Introduction
According to the 2020 Global Cancer Statistics,
liver cancer ranks 6th in terms of global morbidity and 3rd among
causes of cancer-associated mortality worldwide (1). Hepatocellular carcinoma (HCC) is the
most frequent subtype of primary liver cancer that occurs in 75–85%
of patients with liver cancer (2).
Despite the great progress made in anti-HCC therapy, HCC is
associated with a poor prognostic outcome (3). Tumor metastasis is an important factor
contributing to the poor HCC prognosis (4). Tumor budding (TB) is defined as
dissociation of isolated cancer cells and/or discrete clusters
(<5 cancer cells), and its prognostic role was first reported in
colorectal cancer (CRC) (5). In
addition to CRC, TB has been observed in various cancer types and
predicts a poor prognosis, including in esophageal (6), nasopharyngeal (7), lung (8) and pancreatic cancer (9). Furthermore, TB has been reported to be
associated with epithelial-mesenchymal transition (EMT), which is a
critical event that supports tumor migration and invasion (10,11).
However, the impact of TB on HCC remains poorly understood. Only
two studies have analyzed the prognostic value of TB in HCC
(12,13), while the mechanistic basis of TB in
HCC remains unclear.
A disintegrin and metalloproteinase domain with
thrombospondin motifs 16 (ADAMTS16) belongs to the ADAMTS family.
Its role has been investigated in esophageal squamous cell
carcinoma (14) and CRC (15,16).
However, to the best of our knowledge, the role of ADAMTS16 in HCC
has not yet been studied. Bone morphogenetic protein 2 (BMP2),
which belongs to the transforming growth factor-β superfamily,
participates in different cancer occurrence and development
processes (17–19). Certain studies have indicated that
BMP2 enhances HCC cell growth, invasion and migration (20,21).
However, the relationship between BMP2 and TB in HCC remains to be
studied.
The aim of this study was to identify differentially
expressed genes (DEGs) in TB-positive (TB-pos) HCC tissue samples
relative to TB-negative (TB-neg) HCC tissue samples through
bioinformatics analysis, and then explore the potential mechanism
of TB in HCC. In the present study, it was demonstrated that
ADAMTS16 and BMP2 levels were significantly increased in the TB-pos
HCC samples, and BMP2 expression was significantly increased in
budding cells compared with the tumor center. Additionally, it was
demonstrated that overexpression (OE) of ADAMTS16 and BMP2 promoted
invasion of HepG2 cells, which implied that ADAMTS16 and BMP2 may
regulate TB in liver cancer. The association of ADAMTS16 and BMP2
expression with clinicopathological characteristics and patient
survival time were also analyzed. The findings of the present study
provide novel mechanistic insights into the role of ADAMTS16 and
BMP2 in HCC.
Materials and methods
Patients and tissues
Paraffin-embedded tumor specimens from patients with
HCC (n=308) were retrospectively collected from The Affiliated
Hospital of Jining Medical University (Jining, China) between June
2013 and August 2019. The clinical and pathological details of the
patients were retrospectively reviewed by accessing the hospital's
existing electronic medical record system. Ultimately, 266/308
(86%) of patients with HCC with complete clinical information were
selected for the subsequent clinical significance analysis. In
addition, of the 308 patients, 66 were excluded from the survival
analysis due to a lack of follow-up. And additional 40 HCC tissue
samples were prospectively collected from The Affiliated Hospital
of Jining Medical University between February 2020 and June 2021
for transcriptome sequencing. All samples were histopathologically
and clinically confirmed as HCC tissues. The histology, diagnostic
methods, and α-fetoprotein (AFP) levels were reviewed for each case
of HCC identified to eliminate cases of metastatic cancer or other
primary liver cancer types. Microscopically, the HCC cells were
arranged in solid nests, trabeculae, acinar or pseudoglandular
structures. Papillary structures were occasionally seen. Abundant
sinusoidal capillaries could be seen between the tumor alveolus,
with steatosis and eosinophilic bodies. Ethics approval was
provided by The Ethics Committee of The Affiliated Hospital of
Jining Medical University (Jining, China; ethical approval no.
2021C145). Each participant (>18 years of age) in the study
provided written informed consent and signed informed consent
forms.
Cell culture and lentiviral
infection
The human hepatoblastoma cell line, HepG2, was
purchased from ATCC via the Shanghai Huiying Biological Technology
Co., Ltd. The cell line was authenticated by human STR profiling
and confirmed to be mycoplasma free. HepG2 cells were cultured in
DMEM (Nanjing KeyGen Biotech Co., Ltd.) containing 10% fetal bovine
serum (FBS; Biological Industries) and 1% penicillin-streptomycin
(Nanjing KeyGen Biotech Co., Ltd.), and incubated in a humidified
5% CO2 incubator at 37°C.
The lentiviral vectors, GV513-ADAMTS16
(Ubi-MCS-CBh-gcGFP-IRES-puromycin) and GV358-BMP2
(Ubi-MCS-3FLAG-SV40-EGFP-IRES-puromycin), and the corresponding
control lentiviruses containing the empty vector, CON335 and
CON238, respectively, were purchased from Shanghai GeneChem Co.,
Ltd. Accession numbers for the two genes used in the present study
are as follows: ADAMTS16, NM139056; BMP2, NM001200. For lentiviral
infection, HepG2 cells were incubated with lentivirus at a MOI of
10 for 16 h, and the stable cell line was selected using 2 µg/ml
puromycin for a week, followed by 2 µg/ml puromycin maintenance.
The overexpression efficiency of ADAMTS16 and BMP2 were assessed
through reverse transcription-quantitative PCR (RT-qPCR).
Transcriptome sequencing and
analysis
In total, 40 HCC tissue samples were divided into
TB-pos and TB-neg groups by two independent pathologists on the
basis of hematoxylin-eosin (HE) staining for transcriptome
sequencing, which was conducted by Berry Genomics Co., Ltd.
Briefly, using the NEBNext® UltraTM RNA Library Prep Kit
for Illumina® (E7770S, New England Biolabs, Inc.), total
RNA (≥1 µg) from each sample was used for generating sequencing
libraries in-line with the manufacturer's instructions. Oligo (dT)
magnetic beads were used to enrich and purify poly A-containing
mRNA, followed by random interruption of mRNA into short fragments,
which served as templates for random-primed cDNA synthesis
performed using reverse transcriptase. Subsequently, purified
double-stranded cDNA was subjected to end-repair, A-tailing and
adapter ligation. Moreover, AMPure XP beads (Beckman Coulter,
Beverly, USA) were used to purify cDNA library fragments for
selecting the 250-300 bp cDNA fragments. Lastly, PCR amplification
was performed by ABI Q3TMReal-Time PCR System, followed by
purification of PCR products using the AMPure XP beads for
obtaining the cDNA library.
Following library construction, the Qubit 2.0
Fluorometer (Thermo Fisher Scientific, Inc.) was used to quantify
the library before diluting to 1.5 ng/µl. Then, the Agilent 2100
Bioanalyzer was used to assess the library insert size. When the
expected insert size was obtained, qPCR was conducted to precisely
quantify library effective concentration (>2 nM) for ensuing
library quality. After the library quality was confirmed, the
Illumina platform was used for sequencing to generate 150 bp paired
end reads.
The edgeR software package (version 3.28.1;
http://bioconductor.org/packages/release/bioc/html/edgeR.html)
(22) in the R/Bioconductor
environment (Release 3.6.1) was used for differential expression
analysis. To control the false discovery rate, the resulting
P-values were adjusted according to the Benjamini and Hochberg's
approach (23). Genes with log2
(Fold Change) >1 and Q<0.05 were assigned as differentially
expressed. To annotate the function of these DEGs, Gene ontology
(GO) enrichment analysis of DEG sets were implemented in topGO
(version 2.38.1; http://www.bioconductor.org/packages/release/bioc/html/topGO.html)
in the R/Bioconductor environment (Release 3.6.1). GO terms with
adjusted P<0.05 were considered as significantly enriched by
DEGs.
RT-qPCR
Total RNA from the HepG2 cells was extracted using
TRIzol reagent (Ambion; Thermo Fisher Scientific, Inc.). Reverse
transcription was conducted using the HiScript III RT SuperMix for
qPCR (+gDNA wiper) (Vazyme Biotech Co., Ltd.) according to the
manufacturer's protocol. The reverse transcription reactions were
performed at 42°C for 2 min, followed by 37°C for 15 min and 85°C
for 5 sec. qPCR was performed using ChamQ Universal SYBR qPCR
Master Mix (Vazyme Biotech Co., Ltd.) in a CFX Connect Real-time
System (Bio-Rad Laboratories Inc.). GAPDH was employed as the
internal reference and the 2−∆∆Cq method was used for
quantification (24). Primer
sequences for GAPDH, ADAMTS16 and BMP2 were as follows: GAPDH,
5′-CTGACTTCAACAGCGACACC-3′ (forward) and
5′-TGCTGTAGCCAAATTCGTTGT-3′ (reverse); ADAMTS16,
5′-CCGGCCGGTACAAATTTTCG-3′ (forward) and
5′-AACAGCAGCTCCACAATCAGT-3′ (reverse); BMP2,
5′-AGAATGCAAGCAGGTGGGAA-3′ (forward) and 5′-TCTTGGTGCAAAGACCTGCT-3′
(reverse).
Inverse Matrigel invasion assays
The inverse Matrigel invasion assays were performed
as reported previously (25). To
evaluate the TB capacity of HepG2 cells, 8-µm pore sized Transwell
chambers (Corning, Inc.) were used. Briefly, the Transwell chambers
were hydrated using serum-free DMEM medium for 30 min. Then, the
Matrigel (BD Biosciences) was added to each chamber and solidified
in an incubator at 37°C for 1 h. Afterwards, the chambers were
inverted and the cells (2×105/well) were seeded onto the
upward facing underside of the chamber. Following cell attachment,
serum-free DMEM medium was placed in the lower chamber and DMEM
medium containing 20% FBS (serving as a chemoattractant) was added
to the upper chamber. The cells were allowed to invade through the
Matrigel for 3-5 days in a 37°C incubator. Finally, the 3D
reconstruction of cell invasion was obtained through Zeiss 800
laser confocal microscopy. The ratio of the fluorescence values of
GFP-positive budding cells/total fluorescence value of all
GFP-positive cells × 100% was calculated using Image J software
(version 1.8.0) to quantify the invaded cells (TB rate).
Spheroid-based sprouting assays
Spheroid-based sprouting assays were performed as
previously described (26), with
some optimization. HepG2 spheroids were obtained using the hanging
drop method. HepG2 cells were suspended at a density of
1×104 cells/45 µl and then seeded on the lid of 48-well
culture plates for 2 days in an incubator at 37°C. Then, all
spheroids were harvested and embedded in Matrigel for 2 days in a
37°C incubator. Finally, images were captured using a light
microscope (OPTIKA).
Immunohistochemistry (IHC)
In total, 308 formalin-fixed, paraffin-embedded
tissue samples from patients with HCC were used to establish HCC
tissue microarrays (TMAs) with a 2.0-mm diameter per core for IHC.
In brief, TMAs were dewaxed, hydrated and antigen repaired by EDTA
antigen repair buffer (PH 8.0), and the endogenous peroxidase
activity was then blocked. TMAs were blocked in goat serum (OriGene
Technologies, Inc.) at room temperature for 30 min to block
non-specific staining. Primary antibodies were incubated with the
TMAs overnight at 4°C, including ADAMTS16 (1:200; cat. no. DF9173;
Affinity Biosciences) and BMP2 (1:200; cat. no. A0231; ABclonal
Biotech Co., Ltd.). Subsequently, a horseradish peroxidase-labeled
secondary antibody (KIT-5020; Maxim Co., Ltd., Fuzhou, China) was
incubated with the TMAs for 30 min at room temperature. Finally, a
chromogenic reaction was developed with DAB kit (DAB-0031 (20×);
Fuzhou Maixin Biotech Co., Ltd.), followed by counterstaining with
hematoxylin (G1120; Beijing Solarbio Science & Technology Co.,
Ltd.) for 10-30 sec at room temperature. All IHC staining analyses
were assessed using a semi-quantitative scoring approach by two
senior pathologists. IHC scores were calculated by multiplying
staining intensity with the stained area, in which the staining
intensity was scored as follows: 0 (no staining), 1 (light yellow
staining), 2 (yellow-brown staining) and 3 (brown staining). The
staining area was scored as follows: 1 (1–25%), 2 (26–50%), 3
(51–75%) and 4 (76–100%), according to the percentage of stained
area in the field of vision. The median IHC score of ADAMTS16 or
BMP2 was used as a cut-off to divide the samples into the low
expression and high expression groups. ‘High’ was defined an IHC
score higher than the cut-off value, and ‘low’ was defined as an
IHC score lower than or equal the cut-off value.
Statistical analysis
SPSS 25.0 (IBM Corp.) was used for statistical
analyses. Differences between two groups were analyzed using the
Wilcoxon rank-sum test or unpaired student's t-test. The
association of ADAMTS16 and BMP2 expressions with
clinicopathological features was analyzed using the chi-square test
or Fisher's exact test as appropriate. Kaplan-Meier (KM) curves
were generated using the ‘survfit’ function in the survival package
of R software (version 3.5.3), while significant differences in
survival were compared using the log-rank test or Cramer-von Mises
test as appropriate. Cramer-von Mises test was performed to
generate the P-values when KM curves crossed over. P<0.05 was
considered to indicate a statistically significant difference.
Results
Transcriptome sequencing analysis
A total of 40 surgical HCC specimens were divided
into two groups: TB-pos group (n=21) and TB-neg group (n=19),
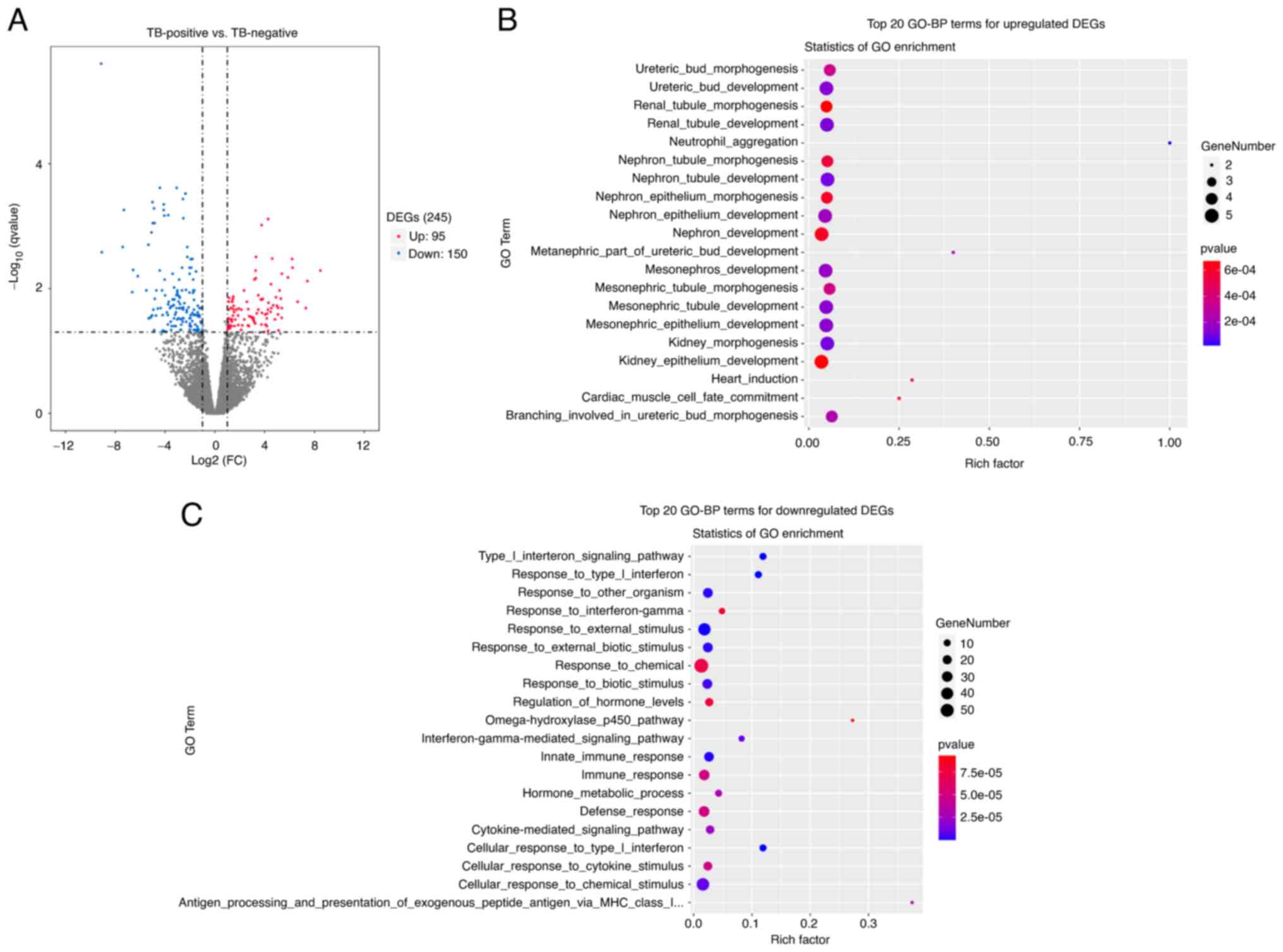
followed by transcriptome sequencing analysis. As shown in the
volcano plot in Fig. 1A, 245 DEGs
including 95 upregulated DEGs and 150 downregulated DEGs were
obtained for the TB-pos group compared with the TB-neg group. GO
enrichment analysis was subsequently performed for the 95
upregulated DEGs. The top 20 upregulated biological processes (BPs)
are shown in Fig. 1B. The
predominant BPs of the upregulated DEGs are associated with
embryonic kidney development.
Results of GO (BP) enrichment analysis for
downregulated genes are shown in Fig.
1C. It was noted that downregulated genes were mainly involved
in immune-related processes, such as immune response, innate immune
response and response to type I interferon. Certain studies have
illustrated that the downregulated immune-related genes are
associated with tumor immune escape (27–29).
The details of BP enrichment of upregulated genes in
the GO analysis are provided in Table
SI. In the present study, BPs of embryonic kidney
development-related processes related to the upregulated genes of
HCC with TB was made the focus as this is a novel area. Genes
involved in the BPs associated with embryonic kidney development
include ADAMTS16, BMP2, CALB1, FOXD1 and WT1. Furthermore, we found
that there were 4 common upregulated genes in these embryonic
kidney development-related processes, including ADAMTS16, BMP2, WT1
and FOXD1. ADAMTS16 and BMP2 were selected for further
investigation. In addition, we noted that S100A9 which involved in
neutrophil aggregation, was a poor prognosis-related gene. The
survival analysis result of S100A9 based on TCGA data showed that
the higher expression level of S100A9 was also linked with a worse
prognosis of HCC patients (P=0.013) (Fig. S1). It was also reported in the
literature that S100A9 expression could potentially serve as an
independent prognostic marker for HCC (30).
High ADAMTS16 and BMP2 expression
levels are associated with TB in HCC
To identify ADAMTS16 and BMP2 expression profiles
within HCC tissues with different TB statuses, IHC staining was
performed using TMAs of 308 paraffin-embedded HCC tissues. In
addition, the DEGs of the budding cells and cancer center were
compared. The statistical analysis results for the IHC staining
scores of ADAMTS16 and BMP2 expression are summarized in Tables I and II. As shown in Table I, the staining scores of ADAMTS16
expression in the TB-pos HCC tissues were significantly higher than
those in the TB-neg HCC tissues (P=0.005). However, the staining
scores of ADAMTS16 expression demonstrated no significant
difference between the tumor center and budding cells (P=0.174). As
shown in Table II, the staining
scores of BMP2 expression were significantly higher in TB-pos group
compared with that in TB-neg group (P=0.015), and the significantly
higher staining scores in budding cells than in tumor center
(P=0.042) were observed. The representative results for IHC
staining of ADAMTS16 and BMP2 expressions are illustrated in
Fig. 2. These results demonstrated
that upregulation of ADAMTS16 and BMP2 expression may be related to
TB in HCC. Moreover, BMP2 expression was significantly increased in
the budding cells compared with the tumor center.
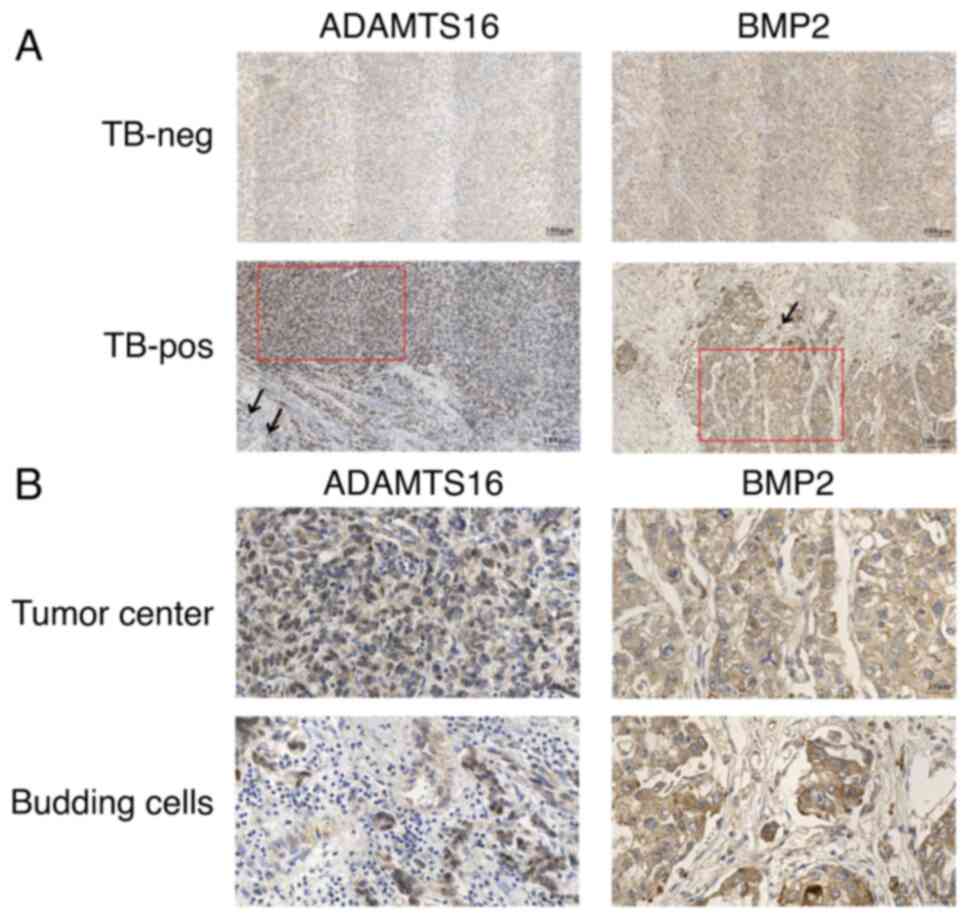 | Figure 2.Immunohistochemical analysis of
ADAMTS16 and BMP2 in patients with HCC with different TB statuses.
(A) Representative IHC staining images of ADAMTS16 and BMP2 in
TB-neg and TB-pos HCC tissues (magnification, ×100). Red boxes
represent the tumor center and black arrows indicate budding tumor
cells. (B) Representative IHC images of ADAMTS16 and BMP2 levels in
the tumor center and budding cells (magnification, ×400), which are
magnified views of the areas indicated by the red boxes and black
arrows in (A), respectively. ADAMTS16, a metalloproteinase domain
with thrombospondin motifs 16; BMP2, bone morphogenetic protein 2;
HCC, hepatocellular carcinoma; IHC, immunohistochemistry; neg,
negative; pos, positive; TB, tumor budding. |
 | Table I.Association between a
metalloproteinase domain with thrombospondin motifs 16 expression
and TB in hepatocellular carcinoma tissues (n=308). |
Table I.
Association between a
metalloproteinase domain with thrombospondin motifs 16 expression
and TB in hepatocellular carcinoma tissues (n=308).
|
|
|
| Two-sample Wilcoxon
rank-sum test |
|---|
|
|
|
|
|
|---|
| Group | Median (P25,
P75) | Median difference
(95% CI) | Z value | P-value |
|---|
| TB |
| −1.000
(−1.000-0.000) | −2.802 | 0.005a |
|
TB-neg | 2 (1, 4) |
|
|
|
|
TB-pos | 3 (1, 6) |
|
|
|
| Area |
| 0.000
(−2.000-0.000) | −1.359 | 0.174 |
| Tumor
center | 3 (2, 6) |
|
|
|
| Budding
cells | 4 (3, 8) |
|
|
|
 | Table II.Association between bone
morphogenetic protein 2 expression and TB in hepatocellular
carcinoma tissues (n=308). |
Table II.
Association between bone
morphogenetic protein 2 expression and TB in hepatocellular
carcinoma tissues (n=308).
|
|
|
| Two-sample Wilcoxon
rank-sum test |
|---|
|
|
|
|
|
|---|
| Group | Median (P25,
P75) | Median difference
(95% CI) | Z value | P-value |
|---|
| TB |
| 0.000
(0.000-0.000) | −2.434 | 0.015a |
|
TB-neg | 8 (8, 9) |
|
|
|
|
TB-pos | 8 (8, 12) |
|
|
|
| Area |
| 0.000
(−2.000-0.000) | −2.038 | 0.042a |
| Tumor
center | 8 (8, 12) |
|
|
|
| Budding
cells | 12 (8, 12) |
|
|
|
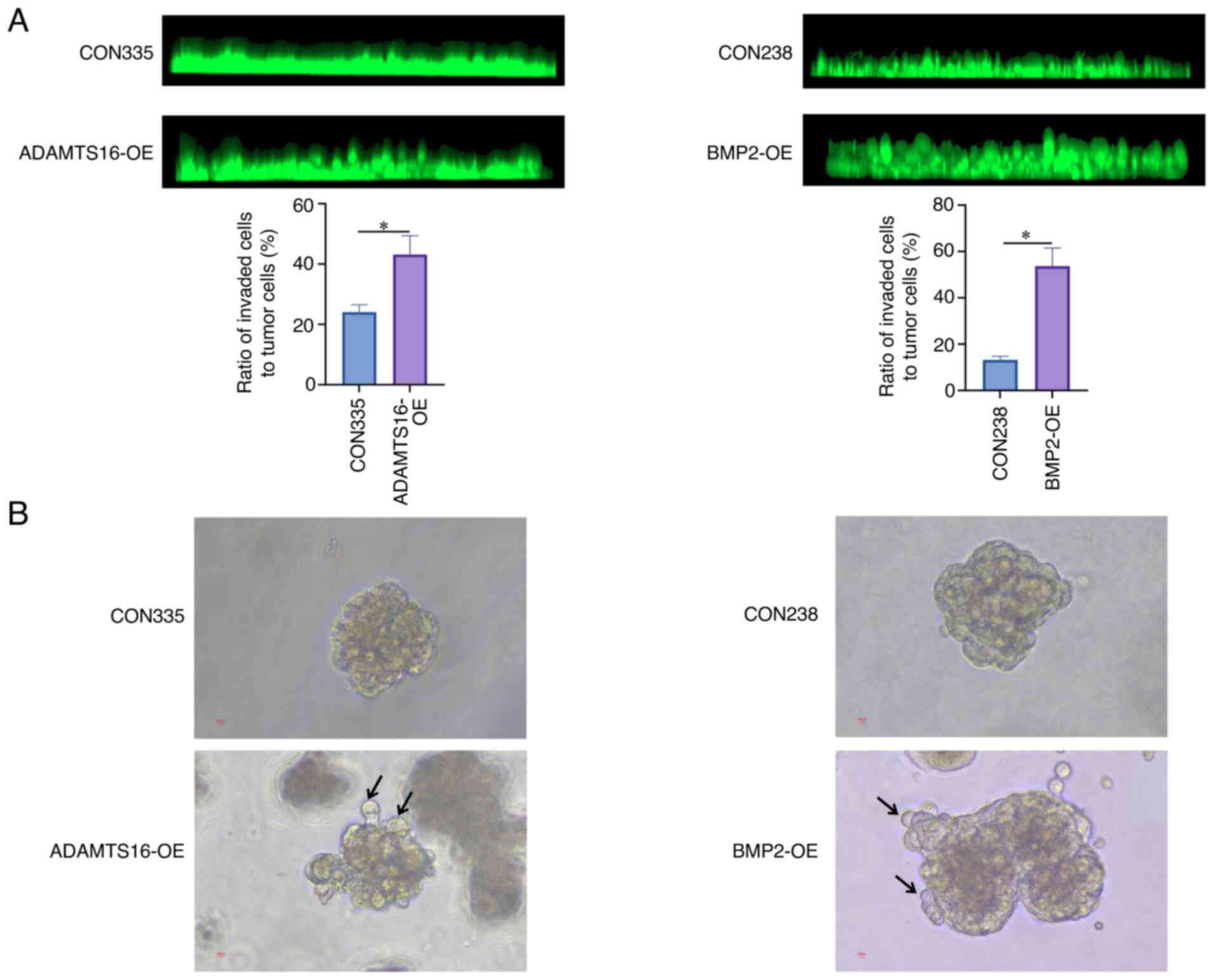
ADAMTS16 and BMP2 promote the TB of
liver cancer in vitro
To explore whether ADAMTS16 and BMP2 play a role in
the TB of liver cancer, HepG2 cells with stable ADAMTS16-OE or
BMP2-OE were constructed using lentiviral vectors. The
overexpression efficiency was assessed through RT-qPCR assays
(Fig. S2). Subsequently, the TB
ability of HepG2 cells was evaluated using in vitro inverse
Matrigel invasion and spheroid-based sprouting assays. As shown in
Fig. 3A, the ratio of TB in
ADAMTS16-OE or BMP2-OE HepG2 cells increased significantly relative
to their respective controls. Similarly, the results of the
spheroid-based sprouting assays demonstrated that ADAMTS16-OE or
BMP2-OE in HepG2 cells resulted in more budding cells compared with
the control cells (Fig. 3B). These
results demonstrated that ADAMTS16 and BMP2 may be involved in the
regulation of TB of liver cancer.
Association of ADAMTS16 and BMP2
expression levels with clinicopathological characteristics in
HCC
To analyze the association of ADAMTS16 and BMP2
expression levels with HCC clinicopathological characteristics, 266
HCC cases with complete available clinicopathological information
for analysis were enrolled. These 266 cases were classified into
the low- or high-expression groups by considering the respective
median IHC scores for ADAMTS16 and BMP2 expression as the cut-off.
The relationship between the clinicopathological characteristics of
patients and gene expression was assessed using the chi-square test
and Fisher's exact test. Consequently, ADAMTS16 expression was
found to be significantly associated with necrosis (P=0.023) and
cholestasis (P=0.011) (Table
III). As shown in Table IV,
BMP2 expression level had a significant association with the
Barcelona Clinic Liver Cancer (BCLC) stage (B-C vs. 0-A; P=0.003)
and the vessels encapsulating tumor cluster (VETC; P=0.014), which
is one of the vessel types in HCC.
 | Table III.Association between ADAMTS16
expression and the clinicopathological characteristics in patients
with hepatocellular carcinoma (n=266). |
Table III.
Association between ADAMTS16
expression and the clinicopathological characteristics in patients
with hepatocellular carcinoma (n=266).
|
| ADAMTS16 |
|
|---|
|
|
|
|
|---|
| Clinicopathological
characteristics | Low, n (n=131) | High, n
(n=135) | P-value |
|---|
| Age, years |
|
| 0.848 |
|
>60 | 48 | 51 |
|
|
≤60 | 83 | 84 |
|
| Sex |
|
| 0.359 |
|
Male | 105 | 114 |
|
|
Female | 26 | 21 |
|
| HBV infection |
|
| 0.193 |
|
Positive | 116 | 112 |
|
|
Negative | 15 | 23 |
|
| HCV infection |
|
| 0.122 |
|
Positive | 0 | 4 |
|
|
Negative | 131 | 131 |
|
| AFP serum level,
ng/ml |
|
| 0.407 |
|
>400 | 43 | 38 |
|
|
≤400 | 88 | 97 |
|
| Liver cirrhosis
(Yes vs. No) |
|
| 0.359 |
|
Yes | 106 | 103 |
|
| No | 25 | 32 |
|
| BCLC stage |
|
| 0.325 |
|
B-C | 21 | 16 |
|
|
0-A | 110 | 119 |
|
| Tumor size, cm |
|
| 0.969 |
|
>5 | 55 | 57 |
|
| ≤5 | 76 | 78 |
|
| Tumor number |
|
| 0.114 |
|
Multiple | 12 | 21 |
|
|
Single | 119 | 114 |
|
| Intrahepatic
metastasis |
|
| 0.114 |
|
Yes | 12 | 21 |
|
| No | 119 | 114 |
|
| Collective
invasion |
|
| 0.693 |
|
Yes | 9 | 11 |
|
| No | 122 | 124 |
|
| Ki-67, % |
|
| 0.236 |
|
>30 | 31 | 24 |
|
|
≤30 | 100 | 111 |
|
| Necrosis |
|
| 0.023a |
|
Yes | 28 | 15 |
|
| No | 103 | 120 |
|
| Vessel carcinoma
embolus |
|
| 0.285 |
|
Yes | 17 | 12 |
|
| No | 114 | 123 |
|
| Microtrabecular
pattern |
|
| 0.735 |
|
Yes | 106 | 107 |
|
| No | 25 | 28 |
|
| Macrotrabecular
pattern |
|
| 0.780 |
|
Yes | 75 | 75 |
|
| No | 56 | 60 |
|
| Pseudoglandular
pattern |
|
| 0.130 |
|
Yes | 23 | 34 |
|
| No | 108 | 101 |
|
| Compact
pattern |
|
| 0.865 |
|
Yes | 53 | 56 |
|
| No | 78 | 79 |
|
| Cholestasis |
|
| 0.011a |
|
Yes | 17 | 34 |
|
| No | 114 | 101 |
|
| Hyaline bodies |
|
| 0.273 |
|
Yes | 23 | 31 |
|
| No | 108 | 104 |
|
| Steatosis |
|
| 0.908 |
|
Yes | 24 | 24 |
|
| No | 107 | 111 |
|
| Edmondson
grade |
|
| 0.136 |
|
III–IV | 46 | 36 |
|
|
I–II | 85 | 99 |
|
| VETC |
|
| 0.497 |
|
Yes | 33 | 39 |
|
| No | 98 | 96 |
|
 | Table IV.Association between BMP2 expression
and the clinicopathological characteristics in patients with
hepatocellular carcinoma (n=266). |
Table IV.
Association between BMP2 expression
and the clinicopathological characteristics in patients with
hepatocellular carcinoma (n=266).
|
| BMP2 |
|
|---|
|
|
|
|
|---|
| Clinicopathological
characteristics | Low, n (n=172) | High, n (n=94) | P-value |
|---|
| Age, years |
|
| 0.290 |
|
>60 | 68 | 31 |
|
|
≤60 | 104 | 63 |
|
| Sex |
|
| 0.588 |
|
Male | 140 | 79 |
|
|
Female | 32 | 15 |
|
| HBV infection |
|
| 0.105 |
|
Positive | 143 | 85 |
|
|
Negative | 29 | 9 |
|
| HCV infection |
|
| 0.301 |
|
Positive | 4 | 0 |
|
|
Negative | 168 | 94 |
|
| AFP serum level,
ng/ml |
|
| 0.508 |
|
>400 | 50 | 31 |
|
|
≤400 | 122 | 63 |
|
| Liver cirrhosis
(Yes vs. No) |
|
| 0.789 |
|
Yes | 136 | 73 |
|
| No | 36 | 21 |
|
| BCLC stage |
|
| 0.003a |
|
B-C | 16 | 21 |
|
|
0-A | 156 | 73 |
|
| Tumor size, cm |
|
| 0.054 |
|
>5 | 65 | 47 |
|
| ≤5 | 107 | 47 |
|
| Tumor number |
|
| 0.091 |
|
Multiple | 17 | 16 |
|
|
Single | 155 | 78 |
|
| Intrahepatic
metastasis |
|
| 0.091 |
|
Yes | 17 | 16 |
|
| No | 155 | 78 |
|
| Collective
invasion |
|
| 0.604 |
|
Yes | 14 | 6 |
|
| No | 158 | 88 |
|
| Ki-67, % |
|
| 0.417 |
|
>30 | 33 | 22 |
|
|
≤30 | 139 | 72 |
|
| Necrosis |
|
| 0.530 |
|
Yes | 26 | 17 |
|
| No | 146 | 77 |
|
| Vessel carcinoma
embolus |
|
| 0.123 |
|
Yes | 15 | 14 |
|
| No | 157 | 80 |
|
| Microtrabecular
pattern |
|
| 0.683 |
|
Yes | 139 | 74 |
|
| No | 33 | 20 |
|
| Macrotrabecular
pattern |
|
| 0.302 |
|
Yes | 93 | 57 |
|
| No | 79 | 37 |
|
| Pseudoglandular
pattern |
|
| 0.964 |
|
Yes | 37 | 20 |
|
| No | 135 | 74 |
|
| Compact
pattern |
|
| 0.511 |
|
Yes | 73 | 36 |
|
| No | 99 | 58 |
|
| Cholestasis |
|
| 0.750 |
|
Yes | 32 | 19 |
|
| No | 140 | 75 |
|
| Hyaline bodies |
|
| 0.352 |
|
Yes | 32 | 22 |
|
| No | 140 | 72 |
|
| Steatosis |
|
| 0.186 |
|
Yes | 35 | 13 |
|
| No | 137 | 81 |
|
| Edmondson
grade |
|
| 0.264 |
|
III–IV | 49 | 33 |
|
|
I–II | 123 | 61 |
|
| VETC |
|
| 0.014a |
|
Yes | 38 | 34 |
|
| No | 134 | 60 |
|
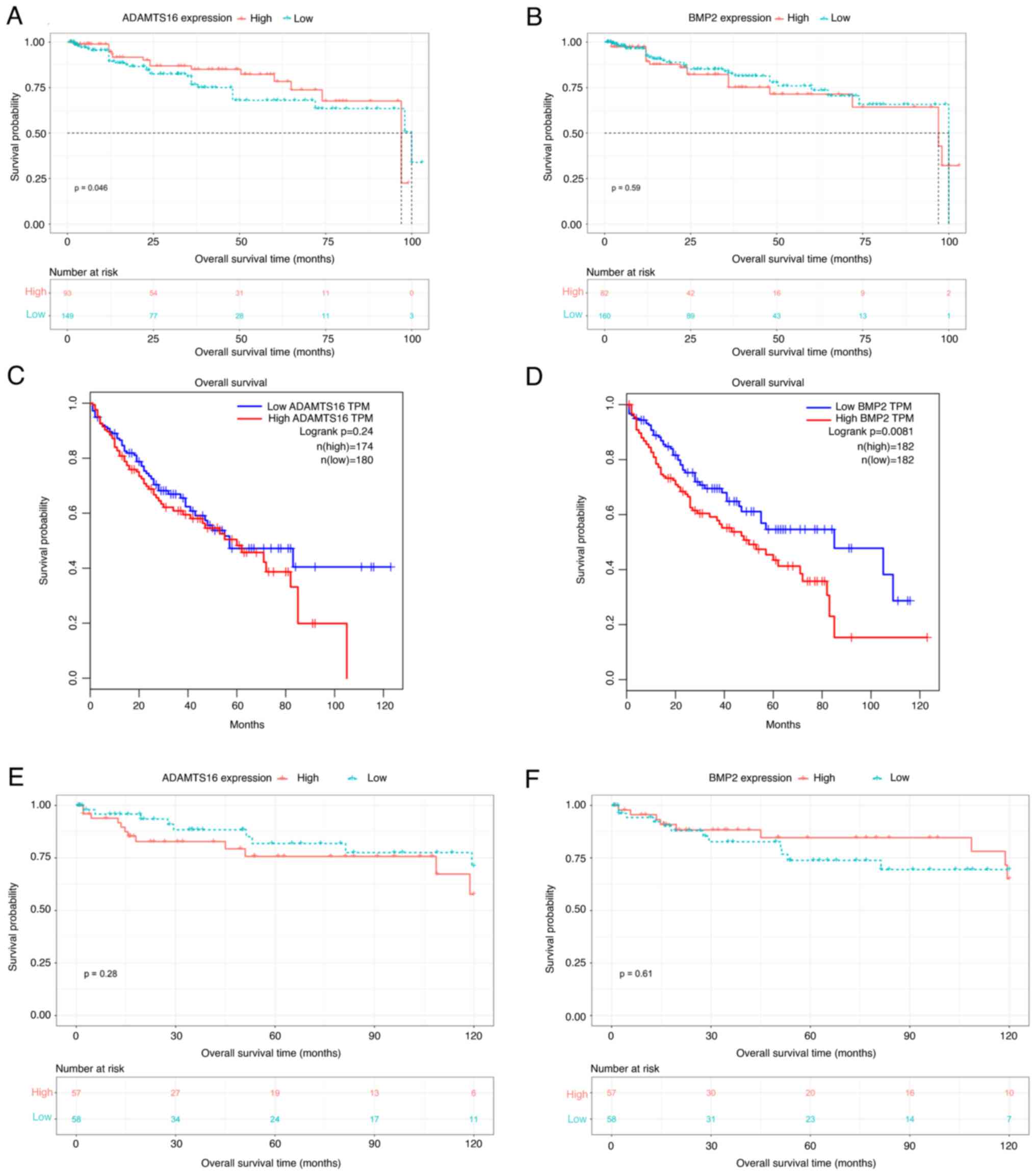
Relationship between ADAMTS16 and BMP2
levels and the prognosis of patients with HCC
A total of 242 HCC cases with available follow-up
data were enrolled for survival analysis. These 242 HCC cases were
divided into two groups (low or high expression) based on the
median IHC scores for ADAMTS16 or BMP2. As shown in Fig. 4A and B, the KM survival curves
demonstrated that ADAMTS16 expression was statistically
significantly associated with overall survival (OS; ADAMTS16,
P=0.046) in patients with HCC. In addition, BMP2 expression was not
associated with the OS of patients with HCC (BMP2, P=0.59).
Subsequently, the survival curves of these two genes were
downloaded from the GEPIA database (http://gepia.cancer-pku.cn/detail.php). As shown in
Fig. 4C and D, the survival time of
patients with HCC with high BMP2 expression significantly decreased
compared with that of patients with low BMP2 expression (P=0.0081).
ADAMTS16 expression did not differ significantly between the two
groups (P=0.24). The GEO database (accession no. GSE76427;
http://www.ncbi.nlm.nih.gov/geo/) was
also utilized to perform survival analyses comparing expression
levels of ADAMTS16 and BMP2 in HCC. The results demonstrated that
neither ADAMTS16 (P=0.28) nor BMP2 (P=0.61) expression were
associated with the OS time of patients with HCC (Fig. 4E and F). The potential reasons for
this discrepancy were elucidated in the discussion.
Discussion
TB, a histological phenomenon observed in the tumor
invasive edge, has been recognized as the initial stage of cancer
invasion and metastasis (31,32).
TB has been reported to portend lymphatic/vascular invasion, lymph
node metastasis, distant metastasis, failure to respond to
neoadjuvant chemoradiotherapy, locoregional relapse and poor
survival (6,7,33–37).
Certain studies have demonstrated an association between TB and low
survival rates in numerous types of solid cancers, including
nasopharyngeal, lung, pancreatic and esophageal cancer (6–9).
However, little is known about the effect of TB on HCC. Moreover,
TB can only be definitively determined following surgery and based
on detailed pathological and immunohistochemical examinations, thus
limiting its role in selecting the therapeutic options for HCC.
Therefore, exploring the role of TB in HCC and its molecular
mechanisms is crucial for identifying new therapeutic targets for
combating HCC.
In the present study, the expression profiles of 40
HCC tissues were analyzed using transcriptome sequencing
technology. Functional annotation indicated that upregulated DEGs,
obtained by comparing the gene expression of TB-pos and TB-neg HCC
tissues, were mainly involved in embryonic kidney development,
including nephron tubule development, kidney morphogenesis, renal
tubule development and ureteric bud development. With advancements
in developmental and cancer biology, numerous studies have
suggested that cancer invasion and metastasis are similar to normal
embryonic development (38–40). Hence, the TB process may be
considered to simulate embryonic kidney development. Furthermore,
ADAMTS16 and BMP2 were selected from the enriched genes associated
with embryonic kidney development for follow-up studies. ADAMTS16
belongs to the ADAMTS family and it has been demonstrated to
promote the proliferation and invasion of gastric cancer (41) and esophageal cancer (15) cells. However, to the best of our
knowledge, the function of ADAMTS16 has not yet been studied in
HCC. BMP2, which belongs to the transforming growth factor-β
superfamily, participates in different cancer occurrence and
development processes (17–19). Certain studies have indicated that
BMP2 may enhance HCC cell growth, invasion and migration (20,21).
However, to the best of our knowledge, no study has discussed the
relationship between BMP2 and TB in HCC.
The IHC results in the present study indicated an
upregulation of ADAMTS16 and BMP2 expression in TB-pos HCC tissues
compared with TB-neg HCC tissues. BMP2 expression in budding cells
was also increased compared with that in the tumor center.
Moreover, through the inverse Matrigel invasion and spheroid-based
sprouting assays, the present study revealed that ADAMTS16 and BMP2
may regulate the TB process of liver cancer. Therefore, we
hypothesize that these two genes may be associated with the
malignant progression of liver cancer. Furthermore, the association
of ADAMTS16 and BMP2 expression with clinicopathological
characteristics of patients with HCC was analyzed, and an
association between ADAMTS16 expression and necrosis and
cholestasis was confirmed in. In addition, BMP2 expression was
significantly associated with the BCLC stage and VETC.
It is well known that rapidly growing tumor cells,
which represent an important malignant behavior of tumors, require
an adequate supply of oxygen and nutrients, and the blood supply
cannot meet this need of rapid tumor growth, eventually resulting
in tumor necrosis (42). Therefore,
the significant association between ADAMTS16 and necrosis in HCC
observed in the present study may be caused by the ability of
ADAMTS16 to promote tumor malignant progression of HCC. To the best
of our knowledge, no study has reported the association of ADAMTS
family members with cholestasis. However, a close relative of the
ADAMTS family, ADAM17, has been reported to be related to
cholestasis (43). Increased ADAM17
expression was found in patients with two important cholestatic
liver diseases, including primary biliary cholangitis and primary
sclerosing cholangitis (43). Given
the functional similarity of these two genes, the result regarding
the association of ADAMTS16 with cholestasis in HCC is reasonable
and reliable.
As for BMP2, in the present study, patients with
BCLC stage B-C had significantly higher BMP2 expression levels than
patients with BCLC stage 0-A, suggesting the involvement of BMP2 in
the progression and metastasis of HCC. Previous studies have
demonstrated that BMP2 may promote HCC cell proliferation and
invasion, thereby promoting malignant progression of HCC (20,21,44).
As such, the results of the present study are in accordance with
previously reported results. Certain studies have also demonstrated
that BMP2 promotes angiogenesis of solid tumors, including HCC
(45,46). In addition, VETC, a novel vascular
pattern distinct from microvascular invasion, has become a powerful
predictor of aggressive HCC (47).
To the best of our knowledge, the present study is the first to
report that BMP2 is significantly associated with VETC in HCC,
which may explain the mechanism of this specific angiogenesis
pattern in HCC. Tumor malignancy is closely associated with the
invasiveness and metastasis of tumor which depends upon EMT
(48,49). And TB is considered to be an
EMT-like process (10). It has been
documented that ADAMTS16 may promote cell migration and invasion
through the NF-κB pathway (41). In
addition, BMP2 also enhances the migration, invasion and EMT of
tumor cells through the m-TOR signaling pathway (45,50).
Hence, it is speculated that a contributing mechanism to TB in HCC
may involve the m-TOR and/or NF-κB pathways.
The overall survival analysis of 242 patients with
HCC indicated that ADAMTS16 expression was associated with HCC
prognosis whereas BMP2 expression was not associated with HCC
prognosis. In addition, the survival data obtained in the present
study is inconsistent with the results predicted using the GSE76427
dataset and the results predicted using GEPIA, which is a The
Cancer Genome Atlas (TCGA)-based online tool. The results of the
GEPIA analysis demonstrated that BMP2 upregulation was
significantly associated with poor OS. The results of the GSE76427
dataset showed that neither ADAMTS16 expression nor BMP2 expression
was associated with HCC prognosis. The possible reasons for this
discrepancy are as follows: Firstly, sample size varied widely
across these three cohorts. which may cause the inconsistent
results. And compared with the large sample size within the TCGA
cohort, the retrospective cohort of the present study and the
GSE76427 cohort have relatively small sample sizes. Secondly, the
sources of the tumor samples in the three cohorts were different.
The majority of patients from TCGA database are white. In GSE76427
cohort, all of the patients were derived from Singapore. And our
study is based on data collected from a single center (Affiliated
Hospital of Jining Medical University, China). Thirdly, as patient
characteristics, surgical skills and treatment regimens are
different among countries, the final outcome of patients with HCC
could be affected. The absence of animal models of HCC is also a
limitation of this study, animal models of HCC will be constructed
for further study validation in the future.
Regardless of these limitations, to the best of our
knowledge, the present study was the first to investigate the
TB-related molecular mechanism in HCC. The findings of the present
study provide evidence for mechanism studies of TB. Moreover, the
present study provides a basis for the potential application of
ADAMTS16 and BMP2 as predictive diagnosis markers and treatment
targets for HCC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural Science
Foundation of China (grant no. 81972629), the Taishan Scholars
Program of Shandong Province (grant no. tsqn201909193), the
Shandong Youth Innovation and Technology program (grant no.
2020KJL003), the Jining Research and Development Program (grant
nos. 2021YXNS065 and 2021YXNS075), and the Research Fund for Lin He
Academician New Medicine (grant no. JYHL2021FMS12).
Availability of data and materials
The RNA-Seq datasets generated and/or analyzed
during the current study are available in the GEO repository, the
accession number is GSE227335. All other data generated or analyzed
during this study are included in this published article.
Authors' contributions
DJ performed the experiments, data analysis and
collection of clinical specimens, and drafted the manuscript. SX
assisted with the data analysis and reverse
transcription-quantitative PCR. CZ performed the bioinformatics
analysis of the GEO database. CZ, CH and LL contributed to the
analysis and interpretation of the data. MZ and HW contributed to
the acquisition and interpretation of the data. DY and YL conceived
and designed the study. YL revised the manuscript. DJ and SX
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients. The study was approved by The Ethics Committee of the
Affiliated Hospital of Jining Medical University (ethical approval
no. 2021C145; Jining, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang JD, Hainaut P, Gores GJ, Amadou A,
Plymoth A and Roberts LR: A global view of hepatocellular
carcinoma: Trends, risk, prevention and management. Nat Rev
Gastroenterol Hepatol. 16:589–604. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Turajlic S and Swanton C: Metastasis as an
evolutionary process. Science. 352:169–175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hase K, Shatney C, Johnson D, Trollope M
and Vierra M: Prognostic value of tumor ‘budding’ in patients with
colorectal cancer. Dis Colon Rectum. 36:627–635. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Koike M, Kodera Y, Itoh Y, Nakayama G,
Fujiwara M, Hamajima N and Nakao A: Multivariate analysis of the
pathologic features of esophageal squamous cell cancer: Tumor
budding is a significant independent prognostic factor. Ann Surg
Oncol. 15:1977–1982. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Luo WR, Gao F, Li SY and Yao KT: Tumour
budding and the expression of cancer stem cell marker aldehyde
dehydrogenase 1 in nasopharyngeal carcinoma. Histopathology.
61:1072–1081. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Masuda R, Kijima H, Imamura N, Aruga N,
Nakamura Y, Masuda D, Takeichi H, Kato N, Nakagawa T, Tanaka M, et
al: Tumor budding is a significant indicator of a poor prognosis in
lung squamous cell carcinoma patients. Mol Med Rep. 6:937–943.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Karamitopoulou E, Zlobec I, Born D,
Kondi-Pafiti A, Lykoudis P, Mellou A, Gennatas K, Gloor B and Lugli
A: Tumour budding is a strong and independent prognostic factor in
pancreatic cancer. Eur J Cancer. 49:1032–1039. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Grigore AD, Jolly MK, Jia D, Farach-Carson
MC and Levine H: Tumor budding: The name is EMT. Partial EMT. J
Clin Med. 5:512016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
De Smedt L, Palmans S, Andel D, Govaere O,
Boeckx B, Smeets D, Galle E, Wouters J, Barras D, Suffiotti M, et
al: Expression profiling of budding cells in colorectal cancer
reveals an EMT-like phenotype and molecular subtype switching. Br J
Cancer. 116:58–65. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kairaluoma V, Kemi N, Pohjanen VM, Saarnio
J and Helminen O: Tumour budding and tumour-stroma ratio in
hepatocellular carcinoma. Br J Cancer. 123:38–45. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wei L, Delin Z, Kefei Y, Hong W, Jiwei H
and Yange Z: A classification based on tumor budding and immune
score for patients with hepatocellular carcinoma. Oncoimmunology.
9:16724952019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang D, Qian C, Wei H and Qian X:
Identification of the prognostic value of tumor
microenvironment-related genes in esophageal squamous cell
carcinoma. Front Mol Biosci. 7:5994752020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sakamoto N, Oue N, Noguchi T, Sentani K,
Anami K, Sanada Y, Yoshida K and Yasui W: Serial analysis of gene
expression of esophageal squamous cell carcinoma: ADAMTS16 is
upregulated in esophageal squamous cell carcinoma. Cancer Sci.
101:1038–1044. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mersakova S, Janikova K, Kalman M,
Marcinek J, Grendar M, Vojtko M, Kycina R, Pindura M, Janik J,
Mikolajcik P, et al: Cancer stem cell marker expression and
methylation status in patients with colorectal cancer. Oncol Lett.
24:2312022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Raida M, Clement JH, Ameri K, Han C, Leek
RD and Harris AL: Expression of bone morphogenetic protein 2 in
breast cancer cells inhibits hypoxic cell death. Int J Oncol.
26:1465–1470. 2005.PubMed/NCBI
|
|
18
|
Yang S, Pham LK, Liao CP, Frenkel B, Reddi
AH and Roy-Burman P: A novel bone morphogenetic protein signaling
in heterotypic cell interactions in prostate cancer. Cancer Res.
68:198–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hsu YL, Huang MS, Yang CJ, Hung JY, Wu LY
and Kuo PL: Lung tumor-associated osteoblast-derived bone
morphogenetic protein-2 increased epithelial-to-mesenchymal
transition of cancer by Runx2/Snail signaling pathway. J Biol Chem.
286:37335–37346. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu G, Huang F, Chen Y, Zhuang Y, Huang Y
and Xie Y: High levels of BMP2 promote liver cancer growth via the
activation of myeloid-derived suppressor cells. Front Oncol.
10:1942020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu Z, Sun J, Wang X and Cao Z:
MicroRNA-129-5p promotes proliferation and metastasis of
hepatocellular carcinoma by regulating the BMP2 gene. Exp Ther Med.
21:2572021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate-a practical and powerful approach to
multiple testing. J Roy Statist Soc B. 57:289–300. 1995.
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Scott RW, Crighton D and Olson MF:
Modeling and imaging 3-dimensional collective cell invasion. J Vis
Exp. 35252011.PubMed/NCBI
|
|
26
|
Pfisterer L and Korff T: Spheroid-based in
vitro angiogenesis model. Methods Mol Biol. 1430:167–177. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Muenst S, Läubli H, Soysal SD, Zippelius
A, Tzankov A and Hoeller S: The immune system and cancer evasion
strategies: Therapeutic concepts. J Intern Med. 279:541–562. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aust S, Felix S, Auer K, Bachmayr-Heyda A,
Kenner L, Dekan S, Meier SM, Gerner C, Grimm C and Pils D: Absence
of PD-L1 on tumor cells is associated with reduced MHC I expression
and PD-L1 expression increases in recurrent serous ovarian cancer.
Sci Rep. 7:429292017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baloche V, Rivière J, Tran TBT, Gelin A,
Bawa O, Signolle N, Diop MBK, Dessen P, Beq S, David M and Busson
P: Serial transplantation unmasks galectin-9 contribution to tumor
immune escape in the MB49 murine model. Sci Rep. 11:52272021.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liao J, Li JZ, Xu J, Xu Y, Wen WP, Zheng L
and Li L: High S100A9+ cell density predicts a poor
prognosis in hepatocellular carcinoma patients after curative
resection. Aging (Albany NY). 13:16367–16380. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liang F, Cao W, Wang Y, Li L, Zhang G and
Wang Z: The prognostic value of tumor budding in invasive breast
cancer. Pathol Res Pract. 209:269–275. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Prall F: Tumour budding in colorectal
carcinoma. Histopathology. 50:151–162. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Morodomi T, Isomoto H, Shirouzu K,
Kakegawa K, Irie K and Morimatsu M: An index for estimating the
probability of lymph node metastasis in rectal cancers. Lymph node
metastasis and the histopathology of actively invasive regions of
cancer. Cancer. 63:539–543. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ueno H, Murphy J, Jass JR, Mochizuki H and
Talbot IC: Tumour ‘budding’ as an index to estimate the potential
of aggressiveness in rectal cancer. Histopathology. 40:127–132.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mitrovic B, Schaeffer DF, Riddell RH and
Kirsch R: Tumor budding in colorectal carcinoma: Time to take
notice. Mod Pathol. 25:1315–1325. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Satoh K, Nimura S, Aoki M, Hamasaki M,
Koga K, Iwasaki H, Yamashita Y, Kataoka H and Nabeshima K: Tumor
budding in colorectal carcinoma assessed by cytokeratin
immunostaining and budding areas: Possible involvement of c-Met.
Cancer Sci. 105:1487–1495. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rogers AC, Gibbons D, Hanly AM, Hyland JM,
O'Connell PR, Winter DC and Sheahan K: Prognostic significance of
tumor budding in rectal cancer biopsies before neoadjuvant therapy.
Mod Pathol. 27:156–162. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li A and Machesky LM: Melanoblasts on the
move: Rac1 sets the pace. Small GTPases. 3:115–119. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gupta S and Maitra A: EMT: Matter of life
or death? Cell. 164:840–842. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yu M, Bardia A, Wittner BS, Stott SL, Smas
ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, et al:
Circulating breast tumor cells exhibit dynamic changes in
epithelial and mesenchymal composition. Science. 339:580–584. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li T, Zhou J, Jiang Y, Zhao Y, Huang J, Li
W, Huang Z, Chen Z, Tang X, Chen H and Yang Z: The novel protein
ADAMTS16 promotes gastric carcinogenesis by targeting IFI27 through
the NF-κb signaling pathway. Int J Mol Sci. 23:110222022.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu CX, Lin GS, Lin ZX, Zhang JD, Chen L,
Liu SY, Tang WL, Qiu XX and Zhou CF: Peritumoral edema on magnetic
resonance imaging predicts a poor clinical outcome in malignant
glioma. Oncol Lett. 10:2769–2776. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Almishri W, Swain LA, D'Mello C, Le TS,
Urbanski SJ and Nguyen HH: ADAM metalloproteinase domain 17
regulates cholestasis-associated liver injury and sickness behavior
development in mice. Front Immunol. 12:7791192022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Guo J, Guo M and Zheng J: Inhibition of
BONE MORPHOGENETIC PROtein 2 suppresses the stemness maintenance of
cancer stem cells in hepatocellular carcinoma via the MAPK/ERK
pathway. Cancer Manag Res. 13:773–785. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zuo WH, Zeng P, Chen X, Lu YJ, Li A and Wu
JB: Promotive effects of bone morphogenetic protein 2 on
angiogenesis in hepatocarcinoma via multiple signal pathways. Sci
Rep. 6:374992016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Raida M, Clement JH, Leek RD, Ameri K,
Bicknell R, Niederwieser D and Harris AL: Bone morphogenetic
protein 2 (BMP-2) and induction of tumor angiogenesis. J Cancer Res
Clin Oncol. 131:741–750. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Renne SL, Woo HY, Allegra S, Rudini N,
Yano H, Donadon M, Viganò L, Akiba J, Lee HS, Rhee H, et al:
Vessels encapsulating tumor clusters (VETC) is a powerful predictor
of aggressive hepatocellular carcinoma. Hepatology. 71:183–195.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Rodriguez MI, Peralta-Leal A, O'Valle F,
Rodriguez-Vargas JM, Gonzalez-Flores A, Majuelos-Melguizo J, López
L, Serrano S, de Herreros AG, Rodríguez-Manzaneque JC, et al:
PARP-1 regulates metastatic melanoma through modulation of
vimentin-induced malignant transformation. PLoS Genet.
9:e10035312013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang MH, Zhou XM, Zhang MY, Shi L, Xiao
RW, Zeng LS, Yang XZ, Zheng XFS, Wang HY and Mai SJ: BMP2 promotes
proliferation and invasion of nasopharyngeal carcinoma cells via
mTORC1 pathway. Aging (Albany NY). 9:1326–1340. 2017. View Article : Google Scholar : PubMed/NCBI
|