
with LUAD. Multivariate Cox regression analysis pointed toward
hnRNPAB as an independent prognostic marker. These results
suggested that hnRNPAB may play an important role in NSCLC
progression.
Metastasis is a main cause of mortality in cancer
patients (32). EMT is often
activated during tumor metastasis (32). EMT is a reversible biological
cellular process in which cells transform from an epithelial form
to a mesenchymal form (7). In the
process of EMT, the expressions of many mesenchymal related
proteins are often increased, especially Vimentin (33). Cells in EMT acquire stem cell-like
properties, in which the protein expression changed from E-cadherin
to N-cadherin, ZEB1 and SNAI1, of which ZEB1 and SNAI1 directly
suppressed the expression of E-cadherin by binding to its promoter
(34–36). Our results showed that hnRNPAB
knockdown increased the expression of E-cadherin and reduced the
expressions of ZEB1, N-cadherin, Vimentin, and SNAI1, indicating
EMT was reversed. In general, cancer cells obtain stronger
proliferation, migration, and invasion abilities after EMT
(37,38). Consistently, hnRNPAB silencing
inhibited the proliferation, migration, and invasion of NSCLC
cells. These observations suggested that hnRNPAB contributes to
tumor progression, probably via the process of tumor proliferation
and metastasis.
To explore the molecular mechanism of hnRNPAB
regulating the proliferation and metastasis of NSCLC cells, 10
genes positively correlated with hnRNPAB expression were selected
for verification. The results showed that the expressions of NOP16,
NPM1, TCOF1, CENPH and CCNB1 were not only positively correlated
with the expression of hnNRPAB in LUAD and LUSC cases but also
down-regulated in hnRNPAB silenced cells. Previous studies have
shown that these 5 genes were highly expressed in a variety of
tumor tissues, which promoted the malignant transformation of tumor
cells (15–18,21).
Increased rRNA production is a hallmark of cancer (15,39).
It is verified that NOP16 and TCOF1 could coordinate the production
of rRNA to promote HCC and breast cancer tumorigenesis (15,39).
NPM1 is a well-known nucleocytoplasmic shuttling protein and its
overexpression, translocation, mutation, or loss of function can
lead to cancer and tumorigenesis (16). CENPH is highly expressed in NSCLC
and may be used as a prognostic biomarker for patients with NSCLC
(40). Likewise, CCNB1 is
overexpressed in LUAD and involved in the proliferation and
metastasis of tumor cells (21).
These analyses indicate that hnRNPAB may promote the cell
proliferation and metastasis of NSCLC by up-regulating the
expressions of NOP16, NPM1, TCOF1, CENPH and CCNB1.
In conclusion, this study identified hnRNPAB as a
useful biomarker in the prognosis of patients with LUAD. The
overexpression of hnRNPAB promoted the proliferation and metastasis
of NSCLC cells partially by up-regulating the expressions of NOP16,
NPM1, TCOF1, CENPH and CCNB1. Our results may offer a potential
novel target for the treatment of NSCLC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science
Foundation of China (grant no. 81660474), the Guizhou Provincial
Science and Technology Project [grant no. ZK(2022)041], the
Academic Seedling Project of Guizhou Medical University (grant no.
19NSP009) and the Natural Science Foundation of Guizhou Provincial
Health Commission (grant no. gzwkj2023-254).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QW and KS designed the research and confirmed the
authenticity of all the raw data. XG performed experiments. XG, LL
and HY performed statistical analysis. QW wrote the manuscript. QW
and XG prepared figures. TZ, YZ, YX and JZ interpreted the data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Oliver AL: Lung cancer: Epidemiology and
screening. Surg Clin North Am. 102:335–344. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Passiglia F, Bertaglia V, Reale ML,
Delcuratolo MD, Tabbò F, Olmetto E, Capelletto E, Bironzo P and
Novello S: Major breakthroughs in lung cancer adjuvant treatment:
Looking beyond the horizon. Cancer Treat Rev. 101:1023082021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Passiglia F, Pilotto S, Facchinetti F,
Bertolaccini L, Del Re M, Ferrara R, Franchina T, Malapelle U,
Menis J, Passaro A, et al: Treatment of advanced non-small-cell
lung cancer: The 2019 AIOM (Italian Association of Medical
Oncology) clinical practice guidelines. Crit Rev Oncol Hematol.
146:1028582020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bakir B, Chiarella AM, Pitarresi JR and
Rustgi AK: EMT, MET, plasticity, and tumor metastasis. Trends Cell
Biol. 30:764–776. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
O'Leary K, Shia A and Schmid P: Epigenetic
regulation of EMT in non-small cell lung cancer. Curr Cancer Drug
Targets. 18:89–96. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yuan X, Wu H, Han N, Xu H, Chu Q, Yu S,
Chen Y and Wu K: Notch signaling and EMT in non-small cell lung
cancer: Biological significance and therapeutic application. J
Hematol Oncol. 7:872014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mittal V: Epithelial mesenchymal
transition in tumor metastasis. Annu Rev Pathol. 13:395–412. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Geuens T, Bouhy D and Timmerman V: The
hnRNP family: Insights into their role in health and disease. Hum
Genet. 135:851–867. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Han N, Li W and Zhang M: The function of
the RNA-binding protein hnRNP in cancer metastasis. J Cancer Res
Ther. 9 (Suppl):S129–S134. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou ZJ, Dai Z, Zhou SL, Hu ZQ, Chen Q,
Zhao YM, Shi YH, Gao Q, Wu WZ, Qiu SJ, et al: HNRNPAB induces
epithelial-mesenchymal transition and promotes metastasis of
hepatocellular carcinoma by transcriptionally activating SNAIL.
Cancer Res. 74:2750–2762. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang Y, Chen Q, Piao HY, Wang B, Zhu GQ,
Chen EB, Xiao K, Zhou ZJ, Shi GM, Shi YH, et al: HNRNPAB-regulated
lncRNA-ELF209 inhibits the malignancy of hepatocellular carcinoma.
Int J Cancer. 146:169–180. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cao Y, Zhang W, Jin YT and Zou Q: Mining
the prognostic value of HNRNPAB and its function in breast
carcinoma. Int J Genomics. 2020:37506732020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pan T, Yu Z, Jin Z, Wu X, Wu A, Hou J,
Chang X, Fan Z, Li J, Yu B, et al: Tumor suppressor lnc-CTSLP4
inhibits EMT and metastasis of gastric cancer by attenuating
HNRNPAB-dependent Snail transcription. Mol Ther Nucleic Acids.
23:1288–1303. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen ZQ, Yuan T, Jiang H, Yang YY, Wang L,
Fu RM, Luo SQ, Zhang T, Wu ZY and Wen KM: MicroRNA-8063 targets
heterogeneous nuclear ribonucleoprotein AB to inhibit the
self-renewal of colorectal cancer stem cells via the Wnt/β-catenin
pathway. Oncol Rep. 46:2192021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang C, Yin C, Wang L, Zhang S, Qian Y,
Ma J, Zhang Z, Xu Y and Liu S: HSPC111 governs breast cancer growth
by regulating ribosomal biogenesis. Mol Cancer Res. 12:583–594.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Karimi Dermani F, Gholamzadeh Khoei S,
Afshar S and Amini R: The potential role of nucleophosmin (NPM1) in
the development of cancer. J Cell Physiol. 236:7832–7852. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu J, Lai Y, Huang H, Ramakrishnan S, Pan
Y, Ma VWS, Cheuk W, So GYK, He Q, Geoffrey Lau C, et al: TCOF1
upregulation in triple-negative breast cancer promotes stemness and
tumour growth and correlates with poor prognosis. Br J Cancer.
126:57–71. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu G, Shan T, He S, Ren M, Zhu M, Hu Y, Lu
X and Zhang D: Overexpression of CENP-H as a novel prognostic
biomarker for human hepatocellular carcinoma progression and
patient survival. Oncol Rep. 30:2238–2244. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He WL, Li YH, Yang DJ, Song W, Chen XL,
Liu FK, Wang Z, Li W, Chen W, Chen CY, et al: Combined evaluation
of centromere protein H and Ki-67 as prognostic biomarker for
patients with gastric carcinoma. Eur J Surg Oncol. 39:141–149.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu X, Lin Y, Shi L, Huang Y, Lai C, Wang
Y, Zhang M, Wang S, Heng B, Yu G, et al: Upregulation of centromere
protein H is associated with progression of renal cell carcinoma. J
Mol Histol. 46:377–385. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bao B, Yu X and Zheng W: MiR-139-5p
targeting CCNB1 modulates proliferation, migration, invasion and
cell cycle in lung adenocarcinoma. Mol Biotechnol. 64:852–860.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guan Y, Zhu X, Liang J, Wei M, Huang S and
Pan X: Upregulation of HSPA1A/HSPA1B/HSPA7 and downregulation of
HSPA9 were related to poor survival in colon cancer. Front Oncol.
11:7496732021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen L, Kang Y, Jiang Y, You J, Huang C,
Xu X and Chen F: Overexpression of SFXN1 indicates poor prognosis
and promotes tumor progression in lung adenocarcinoma. Pathol Res
Pract. 237:1540312022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen D, Zhao Z, Chen L, Li Q, Zou J and
Liu S: PPM1G promotes the progression of hepatocellular carcinoma
via phosphorylation regulation of alternative splicing protein
SRSF3. Cell Death Dis. 12:7222021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han GH, Yun H, Chung JY, Kim JH and Cho H:
TMED9 expression level as a biomarker of epithelial ovarian cancer
progression and prognosis. Cancer Genomics Proteomics. 19:692–702.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Weinreb JT and Bowman TV: Clinical and
mechanistic insights into the roles of DDX41 in haematological
malignancies. FEBS Lett. 596:2736–2745. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wenk MR and Choi H: Abundant circulating
lipids-a new opportunity for NSCLC detection? Nat Rev Clin Oncol.
19:361–362. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xie W, Zhu H, Zhao M, Wang L, Li S, Zhao
C, Zhou Y, Zhu B, Jiang X, Liu W and Ren C: Crucial roles of
different RNA-binding hnRNP proteins in Stem Cells. Int J Biol Sci.
17:807–817. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lu J, Zhong C, Luo J, Shu F, Lv D, Liu Z,
Tan X, Wang S, Wu K, Yang T, et al: HnRNP-L-regulated
circCSPP1/miR-520h/EGR1 axis modulates autophagy and promotes
progression in prostate cancer. Mol Ther Nucleic Acids. 26:927–944.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Torosyan Y, Dobi A, Glasman M, Mezhevaya
K, Naga S, Huang W, Paweletz C, Leighton X, Pollard HB and
Srivastava M: Role of multi-hnRNP nuclear complex in regulation of
tumor suppressor ANXA7 in prostate cancer cells. Oncogene.
29:2457–2466. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu Y, Wang X, Gu Q, Wang J, Sui Y, Wu J
and Feng J: Heterogeneous nuclear ribonucleoprotein A/B: An
emerging group of cancer biomarkers and therapeutic targets. Cell
Death Discov. 8:3372022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fares J, Fares MY, Khachfe HH, Salhab HA
and Fares Y: Molecular principles of metastasis: A hallmark of
cancer revisited. Signal Transduct Target Ther. 5:282020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ostrowska-Podhorodecka Z and McCulloch CA:
Vimentin regulates the assembly and function of matrix adhesions.
Wound Repair Regen. 29:602–612. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang Y, Zhao B, Lv L, Yang Y, Li S and Wu
H: FBXL10 promotes EMT and metastasis of breast cancer cells via
regulating the acetylation and transcriptional activity of SNAI1.
Cell Death Discov. 7:3282021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
López-Moncada F, Torres MJ, Lavanderos B,
Cerda O, Castellón EA and Contreras HR: SPARC induces E-cadherin
repression and enhances cell migration through Integrin αvβ3 and
the transcription factor ZEB1 in prostate cancer cells. Int J Mol
Sci. 23:58742022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xiong Y, Lai X, Xiang W, Zhou J, Han J, Li
H, Deng H, Liu L, Peng J and Chen L: Galangin (GLN) suppresses
proliferation, migration, and invasion of human glioblastoma cells
by targeting Skp2-induced epithelial-mesenchymal transition (EMT).
Onco Targets Ther. 13:9235–9244. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li JL, Wang ZQ and Sun XL: MYL6B drives
the capabilities of proliferation, invasion, and migration in
rectal adenocarcinoma through the EMT process. Open Life Sci.
15:522–531. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu C, Xia D, Wang D, Wang S, Sun Z, Xu B
and Zhang D: TCOF1 coordinates oncogenic activation and rRNA
production and promotes tumorigenesis in HCC. Cancer Sci.
113:553–564. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liao WT, Wang X, Xu LH, Kong QL, Yu CP, Li
MZ, Shi L, Zeng MS and Song LB: Centromere protein H is a novel
prognostic marker for human nonsmall cell lung cancer progression
and overall patient survival. Cancer. 115:1507–1517. 2009.
View Article : Google Scholar : PubMed/NCBI
|