Introduction
Cervical cancer (CC) is the fourth most common
cancer and the fourth leading cause of cancer mortality in women
(1). In 2020, 604,000 new diagnoses
and 342,000 CC-related deaths were estimated worldwide, accounting
for ~3.2% of all new cancer diagnoses and 3.4% of all cancer deaths
(1). Given the high incidence and
mortality of CC, the World Health Organization has assembled the CC
Elimination Modelling Consortium to eliminate CC (2,3).
Despite increases in human papillomavirus vaccination coverage, and
advances in the screening approach of CC in recent years, the
burden of CC is heavy in low-income countries (3). Therefore, it remains necessary to
elucidate the molecular mechanisms underlying the progression of CC
as it would help to identify effective therapeutic strategies.
The SHC binding and spindle associated 1 (SHCBP1)
gene is located on chromosome 16q11.2; SHCBP1 is a member of the
Src homolog and collagen homolog (Shc) family, and it was first
characterized as a protein which bound to the Shc adaptor protein,
Shc A (4). SHCBP1 is able to
regulate cell proliferation, differentiation and wound inflammation
(4,5). Increasing evidence has suggested that
SHCBP1 is often dysregulated in cancer, and acts as a key regulator
of tumor progression and metastasis in various types of cancer,
such as gastric carcinoma (6),
synovial sarcoma (7) and non-small
cell lung carcinoma (8).
Nevertheless, the functional and molecular mechanisms of SHCBP1 in
CC remain unexplored.
As a transcription factor, NF-κB has been linked to
inflammatory responses in various human diseases (9). In mammals, the NF-κB family is
composed of five members, namely p65, c-Rel, RelB, p50 and p52, all
of which share an N-terminal DNA-binding/dimerization domain
referred to as the REL homology domain (10). NF-κB proteins form homo- and
heterodimers that can bind to DNA sequences of target genes and
ultimately regulate gene transcription (10). A strong association has been
established between the activation of NF-κB and cancer, including
CC (11–13). Zhou et al (14) confirmed the positive regulation of
NF-κB signaling pathway activation and cell mobility in glioma
induced by SHCBP1 by performing gain-/loss-of-function experiments.
The link between SHCBP1 and NF-κB signaling in CC remains to be
fully elucidated.
Eukaryotic translation initiation factor 5A (EIF5A),
a key factor involved in translation elongation, exerts oncogenic
roles in a variety of cancer types, including colorectal cancer
(15), pancreatic cancer (16) and CC (17,18).
EIF5A silencing has been reported to inhibit CC cell proliferation
and promote NF-κB inhibitor (IκB) expression in CC cells (17). Furthermore, small interfering RNA
(siRNA)-mediated inhibition of EIF5A has been shown to reverse the
phosphorylation of p65 in multiple myeloma cells (19), highlighting the modulation of NF-κB
activation by EIF5A. A dataset (GSE154307) downloaded from the Gene
Expression Omnibus (GEO) database indicates that SHCBP1 short
hairpin RNA (shRNA) reduced EIF5A mRNA expression in papillary
thyroid cancer cells (20).
Therefore, it was hypothesized that SHCBP1 may be involved in the
activation of the NF-κB signaling pathway in CC cells via
EIF5A.
In the present study, gain- and/or loss-of-function
experiments were performed to explore the role of SHCBP1 in the
proliferation and stemness of CC cells, and the in vitro
activation of the NF-κB signaling pathway. In addition, the
molecular mechanism of SHCBP1 in the progression of CC was
discussed.
Materials and methods
Screening of differentially expressed
genes (DEGs)
The CC microarray datasets (GSE9750, GSE63514 and
GSE7803) (21–23) were downloaded from the GEO database
(https://www.ncbi.nlm.nih.gov/geo/).
Genes with P<0.05 and log2(fold change) ≥1 or ≤-1
were considered to be DEGs between healthy control cervical tissues
and CC tissues.
Bioinformatics analysis
The Gene Expression Profiling Interactive Analysis
(GEPIA, http://gepia.cancer–pku.cn/index.html) database was
used to profile the expression level of SHCBP1 in tumor tissues
from 31 types of cancer and the corresponding healthy control
tissues based on the TCGA and GTEx databases. The mutual DEGs among
the CC microarray datasets (GSE9750, GSE63514 and GSE7803) were
identified using the online tool Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/index.html).
The Database for Annotation, Visualization and Integrated Discovery
(https://david.ncifcrf.gov/) was utilized
to perform Gene Ontology (GO, http://geneontology.org/) and Kyoto Encyclopedia of
Genes and Genomes (KEGG, http://www.genome.jp/kegg/) pathway enrichment
analyses for the mutual DEGs.
Cell culture and transfection
Human CC cell lines were obtained from iCell
Bioscience, Inc. (HeLa, cat. no. iCell-h088) and Procell Life
Science & Technology Co., Ltd. (SiHa,C-33A and CaSki). HeLa,
SiHa and C-33A cells were cultured in minimum essential medium
(MEM, Beijing Solarbio Science & Technology Co., Ltd.)
supplemented with 10% FBS (Zhejiang Tianhang Biotechnology Co.,
Ltd.), 100 U/ml penicillin and 0.1 mg/ml streptomycin with 5%
CO2 at 37°C. CaSki cells were maintained in RPMI-1640
medium (Beijing Solarbio Science & Technology Co., Ltd.)
supplemented with 10% FBS (Zhejiang Tianhang Biotechnology Co.,
Ltd.), 100 U/ml penicillin and 0.1 mg/ml streptomycin with 5%
CO2 at 37°C.
For in vitro functional experiments, plasmids
(2.5 µg) expressing shRNAs targeting SHCBP1 or non-targeting shRNAs
(negative control, NC) were transfected into SiHa cells and
plasmids (2.5 µg) expressing SHCBP1 coding DNA sequence (CDS) or
empty vectors were transfected into CaSki cells using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions at
37°C, and the medium was changed after 8 h. The sequences of shRNAs
were as follows: SHCBP1 shRNA-1,
5′-GGATTGTATGTGTTTGGTTATTCAAGAGATAACCAAACACATACAATCTTTTT-3′; SHCBP1
shRNA-2,
5′-GGGTTATCAGAAGAATTCTATTCAAGAGATAGAATTCTTCTGATAACCTTTTT-3′; and
non-targeting shRNA,
5′-GTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTT-3′. The
mock control represents the cells without any transfection. The
shRNA was inserted into the pRNA-H1.1/Neo vector (cat. no. SD1203;
GenScript) between BamHI and HindIII. The SHCBP1 CDS
(General Biology (Anhui) Co., Ltd.) was inserted into the
pcDNA3.1(+) vector (cat. no. V79020; Invitrogen; Thermo Fisher
Scientific, Inc.) between HindIII and XhoI. At 24 h
post-transfection, the SiHa cells stably silencing SHCBP1 were
selected in complete MEM containing 350 µg/ml geneticin (Beijing
Solarbio Science & Technology Co., Ltd.) for 1–2 weeks, and the
CaSki cells stably overexpressing SHCBP1 were selected in complete
RPMI-1640 medium containing 300 µg/ml geneticin for 1–2 weeks, and
the medium was changed every 2–3 days. A large number of cells died
after 1- or 2-week geneticin selection, and then the surviving
cells were cultured in complete MEM or RPMI-1640 medium containing
geneticin (SiHa: 175 µg/ml; CaSki: 150 µg/ml) for 2 weeks.
In addition, stable SHCBP1-overexpressing CaSki
cells or blank CaSki cells (without any manipulation of SHCBP1
expression) were transfected with siRNA-targeting EIF5A (75 pmol;
General Biology (Anhui) Co., Ltd.) using Lipofectamine®
3000 (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C, and the
medium was changed after 8 h. The sequences of siRNAs were as
follows: EIF5A siRNA sense, 5′-GCAAGGAGAUUGAGCAGAATT-3′ and
anti-sense 5′-UUCUGCUCAAUCUCCUUGCTT-3′; and non-targeting siRNA
sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and anti-sense,
5′-ACGUGACACGUUCGGAGAATT-3′. At 48 h post-transfection, the cells
were used for subsequent experiments.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the four CC cell lines
with TRIpure reagent (BioTeke Corporation). First-strand cDNA was
synthesized using BeyoRT II M-MLV reverse transcriptase (Beyotime
Institute of Biotechnology) with dNTPs, 5× buffer, Rnase inhibitor
(BioTeke Corporation), oligo (dT)15 and random primers
at 25°C for 10 min, at 42°C for 50 min and at 80°C for 10 min.
Relative mRNA expression was determined by RT-qPCR on an Exicycler™
96 Real-Time PCR system (Bioneer Corporation) using SYBR Green 2×
Taq PCR Master Mix (Beijing Solarbio Science & Technology Co.,
Ltd.) and gene-specific primers (General Biology (Anhui) Co.,
Ltd.). The thermocycling conditions were listed as follows: 94°C
for 5 min (initial denaturation), 40 cycles of 94°C for 10 sec,
60°C for 20 sec and 72°C for 30 sec, followed by a single final
extension for 2.5 min at 72°C. GAPDH was used to normalize gene
expression. The relative expression levels of mRNAs were calculated
using the 2−ΔΔCq method (24). The sequences of the gene-specific
primers used in RT-qPCR are listed in Table I.
 | Table I.Sequences of primers used in reverse
transcription-quantitative PCR analysis. |
Table I.
Sequences of primers used in reverse
transcription-quantitative PCR analysis.
| Gene | Sequences
(5′-3′) |
|---|
| Human SHCBP1 | Forward:
ATCCAAGCAAAGGGTGTC |
|
| Reverse:
CAAAGCCTGTCCGTAAGC |
| Human EIF5A | Forward:
TAAGAATGGCTTTGTGGTG |
|
| Reverse:
TGTCCTGGAGCAGTGATAG |
| Human CD133 | Forward:
GCACTCTATACCAAAGCGTCAA |
|
| Reverse:
CTCCCATACTTCTTAGTTTCCTCA |
| Human CD44 | Forward:
ATCCCTGCTACCACTTTG |
|
| Reverse:
TTCTTCATTTGGCTCCC |
| Human NANOG | Forward:
CACCTATGCCTGTGATTT |
|
| Reverse:
CAGAAGTGGGTTGTTTGC |
| Human OCT4 | Forward:
GTGGAGGAAGCTGACAACAATG |
|
| Reverse:
CTCACTCGGTTCTCGATACTGG |
| Human GAPDH | Forward:
GACCTGACCTGCCGTCTAG |
|
| Reverse:
AGGAGTGGGTGTCGCTGT |
Western blotting
Protein was extracted from the four CC cell lines
with RIPA buffer (Beijing Solarbio Science & Technology Co.,
Ltd.) containing 1 mM phenylmethanesulfonyl fluoride (Beijing
Solarbio Science & Technology Co., Ltd.). The concentration of
protein was measured using BCA protein assay kits (Beijing Solarbio
Science & Technology Co., Ltd.) according to the manufacturer's
protocols. Protein samples (10–20 µg/lane) were separated by
SDS-PAGE on 10 or 14% gels and transferred to PVDF membranes
(MilliporeSigma). The membranes were blocked with 5% BSA (Biosharp
Life Sciences) at room temperature for 1 h, followed by incubation
with anti-SHCBP1 (dilution, 1:500; cat no. 12672-1-AP, Proteintech
Group, Inc.), anti-cyclin D1 (dilution, 1:500; cat no. A19038,
ABclonal Biotech Co., Ltd.), anti-p21 (dilution, 1:500; cat no.
A19094, ABclonal Biotech Co., Ltd.), anti-phosphorylated (p)-IκBα
(dilution, 1:500; Ser-32/Ser-36, cat no. AF2002, Affinity
Biosciences, Ltd.), anti-IκBα (dilution, 1:1,000; cat no. AF5002,
Affinity Biosciences, Ltd.), anti-p-p65 (dilution, 1:400; Ser-536,
cat no. AF2006, Affinity Biosciences, Ltd.), anti-p65 (dilution,
1:1,000; cat no. AF5006, Affinity Biosciences, Ltd.), anti-EIF5A
(dilution, 1:1,000; cat no. 11309-1-AP, Proteintech Group, Inc.),
and anti-GAPDH (dilution, 1:10,000; cat no. 60004-1-Ig, Proteintech
Group, Inc.) antibodies overnight at 4°C. The following day,
membranes were incubated with HRP-conjugated goat anti-rabbit IgG
(dilution, 1:3,000; cat no. SE134, Beijing Solarbio Science &
Technology Co., Ltd.) or goat anti-mouse IgG (dilution, 1:3,000;
cat no. SE131, Beijing Solarbio Science & Technology Co., Ltd.)
for 1 h at 37°C. The immunoreactive protein bands were visualized
using an ECL kit (Beijing Solarbio Science & Technology Co.,
Ltd.). GAPDH was used as a loading control to normalize protein
expression.
Cell proliferation assays
MTT assays were used to assess cell proliferation.
In brief, CC cells were seeded into 96-well culture plates at a
density of 4×103 cells/well and cultured for 0, 12, 24,
36 and 48 h. Cell proliferation was measured using MTT assay kits
(Nanjing KeyGen Biotech Co., Ltd.) according to the manufacturer's
instructions. DMSO (Nanjing KeyGen Biotech Co., Ltd.) was used to
dissolve formazan and the optical density was measured at 490
nm.
Immunofluorescence assays (IFAs)
SiHa and CaSki cells were grown on glass coverslips
(1×105 cells per well) in 24-well plates, fixed in 4%
paraformaldehyde (Sinopharm Chemical Reagent Co., Ltd.) for 15 min
at room temperature and permeabilized with 0.1% Triton X-100
(Beyotime Institute of Biotechnology) for 30 min at room
temperature. After blocking with 1% BSA (Sangon Biotech Co., Ltd.)
for 15 min at room temperature, the coverslips were incubated with
anti-ki67 (dilution, 1:200; cat no. A11390, ABclonal Biotech Co.,
Ltd.) or anti-p65 antibodies (dilution, 1:200; cat no. A19653,
ABclonal Biotech Co., Ltd.) at 4°C overnight, followed by
incubation with the Alexa Fluor™ 555-conjugated goat anti-rabbit
(dilution, 1:200; cat no. A27039, Invitrogen; Thermo Fisher
Scientific, Inc.) at room temperature for 1 h. Cell nuclei were
stained with DAPI (Shanghai Aladdin Biochemical Technology Co.,
Ltd.) at room temperature for 5 min and observed under a
fluorescence microscope. Total p65 and nuclear p65 levels were
quantified. Relative p65 nuclear localization was expressed as the
ratio of the fluorescence density of nuclear p65 staining to the
fluorescence density of total p65 cellular staining.
Cell cycle analysis
Cell cycle progression was examined using cell cycle
analysis kits (cat no. BL114A, Biosharp Life Sciences). SiHa and
CaSki cells (5×105 cells) were collected by
centrifugation (106 × g for 5 min) at 4°C and fixed with ice-cold
70% ethanol for 2 h at 4°C. The cell pellets were washed with PBS
twice and the staining buffer, PI (20×) and RNase A (50×) (cat no.
BL114A, Biosharp Life Sciences) were added. After incubation at
37°C for 30 min, the cells were analyzed by a flow cytometer
(NovoCyte 2040R; ACEA Bioscience, Inc.; Agilent Technologies, Inc.)
using the NovoExpress software (version 1.4.1; ACEA Bioscience,
Inc.; Agilent Technologies, Inc.).
Sphere formation assays
CC cells were seeded into 6-well ultra-low plates at
a density of 3×103 cells/ml and cultured in stem cell
medium for 2 weeks with 5% CO2 at 37°C. The medium was
changed every 2 days. The stem cell medium was DMEM/F12 (Procell
Life Science & Technology Co., Ltd.) supplemented with 1% B-27
(Gibco; Thermo Fisher Scientific, Inc.), 1% N-2 (Gibco; Thermo
Fisher Scientific, Inc.), 20 ng/ml EGF (Sino Biological Inc.) and
10 ng/ml FGF (Sino Biological Inc.). Spheres were observed under an
inverted phase contrast microscope and analyzed using Image-Pro
Plus software (version 6.0, Media Cybernetics, Inc.). Spherical
cell clusters were considered to be spheres when their diameter was
>75 µm. The spheres were formed based on the cancer stem cell
(CSC)-like properties of cancer cells.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism (version 8.0.2, GraphPad Software; Dotmatics). Results are
presented as the mean ± SD from three independent experiments.
One-way and two-way ANOVA followed by Tukey or Sidak tests,
unpaired t-test and Mann Whitney U test were used to analyze
statistical significance. P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of DEGs in CC
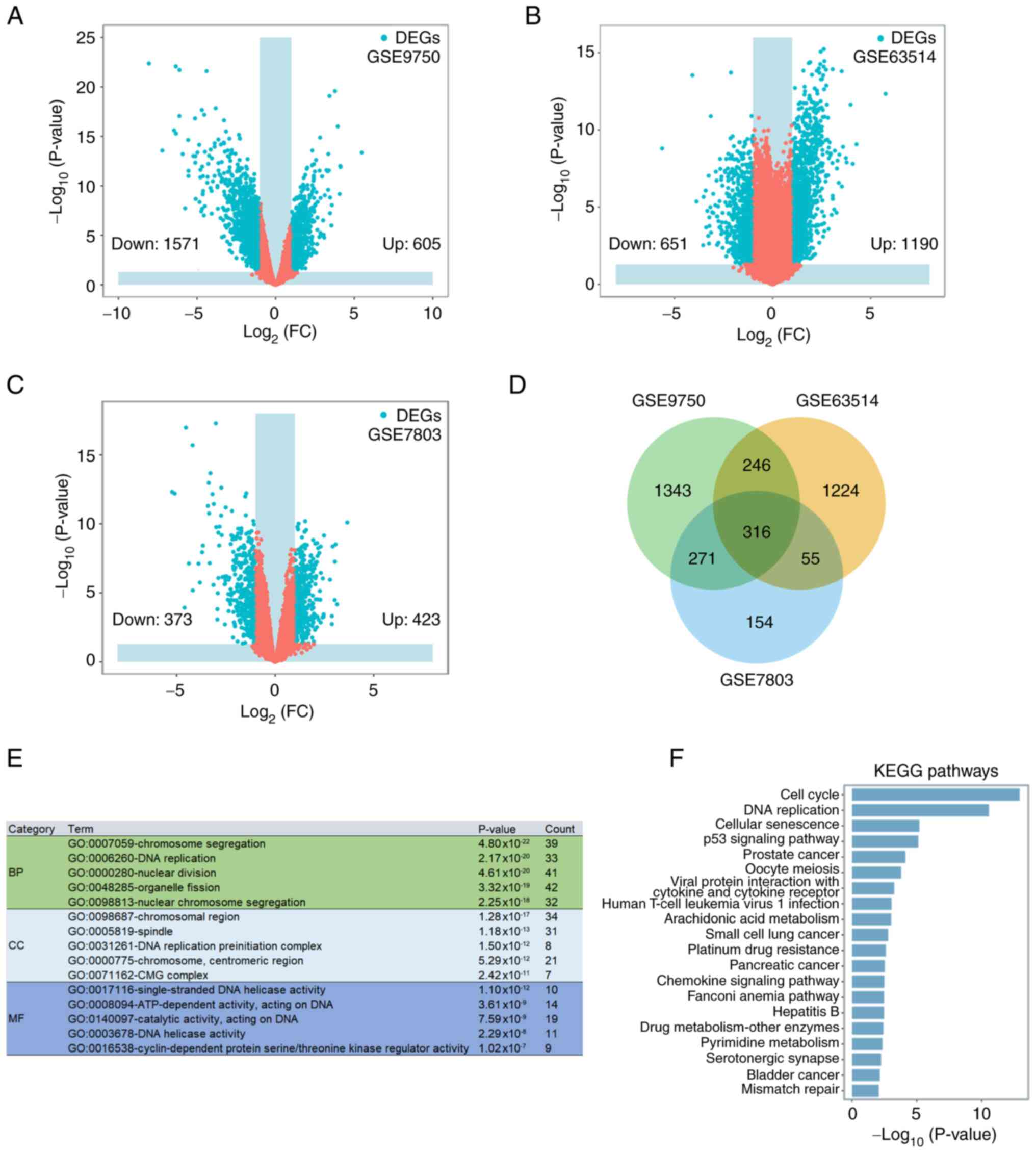
The GSE9750, GSE63514 and GSE7803 datasets indicated
2,176, 1,841 and 796 DEGs in CC compared with the healthy control
cervical tissues, respectively (Fig.
1A-C). The Venn diagram of DEGs in the three datasets indicated
a total of 316 mutual DEGs (Fig.
1D). The function of the mutual DEGs was examined by performing
GO (Fig. 1E) and KEGG pathway
(Fig. 1F) enrichment analyses. GO
analysis suggested that the mutual DEGs were mainly linked to
‘chromosome segregation’ [biological process (BP)], ‘DNA
replication’ (BP), ‘nuclear division’ (BP), ‘chromosomal region’
[cell component (CC)], ‘spindle’ (CC), ‘DNA replication
preinitiation complex’ (CC), ‘single-stranded DNA helicase
activity’ [molecular function (MF)], ‘ATP-dependent activity,
acting on DNA’ (MF) and ‘catalytic activity, acting on DNA’ (MF).
The three most enriched pathways in the KEGG analysis were ‘cell
cycle’, ‘DNA replication’ and ‘cellular senescence’.
Upregulation of SHCBP1 mRNA levels in
CC
The mRNA expression level of SHCBP1 was upregulated
in CC compared with the healthy control cervix tissues in the
GSE9750, GSE63514 and GSE7803 datasets, and the expression of
SHCBP1 in the tumor and healthy control cervical tissues in the
three datasets was shown in Fig.
2A. The Gene Expression Profiling Interactive Analysis database
showed that SHCBP1 expression was significantly upregulated in 21
human cancer types, particularly lymphoid neoplasm diffuse large
B-cell lymphoma and cervical squamous cell carcinoma and
endocervical adenocarcinoma (Fig.
2B). The highest median value of SHCBP1 expression was observed
in the healthy control for acute myeloid leukemia, which indicated
a high level of SHCBP1 mRNA in bone marrow (Fig. 2B). There was not a statistically
significant difference between the bone marrow from the healthy
control and acute myeloid leukemia patients (Fig. 2B).
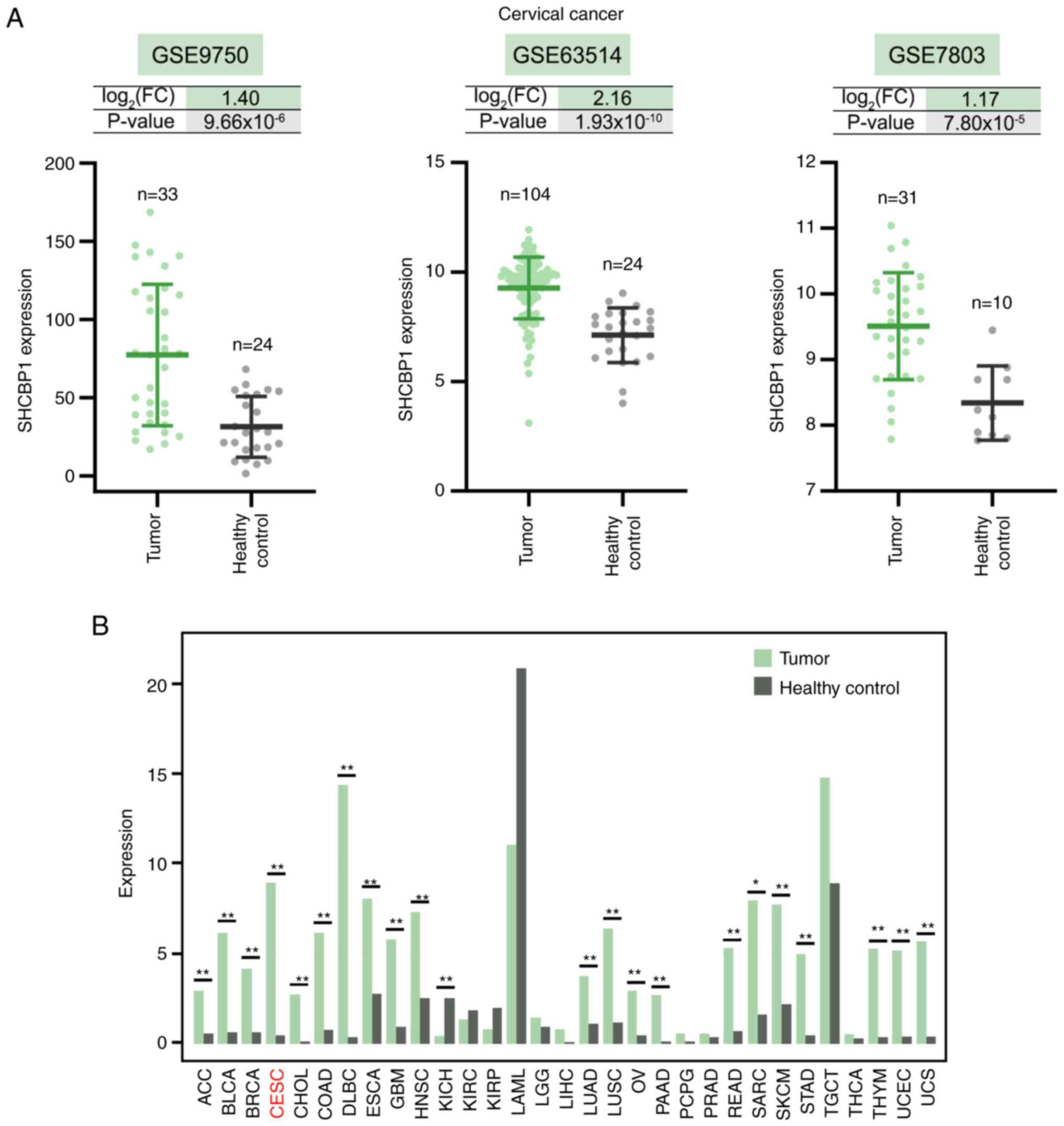 | Figure 2.SHCBP1 expression in cervical cancer
and other cancer types. (A) SHCBP1 expression in the GSE9750,
GSE63514 and GSE7803 datasets downloaded from the Gene Expression
Omnibus database. (B) Pan-cancer expression profiling analysis of
SHCBP1 using the Gene Expression Profiling Interactive Analysis
database. *P<0.05 and **P<0.01. SHCBP1, SHC binding and
spindle associated 1; FC, fold change; ACC, adrenocortical
carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast
invasive carcinoma; CESC, cervical squamous cell carcinoma and
endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon
adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell
lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme;
HNSC, head and neck squamous cell carcinoma; KICH, kidney
chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney
renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG,
brain lower grade glioma; LIHC, liver hepatocellular carcinoma;
LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV,
ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma;
PCPG, pheochromocytoma and paraganglioma; PRAD, prostate
adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM,
skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT,
testicular germ cell tumors; THCA, thyroid carcinoma; THYM,
thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine
carcinosarcoma. |
Pro-proliferative and pro-stemness
role of SHCBP1 in CC cells in vitro
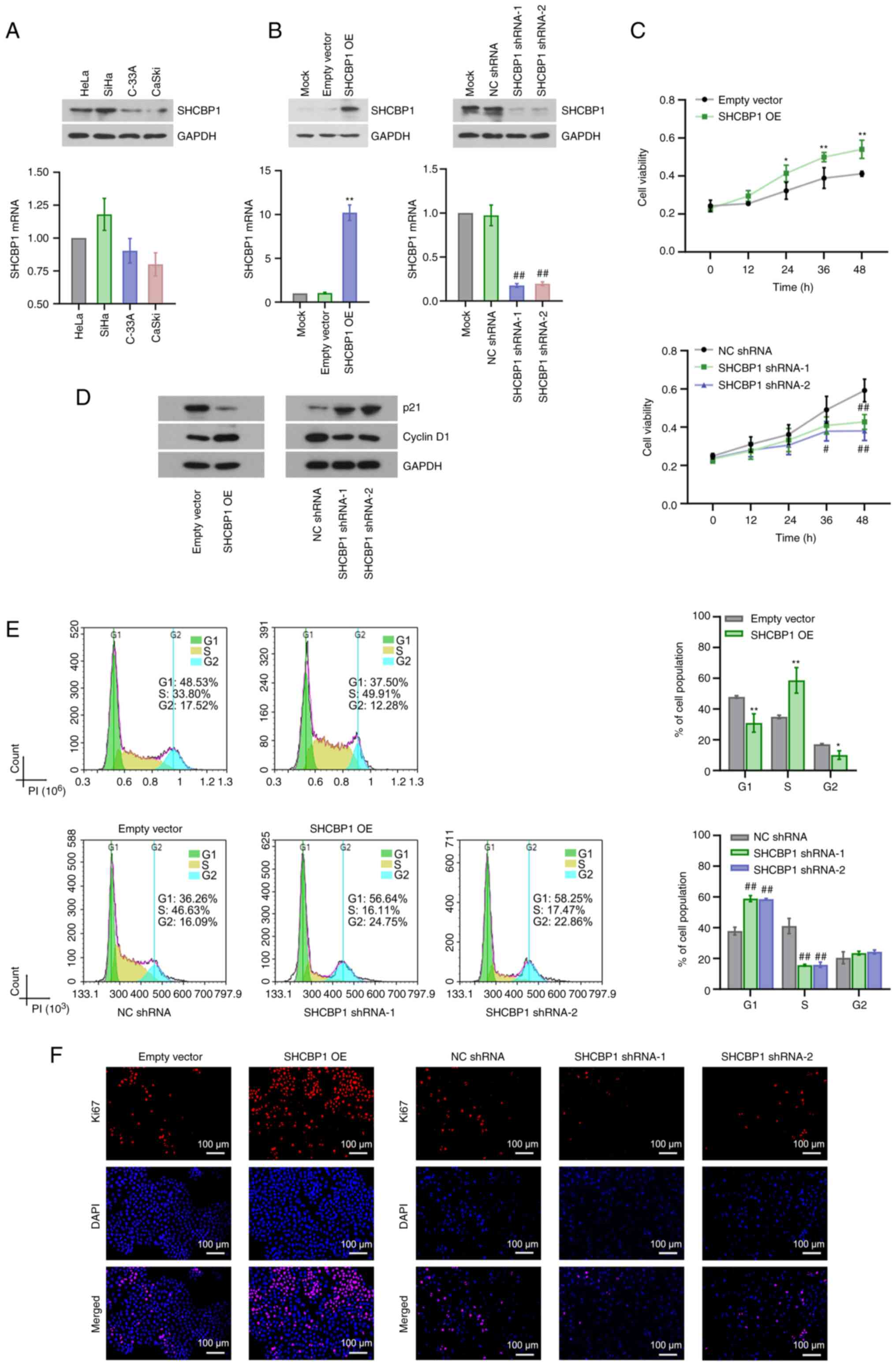
The basal expression levels of SHCBP1 in HeLa, SiHa,
C-33A and CaSki CC cell lines were determined by RT-qPCR and
western blotting and the results demonstrated a markedly higher
expression of SHCBP1 in SiHa cells and a markedly lower expression
of SHCBP1 in CaSki cells than that in other CC cell lines (Fig. 3A). Stable SHCBP1-overexpressing
CaSki cells and stable SHCBP1-silenced SiHa cells were successfully
established (Fig. 3B). MTT assays
demonstrated that SHCBP1 overexpression promoted but SHCBP1
knockdown inhibited cell proliferation in vitro (Fig. 3C). Proteins involved in cell cycle
progression were detected by western blotting. SHCBP1
overexpression promoted cyclin D1 expression and inhibited p21
expression in CaSki cells, yet SHCBP1 knockdown induced an opposite
effect on SiHa cells (Fig. 3D).
Flow cytometric analysis suggested that SHCBP1 overexpression
enhanced entry into the S phase but SHCBP1 knockdown induced arrest
at the G1 phase of the cell cycle (Fig.
3E). In addition, SHCBP1 overexpression decreased the number of
cells at the G2 phase, which might be the result of the extreme
increases in cell density induced by SHCBP1 overexpression
(Fig. 3E). IFA of ki67 showed
increased ki67 expression induced by SHCBP1 overexpression and
decreased ki67 expression induced by SHCBP1 knockdown, which
indicated the pro-proliferative roles of SHCBP1 (Fig. 3F). Subsequently, the effect of
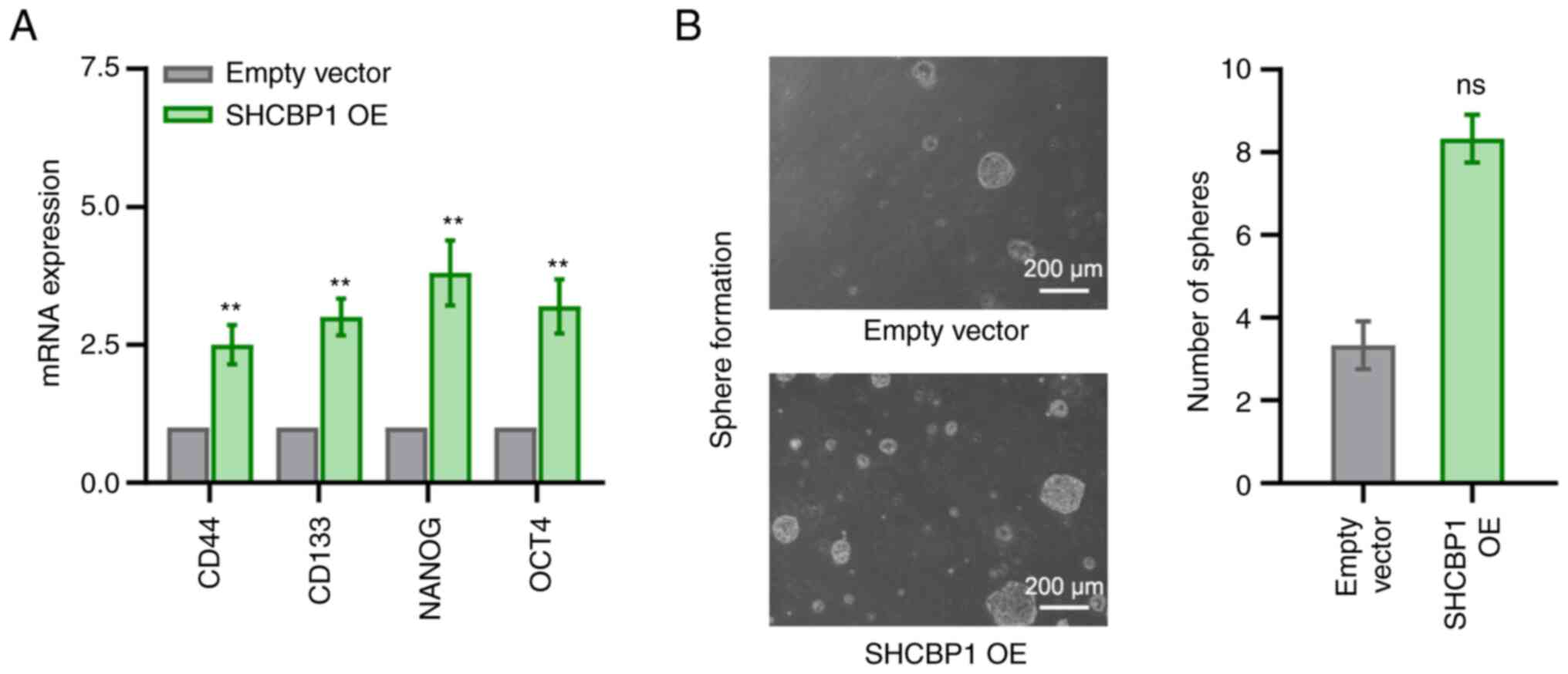
SHCBP1 on the CSC-like properties of CC cells was investigated. The
mRNA expression levels of stemness-related markers, including CD44,
CD133, NANOG and OCT4, were analyzed, and the results demonstrated
that SHCBP1 overexpression significantly increased the mRNA
expression levels of these markers (Fig. 4A). Furthermore, SHCBP1
overexpression tended to upregulate the sphere-forming ability of
CC cells, although there was no statistically significant
difference between the empty vector and the SHCBP1 OE group
(Fig. 4B). Taken together, SHCBP1
exerted a key role in maintaining the malignant phenotype and
self-renewal of CC cells in vitro.
Activation of the NF-κB signaling
pathway by SHCBP1 in CC cells
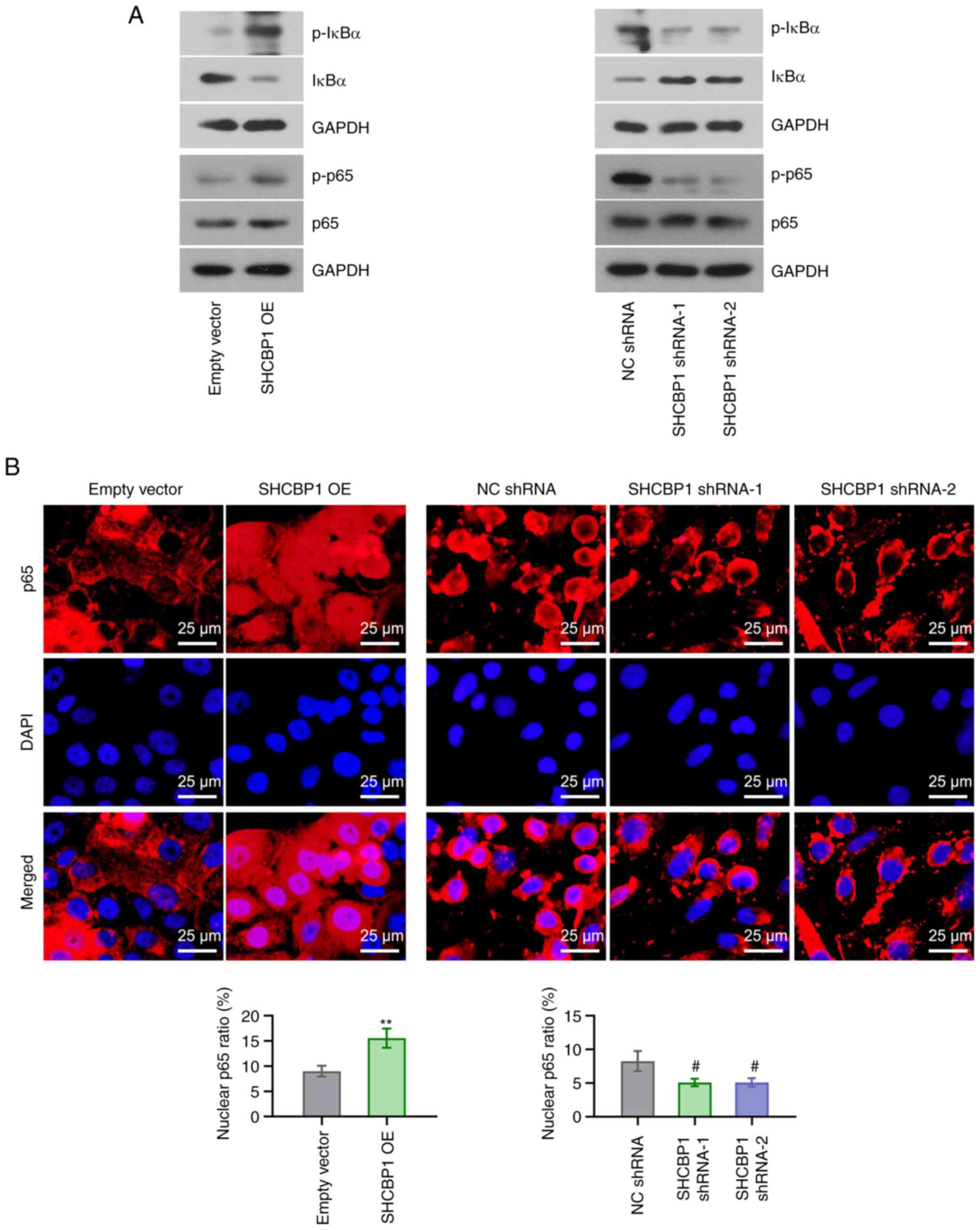
Activation of the NF-κB pathway serves a key role in
cancer (25). The phosphorylation
of IκBα and p65 was assessed by western blotting. SHCBP1
overexpression increased p-IκBα (Ser-32/Ser-36) and p-p65 (Ser-536)
expressions but decreased total IκBα expression, however, SHCBP1
knockdown exerted the opposite effect (Fig. 5A). IFA for p65 demonstrated that a
significantly higher number of positive signals of p65 was detected
in the nucleus after SHCBP1 overexpression; however, SHCBP1
knockdown significantly reduced nuclear signals of p65 (Fig. 5B). These findings suggested that
SHCBP1 was involved in the activation of the NF-κB signaling
pathway in CC cells.
SHCBP1 depends on EIF5A to exert
pro-proliferative and self-renewal effects and activate the NF-κB
signaling pathway in CC cells
EIF5A exerts oncogenic roles in CC (17,18). A
GEO dataset (GSE154307) indicates downregulated EIF5A mRNA
expression by SHCBP1 shRNA in papillary thyroid cancer cells
(20). The results of the present
study demonstrated that EIF5A mRNA expression was significantly
promoted by SHCBP1 overexpression (Fig. S1A), yet significantly inhibited by
SHCBP1 knockdown (Fig. S1B). To
investigate whether EIF5A mediated the biological function of
SHCBP1 in CC cells, untransfected CaSki cells and stable
SHCBP1-overexpressing CaSki cells were transiently transfected with
EIF5A siRNA. The knockdown efficiency of EIF5A was verified in
untransfected CaSki cells and stable SHCBP1-overexpressing CaSki
cells by RT-qPCR and western blotting (Fig. 6A, B and E). The stemness-related
markers including NANOG and OCT4 were confirmed to be regulated by
EIF5A (26). EIF5A siRNA inhibited
the SHCBP1-induced increases in NANOG and OCT4 expression (Fig. 6B). In addition, EIF5A siRNA reversed
the pro-proliferative effects (Fig. 6C,
D) and the activation of the NF-κB signaling pathway induced by
SHCBP1 overexpression (Fig. 6E).
These results confirmed that SHCBP1 enhanced the malignant
phenotype and self-renewal of CC cells, and activated the NF-κB
signaling pathway in vitro via EIF5A. The potential
mechanism of SHCBP1 in CC cells is shown in Fig. 6F. Based on the findings of the
present study, it was hypothesized that SHCBP1 increased the
nuclear translocation of p65 via EIF5A, thereby enhancing the
NF-κB-mediated gene transcription and promoting cell proliferation
and cell stemness.
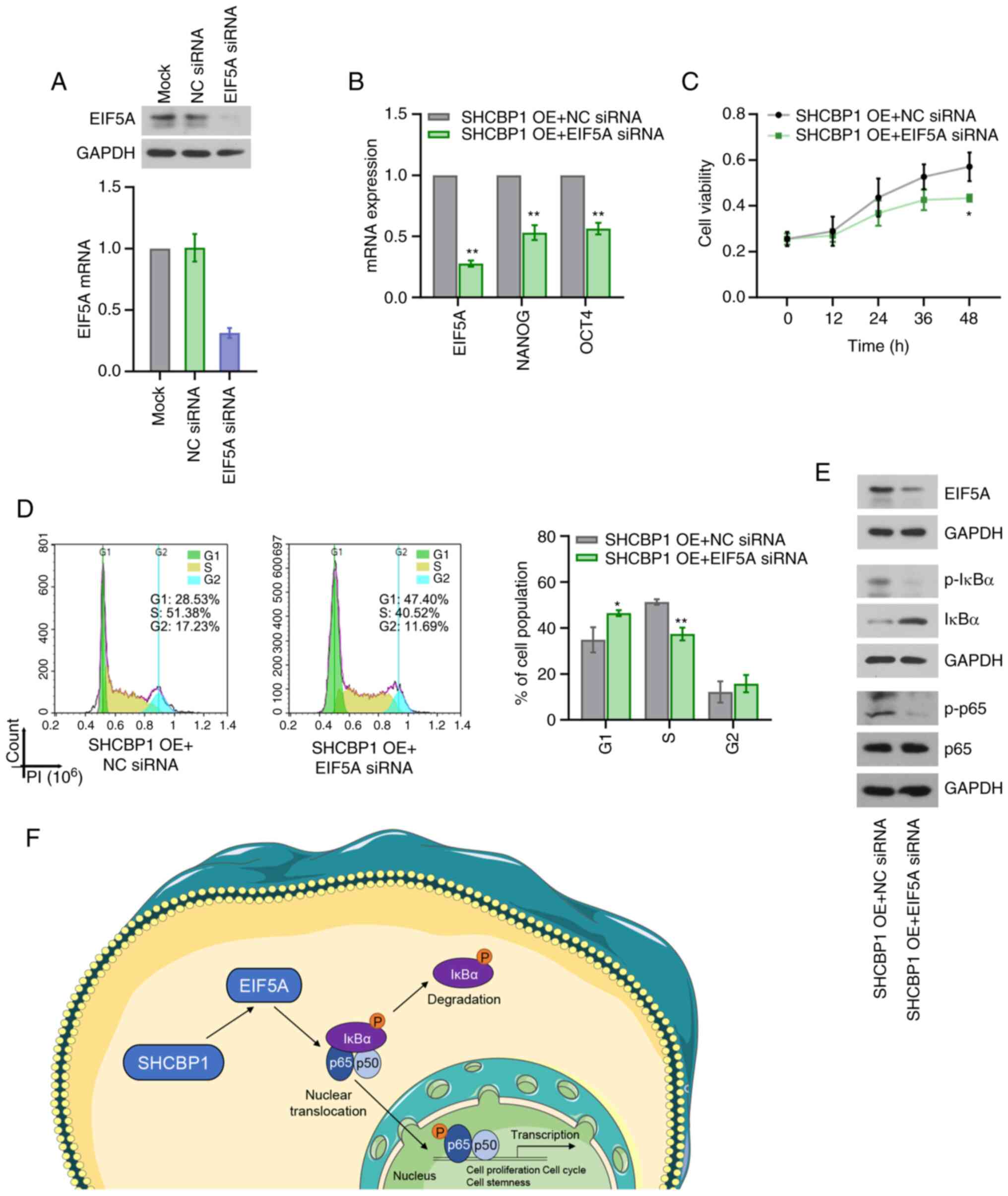 | Figure 6.Effect of EIF5A on cell
proliferation, cell stemness and activation of the NF-κB signaling
pathway induced by SHCBP1 in CC. CaSki cells were transiently
transfected with EIF5A siRNA. (A) Knockdown efficiency was
evaluated by measuring mRNA and protein expression levels of EIF5A
using RT-qPCR and western blotting. Data were analyzed with one-way
ANOVA followed by Tukey's test. **P<0.01 vs. NC siRNA group. (B)
CaSki cells stably overexpressing SHCBP1 were transiently
transfected with EIF5A siRNA. The mRNA expression levels of EIF5A,
NANOG and OCT4 were examined by RT-qPCR. Data were analyzed with
unpaired t-test. (C) Cell proliferation was measured using an MTT
assay. Data were analyzed with two-way ANOVA followed by Sidak's
test. (D) Flow cytometric analysis of cell cycle distribution. Data
were analyzed with unpaired t-test. (E) Western blot analysis of
EIF5A, p-IκBα, IκBα, p-p65 and p65. (F) Schematic overview of the
molecular mechanism underlying the function of SHCBP1 in CC cells.
Data are presented as the mean ± SD. *P<0.05 and **P<0.01 vs.
SHCBP1-OE + NC siRNA group. n=3. CC, cervical cancer; EIF5A,
eukaryotic translation initiation factor 5A; IκBα, NF-κB inhibitor
α; NC, negative control; OE, overexpression; p-, phosphorylated;
RT-qPCR, reverse transcription-quantitative PCR; SHCBP1, SHC
binding and spindle associated 1; siRNA, small interfering RNA. |
Discussion
Unchecked proliferation is a fundamental hallmark of
cancer cells (27). The accurate
transition from the G1 phase to the S phase of the cell cycle is
crucial for the control of cell proliferation, and its
disorganization results in uncontrolled cell proliferation
(28). SHCBP1 was first identified
in proliferating cells and it was absent in quiescent tissues and
growth-arrested cells (4). SHCBP1
mRNA is expressed during entry into S phase and its expression was
higher in cells treated with a stimulator of cell cycle progression
(4). These evidence indicate the
important role of SHCBP1 in regulating cell proliferation and cell
cycle progression (4). The present
study revealed that SHCBP1 increased CC cell proliferation, ki67
expression and cell cycle transition from the G1 phase to the S
phase of the cell cycle. The pro-proliferative effect of SHCBP1 has
also been observed in cell lines from other types of cancer
(20,29–32),
highlighting the important role of SHCBP1 in cancer cell
proliferation. Therefore, SHCBP1 inhibition may be a potential
strategy for the control of tumor progression.
CSCs, a key population of tumor cells, are
pluripotent, self-renewing and they contribute to tumor initiation,
metastasis, recurrence and resistance to therapies (33). Cumulative reports have highlighted
the importance of CSCs in CC (34–36).
SHCBP1 knockdown induced inhibition of stem cell-like properties of
non-small cell lung carcinoma (8)
and papillary thyroid carcinoma (20) cells, including generation of tumor
cell spheres and the expression of stemness-related genes and
proteins. In line with previous reports (8,20), the
present study suggested that SHCBP1 activated CSC-related genes and
promoted the self-renewal of CC cells. These findings further
indicated the significance of SHCBP1 in CC.
The NF-κB signaling pathway is one of the important
pathways involved in cell cycle regulation (37). NF-κB can activate the transcription
of cell cycle-related genes, including cyclin D1, cyclin E1, CDK2,
CDK2 and Myc, thereby modulating cell cycle distribution (38–41).
The NF-κB pathway also contributes to maintaining CSC-like
properties in multiple types of cancer (42–44).
Overexpression of p65 enhances the expression of CSC
markers, such as CD44 and CD133, in CC cells (45). Bay-11-7082, an inhibitor of NF-κB,
has been demonstrated to reduce CD44 expression in breast cancer
cells (46) and NANOG and SOX2
expression in CD44-positive and myeloid differentiation primary
response 88-positive ovarian cancer cells (47). Knockdown of p65 and/or RelB
decreases CD133-positive skin cancer cells, and their self-renewal
abilities (48). The present study
demonstrated that SHCBP1 activated the NF-κB signaling pathway in
CC cells. Given the importance of NF-κB in cell proliferation and
stemness maintenance in cancer cells, it is hypothesized that the
pro-proliferative and pro-stemness effects of SHCBP1 may be
attributed to its induction of NF-κB activation.
The positive regulation of NF-κB activation by EIF5A
has been indicated in previous studies (17,19).
The inhibitory effect of SHCBP1 shRNA transfection on EIF5A mRNA
was confirmed in the present study. Similar effects have been
observed in papillary thyroid cancer cells (20), and it was hypothesized that EIF5A
may mediate the biological function of SHCBP1 and the regulation of
the NF-κB signaling pathway by SHCBP1 in CC. The findings of the
present study confirm this hypothesis. The regulatory mechanism of
SHCBP1 on EIF5A remains unclear, which is one of the limitations of
the present study. SHCBP1 could enhance the interaction between
CREB-binding protein (CBP) and β-catenin, thereby activating the
transcription of genes driven by β-catenin (49). The interaction between CBP and KLF5
is of importance for KLF5 transactivation activities (50). There are KLF5 binding sites at the
promoter of EIF5A. KLF5 has been confirmed to bind to EIFA5 and
activate its transcription (51).
Based on previous studies (49–51),
it was hypothesized that the regulation of EIF5A mRNA by SHCBP1 may
be associated with the enhanced interaction between CBP and KLF5
triggered by SHCBP1, which needs to be explored in future work. In
addition, the effect of SHCBP1 on tumor growth in a xenograft model
of CC was not investigated, which may be an additional limitation
of the present study.
In summary, the role of SHCBP1 in CC was identified
in vitro. Mechanistically, SHCBP1 facilitated CC cell
proliferation, stemness and activation of the NF-κB signaling
pathway through EIF5A in vitro. The present study suggested
a novel molecular mechanism underlying the progression of CC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science
Foundation of China (grant no. 81902633).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BYD conceptualized the research project, devised
methodology, completed formal analysis and data curation, prepared
and wrote the original draft manuscript. ALL supervised the study,
performed data analysis and reviewed and edited the manuscript. YZ,
YYZ and JF performed experiments and validated the data. YM
performed interpretation and visualization of the data. BYD, ALL
and YM confirm the authenticity of all the raw data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brisson M and Drolet M: Global elimination
of cervical cancer as a public health problem. Lancet Oncol.
20:319–321. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brisson M, Kim JJ, Canfell K, Drolet M,
Gingras G, Burger EA, Martin D, Simms KT, Bénard É, Boily MC, et
al: Impact of HPV vaccination and cervical screening on cervical
cancer elimination: A comparative modelling analysis in 78
low-income and lower-middle-income countries. Lancet. 395:575–590.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schmandt R, Liu SK and McGlade CJ: Cloning
and characterization of mPAL, a novel Shc SH2 domain-binding
protein expressed in proliferating cells. Oncogene. 18:1867–1879.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li D, Peng H, Qu L, Sommar P, Wang A, Chu
T, Li X, Bi X, Liu Q, Sérézal IG, et al: MiR-19a/b and miR-20a
promote wound healing by regulating the inflammatory response of
keratinocytes. J Invest Dermatol. 141:659–671. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shi W, Zhang G, Ma Z, Li L, Liu M, Qin L,
Yu Z, Zhao L, Liu Y, Zhang X, et al: Hyperactivation of
HER2-SHCBP1-PLK1 axis promotes tumor cell mitosis and impairs
trastuzumab sensitivity to gastric cancer. Nat Commun. 12:28122021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peng C, Zhao H, Song Y, Chen W, Wang X,
Liu X, Zhang C, Zhao J, Li J, Cheng G, et al: SHCBP1 promotes
synovial sarcoma cell metastasis via targeting TGF-beta1/Smad
signaling pathway and is associated with poor prognosis. J Exp Clin
Cancer Res. 36:1412017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu L, Yang Y, Liu S, Tao T, Cai J, Wu J,
Guan H, Zhu X, He Z, Li J, et al: EGF-induced nuclear localization
of SHCBP1 activates beta-catenin signaling and promotes cancer
progression. Oncogene. 38:747–764. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wullaert A, Bonnet MC and Pasparakis M:
NF-κB in the regulation of epithelial homeostasis and inflammation.
Cell Res. 21:146–158. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ghosh S and Hayden MS: New regulators of
NF-kappaB in inflammation. Nat Rev Immunol. 8:837–848. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu Z, Peng X, Li J, Zhang Y and Hu L:
Constitutive activation of nuclear factor kappaB contributes to
cystic fibrosis transmembrane conductance regulator expression and
promotes human cervical cancer progression and poor prognosis. Int
J Gynecol Cancer. 23:906–915. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang J, Ou J, Guo Y, Dai T, Li X, Liu J,
Xia M, Liu L and He M: TBLR1 is a novel prognostic marker and
promotes epithelial-mesenchymal transition in cervical cancer. Br J
Cancer. 111:112–124. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang W, Li X, Xu Y, Guo W, Yu H, Zhang L,
Wang Y and Chen X: Acetylation-stabilized chloride intracellular
channel 1 exerts a tumor-promoting effect on cervical cancer cells
by activating NF-kappaB. Cell Oncol (Dordr). 44:557–568. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou Y, Tan Z, Chen K, Wu W, Zhu J, Wu G,
Cao L, Zhang X, Zeng X, Li J and Zhang W: Overexpression of SHCBP1
promotes migration and invasion in gliomas by activating the
NF-kappaB signaling pathway. Mol Carcinog. 57:1181–1190. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Coni S, Serrao SM, Yurtsever ZN, Magno LD,
Bordone R, Bertani C, Licursi V, Ianniello Z, Infante P, Moretti M,
et al: Blockade of EIF5A hypusination limits colorectal cancer
growth by inhibiting MYC elongation. Cell Death Dis. 11:10452020.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Z, Jiang J, Qin T, Xiao Y and Han L:
EIF5A regulates proliferation and chemoresistance in pancreatic
cancer through the sHH signalling pathway. J Cell Mol Med.
23:2678–2688. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mémin E, Hoque M, Jain MR, Heller DS, Li
H, Cracchiolo B, Hanauske-Abel HM, Pe'ery T and Mathews MB:
Blocking eIF5A modification in cervical cancer cells alters the
expression of cancer-related genes and suppresses cell
proliferation. Cancer Res. 74:552–562. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu Z, Teng L, Gao L, Wang H, Su Y and Li
J: The role of eukaryotic translation initiation factor 5A-1
(eIF5A-1) gene in HPV 16 E6 induces cell growth in human cervical
squamous carcinoma cells. Biochem Biophys Res Commun. 504:6–12.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Taylor CA, Liu Z, Tang TC, Zheng Q,
Francis S, Wang TW, Ye B, Lust JA, Dondero R and Thompson JE:
Modulation of eIF5A expression using SNS01 nanoparticles inhibits
NF-κB activity and tumor growth in murine models of multiple
myeloma. Mol Ther. 20:1305–1314. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Geng H, Guo M, Xu W, Zang X, Wu T, Teng F,
Wang Y, Liu X, Wang X, Sun Q and Liang J: SHCBP1 promotes papillary
thyroid carcinoma carcinogenesis and progression through promoting
formation of integrin and collagen and maintaining cell stemness.
Front Endocrinol (Lausanne). 11:6138792021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Scotto L, Narayan G, Nandula SV,
Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright
JD, Pothuri B, Mansukhani M and Murty VV: Identification of copy
number gain and overexpressed genes on chromosome arm 20q by an
integrative genomic approach in cervical cancer: Potential role in
progression. Genes Chromosomes Cancer. 47:755–765. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
den Boon JA, Pyeon D, Wang SS, Horswill M,
Schiffman M, Sherman M, Zuna RE, Wang Z, Hewitt SM, Pearson R, et
al: Molecular transitions from papillomavirus infection to cervical
precancer and cancer: Role of stromal estrogen receptor signaling.
Proc Natl Acad Sci USA. 112:E3255–E3264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhai Y, Kuick R, Nan B, Ota I, Weiss SJ,
Trimble CL, Fearon ER and Cho KR: Gene expression analysis of
preinvasive and invasive cervical squamous cell carcinomas
identifies HOXC10 as a key mediator of invasion. Cancer Res.
67:10163–10172. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mirzaei S, Saghari S, Bassiri F, Raesi R,
Zarrabi A, Hushmandi K, Sethi G and Tergaonkar V: NF-κB as a
regulator of cancer metastasis and therapy response: A focus on
epithelial-mesenchymal transition. J Cell Physiol. 237:2770–2795.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Strnadel J, Choi S, Fujimura K, Wang H,
Zhang W, Wyse M, Wright T, Gross E, Peinado C, Park HW, et al:
eIF5A-PEAK1 signaling regulates YAP1/TAZ protein expression and
pancreatic cancer cell growth. Cancer Res. 77:1997–2007. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tian M, Neil JR and Schiemann WP:
Transforming growth factor-β and the hallmarks of cancer. Cell
Signal. 23:951–962. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bertoli C, Skotheim JM and de Bruin RA:
Control of cell cycle transcription during G1 and S phases. Nat Rev
Mol Cell Biol. 14:518–528. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mo M, Tong S, Yin H, Jin Z, Zu X and Hu X:
SHCBP1 regulates STAT3/c-Myc signaling activation to promote tumor
progression in penile cancer. Am J Cancer Res. 10:3138–3156.
2020.PubMed/NCBI
|
|
30
|
Yang C, Hu JF, Zhan Q, Wang ZW, Li G, Pan
JJ, Huang L, Liao CY, Huang Y, Tian YF, et al: SHCBP1 interacting
with EOGT enhances O-GlcNAcylation of NOTCH1 and promotes the
development of pancreatic cancer. Genomics. 113:827–842. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu N, Wu YP, Yin HB, Chen SH, Li XD, Xue
XY and Gou X: SHCBP1 promotes tumor cell proliferation, migration,
and invasion, and is associated with poor prostate cancer
prognosis. J Cancer Res Clin Oncol. 146:1953–1969. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Feng W, Li HC, Xu K, Chen YF, Pan LY, Mei
Y, Cai H, Jiang YM, Chen T and Feng DX: SHCBP1 is over-expressed in
breast cancer and is important in the proliferation and apoptosis
of the human malignant breast cancer cell line. Gene. 587:91–97.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Batlle E and Clevers H: Cancer stem cells
revisited. Nat Med. 23:1124–1134. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tyagi A, Vishnoi K, Mahata S, Verma G,
Srivastava Y, Masaldan S, Roy BG, Bharti AC and Das BC: Cervical
cancer stem cells selectively overexpress HPV oncoprotein E6 that
controls stemness and self-renewal through upregulation of HES1.
Clin Cancer Res. 22:4170–4184. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Feng Q, Li S, Ma HM, Yang WT and Zheng PS:
LGR6 activates the Wnt/beta-catenin signaling pathway and forms a
β-catenin/TCF7L2/LGR6 feedback loop in LGR6(high) cervical cancer
stem cells. Oncogene. 40:6103–6114. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Low HY, Lee YC, Lee YJ, Wang HL, Chen YI,
Chien PJ, Li ST and Chang WW: Reciprocal regulation between
indoleamine 2,3-dioxigenase 1 and notch1 involved in radiation
response of cervical cancer stem cells. Cancers (Basel).
12:15472020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ledoux AC and Perkins ND: NF-κB and the
cell cycle. Biochem Soc Trans. 42:76–81. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Guttridge DC, Albanese C, Reuther JY,
Pestell RG and Baldwin AS Jr: NF-kappaB controls cell growth and
differentiation through transcriptional regulation of cyclin D1.
Mol Cell Biol. 19:5785–5799. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cheng SH, Hsia C, Leone G and Liou HC:
Cyclin E and Bcl-x(L) cooperatively induce cell cycle progression
in c-Rel(−/-) B cells. Oncogene. 22:8472–8486. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhu SM, Al-Mathkour M, Cao L, Khalafi S,
Chen Z, Poveda J, Peng D, Lu H, Soutto M, Hu T, et al: CDK1 bridges
NF-κB and β-catenin signaling in response to H. pylori infection in
gastric tumorigenesis. Cell Rep. 42:1120052023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu JL, Ma HP, Lu XL, Sun SH, Guo X and Li
FC: NF-κB induces abnormal centrosome amplification by upregulation
of CDK2 in laryngeal squamous cell cancer. Int J Oncol. 39:915–924.
2011.PubMed/NCBI
|
|
42
|
Marquardt JU, Gomez-Quiroz L, Camacho LO,
Pinna F, Lee YH, Kitade M, Domínguez MP, Castven D, Breuhahn K,
Conner EA, et al: Curcumin effectively inhibits oncogenic NF-κB
signaling and restrains stemness features in liver cancer. J
Hepatol. 63:661–669. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang M, Wang L, Yue Y, Zhang L, Liu T,
Jing M, Liang X, Ma M, Xu S, Wang K, et al: ITPR3 facilitates tumor
growth, metastasis and stemness by inducing the NF-ĸB/CD44 pathway
in urinary bladder carcinoma. J Exp Clin Canc Res. 40:652021.
View Article : Google Scholar
|
|
44
|
Gonzalez-Torres C, Gaytan-Cervantes J,
Mandujano-Tinoco EA, Ceballos-Cancino G, Garcia-Venzor A, Zampedri
C, Sanchez-Maldonado P, Mojica-Espinosa R, Jimenez-Hernandez LE and
Maldonado V: NF-κB participates in the stem cell phenotype of
ovarian cancer cells. Arch Med Res. 48:343–351. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dong W, Sun S, Cao X, Cui Y, Chen A, Li X,
Zhang J, Cao J and Wang Y: Exposure to TNFalpha combined with
TGFbeta induces carcinogenesis in vitro via NF-kappaB/Twist axis.
Oncol Rep. 37:1873–1882. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Smith SM, Lyu YL and Cai L: NF-κB affects
proliferation and invasiveness of breast cancer cells by regulating
CD44 expression. PLoS One. 9:e1069662014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chefetz I, Alvero AB, Holmberg JC,
Lebowitz N, Craveiro V, Yang-Hartwich Y, Yin G, Squillace L,
Soteras MG, Aldo P and Mor G: TLR2 enhances ovarian cancer stem
cell self-renewal and promotes tumor repair and recurrence. Cell
Cycle. 12:511–521. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Quan XX, Hawk NV, Chen W, Coupar J, Lee
SK, Petersen DW, Meltzer PS, Montemarano A, Braun M, Chen Z and Van
Waes C: Targeting Notch1 and IKKalpha enhanced NF-kappaB activation
in CD133(+) skin cancer stem cells. Mol Cancer Ther. 17:2034–2048.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sun Y, Pan H, He Y, Hu C and Gu Y:
Functional roles of the SHCBP1 and KIF23 interaction in modulating
the cell-cycle and cisplatin resistance of head and neck squamous
cell carcinoma. Head Neck. 44:591–605. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang Z and Teng CT: Phosphorylation of
Kruppel-like factor 5 (KLF5/IKLF) at the CBP interaction region
enhances its transactivation function. Nucleic Acids Res.
31:2196–2208. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ma D, Zheng B, Liu HL, Zhao YB, Liu X,
Zhang XH, Li Q, Shi WB, Suzuki T and Wen JK: Klf5 down-regulation
induces vascular senescence through eIF5a depletion and
mitochondrial fission. PLoS Biol. 18:e30008082020. View Article : Google Scholar : PubMed/NCBI
|