Introduction
MicroRNAs (miRNAs) consist of 20–30 bases and play
an important role in fostering communication among cells in various
organisms (1). Tumor cells also
affect the surrounding normal cells via miRNAs, encouraging changes
in the tumor microenvironment to facilitate tumor progression
(2–6). These miRNAs have been reported to be
transmitted to the target cells via tumor-derived small
extracellular vesicles (3–6). Thus, these miRNAs in circulating blood
may be useful as prognostic or diagnostic markers of tumors
(7–11).
Sarcoma is a rare cancer, and osteosarcoma typically
occurs in children, adolescents and young adults (12,13).
Thus far, the diagnosis of osteosarcoma has generally depended on
the histological findings of a biopsy specimen. The prognosis of
these patients has been reported to be associated with the presence
of metastasis at presentation, a delayed initiation of treatment, a
poor response to chemotherapy and small amounts of mature
osteoclasts in biopsy specimens (14–19).
Compared with these factors, however, circulating biomarkers
assessed using blood tests are simpler and easier to evaluate,
especially in children. Previously, several serum miRNAs associated
with the prognosis or diagnosis of osteosarcoma have been reported,
including circulating miR-663a, miR-25-3p, miR-487a, miR-493-5p,
miR-501-3p and miR-502-5p (20–25).
In our previous study, osteosarcoma advanced locally
and metastasis was promoted through the secretion of large number
of small extracellular vesicles, followed by the suppression of
osteoclastogenesis via the upregulation of miR-146a-5p (26). Several other miRNAs were also
identified (miR-1260a, miR-487b-3p, miR-6720-3p, miR-1260b,
miR-4758-3p, miR-4690-3p, miR-4286, miR-6765-3p, miR-1261,
miR-7975, miR-4664-5p and miR-3907) that were included in small
extracellular vesicles derived from high-grade osteosarcoma cells
with the capacity to metastasize at rates ≥6× than in those with
low metastatic potential (26).
However, to the best of our knowledge, the utility of the 13
identified miRNAs for determining the prognosis or diagnosis of
high-grade osteosarcoma has not yet been validated in the clinical
setting.
Therefore, in the present study, the association of
these miRNAs with the prognosis of patients with osteosarcoma and
their potential utility in the diagnosis of osteosarcoma and
differentiation from other bone tumors was investigated.
Materials and methods
Patient enrollment
A total of 43 patients with osteosarcoma were
treated at The National Cancer Center Hospital (NCCH) in Tokyo,
Japan, from January 2007 to June 2013. Among these patients, 13
were excluded due to a previous history of chemotherapy or surgical
treatment at another institution (n=7), metastatic lesions at
presentation (n=2), the histology of dedifferentiated osteosarcoma
from low-grade osteosarcoma (n=2) and absence of microarray data
(n=2). The remaining 30 patients who had no previous history of
treatment for the histology of primary osteosarcoma without
metastatic lesions at presentation and whose microarray data were
sufficiently obtained from their serum samples, were
retrospectively reviewed in the present study. A total of 27
patients underwent standard treatment with the combination of
chemotherapy and surgery in accordance with the National
Comprehensive Cancer Network (NCCN) guidelines (27) for osteosarcoma during the follow-up
period at the NCCH. In total, 3 patients did not undergo either
neoadjuvant or adjuvant chemotherapy. Of these 3 patients, 1
patient was assumed to have a low-grade malignant bone tumor before
surgery, while another patient did not receive adjuvant
chemotherapy due to the patient's refusal to receive further
treatment after surgery. The other patient opted to receive surgery
and radiotherapy instead of chemotherapy.
Patients with a histological diagnosis of
osteochondroma (n=15), enchondroma (n=11) and fibrous dysplasia
(n=10) as benign tumors, giant cell tumors of the bone (n=28),
synovial chondromatosis (n=5) and osteoblastoma (n=3) as
intermediate-grade tumors, Ewing sarcoma (n=26), chordoma (n=8) and
chondrosarcoma (grade 2–3) (n=6) as high-grade tumors [according to
the histological grade in the 2020 WHO classification (12)], who were treated at the NCCH from
January 2007 to June 2013 and whose microarray data were
sufficiently obtained from their serum samples at presentation
before any treatment, were also included in the study.
The present study was conducted in accordance with
the 1975 Declaration of Helsinki. It was approved by The NCCH
Institutional Review Board (Tokyo, Japan; approval nos. 2004-050,
2013-111 and 2015-266). Written informed consent was obtained from
each participant and/or their family.
Data collection on patient and control
characteristics
The age (mean and range), sex, tumor location (limb
or trunk), tumor size (mean and range), staging [using the American
Joint Committee on Cancer (AJCC) 8th edition] (28), surgery, chemotherapy, radiation
therapy, distant metastasis, local recurrence, follow-up periods
(mean and range), oncological outcomes at the final follow-up time
and the levels of 13 serum miRNAs (miR-146a-5p, miR-1260a,
miR-487b-3p, miR-6720-3p, miR-1260b, miR-4758-3p, miR-4690-3p,
miR-4286, miR-6765-3p, miR-1261, miR-7975, miR-4664-5p and
miR-3907) at the first visit were retrospectively investigated in
all patients with osteosarcoma selected for study using medical
records. The age (mean and range), sex and the level of the 13
serum miRNAs at the first visit in patients with other types of
bone tumors (osteochondroma, enchondroma, fibrous dysplasia, giant
cell tumors of the bone, synovial chondromatosis, osteoblastoma,
Ewing sarcoma, chordoma and chondrosarcoma) and healthy controls
were also reviewed. The 13 serum miRNA levels in all patients in
the present study were used in microarray experiments in a previous
study conducted at the NCCH, which are publicly available (25). A microarray analysis was not newly
conducted in the present study.
miRNA mimics and inhibitor
transfection
To determine the effects of miR-1261, 70 pmol of
miRNA hairpin-inhibitor transfection control with Dy547 (miR
hairpin inhibitor-Ctrl; cat. no. IP-004500-01-05) or hsa-miR-1261
hairpin-inhibitor (cat. no. IH-301388-01-0002) (both from Horizon
Discovery Ltd.; PerkinElmer, Inc.) was transfected into 143B cells
[highly aggressive, k-ras activated, human osteosarcoma cells (OS)]
obtained from the ATCC using Lipofectamine™ RNAiMAX (Thermo Fisher
Scientific, Inc.), and 70 pmol of miRNA mimic transfection control
with Dy547 (miR mimic-Ctrl; cat. No. CP-004500-01-05) or
hsa-miR-1261 mimic (cat. no. C-301388-00-0002) (both from Horizon
Discovery Ltd.; PerkinElmer, Inc.) was transfected into HOS cells
(non-aggressive, wild-type k-ras, human OS) obtained from the ATCC
using Lipofectamine RNAiMAX. These two cell lines were cultured at
37°C in DMEM (Nacalai Tesque, Inc.) with 10% heat inactivated FBS
(Biowest) and 100 units/ml Penicillin G and 100 µg/ml Streptomycin
(FUJIFILM Wako Pure Chemical Corporation) for 1 day before
transfection. Both cells were cultured at 37°C in DMEM after
transfection for 2 days. To confirm the transfection efficacy of
the cells, uptake of the Dy547-labelled control in each cell line
was observed using a BZ-9000 phase-contrast and fluorescence
microscope (Keyence Corporation) and the positive cells were
counted by a BZ-II Analyzer, version 1.42 (Keyence Corporation)
(Fig. S1).
Cell viability and cell proliferation
assays
143B and HOS cells were seeded at 3×103
cells/200 µl/96-well (n=3) after the aforementioned miRNA
transfection. After incubating for 2 days, cellular viability was
evaluated using WST-8 assay reagent (Nacalai Tesque, Inc.) for an
hour.
After the culture of cells (5×104
cells/500 µl/4-well glass slide) for 2 days, the cells were fixed
using 4% paraformaldehyde 500 µl/well at room temperature for 20
min. Cells were then blocked using serum-free unconjugated blocking
liquid (cat. no. X0909; EnVision Systems; Dako; Agilent
Technologies, Inc.) at room temperature for 10 min. For
immunocytochemistry, the primary anti-rabbit polyclonal antibody
for Ki-67 (1:200; cat. no. NB500-170; Novus Biologicals, LLC) was
incubated with the cells at 4°C for 1 day. Anti-mouse IgG
conjugated with peroxidase-labeled polymers (cat. no. K4001;
EnVision Systems; Dako; Agilent Technologies, Inc.) was incubated
with the cells at room temperature for 30 min as the secondary
antibody. DAB-Chromogen staining was conducted for 3 min at room
temperature (cat. no. GV825; EnVision Systems; Dako; Agilent
Technologies, Inc.). The percentage of the Ki-67+ cells
per total cells was measured by phase-contrast and fluorescence
microscopy using a BZ-9000 and BZ-II Analyzer version 1.42.
(KEYENCE).
Reverse transcription-quantitative PCR
(RT-qPCR)
To detect mRNA, total RNA extraction was performed
on 143B and HOS cells that had been seeded at 6×105
cells/3 ml/60-mm dish after miRNA transfection and cultured at 37°C
for 2 days, using a Fast Gene RNA Basic Kit (Nippon Gene Co.,
Ltd.). The mRNA was reverse transcribed using 100 ng RNA and 10 µl
reaction mix from a ReverTra Ace qPCR RT Kit (Toyobo Life Science)
according to the manufacturer's protocol (37°C for 15 min, 50°C for
5 min, 98°C for 5 min and held at 4°C). After diluting the cDNA
with 90 µl dH2O, 4 µl was incubated with 16 µl Fast SYBR Green
Master Mix (Thermo Fisher Scientific, Inc.) in a LightCycler96
system (Roche Diagnostics K.K.). The thermocycler conditions were
as follows: 95°C for 20 sec, 45 cycles of 95°C for 3 sec
(denaturation) and 60°C for 30 sec (combination of annealing and
extension). At the end of the qPCR, specificity was confirmed using
melting peak analysis. Paired primers are listed in Table SI. mRNA expression of each gene was
normalized to GAPDH using 2−ΔΔCq (29).
Statistical analysis
Using receiver operating characteristic (ROC)
curves, the optimal threshold values for miR-146a-5p, miR-1260a,
miR-487b-3p, miR-6720-3p, miR-1260b, miR-4758-3p, miR-4690-3p,
miR-4286, miR-6765-3p, miR-1261, miR-7975, miR-4664-5p and miR-3907
in patients with osteosarcoma were obtained when the Youden index
was maximal, from which death from disease, distant metastasis and
freedom from disease were predicted. The 27 patients with
osteosarcoma who underwent standard treatment with a combination of
chemotherapy and surgery in accordance with the NCCN guidelines
were divided into two groups based on the area under the curve
values (low vs. high level) determined by ROC curves analyses. The
overall survival rate, distant metastasis-free survival rate and
disease-free survival rate were plotted using the Kaplan-Meier
method for each group and compared by the log-rank test. P<0.05
was considered to indicate a statistically significant difference.
The overall survival was defined as the period until the death of
the patient since the osteosarcoma diagnosis was made. The distant
metastasis-free survival was defined as the period until a distant
metastasis was detected by an imaging examination. The disease-free
survival was defined as the period of no evidence of disease.
For differentiation of osteosarcoma, serum miRNA
levels at presentation in 30 patients with osteosarcoma were
compared with those in patients with other types of bone tumors
(benign, intermediate and high-grade) and healthy controls. Using
the level of each miRNA (miR-146a-5p, miR-1260a, miR-487b-3p,
miR-6720-3p, miR-1260b, miR-4758-3p, miR-4690-3p, miR-4286,
miR-6765-3p, miR-1261, miR-7975, miR-4664-5p and miR-3907) in the
10 bone tumor groups and the osteosarcoma group, the Kruskal-Wallis
test was performed to differentiate osteosarcoma and, as a post-hoc
test, all 11 groups (including the healthy control group) were
compared by Steel-Dwass multiple tests. P<0.05 was considered to
indicate a statistically significant difference.
Cell viability using the WST-8 assay, the percentage
of Ki-67+ cells, and the change in miRNA expression
levels for miR-1261 target genes between HOS cells transfected with
hsa-miR-1261 mimic and HOS cells transfected with miRNA mimic
control, or between 143B cells transfected with hsa-miR-1261
hairpin-inhibitor and 143B cells transfected with miRNA
hairpin-inhibitor control were compared by unpaired Student's
t-test. P<0.05 was considered to indicate statistically
significant difference.
All statistical analyses were performed using EZR
version 1.61 (Saitama Medical Center, Jichi Medical University),
which is a graphical user interface for the R software program (The
R Foundation for Statistical Computing). More precisely, it is a
modified version of R commander designed to add statistical
functions frequently used in biostatistics (30,31).
Results
Patient characteristics
The osteosarcoma study population included 20 male
and 10 female patients with a mean age of 24 (range, 10–68) years
old. The histological findings were high-grade osteosarcoma in all
cases. Tumors were located in the pelvis in 3 patients and in the
appendicular skeleton in 27 patients. No distant metastasis was
observed at presentation in all cases. The mean tumor size was 9.7
cm in the greatest dimension (range, 6–18 cm). The tumor stage was
IIA in 13 patients, IIB in 14 patients and III in 3 patients
according to the AJCC 8th staging classification. In total, 27
patients underwent surgical treatment after neoadjuvant
chemotherapy, and thereafter received adjuvant chemotherapy. The
mean follow-up period was 108 (range, 26–182) months. Local
recurrence was observed in 1 patient at 48 months after surgery,
and distant metastasis was observed in 11 patients at a mean
follow-up period of 23 (range, 10–37) months. In total, 6 patients
died of disease at a mean follow-up period of 49 (range, 26–90)
months. At the final follow-up, a total of 19 patients were free
from disease (Table I).
 | Table I.Patient characteristics (n=30). |
Table I.
Patient characteristics (n=30).
| Patient
characteristic | Value |
|---|
| Mean age (range),
years | 24 (10–68) |
| Sex, n |
|
|
Male | 20 |
|
Female | 10 |
| Mean follow-up
period (range), months | 108 (26–182) |
| Tumor location,
n |
|
|
Pelvis | 3 |
|
Femur | 17 |
|
Tibia | 10 |
| Mean tumor size
(range), cm | 9.7 (6–18) |
| Tumor stage, n |
|
|
IIA | 13 |
|
IIB | 14 |
|
III | 3 |
| Treatment, n |
|
|
Surgery | 30 |
|
Chemotherapy (neoadjuvant +
adjuvant) | 27 |
|
Neoadjuvant | 28 |
|
Doxorubicin +
cisplatin + MTX | 19 |
|
Doxorubicin +
cisplatin + ifosfamide | 9 |
|
Adjuvant | 28 |
|
Radiotherapy | 2 |
| Clinical outcome,
n |
|
|
Progression-free | 18 |
| Local
recurrence | 1 |
| Distant
metastasis | 11 |
| Oncological outcome
at final follow-up, n |
|
|
DOD | 6 |
|
AWD | 5 |
| Disease-free | 19 |
The characteristics of the patients with other types
of bone tumors were as follows: i) 8 male and 7 female patients
with a mean age of 27 (range, 6–48) years old with osteochondroma;
ii) 7 male and 4 female patients with a mean age of 42 (range,
11–75) years old with enchondroma; iii) 7 male and 3 female
patients with a mean age of 31 (range, 12–68) years old with
fibrous dysplasia; iv) 15 male and 13 female patients with a mean
age of 35 (range, 10–67) years old with giant cell tumor of the
bone; v) 3 male and 2 female patients with a mean age of 52 (range,
36–63) years old with synovial chondromatosis; vi) 1 male and 2
female patients with a mean age of 16 (range, 15–18) years old with
osteoblastoma; vii) 19 male and 7 female patients with a mean age
of 27 (range, 8–65) years old with Ewing sarcoma; viii) 6 male and
2 female patients with a mean age of 60 (range, 26–92) years old
with chordoma; and ix) 3 male and 3 female patients with a mean age
of 48 (range, 19–76) years old with chondrosaroma [Grade 2–3;
graded using the WHO Classification of Tumors, Soft Tissue and Bone
Tumors, 5th edition (12)]. The
healthy controls were 150 male and 125 female individuals with a
mean age of 52 (range, 35–80) years old.
Survival rates
The optimal cut-off points and areas under the curve
of the miRNAs (miR-146a-5p, miR-1260a, miR-487b-3p, miR-6720-3p,
miR-1260b, miR-4758-3p, miR-4690-3p, miR-4286, miR-6765-3p,
miR-1261, miR-7975, miR-4664-5p and miR-3907) for predicting death
from disease, distant metastasis or freedom from disease are shown
in Table II.
 | Table II.Optimal cut-off values, AUC,
sensitivity and specificity of variables to predict death from
disease, distant metastasis or freedom from disease. |
Table II.
Optimal cut-off values, AUC,
sensitivity and specificity of variables to predict death from
disease, distant metastasis or freedom from disease.
|
| Overall survival
rate | Distant
metastasis-free survival rate | Disease-free
survival rate |
|---|
|
|
|
|
|
|---|
| miRNA | Cut-off value | AUC | Sensitivity, % | Specificity, % | Cut-off value | AUC | Sensitivity, % | Specificity, % | Cut-off value | AUC | Sensitivity, % | Specificity, % |
|---|
| miR-146a-5p | 6.621 | 0.881 | 100 | 67 | 6.693 | 0.718 | 80 | 59 | 6.693 | 0.694 | 80 | 59 |
| miR-1260a | 6.621 | 0.881 | 100 | 62 | 6.693 | 0.753 | 90 | 59 | 6.693 | 0.741 | 90 | 59 |
| miR-487b-3p | 6.621 | 0.889 | 100 | 67 | 6.693 | 0.741 | 90 | 59 | 6.621 | 0.682 | 70 | 65 |
| miR-6720-3p | 6.695 | 0.833 | 100 | 62 | 6.695 | 0.629 | 70 | 59 | 6.695 | 0.747 | 80 | 65 |
| miR-1260b | 7.735 | 0.802 | 100 | 48 | 6.693 | 0.694 | 60 | 82 | 6.693 | 0.724 | 60 | 82 |
| miR-4758-3p | 6.621 | 0.865 | 100 | 62 | 6.693 | 0.735 | 90 | 59 | 6.693 | 0.782 | 90 | 59 |
| miR-4690-3p | 7.849 | 0.633 | 100 | 40 | 7.346 | 0.510 | 78 | 59 | 7.346 | 0.438 | 70 | 56 |
| miR-4286 | 7.378 | 0.579 | 100 | 48 | 7.378 | 0.653 | 90 | 53 | 7.378 | 0.700 | 90 | 53 |
| miR-6765-3p | 6.285 | 0.746 | 67 | 81 | 6.693 | 0.688 | 80 | 59 | 6.693 | 0.612 | 70 | 53 |
| miR-1261 | 9.984 | 0.675 | 83 | 62 | 9.984 | 0.565 | 60 | 59 | 9.984 | 0.665 | 70 | 65 |
| miR-7975 | 6.621 | 0.778 | 100 | 57 | 6.693 | 0.671 | 90 | 53 | 6.693 | 0.724 | 90 | 53 |
| miR-4664-5p | 6.019 | 0.762 | 67 | 81 | 6.621 | 0.612 | 70 | 59 | 6.019 | 0.659 | 50 | 82 |
| miR-3907 | 6.621 | 0.841 | 100 | 57 | 6.693 | 0.700 | 90 | 53 | 6.693 | 0.753 | 90 | 53 |
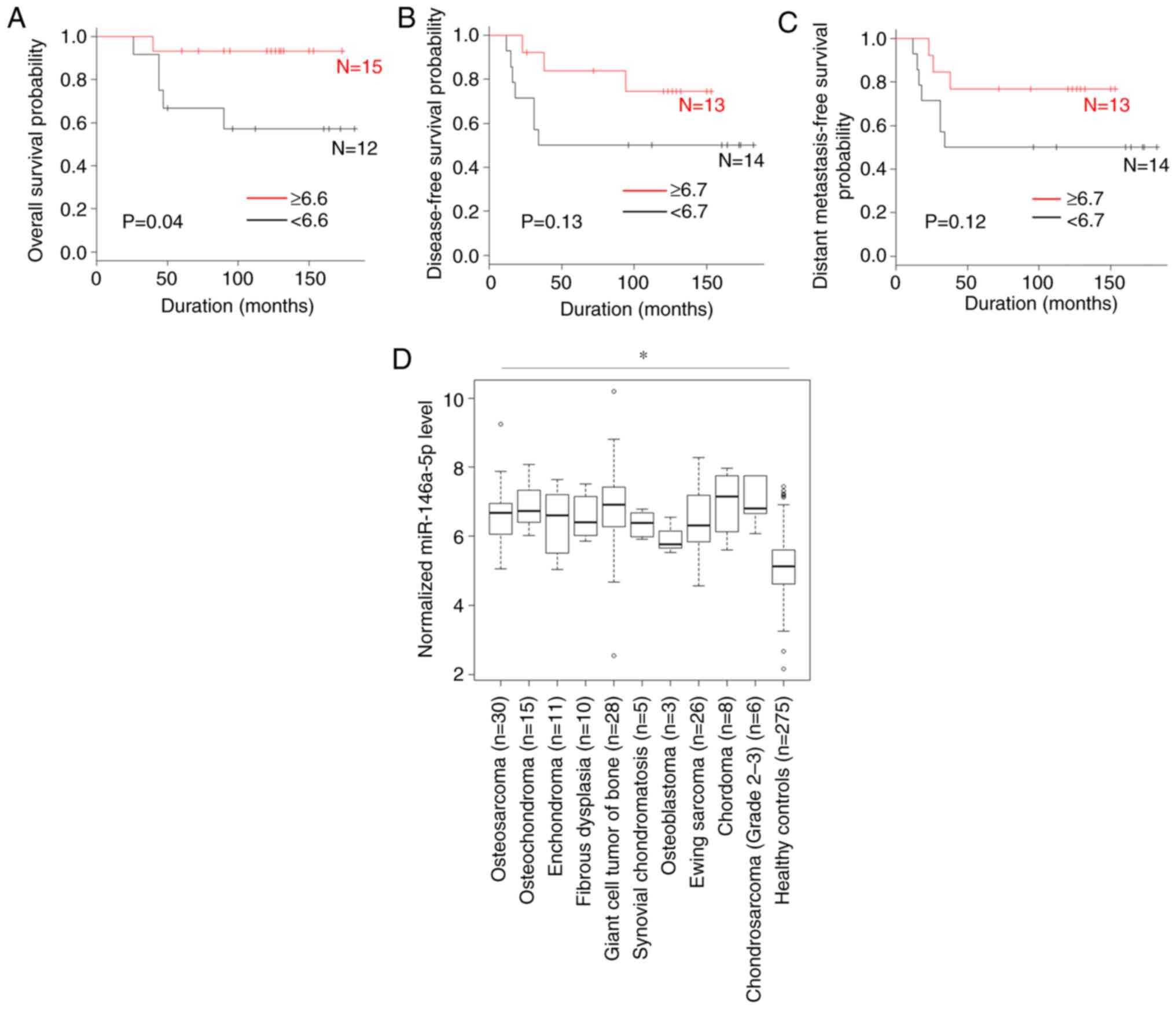
A comparison of the overall survival according to
miRNA levels is shown in Figs. 1A
and 2. The patients with high serum
levels of miR-146a-5p (P=0.04), miR-1260a (P=0.04) and miR-487b-3p
(P=0.04) were significantly associated with an improved overall
survival compared with those with low levels. By contrast, the
serum levels of miR-6720-3p, miR-1260b, miR-4758-3p, miR-4690-3p,
miR-4286, miR-6765-3p, miR-1261, miR-7975, miR-4664-5p and miR-3907
were not associated with the overall survival.
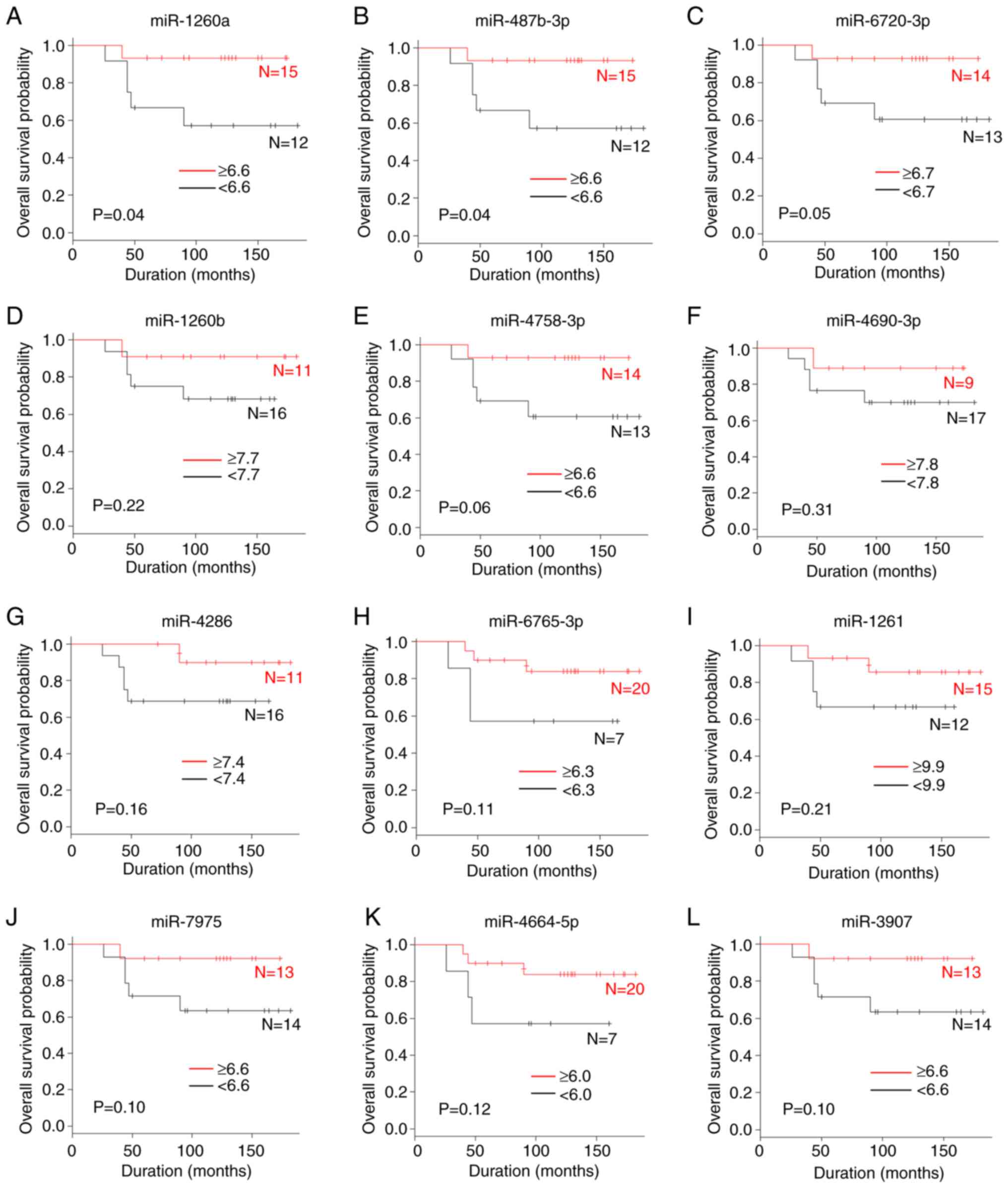 | Figure 2.A comparison of the overall survival
rate according to miRNA levels. (A) miR-1260a, (B) miR-487b-3p, (C)
miR-6720-3p, (D) miR-1260b, (E) miR-4758-3p, (F) miR-4690-3p, (G)
miR-4286, (H) miR-6765-3p, (I) miR-1261, (J) miR-7975, (K)
miR-4664-5p and (L) miR-3907. miR, microRNA. |
A comparison of the distant metastasis-free survival
according to miRNA values is shown in Figs. 1B and 3. The patients with high serum levels of
miR-1260a (P=0.04), miR-487b-3p (P=0.04), miR-1260b (P=0.03) and
miR-4758-3p (P=0.04) were significantly associated with an improved
distant metastasis-free survival compared with those with low
levels. By contrast, the serum levels of miR-146a-5p, miR-6720-3p,
miR-4690-3p, miR-4286, miR-6765-3p, miR-1261, miR-7975, miR-4664-5p
and miR-3907 were not associated with the distant metastasis-free
survival.
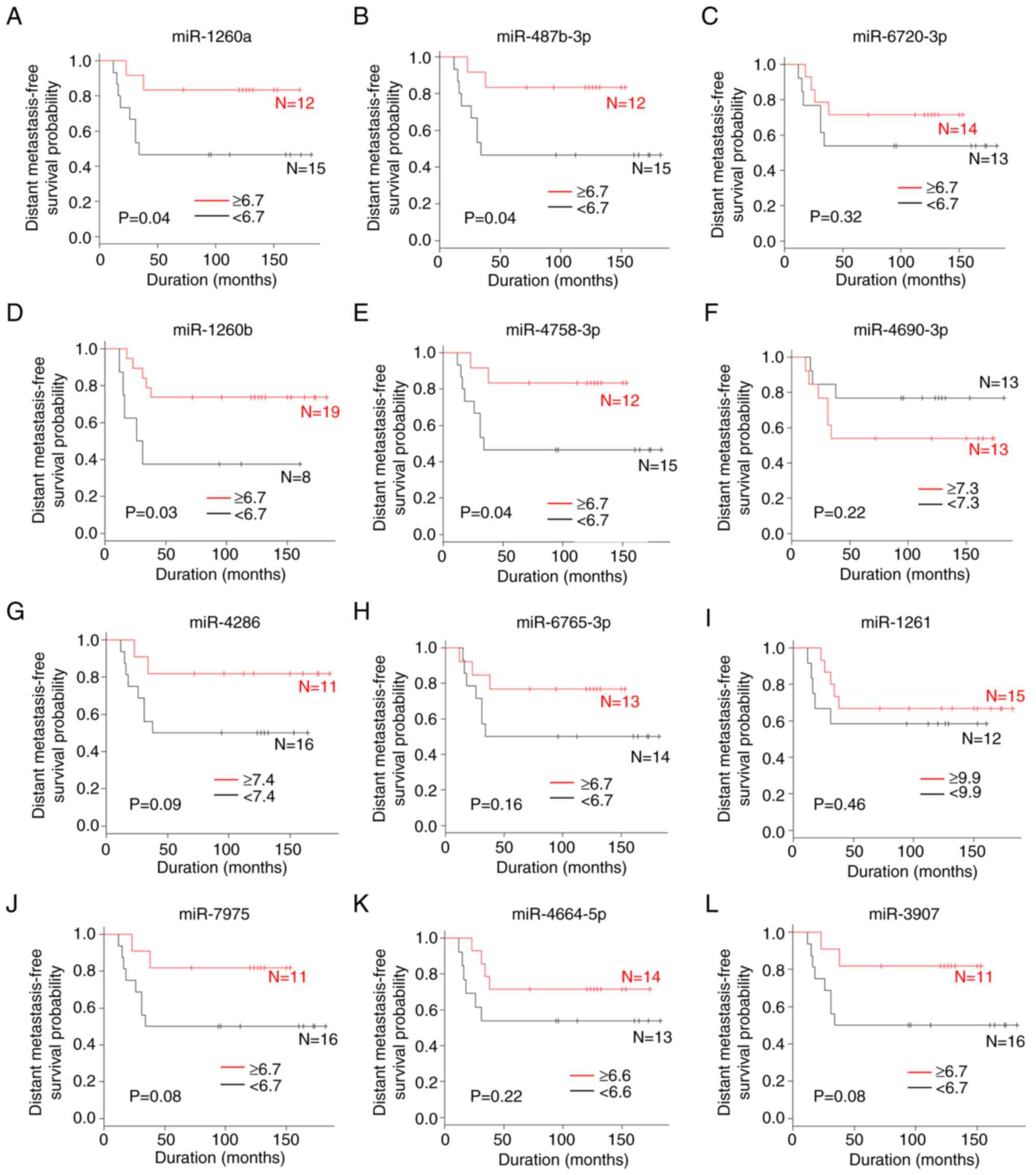 | Figure 3.A comparison of the distant
metastasis-free survival rate according to miRNA levels. (A)
miR-1260a, (B) miR-487b-3p, (C) miR-6720-3p, (D) miR-1260b, (E)
miR-4758-3p, (F) miR-4690-3p, (G) miR-4286, (H) miR-6765-3p, (I)
miR-1261, (J) miR-7975, (K) miR-4664-5p and (L) miR-3907. miR,
microRNA. |
A comparison of the disease-free survival according
to miRNA level is shown in Figs. 1C
and 4. The patients with high serum
levels of miR-1260a (P=0.04), miR-1260b (P=0.02) and miR-4758-3p
(P=0.04) were significantly associated with an improved
disease-free survival compared with those with low levels. By
contrast, the serum levels of miR-146a-5p, miR-487b-3p,
miR-6720-3p, miR-4690-3p, miR-4286, miR-6765-3p, miR-1261,
miR-7975, miR-4664-5p and miR-3907 were not associated with the
disease-free survival.
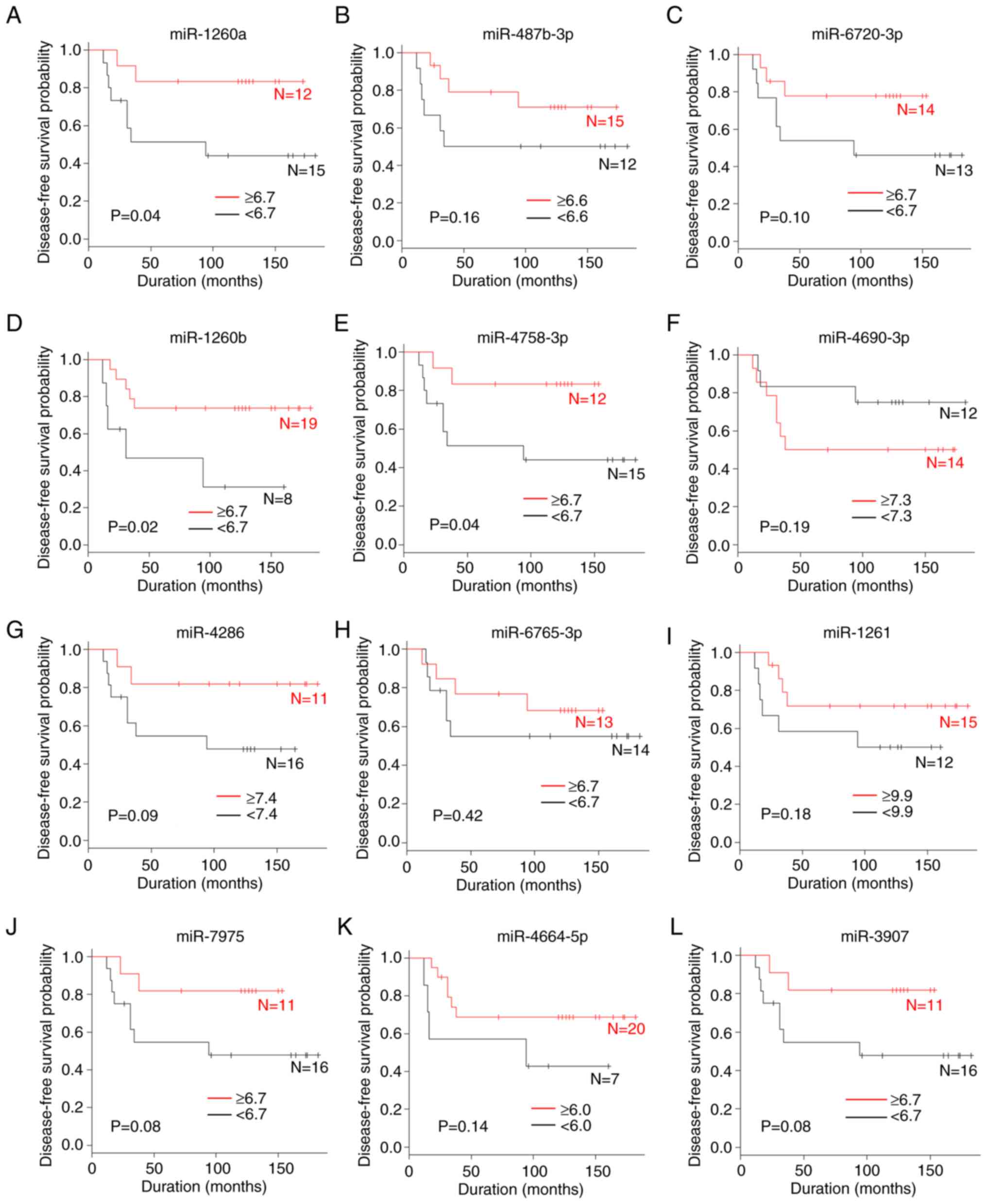 | Figure 4.A comparison of the disease-free
survival rate according to miRNA levels. (A) miR-1260a, (B)
miR-487b-3p, (C) miR-6720-3p, (D) miR-1260b, (E) miR-4758-3p, (F)
miR-4690-3p, (G) miR-4286, (H) miR-6765-3p, (I) miR-1261, (J)
miR-7975, (K) miR-4664-5p and (L) miR-3907. miR, microRNA. |
Differentiation from other types of
bone tumors
A comparison of serum miRNAs levels in patients with
bone tumors and healthy controls is shown in Figs. 1D and 5. The levels of serum miR-1261 in patients
with osteosarcoma were significantly higher than those in the
healthy controls (P<0.05) and patients with benign
(osteochondroma, P<0.05) or intermediate-grade bone tumor (giant
cell tumor of the bone, P<0.05). In addition, the levels of
serum miR-1261 tended to be lower for patients with other benign
bone tumors (enchondroma and fibrous dysplasia) and other
intermediate-grade bone tumors (synovial chondromatosis and
osteoblastoma) compared with patients with osteosarcoma, although
significant differences were not observed. The serum levels of
miR-1261 were not significantly associated with those in patients
with other high-grade bone tumors. By contrast, the serum levels of
miR-146a-5p, miR-1260a, miR-487b-3p, miR-6720-3p, miR-1260b,
miR-4758-3p, miR-4690-3p, miR-4286, miR-6765-3p, miR-7975,
miR-4664-5p and miR-3907 in patients with osteosarcoma were not
markedly different from those in patients with benign bone tumors,
intermediate-grade bone tumors or other high-grade bone tumors,
although the levels were significantly higher than those in healthy
controls (P<0.05).
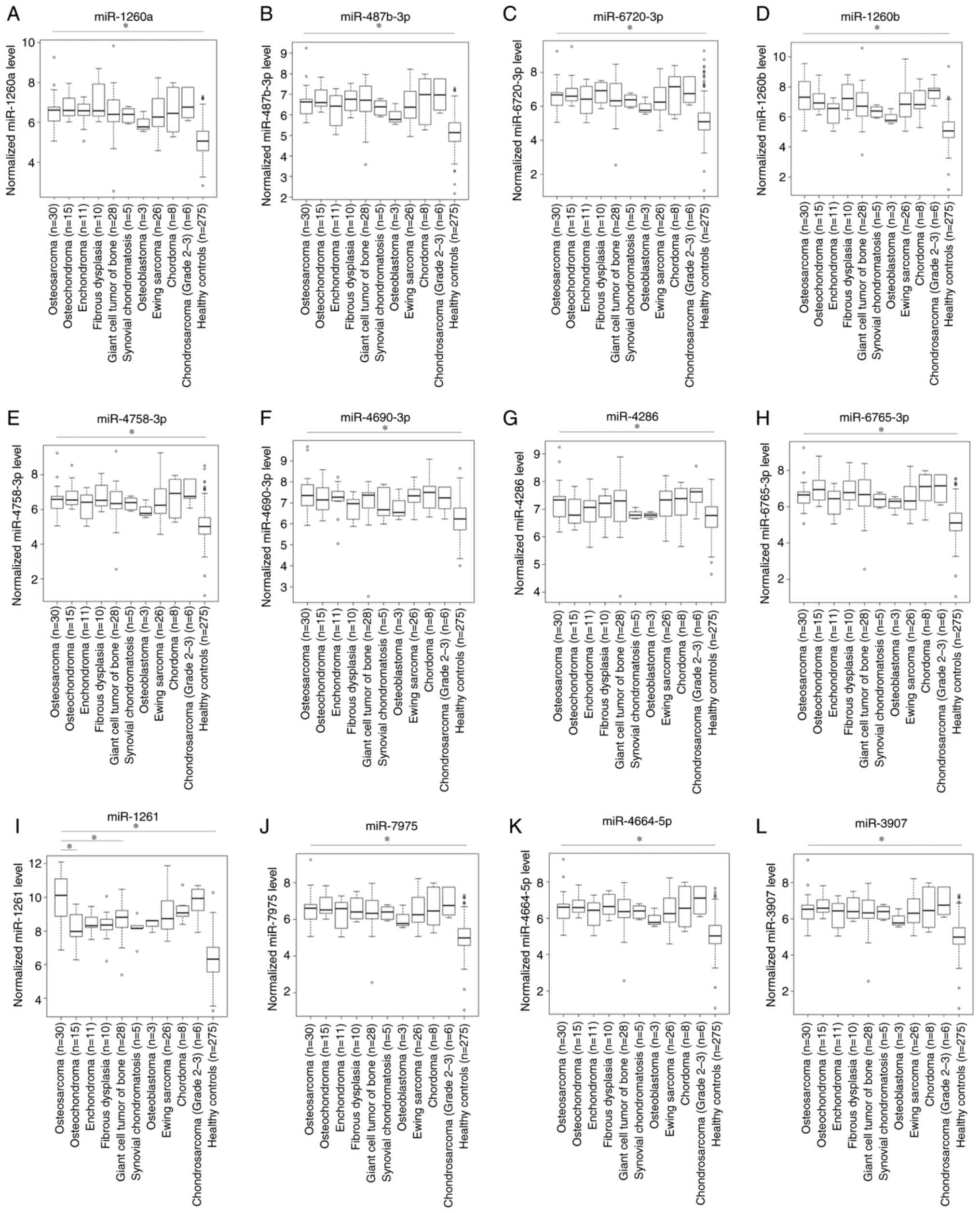 | Figure 5.A comparison of serum miRNAs levels
in patients with bone tumors and healthy controls. (A) miR-1260a,
(B) miR-487b-3p, (C) miR-6720-3p, (D) miR-1260b, (E) miR-4758-3p,
(F) miR-4690-3p, (G) miR-4286, (H) miR-6765-3p, (I) miR-1261, (J)
miR-7975, (K) miR-4664-5p and (L) miR-3907. *P<0.05. miR,
microRNA. |
Effect of miR-1261 on cellular
viability and proliferation
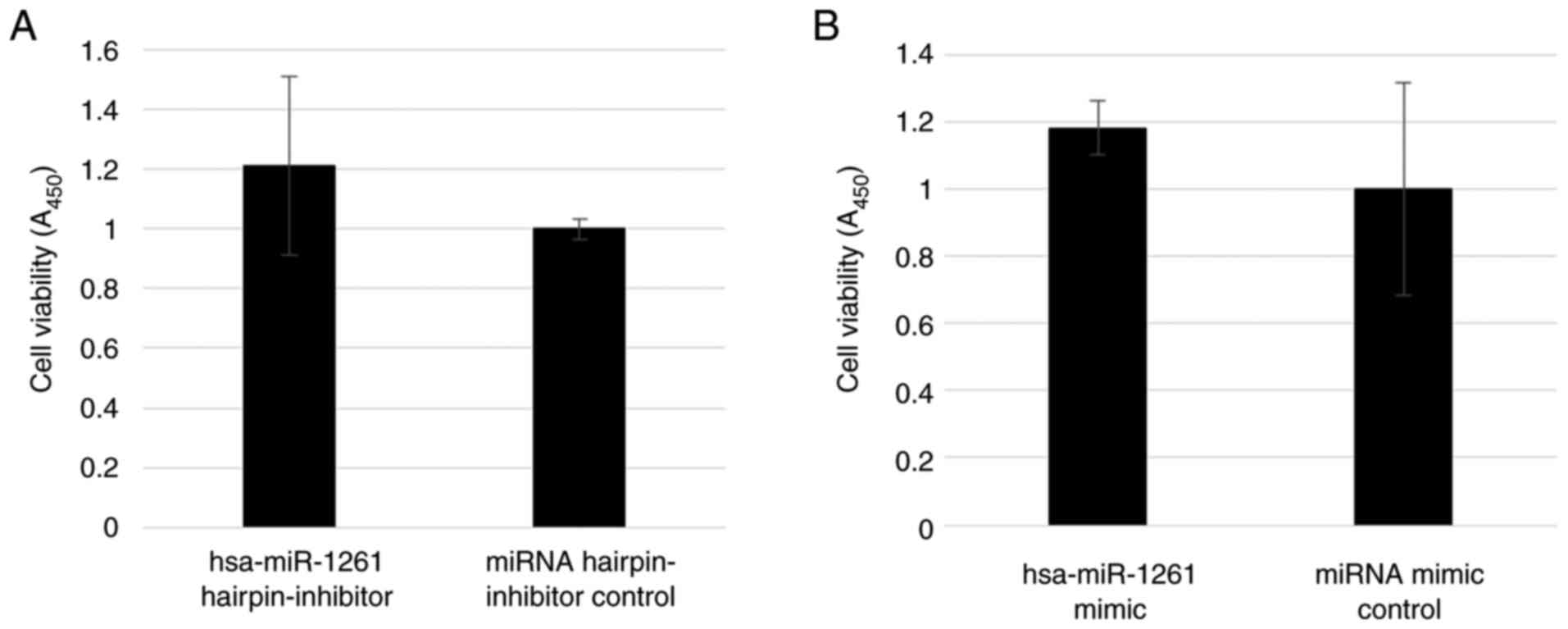
The inhibition of miR-1261 in 143B cells did not
suppress cell viability (P=0.35; Fig.
6A), and the addition of miR-1261 to HOS cells did not promote
cell viability (P=0.42; Fig. 6B).
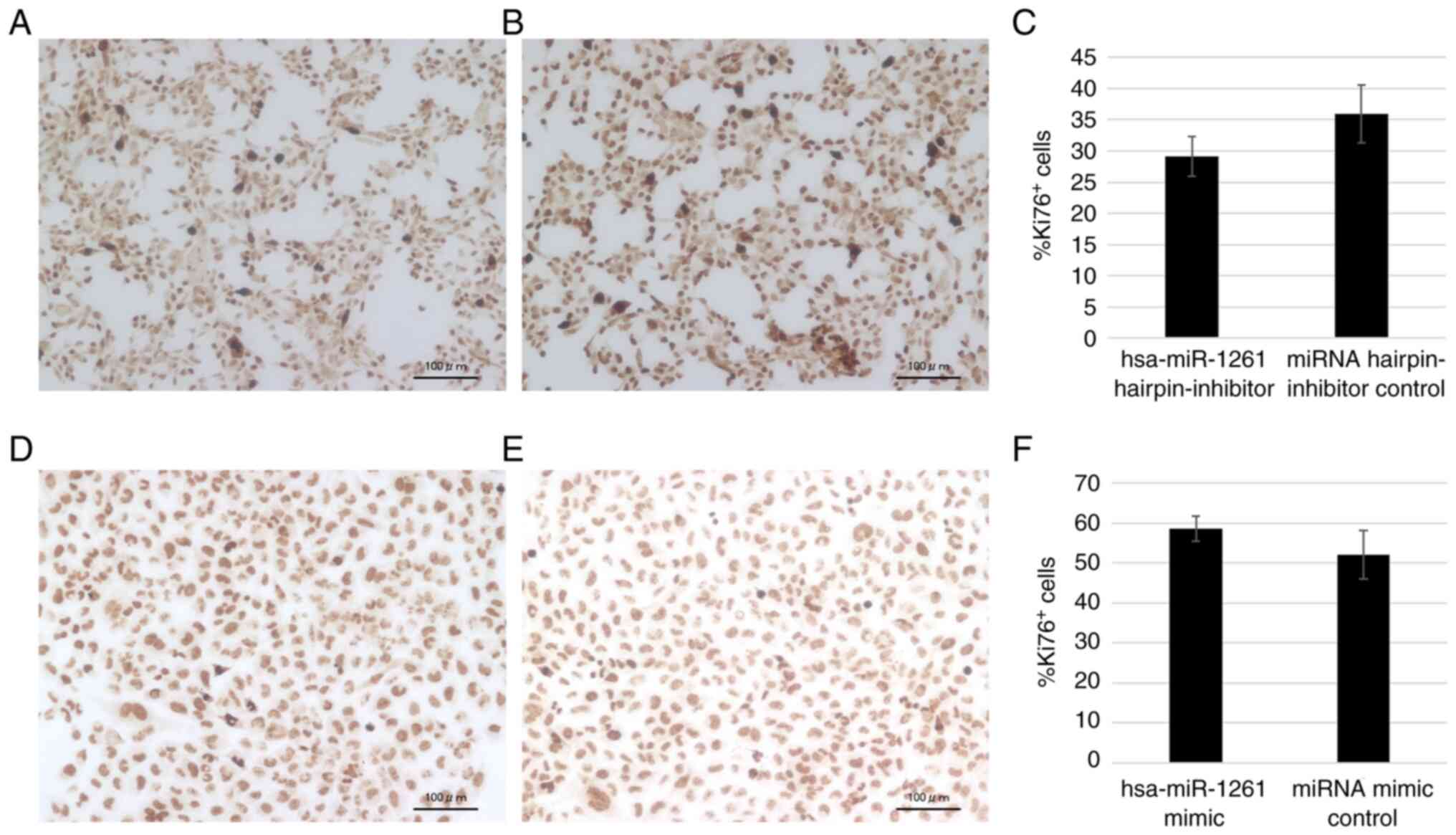
The amount of Ki-67+ 143B cells appeared to decrease
with the inhibition of miR-1261 (Fig.
7A-C), and the amount of Ki-67+ HOS cells appeared
to increase with the transfection of miR-1261 (Fig. 7D-F), although neither assay showed a
statistically significant difference (P=0.11 and P=0.19,
respectively).
Effect of miR-1261 on target
genes
In reference to the list of predicted targets for
hsa-miR-1261 (32), three genes
associated with osteosarcoma were selected for screening, cadherin
6 (CDH6) (33), kinesin family
member 26B (KIF26B) (34) and SET
domain containing 2, histone lysine methyltransferase (SETD2)
(35,36). The inhibition of miR-1261 in 143B
cells significantly increased the mRNA levels of CDH6 and SETD2,
but there was no significant difference in the mRNA level of KIF26B
(Fig. 8A). The addition of miR-1261
mimic to HOS cells significantly decreased the mRNA levels of
KIF26B and SETD2, but there was no significant difference in the
mRNA levels of CDH6 (Fig. 8B).
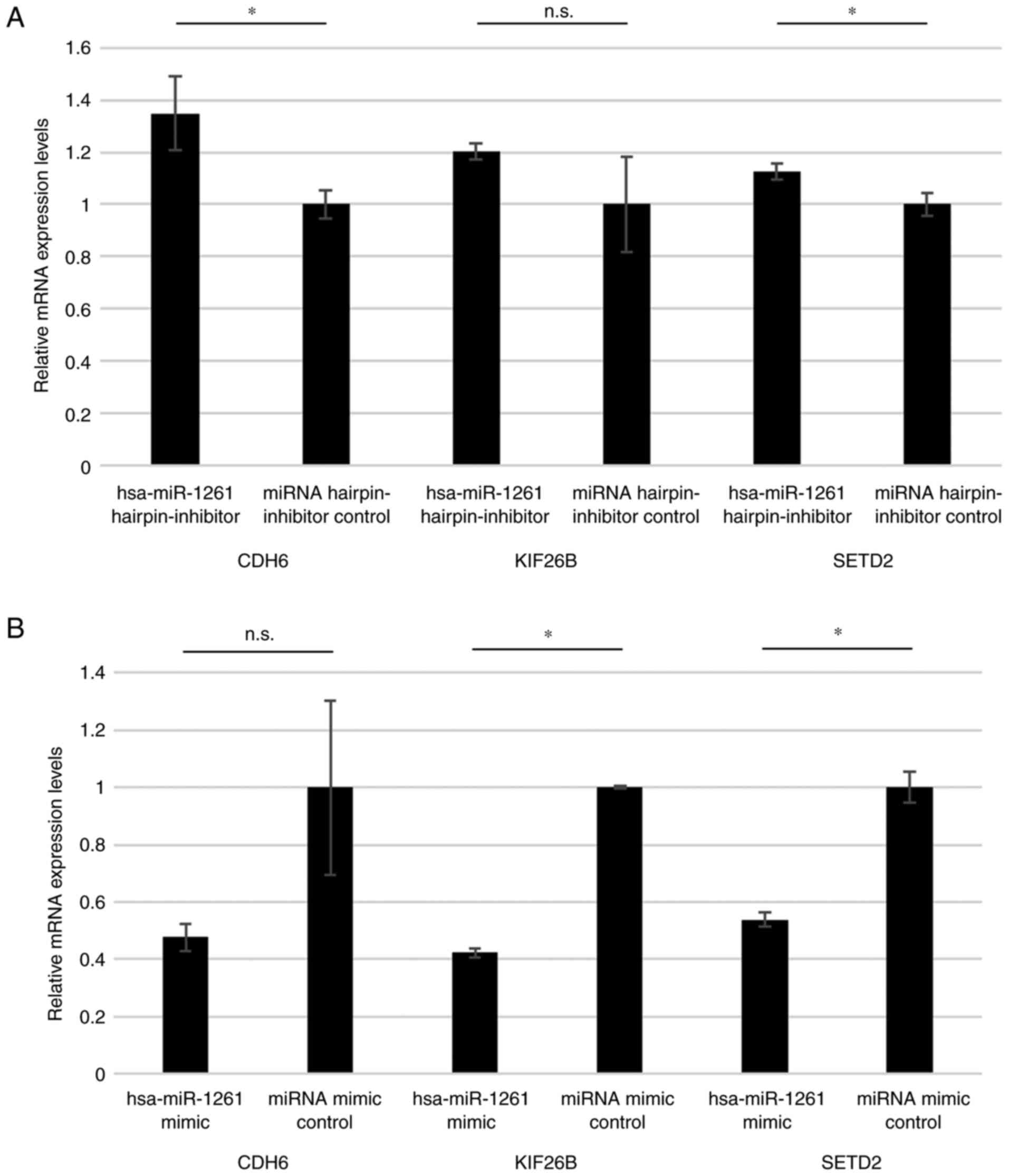 | Figure 8.Reverse transcription-quantitative
PCR of miR-1261 target genes. (A) mRNA levels of CDH6, KIF26B and
SETD2 after the addition of has-miR-1261 hairpin-inhibitor and
miRNA hairpin-inhibitor control in 143B cells. (B) mRNA levels of
CDH6, KIF26B and SETD2 after the addition of has-miR-1261 mimic and
miRNA mimic control in HOS cells. *P<0.05. CDH6, cadherin 6;
KIF26B, kinesin family member 26B; miR, microRNA; n.s., no
significant difference; SETD2, SET domain containing 2, histone
lysine methyltransferase. *P<0.05. miR, microRNA.; n.s., no
significant difference. |
Discussion
The present study is a retrospective observational
study to validate the utility of 13 serum miRNA levels as
prognostic and diagnostic markers in the clinical setting using
pretreatment patient data, as was first noted in our previous basic
study (26). The patients with
osteosarcoma with high serum levels of several types of miRNA
(miR-146a-5p, miR-1260a, miR-487b-3p, miR-1260b and miR-4758-3p)
exhibited an improved survival rate compare with those with low
levels. In particular, patients with high serum levels of miR-1260a
exhibited an improved overall survival rate, metastasis-free
survival rate and disease-free survival rate compared with those
with low levels. Of further note, patients with osteosarcoma
exhibited higher levels of serum miR-1261 than patients with benign
or intermediate-grade bone tumors.
In our previous in vivo and in vitro
study, highly aggressive osteosarcoma advanced locally and promoted
metastasis via the secretion of large number of small extracellular
vesicles (26). 143B cells,
k-ras-activated human osteosarcoma cells with the capacity to
metastasize, secreted greater amounts of small extracellular
vesicles (exosomes) than HOS cells, which are k-ras-wild-type human
osteosarcoma cells with a low metastatic potential. The
tumor-derived exosomes contained various types of miRNAs, including
miR-146a-5p (37,38). miR-146a-5p suppressed
osteoclastogenesis in the tumor bone microenvironment, and patients
with osteosarcoma with few mature osteoclasts on histology of a
biopsy specimen exhibited a worse prognosis and higher rates of
lung metastasis than those with numerous mature osteoclasts
(19,26). Thus, a poor prognosis was expected
in those patients with high levels of miR-146a-5p compared with
those with low levels. The other 12 miRNAs described in the present
study were also identified in the k-ras-activated 143B cells ≥6
times more frequently than in the k-ras-wildtype HOS cells in our
previous study (26). Similarly,
high levels of the other 12 miRNAs were expected to reflect a poor
prognosis.
However, the present study demonstrated unexpectedly
contrary outcomes concerning the prognosis for patients with high
levels of several types of miRNAs (miR-146a-5p, miR-1260a,
miR-487b-3p, miR-1260b and miR-4758-3p). For example, patients with
high serum levels of these miRNAs had an improved survival rate
compared with those with low levels, possibly due to the small
number of patients included in the analysis, the different cut-off
values of miRNAs used, the decomposition of miRNAs in the serum
(39) or the inconsistency in miRNA
levels in the serum and tumor tissues.
The upregulation of miR-1260a was previously
reported to be a biomarker of lung adenocarcinoma (40). However, the serum levels of
miR-1260a were shown to be decreased in ovarian and prostate cancer
and increased in the postoperative state for metastatic melanoma
(41–43). Furthermore, miR-1260a was also
reported to improve osteogenesis and angiogenesis (44), so high serum levels of this miRNA
may contribute to an improved survival rate and have potential
utility as a prognostic marker for some types of cancer, including
osteosarcoma. In a recent study, 143B cells had higher levels of
miR-1260a than HOS cells (26),
however, high-grade malignant cells may also generate some miRNAs
that have a health benefit, not just those that negatively
influence health (45–48). Serum miR-1260a may be a potential
prognostic marker for patients with osteosarcoma, although a
further investigation is required to validate this.
Numerous miRNAs have been reported to be associated
with the diagnosis and prognosis of patients with cancer, but very
few miRNAs are clinically used as specific markers for the disease
(49). One reason for this might be
due to the diverse functions of the miRNAs, depending on the
disease. miR-487b-3p improves chemoresistance or metastasis in
osteosarcoma but impairs osteoblastgenesis, and promotes
tumorigenesis in colon cancer, prostate cancer and glioma (50–54).
miR-1260b promotes tumorigenesis in breast cancer and lung
adenocarcinoma and also inhibits bone loss (55–57).
miR-4758-3p promotes tumorigenesis in glial tumor, gastric cancer
and endometrial tumors (58–60),
but its association with osteosarcoma is unknown.
In the human body, normal cells located around tumor
cells might absorb some tumor-derived miRNAs that negatively
influence cell survival (45). As a
result of this, only a small amount of the tumor-derived miRNAs
that exist in the stromal tissue enter the bloodstream. Some miRNAs
have also been shown to be secreted by normal cells, including
inflammatory cells, in response to the presence of tumor cells
(46–48). Thus, identifying the origin of serum
miRNAs might be difficult, and further investigations are required
to confirm the utility of the 13 miRNAs analyzed in the present
study as prognostic markers in the clinical setting.
miR-1261 has been reported to be associated with
migration and invasion of prostate cancer, progression of lung
adenocarcinoma and as a diagnostic marker of hepatocellular
carcinoma (61–64). However, its association with
osteosarcoma is unclear. In the present study, the serum levels of
miR-1261 in patients with osteosarcoma were significantly higher
than those in healthy controls, patients with benign bone tumors
and patients with intermediate-grade bone tumors, although there
were no significant differences between patients with osteosarcoma
and those of patients with other high-grade bone tumors. The
presence of histological high-grade malignancies, including
osteosarcoma, might be reflected by the high serum levels of
miR-1261 in patients with bone tumors. In the present study, in
vitro experiments were performed to investigate the effect of
miR-1261 on cellular viability and proliferation. Following the
transfection of HOS cells with an miR-1261 mimic and 143B cells
with an miR-1261 hairpin-inhibitor, the cell viabilities were
similar, but cell proliferation appeared to be associated with
miR-1261. Furthermore, the mRNA levels of CDH6, KIF26B and SETD2
[target genes of miR-1261 (32)]
increased by inhibiting miR-1261 and, by contrast, these target
genes decreased upon the addition of an miR-1261 mimic. The SETD2
gene has been described as a tumor suppressor in various types of
cancers, including renal, gastric and lung cancer (65,66).
However, there are only a few studies associated with osteosarcoma,
and the involvement of miR-1261 and SETD2 has not yet been
previously described (35,36,65,66).
Thus, miR-1261 is potentially a novel therapeutic target in
addition to being useful for differentiating whether a bone tumor
is high-grade, although a further investigation is necessary.
Several limitations associated with the present
study warrant mention. Firstly, this was a retrospective,
small-scale, single-institution study. Secondly, the peripheral
blood findings were not compared between the preoperative and
postoperative periods. Changes in the levels of miRNA after surgery
may indicate tumor-derived miRNAs (21–24),
but, in the present study, the postoperative blood findings for
miRNAs were not investigated as the timing of surgery differed
among patients. Thirdly, the characteristics concerning age and sex
in healthy patients and patients with other types of bone tumors
were not compared with those in patients with osteosarcoma since
there was only a small number of patients, and also because the
predominance of age and sex depends on the histological type of
bone tumors (12).
In conclusion, regarding the 13 serum miRNAs
evaluated in the present study, serum miR-1260a may be a potential
prognostic marker for patients with osteosarcoma. Patients with
osteosarcoma had higher levels of serum miR-1261 than those with
benign or intermediate-grade bone tumors, which may be a potential
novel therapeutic target in addition to being useful for
differentiating whether or not a bone tumor is high-grade. A larger
investigation is required to clarify the utility of these miRNAs in
the clinical setting.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Dr Akihiko Yoshida
(Department of Diagnostic Pathology, The National Cancer Center
Hospital, Tokyo, Japan) for performing histological examination of
all the specimens.
Funding
This research was supported by The JSPS Fujita Memorial Fund for
Medical Research (2021) and Grants-in-Aid for Scientific Research
(KAKENHI) from The Ministry of Education, Culture, Sports, Science
and Technology (MEXT; grant no. 22K16712).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HT, NY, AK, NA and YAr conceived and designed the
study. YAr carried out data acquisition. AK and NA provided
assistance for data acquisition. AK and NA managed the patients for
the appropriate treatment and observed them at the follow-up
outpatient clinic. YAr and TY performed the in vitro
experiment. YAr, TY, RH, KH, AT, SMi, KI, TH, KA, HY, SMo and YAs
analyzed and interpretated all the patient's data. HT, NY, JM, TO
and YT contributed to the analysis of the data and critical
appraisal. YAr, TY, NA and NY confirm the authenticity of all the
raw data. YAr wrote the manuscript. KH, AT, SMi, KI, TH, KA, HY,
SMo and YAs supervised the analysis. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
All patients underwent standard treatment during the
follow-up period at The National Cancer Center Hospital in Tokyo,
Japan. The present study was conducted in accordance with the 1975
Declaration of Helsinki. It was approved by The NCCH Institutional
Review Board (Tokyo, Japan; approval nos. 2004-050, 2013-111 and
2015-266). Written informed consent was obtained from each
participant and/or their family.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
miRNA
|
microRNA
|
|
AJCC
|
American Joint Committee on Cancer
|
|
ROC
|
receiver operating characteristic
|
|
NCCH
|
National Cancer Center Hospital
|
|
NCCN
|
National Comprehensive Cancer
Network
|
References
|
1
|
Saliminejad K, Khorram Khorshid HR,
Soleymani Fard S and Ghaffari SH: An overview of microRNAs:
Biology, functions, therapeutics, and analysis methods. J Cell
Physiol. 234:5451–5465. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rupaimoole R, Calin GA, Lopez-Berestein G
and Sood AK: miRNA deregulation in cancer cells and the tumor
microenvironment. Cancer Discov. 6:235–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang
G, Song J, Li Z, Zhang Z and Yuan W: Effect of exosomal miRNA on
cancer biology and clinical applications. Mol Cancer. 17:1472018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pontecorvi G, Bellenghi M, Puglisi R, Carè
A and Mattia G: Tumor-derived extracellular vesicles and microRNAs:
Functional roles, diagnostic, prognostic and therapeutic options.
Cytokine Growth Factor Rev. 51:75–83. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tang Z, Li D, Hou S and Zhu X: The cancer
exosomes: Clinical implications, applications and challenges. Int J
Cancer. 146:2946–2959. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen Q, Li Y, Liu Y, Xu W and Zhu X:
Exosomal Non-coding RNAs-Mediated crosstalk in the tumor
microenvironment. Front Cell Dev Biol. 9:6468642021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McGuire A, Brown JA and Kerin MJ:
Metastatic breast cancer: The potential of miRNA for diagnosis and
treatment monitoring. Cancer Metastasis Rev. 34:145–155. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ishikawa D, Yoshikawa K, Takasu C,
Kashihara H, Nishi M, Tokunaga T, Higashijima J and Shimada M:
Expression level of microRNA-449a predicts the prognosis of
patients with gastric cancer. Anticancer Res. 40:239–244. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Amankwah EK, Devidas M, Teachey DT, Rabin
KR and Brown PA: Six candidate miRNAs associated with early relapse
in pediatric B-cell acute lymphoblastic leukemia. Anticancer Res.
40:3147–3153. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu H, Guan Z, Cuk K, Brenner H and Zhang
Y: Circulating microRNA biomarkers for lung cancer detection in
Western populations. Cancer Med. 7:4849–4862. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J,
Zhou K, Liu X, Ren X, Wang F, et al: Cancer-derived exosomal
miR-25-3p promotes pre-metastatic niche formation by inducing
vascular permeability and angiogenesis. Nat Commun. 9:53952018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
WHO Classification of Tumours Editorial
Board, . WHO Classification of Tumours: Soft Tissue and Bone
Tumours. 5th edition. IARC; Lyon: pp. 403–409. 2020
|
|
13
|
Reed DR, Hayashi M, Wagner L, Binitie O,
Steppan DA, Brohl AS, Shinohara ET, Bridge JA, Loeb DM, Borinstein
SC and Isakoff MS: Treatment pathway of bone sarcoma in children,
adolescents, and young adults. Cancer. 123:2206–2218. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tsukamoto S, Errani C, Angelini A and
Mavrogenis AF: Current treatment considerations for osteosarcoma
metastatic at presentation. Orthopedics. 43:e345–e358. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Araki Y, Yamamoto N, Hayashi K, Takeuchi
A, Miwa S, Igarashi K, Higuchi T, Abe K, Taniguchi Y, Yonezawa H,
et al: Delayed initiation of treatment is associated with
metastasis of malignant bone tumor. Anticancer Res. 41:2993–2999.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hao H, Chen L, Huang D, Ge J, Qiu Y and
Hao L: Meta-analysis of alkaline phosphatase and prognosis for
osteosarcoma. Eur J Cancer Care (Engl). 26:e125362017. View Article : Google Scholar
|
|
17
|
Araki Y, Yamamoto N, Hayashi K, Takeuchi
A, Miwa S, Igarashi K, Higuchi T, Abe K, Taniguchi Y, Yonezawa H,
et al: Pretreatment neutrophil count and platelet-lymphocyte ratio
as predictors of metastasis in patients with osteosarcoma.
Anticancer Res. 42:1081–1089. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ding WZ, Liu K, Li Z and Chen SR: A
meta-analysis of prognostic factors of osteosarcoma. Eur Rev Med
Pharmacol Sci. 24:4103–4112. 2020.PubMed/NCBI
|
|
19
|
Araki Y, Yamamoto N, Hayashi K, Takeuchi
A, Miwa S, Igarashi K, Higuchi T, Abe K, Taniguchi Y, Yonezawa H,
et al: The number of osteoclasts in a biopsy specimen can predict
the efficacy of neoadjuvant chemotherapy for primary osteosarcoma.
Sci Rep. 11:19892021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang J, Liu S, Shi J, Li J, Wang S, Liu H,
Zhao S, Duan K, Pan X and Yi Z: The Role of miRNA in the diagnosis,
prognosis, and treatment of osteosarcoma. Cancer Biother
Radiopharm. 34:605–613. 2019.PubMed/NCBI
|
|
21
|
Raimondi L, De Luca A, Gallo A, Costa V,
Russelli G, Cuscino N, Manno M, Raccosta S, Carina V, Bellavia D,
et al: Osteosarcoma cell-derived exosomes affect tumor
microenvironment by specific packaging of microRNAs.
Carcinogenesis. 41:666–677. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang C, Sun Y, Ma S, Vadamootoo AS, Wang
L and Jin C: Identification of circulating miR-663a as a potential
biomarker for diagnosing osteosarcoma. Pathol Res Pract.
215:1524112019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fujiwara T, Uotani K, Yoshida A, Morita T,
Nezu Y, Kobayashi E, Yoshida A, Uehara T, Omori T, Sugiu K, et al:
Clinical significance of circulating miR-25-3p as a novel
diagnostic and prognostic biomarker in osteosarcoma. Oncotarget.
8:33375–33392. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang C, Wang Q, Ma S, Sun Y, Vadamootoo
AS and Jin C: A four serum-miRNA panel serves as a potential
diagnostic biomarker of osteosarcoma. Int J Clin Oncol. 24:976–982.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Asano N, Matsuzaki J, Ichikawa M, Kawauchi
J, Takizawa S, Aoki Y, Sakamoto H, Yoshida A, Kobayashi E, Tanzawa
Y, et al: A serum microRNA classifier for the diagnosis of sarcomas
of various histological subtypes. Nat Commun. 10:12992019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Araki Y, Aiba H, Yoshida T, Yamamoto N,
Hayashi K, Takeuchi A, Miwa S, Igarashi K, Nguyen TD, Ishii K, et
al: Osteosarcoma-Derived small extracellular vesicles enhance tumor
metastasis and suppress osteoclastogenesis by miR-146a-5p. Front
Oncol. 11:6671092021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
National Comprehensive Cancer Network, .
NCCN Guidelines Bone Cancer. https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1418April
1–2023
|
|
28
|
Amin MB, Edge SB, Greene FL, Compton CC,
Gershenwald JE, Broolland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: AJCC Cancer Staging Manual. 8th edition. Springer;
Chicago, IL: pp. 471–486. 2017, PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ihaka R and Gentleman R: R: A language for
data analysis and graphics. J Comput Graph Stat. 5:299–314. 1996.
View Article : Google Scholar
|
|
31
|
Kanda Y: Investigation of the freely
available easy-to-use software ‘EZR’ for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
miRDB. There are 437 predicted targets for
hsa-miR-1261 in miRDB. https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&searchBox=hsa-miR-1261&full=1April
1–2023
|
|
33
|
Ji Q, Xu X, Song Q, Xu Y, Tai Y, Goodman
SB, Bi W, Xu M, Jiao S, Maloney WJ and Wang Y: miR-223-3p inhibits
human osteosarcoma metastasis and progression by directly targeting
CDH6. Mol Ther. 26:1299–1312. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pu Y, Yi Q, Zhao F, Wang H, Cai W and Cai
S: MiR-20a-5p represses multi-drug resistance in osteosarcoma by
targeting the KIF26B gene. Cancer Cell Int. 16:642016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sakthikumar S, Elvers I, Kim J, Arendt ML,
Thomas R, Turner-Maier J, Swofford R, Johnson J, Schumacher SE,
Alföldi J, et al: SETD2 is recurrently mutated in whole-exome
sequenced canine osteosarcoma. Cancer Res. 78:3421–3431. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jiang C, He C, Wu Z, Li F and Xiao J:
Histone methyltransferase SETD2 regulates osteosarcoma cell growth
and chemosensitivity by suppressing Wnt/β-catenin signaling.
Biochem Biophys Res Commun. 502:382–388. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yurtsever A, Yoshida T, Badami Behjat A,
Araki Y, Hanayama R and Fukuma T: Structural and mechanical
characteristics of exosomes from osteosarcoma cells explored by
3D-atomic force microscopy. Nanoscale. 13:6661–6677. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu X, Yu H and Xu Y: Ras-ERK1/2 signaling
promotes the development of osteosarcoma by regulating H2BK12ac
Through CBP. Cancer Manag Res. 11:9153–9163. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Boele J, Persson H, Shin JW, Ishizu Y,
Newie IS, Søkilde R, Hawkins SM, Coarfa C, Ikeda K, Takayama K, et
al: PAPD5-mediated 3′ adenylation and subsequent degradation of
miR-21 is disrupted in proliferative disease. Proc Natl Acad Sci
USA. 111:11467–11472. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Faversani A, Favero C, Dioni L, Pesatori
AC, Bollati V, Montoli M, Musso V, Terrasi A, Fusco N, Nosotti M,
et al: An EBC/Plasma miRNA Signature discriminates lung
adenocarcinomas from pleural mesothelioma and healthy controls.
Front Oncol. 11:6432802021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen L, Wang K, Li L, Zheng B, Zhang Q,
Zhang F, Chen J and Wang S: Plasma exosomal miR-1260a, miR-7977 and
miR-192-5p as diagnostic biomarkers in epithelial ovarian cancer.
Future Oncol. 18:2919–2931. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mancini M, Grasso M, Muccillo L, Babbio F,
Precazzini F, Castiglioni I, Zanetti V, Rizzo F, Pistore C, De
Marino MG, et al: DNMT3A epigenetically regulates key microRNAs
involved in epithelial-to-mesenchymal transition in prostate
cancer. Carcinogenesis. 42:1449–1460. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Latchana N, DiVincenzo MJ, Regan K, Abrams
Z, Zhang X, Jacob NK, Gru AA, Fadda P, Markowitz J, Howard JH and
Carson WE III: Alterations in patient plasma microRNA expression
profiles following resection of metastatic melanoma. J Surg Oncol.
118:501–509. 2018.PubMed/NCBI
|
|
44
|
Wu D, Chang X, Tian J, Kang L, Wu Y, Liu
J, Wu X, Huang Y, Gao B, Wang H, et al: Bone mesenchymal stem cells
stimulation by magnetic nanoparticles and a static magnetic field:
Release of exosomal miR-1260a improves osteogenesis and
angiogenesis. J Nanobiotechnology. 19:2092021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li K, Rodosthenous RS, Kashanchi F,
Gingeras T, Gould SJ, Kuo LS, Kurre P, Lee H, Leonard JN, Liu H, et
al: Advances, challenges, and opportunities in extracellular RNA
biology: Insights from the NIH exRNA Strategic Workshop. JCI
Insight. 3:e989422018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Engin A: Dark-Side of Exosomes. Adv Exp
Med Biol. 1275:101–131. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Robbins PD and Morelli AE: Regulation of
immune responses by extracellular vesicles. Nat Rev Immunol.
14:195–208. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sinkovics JG: Molecular biology of
oncogenic inflammatory processes. I. Non-oncogenic and oncogenic
pathogens, intrinsic inflammatory reactions without pathogens, and
microRNA/DNA interactions (Review). Int J Oncol. 40:305–349.
2012.PubMed/NCBI
|
|
49
|
Ye H, Hu X, Wen Y, Tu C, Hornicek F, Duan
Z and Min L: Exosomes in the tumor microenvironment of sarcoma:
From biological functions to clinical applications. J
Nanobiotechnology. 20:4032022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cheng M, Duan PG, Gao ZZ and Dai M:
MicroRNA-487b-3p inhibits osteosarcoma chemoresistance and
metastasis by targeting ALDH1A3. Oncol Rep. 44:2691–2700. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
John AA, Prakash R and Singh D:
miR-487b-3p impairs osteoblastogenesis by targeting Notch-regulated
ankyrin-repeat protein (Nrarp). J Endocrinol. 241:249–263. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yi H, Geng L, Black A, Talmon G, Berim L
and Wang J: The miR-487b-3p/GRM3/TGFβ signaling axis is an
important regulator of colon cancer tumorigenesis. Oncogene.
36:3477–3489. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang BL, Dong FL, Guo TW, Gu XH, Huang LY
and Gao DS: MiRNAs Mediate GDNF-Induced proliferation and migration
of glioma cells. Cell Physiol Biochem. 44:1923–1938. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Daniel R, Wu Q, Williams V, Clark G,
Guruli G and Zehner Z: A Panel of MicroRNAs as diagnostic
biomarkers for the identification of prostate cancer. Int J Mol
Sci. 18:12812017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Park S, Kim J, Cho Y, Ahn S, Kim G, Hwang
D, Chang Y, Ha S, Choi Y, Lee MH, et al: Promotion of tumorigenesis
by miR-1260b-targeting CASP8: Potential diagnostic and prognostic
marker for breast cancer. Cancer Sci. 113:2097–2108. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Xia Y, Wei K, Hu LQ, Zhou CR, Lu ZB, Zhan
GS, Pan XL, Pan CF, Wang J, Wen W, et al: Exosome-mediated transfer
of miR-1260b promotes cell invasion through Wnt/β-catenin signaling
pathway in lung adenocarcinoma. J Cell Physiol. 235:6843–6853.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hayashi C, Fukuda T, Kawakami K, Toyoda M,
Nakao Y, Watanabe Y, Shinjo T, Sano T, Iwashita M, Yotsumoto K, et
al: miR-1260b inhibits periodontal bone loss by targeting ATF6β
mediated regulation of ER stress. Front Cell Dev Biol.
10:10612162022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zakrzewska M, Gruszka R, Stawiski K,
Fendler W, Kordacka J, Grajkowska W, Daszkiewicz P, Liberski PP and
Zakrzewski K: Expression-based decision tree model reveals distinct
microRNA expression pattern in pediatric neuronal and mixed
neuronal-glial tumors. BMC Cancer. 19:5442019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Omura T, Shimada Y, Nagata T, Okumura T,
Fukuoka J, Yamagishi F, Tajika S, Nakajima S, Kawabe A and Tsukada
K: Relapse-associated microRNA in gastric cancer patients after S-1
adjuvant chemotherapy. Oncol Rep. 31:613–618. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wu YS, Lin H, Chen D, Yi Z, Zeng B, Jiang
Y and Ren G: A four-miRNA signature as a novel biomarker for
predicting survival in endometrial cancer. Gene. 697:86–93. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
He JH, Li BX, Han ZP, Wang L, Lv YB, Zhou
JB, Cao MR, Li YG and Zhang JZ: Snail-activated long non-coding RNA
PCA3 up-regulates PRKD3 expression by miR-1261 sponging, thereby
promotes invasion and migration of prostate cancer cells. Tumour
Biol. Oct 14–2016.(Epub ahead of print). View Article : Google Scholar
|
|
62
|
Lyu N, Zeng Y, Kong Y, Chen Q, Deng H, Ou
S, Bai Y, Tang H, Wang X and Zhao M: Ferroptosis is involved in the
progression of hepatocellular carcinoma through the
circ0097009/miR-1261/SLC7A11 axis. Ann Transl Med. 9:6752021.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Cao F, Liu S, Li Z, Meng L, Sang M and
Shan B: Activation of circ_0072088/miR-1261/PIK3CA pathway
accelerates lung adenocarcinoma progression. Thorac Cancer.
13:1548–1557. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhu Y, Cao F, Liu F, Liu S, Meng L, Gu L,
Zhao H, Sang M and Shan B: Identification of potential circular RNA
biomarkers in lung adenocarcinoma: A bioinformatics analysis and
retrospective clinical study. Oncol Lett. 23:1442022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chen R, Zhao WQ, Fang C, Yang X and Ji M:
Histone methyltransferase SETD2: A potential tumor suppressor in
solid cancers. J Cancer. 11:3349–3356. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Das S, Idate R, Regan DP, Fowles JS, Lana
SE, Thamm DH, Gustafson DL and Duval DL: Immune pathways and TP53
missense mutations are associated with longer survival in canine
osteosarcoma. Commun Biol. 4:11782021. View Article : Google Scholar : PubMed/NCBI
|