Introduction
SWI/SNF-related, matrix-associated, actin-dependent
regulator of chromatin, subfamily a, member 4 (SMARCA4)- deficient
non-small cell lung cancer (SMARCA4-dNSCLC) is an uncommon yet
notable malignancy originating within the lung. The distinct
characteristics of SMARCA4-dNSCLC were first proposed in 2017 by
Agaimy et al (1) who
highlighted the expression of cytokeratin 7 (CK7) and the loss of
the BRG1 protein as the defining features of the disease.
SMARCA4-dNSCLC is a rare subtype of NSCLC, exhibiting a distinctive
combination of histomorphology, immunophenotype and molecular
genetic attributes. Unlike classical lung adenocarcinoma, the
primary driver genes implicated in SMARCA4-dNSCLC, as revealed by
molecular detection tests, primarily involve SMARCA4, TP53,
KRAS and STK11, while the more commonly associated
driver genes like EGFR, ALK and ROS1 show no marked
association (1).
Drawing from the collective insights of various
retrospective studies (2–4), it is evident that SMARCA4-dNSCLC
predominantly affects individuals ~60 years of age, with a male
predilection and a notable history of prolonged smoking. A
comprehensive literature review underscores the infrequent
occurrence of EGFR mutations in SMARCA4-dNSCLC cases
(1,5,6). In
the present report, a case of SMARCA4-dNSCLC displaying an
EGFR mutation is reported, a finding that carries notable
implications for both pathologists and clinicians. The current case
report further accentuates the intricate landscape of lung cancer
subtypes and highlights the importance of a nuanced understanding
of their molecular underpinnings to guide accurate diagnosis and
tailored therapeutic approaches.
Case report
Case presentation
In January 2023, a 60-year-old female patient was
admitted to Xiaoshan Affiliated Hospital of Wenzhou Medical
University (Hangzhou, China). A ‘mass in the middle lobe of the
right lung’ was found using a chest computed tomography (CT) scan
after undergoing physical and radiological examinations for 2 days.
The patient had undergone breast-conserving surgery >3 years
before the publication of the present case report, and received
postoperative radiotherapy for bilateral breast cancer (5 week
cycles, with 2 Gy, 5 times a week, to a total of 50 Gy; 5 times to
the tumor bed for a total of 30 times).
Physical examination
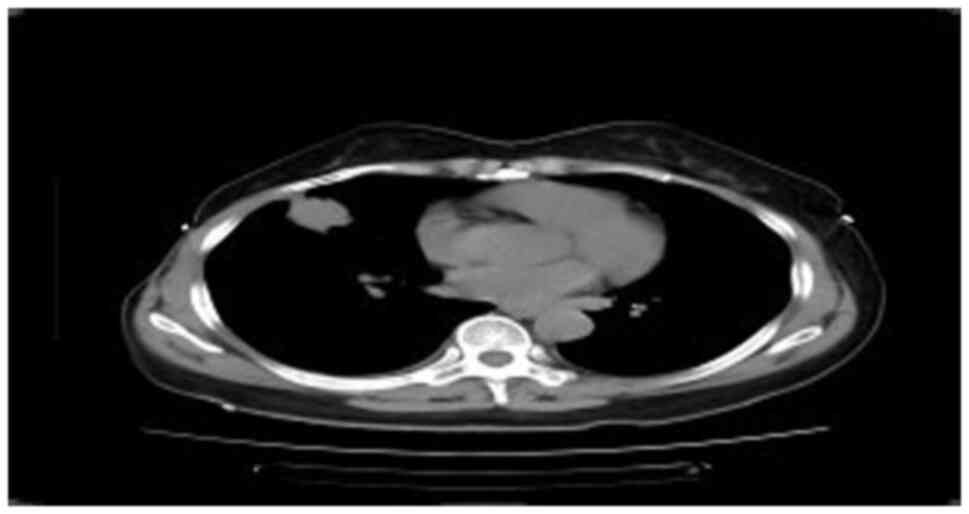
Chest CT (Fig. 1)
revealed a mass in the middle lobe of the right lung, measuring
~3.1×2.8×2.8 cm in size, with small burrs and traction near the
pleura. Multiple small nodules were observed in the right lung, and
the diameter of the largest nodule was ~1.0 cm; no abnormalities of
the cardiac shadow, mediastinum and bilateral hilum were observed.
There was a small effusion in the right thoracic cavity along with
local pleural thickening on the right side. In January 2023,
ultrasound bronchoscopic biopsy was performed on the outer segment
of the right middle lung. Positron emission tomography-CT
examination, also carried out in January 2023, revealed a mass in
the lateral segment of the right middle lobe of the lung, measuring
~3.5×2.5×2.5 cm in size, which was clear with no uniform boundary,
and showed roughly uniform internal density, increased uptake of
fluorodeoxyglucose (FDG), distal bronchus truncation in the lateral
segment of the right middle lobe, and unclear boundary between the
lesion and the adjacent pleura. A few fibrous cord shadows could be
observed in the upper lobe of the lingual and base segments of the
lower lobe of the left lung. No abnormal density lesions, such as
nodules and masses, were observed in the remaining two lung fields,
and no abnormal FDG metabolism was observed. Nodular thickening and
increased FDG uptake were observed in the right pleura, including
the lateral, interlobular fissure and diaphragmatic pleura. A small
effusion in the right thoracic cavity was visible. Slightly larger
lymph nodes were found in the right hilum of the lung and
mediastinum behind the anterior tracheal vena cava and under the
carina. The larger lymph nodes were located behind the anterior
tracheal vena cava, with a diameterof~1.8cm and increased uptake of
FDG.
Macro-examination
Five pieces of gray tissue were obtained with a
total volume 0.3×0.2×0.1 cm. The texture of the tissue was
soft.
Microscopic observations
Tissues were fixed with 4% neutral formalin (12 h at
25°C) and embedded in paraffin. Continuous 4-µm thick tissue
sections were prepared and stained with hematoxylin and eosin (8 h
at 25°C) and EnVision immunohistochemical staining. The sections
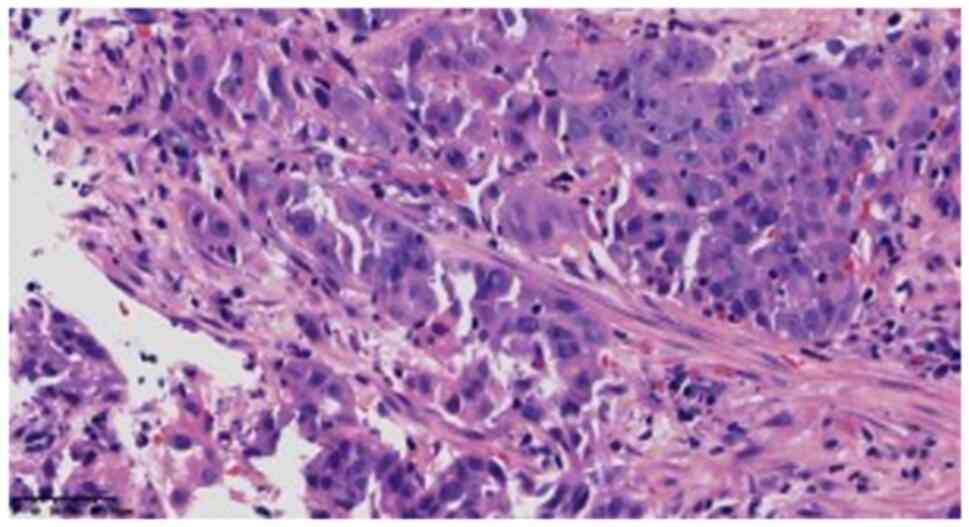
were viewed under a light microscope. In one of the fibrous
tissues, the tumor cells showed solid flaky, nest-like infiltrating
growth, with medium to large cells and some obvious nucleoli
(Fig. 2). In the larger tumor
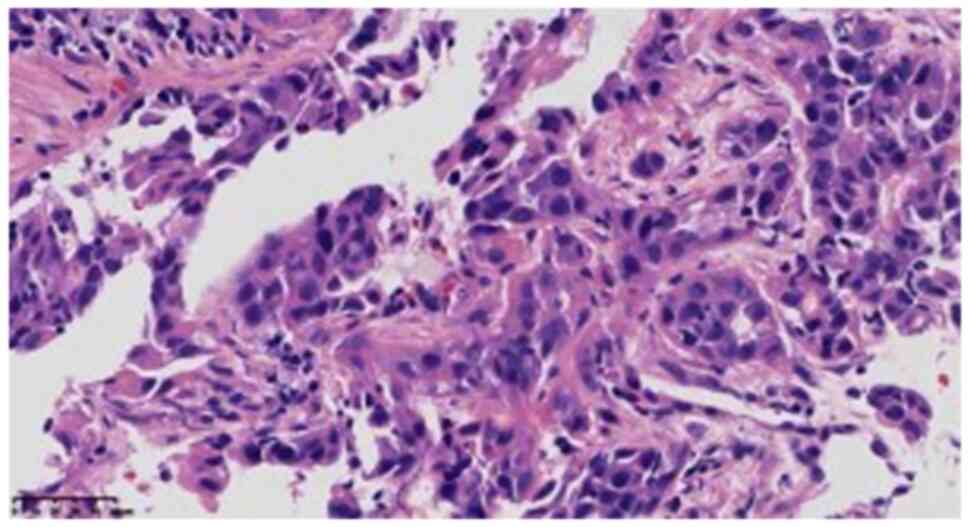
cells, the cytoplasm was rich and partially acidophilus (Fig. 3), cell atypia was obvious, mitotic
image was easy to observe, but no definite necrosis was identified,
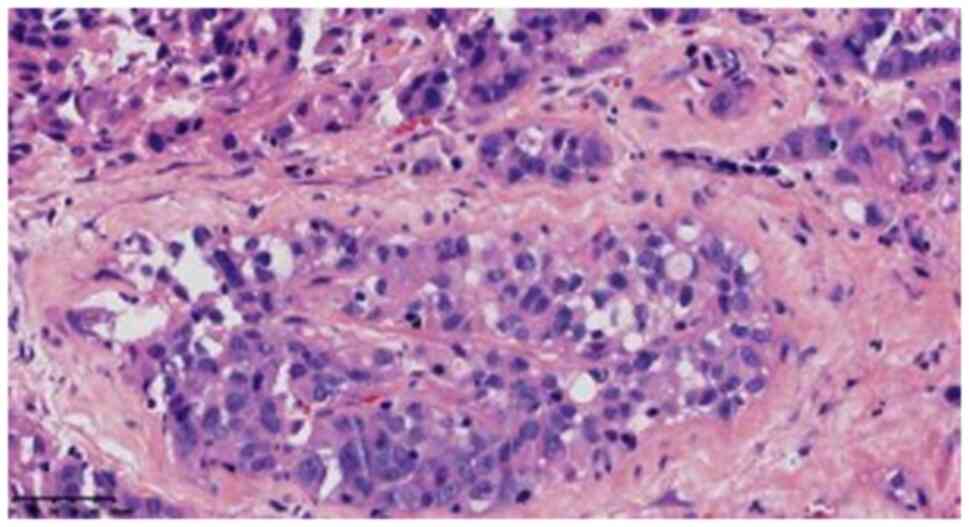
while the cytoplasm of the small focal cells showed light blue
mucus (Fig. 4). Scattered
lymphocyte infiltration was observed in the interstitium.
Immunohistochemistry
Staining was performed using the EnVision Systems
method and the protocol was as follows: Unstained slides were
placed in an oven at 60°C for 120 min, followed by dewaxing in
xylene I, I and III, where Roman numerals indicate different
amounts of 500-ml reagent cylinders, for 10 min per cylinder. The
slides were washed in 100% ethanol I, 100% ethanol II and 95%
ethanol for 3 min per bottle. 85 and 75% ethanol were used for 1
min per bottle, then tissues were rinsed with distilled water to
complete hydration. The slides were placed in EDTA repair solution
(cat. no. MVS-0099; pH 9.0; 1:50; Fuzhou MaixinBiotech Co., Ltd.)
at 100°C for 20 min for antigen repair, washed with water after
natural cooling, treated with 3% hydrogen peroxide solution for 10
min, and then rinsed with PBS. Primary antibody was added and
incubated at room temperature for 40 min, then washed with PBS
three times for 5 min each. Sheep anti-rat/Rabbit IgG polymer
labeled with horseradish peroxidase (HRP)(cat. no. PV8000D; Beijing
Zhongshan Jinqiao Biotechnology Co., Ltd.) was used for incubation
at room temperature for 15 min. Washing was performed with PBS
three times for 5 min each. DAB color developing solution (polymer
method; cat. no. KIT-0014; 1:50; Fuzhou Maixin Biotech Co., Ltd.)
was added and incubated at 25°C for 5–10 min, and then slides were
washed with distilled water. Slides were redyed with hematoxylin
for 1 min, washed with tap water, and turned blue in PBS solution.
Subsequently, the slide was cleaned with 75% ethanol, 2 bottles of
500 ml 95% ethanol and 2 bottles of 500 ml 100% ethanol for 1 min
each, in order to remove excess water and facilitate microscopic
observation. Finally, they were placed in xylene solution I, II and
III for 1 min each, sealed with neutral gum and observed under a
light microscope. Tumor cells showed SMARCA4 (ready-to-use; clone
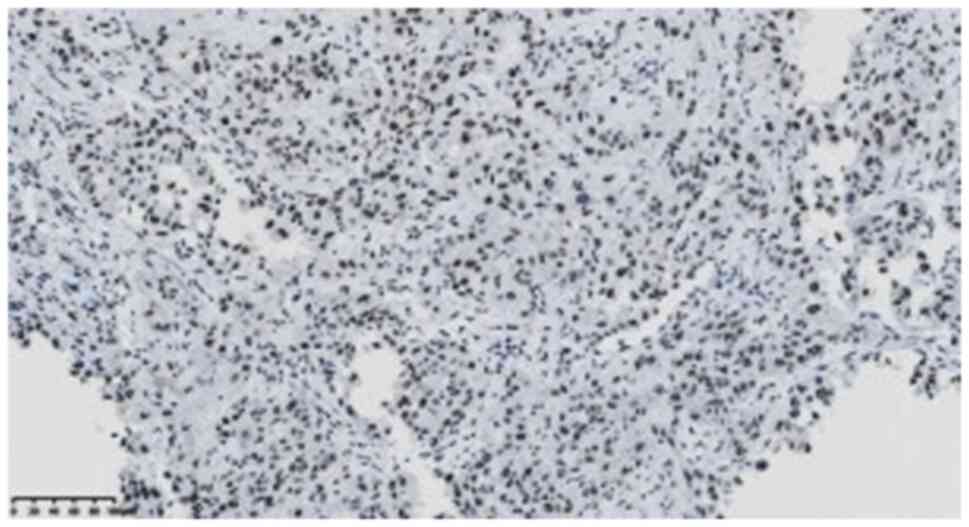
EBV5B; cat. no. ZA-0673) deletion (Fig.
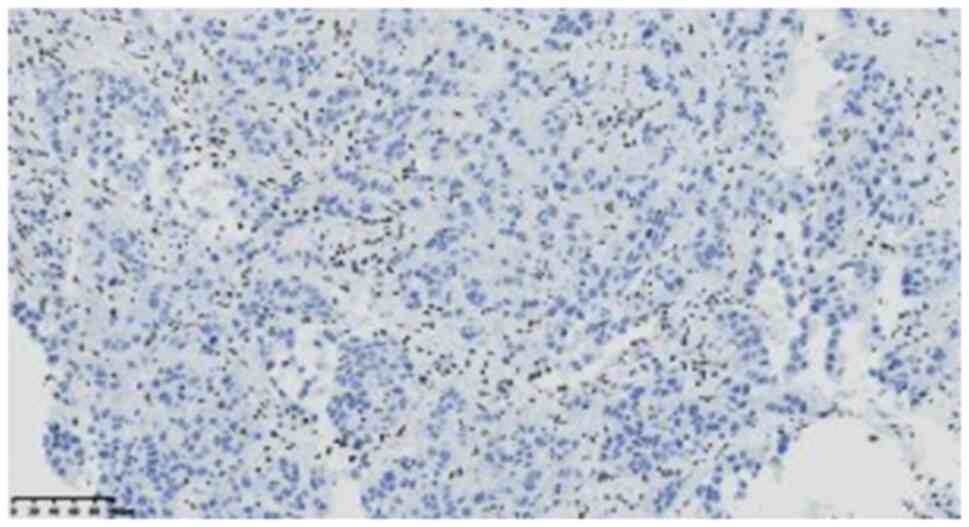
5), and positive results for SMARCB1 (ready-to-use; clone
OTIR4G9; cat. no. ZA-0696) (Fig.
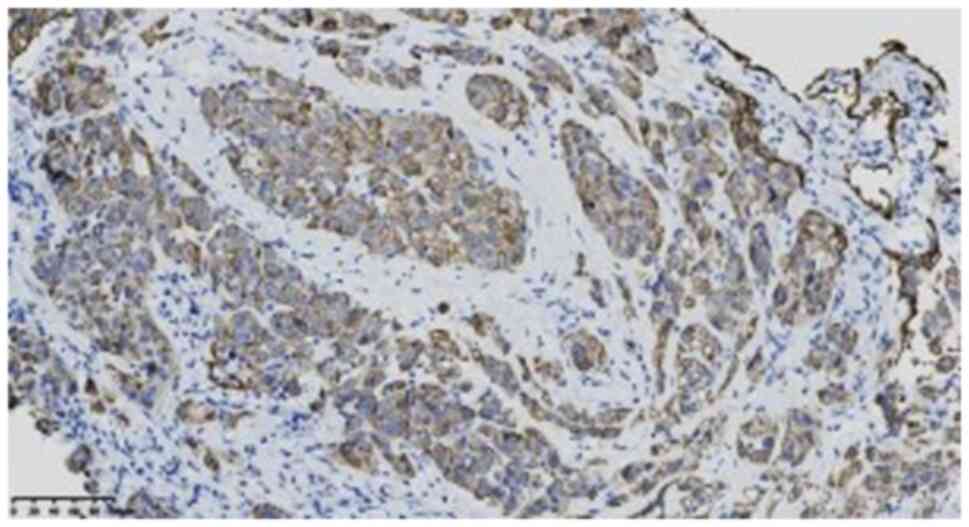
6), CK7 (ready-to-use; clone OV-TL12/30; cat. no. kit-0021)
(Fig. 7), cytokeratin (CAM)5.2
(ready-to-use; clone CAM5.2; cat. no. ZM-0316) (Fig. S1), CK5/6 (ready-to-use; clone
OTI1F8; cat. no. ZM-0313) (Fig.
S2) and calretinin (ready-to-use; clone MX027; cat. no.
MAB-0716) (Fig. S3). However, they
were negative for thyroid transcription factor-1 (TTF-1) (1:200;
clone SPT24; cat. no. ZM-0270) (Fig.
S4), Napsin A (ready-to-use; clone IP64; cat. no. ZM-0473)
(Fig. S5), p40 (ready-to-use;
clone MXR010; cat. no.RMA-1006) (Fig.
S6), Synaptophysin (SYN) (ready-to-use; clone EP158; cat. no.
ZA-0506) (Fig. S7), Chromogranin A
(CGA) (ready-to-use; clone MX018; cat. no. MAB-0707) (Fig. S8), SRY-box transcription factor 2
(SOX2) (ready-to-use; clone EP103; cat. no. ZA-0571) (Fig. S9), CD34 (ready-to-use; clone
QBEnd/10; cat. no. kit-0004) (Fig.
S10), Sal-like protein 4 (SALL4) (ready-to-use; clone 6E3; cat.
no. ZM-0393) (Fig. S11), NUT
(ready-to-use; clone C52B1; cat. no. ZA-0671) (Fig. S12), ALK (ready-to-use; clone 1A4;
cat. no. ZM-0848) (Fig. S13),
podoplanin (D2-40) (ready-to-use; clone D2-40; cat. no. ZM-0465)
(Fig. S14), Wilm's tumor protein
(WT-1) (ready-to-use; clone MX012; cat. no. MAB-0678) (Fig. S15), vimentin (ready-to-use; clone
MX034; cat. no. MAB-0735) (Fig.
S16), GATA binding protein 3 (GATA3) (ready-to-use; clone
EP368; cat. no. ZA-0661) (Fig.
S17) and estrogen receptor (ER) (ready-to-use; clone SP1; cat.
no. kit-0012) (Fig. S18).
Additionally, p63 (ready-to-use; clone UMAB4; cat. no. ZM-0406)
(Fig. S19) was partially weakly
positive, carcinoembryonic antigen (CEA) (ready-to-use; clone
COL-1; cat. no. kit-0008) (Fig.
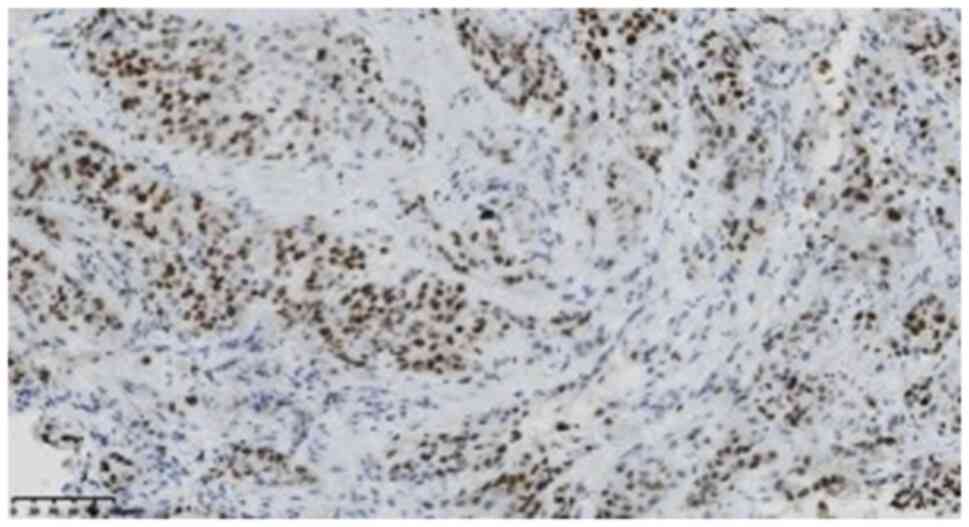
S20) was focally positive, and the Ki-67 (1:200; clone UMAB107;
cat. no. ZM-0166) (Fig. 8)
positivity index was 75%. SMARCA4, SMARCB1, SALL4, SOX2, NUT, ALK,
p63, D2-40, TTF-1, Napsin A, CK5/6, CAM5.2, Ki-67, SYN and GATA3
antibodies were purchased from Beijing Zhongshan Jinqiao
Biotechnology Co., Ltd., and CR, WT-1, CD34, CEA, VIM, ER, CK7, CGA
and p40 antibodies were purchased from Fuzhou Maixin Biotech Co.,
Ltd Molecular detection showed an L858R mutation in exon 21 of
EGFR, mutation abundance of 11.59%, TP53 mutation,
high tumor mutation load, no tumor microsatellite instability and
no mutations in ALK, KRAS, ROS1 and STK11.
Next-generation sequencing (NGS) had been previously performed by
Beijing ACCB Biotech Ltd., and the NGS results were provided by the
patient themselves.
Pathological diagnosis
The final diagnosis was SMARCA4-dNSCLC with an L858R
mutation in exon 21 of EGFR.
Follow-up
After molecular detection, the patient was treated
with osimertinib (80 mg orally, once daily), the targeted drug for
the EGFR mutation. At the publication of the present case
report, the patient was in fair condition.
Discussion
SMARCA4 is a tumor suppressor gene, located
at 19p13.2, encoding the BRG1 protein. This protein is one of the
notable subunits of the SWI/SNF chromatin remodeling complex
(7), which is involved in the
chromatin remodeling process. Therefore, it is involved in marked
cellular processes and in functional regulation, such as gene
expression proliferation and differentiation, and inhibition of
tumorigenesis (7,8). Mutations in the SWI/SNF complex have
been detected in a variety of human tumors, and are most commonly
found in SMARCA4 (9). In 2015, Le
Loarer et al (10) first
reported a group of chest tumors with absence of SMARCA4
expression, characterized by rhabdomyoid histology aggressiveness
and poor prognosis, and the authors proposed the nomenclature
‘thoracicsarcoma with the absence of SMARCA4’. Subsequently,
studies reported that 8–25% of SMARCA4 gene deletions occur
in NSCLC, raising questions about whether these tumors are true
sarcomas, undifferentiated or dedifferentiated carcinomas (2,5,11,12).
Several studies have reported that SMARCA4-dNSCLC ranges from
well-differentiated lung adenocarcinoma to poorly differentiated
lung cancer, and most cases of SMARCA4-dNSCLC are poorly
differentiated (1,3,13,14).
Additionally, it has been reported that SMARCA4-dNSCLC often has
areas of classic NSCLC as well as focal solid areas or a rhabdoid
morphology (15).
In the present case, there was insufficient tissue
for effective examination, and the cells appeared poorly
differentiated under a light microscope (LEICA DM2000), showing
solid flaky and nest-like infiltrating growth. There were no
glandular, tubular and papillary structures, and a light blue mucus
was visible in the cytoplasm of small focal cells, with AB/PAS
positivity. Immunohistochemistry results revealed that CK7, CAM5.2,
CK5/6, and SMARCB1 were positive, while SMARCA4 was negative.
Molecular analysis indicated EGFR and TP53 mutations,
suggesting epithelial differentiation of SMARCA4-dNSCLC, which can
be distinguished from SMARCA4-deficient thoracic tumors.
SMARCA4 deletion has been reported in 10.0, 9.8, 7.0, 3.7
and 2.7% of large cell neuroendocrine carcinomas, adenocarcinomas,
NSCLCs, squamous cell and small cell carcinomas, respectively
(14). Data from several large
retrospective studies indicated that the median age of patients
with SMARCA4-dNSCLC was ~60 years, and that they were predominantly
male with a long history of smoking (2–4). A
review of the literature revealed that EGFR mutations in
SMARCA4-dNSCLC are rare (1,5,6). The
largest number of cases reported is 4,813 from the Memorial
Sloan-Kettering Cancer Center in the United States (5), among which ~8% (n=407) of cases had
the SMARCA4 mutation. Among commonly altered genes in lung
cancer, the most frequent co-occurring mutations with SMARCA4
alterations were in TP53 (56%), KEAP1 (41%),
STK11 (39%) and KRAS (36%). A total of <4% of the
1,140 EGFR mutations were associated with SMARCA4
mutations (5). EGFR
mutations are more common in non-smoking or light smoking Asian
female patients with adenocarcinoma (16). Data from a study show that the
mutation rate of EGFR in patients with lung cancer in China
was 47.6%, which is notably higher than that in Western countries
(16). However, EGFR
mutations in patients with SMARCA4-dNSCLC have not been reported in
China, and it is hypothesized that this finding is related to the
small number of SMARCA4-dNSCLC cases reported in the country;
additionally, most of the cases involve male patients. As the
number of cases increases, reports of related mutated genes may
accumulate. The patient of the present case report was a
60-year-old non-smoking female with a history of double breast
cancer. Molecular analysis revealed an L858R mutation in exon 21 of
EGFR. Whether the EGFR mutation in this patient with
SMARCA4-dNSCLC is related to the bilateral breast cancer history is
unclear. Therefore, more cases need to be reported due to the
limited data currently available.
SMARCA4-dNSCLC should be distinguished from the
following tumors: i) Solid lung adenocarcinoma: Tumor cells have
solid flaky and nest-like structures, it is positive for CK7 and
similar to SMARCA4-dNSCLC, but the former can generally be
identified due to positive expressions of TTF-1 and Napsin A; ii)
epithelioid malignant mesothelioma: Immunohistochemically, it can
be distinguished due to it being WT-1, HMBE1 and D2-40 positive;
iii) large cell carcinoma: The immunohistochemical expression of
squamous cell carcinoma, adenocarcinoma and neuroendocrine
carcinoma is often absent, while CK7 is diffusely positive with
absent SMARCA4 in SMARCA4-dNSCLC; iv) large cell neuroendocrine
carcinoma: It is positive for SYN, CGA, TTF-1 and CD56, and has no
SMARCA4 deletion; v)SMARCA4-deficient thoracic tumor: A new
category of tumors proposed in the 2021 edition of the WHO Thoracic
Tumor Classification (5th edition) (17), defined by SMARCA4 deficiency, but
can also involve expression of CD34, SOX2 and SALL4; CK is only
weakly positive or focal positive, while CD34, SOX2 and SALL4 are
negative, and diffusely strong positive expression of CK7 and
CAM5.2 is also observed; vi) germ cell tumors: Most commonly occur
in children and adolescents, and often express markers such as
SALL4 and OCT3/4; vii) NUT cancer: No NUT expression is observed;
and viii) metastatic SMARCA4 deletion tumors: Metastasis to other
sites should be excluded by clinical examination; for example,
there were no masses in other sites in the present case, Gata3 and
ER were negative, and there was no SMARCA4 deletion in the
primary breast cancer tissue as observed following
immunohistochemical staining, thus, breast cancer metastasis could
be excluded.
In summary, the histomorphological,
immunohistochemical and molecular detection results of the current
case support the diagnosis of SMARCA4-dNSCLC with an EGFR
gene mutation. In terms of treatment, SMARCA4-dNSCLC is a newly
proposed tumor, which usually lacks mutations in common genes such
as EGFR, ALK and ROS1, and there is currently no
unified standardized treatment plan. A study has shown that
inhibitors such as PD-L1/PD1, EZH2 and CDK4/6 may be used for
treatment (3), along with
platinum-based chemotherapy (18).
The patient of the present case report was a non-smoking female
with SMARCA4-dNSCLC accompanied by an L858R mutation in exon 21 of
EGFR. The efficacy of EGFR-targeted drug therapy in
patients with SMARCA4-dNSCLC has not been reported, which may be
related to the small number of reported cases of SMARCA4-dNSCLC,
and most of the reported cases involved male smokers. Since the
patient of the current case report had undergone bilateral breast
cancer treatment 3 years ago, and considering the weakened physical
endurance of the patient, the EGFR-targeted drug osimertinib
was used for treatment for <1 month. The patient survived, but
the evaluation of long-term efficacy of treatment requires
continuous follow-up.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LS and MD conceived the study idea and drafted the
manuscript. JC and QF carried out data collection. LC interpreted
the data and revised the manuscript. LS and MD confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
The patient provided written informed consent for
the case study to be published.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
SMARCA4-dNSCLC
|
SMARCA4-deficient non-small cell lung
cancer
|
|
CT
|
computed tomography
|
|
FDG
|
fluorodeoxyglucose
|
References
|
1
|
Agaimy A, Fuchs F, Moskalev EA, Sirbu H,
Hartmann A and Haller F: SMARCA4-deficient pulmonary
adenocarcinoma: Clinicopathological, immunohistochemical, and
molecular characteristics of a novel aggressive neoplasm with a
consistent
TTF1neg/CK7pos/HepPar-1pos
immunophenotype. Virchows Arch. 471:599–609. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dagogo-Jack I, Schrock AB, Kem M, Jessop
N, Lee J, Ali SM, Ross JS, Lennerz JK, Shaw AT and Mino-Kenudson M:
Clinicopathologic characteristics of BRG1-deficient NSCLC. J Thorac
Oncol. 15:766–776. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Naito T, Udagawa H, Umemura S, Sakai T,
Zenke Y, Kirita K, Matsumoto S, Yoh K, Niho S, Tsuboi M, et al:
Non-small cell lung cancer with loss of expression of the SWI/SNF
complex is associated with aggressive clinicopathological features,
PD-L1-positive status, and high tumor mutation burden. Lung Cancer.
138:35–42. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rekhtman N, Montecalvo J, Chang JC, Alex
D, Ptashkin RN, Ai N, Sauter JL, Kezlarian B, Jungbluth A,
Desmeules P, et al: SMARCA4-deficient thoracic sarcomatoid tumors
represent primarily smoking-related undifferentiated carcinomas
rather than primary thoracic sarcomas. J Thorac Oncol. 15:231–247.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schoenfeld AJ, Bandlamudi C, Lavery JA,
Montecalvo J, Namakydoust A, Rizvi H, Egger J, Concepcion CP, Paul
S, Arcila ME, et al: The Genomic landscape of SMARCA4
alterations and associations with outcomes in patients with lung
cancer. Clin Cancer Res. 26:5701–5708. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sui YX, Jin L, Guo GD, Luo M, Qin XY, You
LS and Chen LF: Clinicopathological analysis of the
SMARCA4-deficient non-small cell lung carcinoma. Zhonghua Bing Li
Xue Za Zhi. 50:1366–1368. 2021.(In Chinese). PubMed/NCBI
|
|
7
|
St Pierre R and Kadoch C: Mammalian
SWI/SNF complexes in cancer: Emerging therapeutic opportunities.
Curr Opin Genet Dev. 42:56–67. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Masliah-Planchon J, Bièche I,
Guinebretière JM, Bourdeaut F and Delattre O: SWI/SNF chromatin
remodeling and human malignancies. Annu Rev Pathol. 10:145–171.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Abou Alaiwi S, Nassar AH, Xie W, Bakouny
Z, Berchuck JE, Braun DA, Baca SC, Nuzzo PV, Flippot R, Mouhieddine
TH, et al: Mammalian SWI/SNF complex genomic alterations and immune
checkpoint blockade in solid tumors. Cancer Immunol Res.
8:1075–1084. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Le Loarer F, Watson S, Pierron G, de
Montpreville VT, Ballet S, Firmin N, Auguste A, Pissaloux D,
Boyault S, Paindavoine S, et al: SMARCA4 inactivation defines a
group of undifferentiated thoracic malignancies transcriptionally
related to BAF-deficient sarcomas. Nat Genet. 47:1200–1205. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fernando TM, Piskol R, Bainer R, Sokol ES,
Trabucco SE, Zhang Q, Trinh H, Maund S, Kschonsak M, Chaudhuri S,
et al: Functional characterization of SMARCA4 variants identified
by targeted exome-sequencing of 131,668 cancer patients. Nat
Commun. 11:55512020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
La Fleur L, Falk-Sörqvist E, Smeds P,
Berglund A, Sundström M, Mattsson JS, Brandén E, Koyi H, Isaksson
J, Brunnström H, et al: Mutation patterns in a population-based
non-small cell lung cancer cohort and prognostic impact of
concomitant mutations in KRAS and TP53 or STK11. Lung Cancer.
130:50–58. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Herpel E, Rieker RJ, Dienemann H, Muley T,
Meister M, Hartmann A, Warth A and Agaimy A: SMARCA4 and SMARCA2
deficiency in non-small cell lung cancer: Immunohistochemical
survey of 316 consecutive specimens. Ann Diagn Pathol. 26:47–51.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nambirajan A, Singh V, Bhardwaj N, Mittal
S, Kumar S and Jain D: SMARCA4/BRG1-deficient non-small cell lung
carcinomas: A case series and review of the literature. Arch Pathol
Lab Med. 145:90–98. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jin YP, Wang L, Wang Y, Wu DY, Zhang H and
Xia QX: [Gastric SWI/SNF complex deletion-associated
undifferentiated carcinoma with rhabdoid phenotype: A
clinicopathological and molecular analysis]. Zhonghua Bing Li Xue
Za Zhi. 51:1229–1234. 2022.(In Chinese). PubMed/NCBI
|
|
16
|
Liu SY, Mok T and Wu YL: Novel targeted
agents for the treatment of lung cancer in China. Cancer. 121
(Suppl 17):S3089–S3096. 2015. View Article : Google Scholar
|
|
17
|
Tsao MS, Nicholson AG, Maleszewski JJ,
Marx A and Travis WD: Introduction to 2021 WHO classification of
thoracic tumors. J Thorac Oncol. 17:e1–e4. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bell EH, Chakraborty AR, Mo X, Liu Z,
Shilo K, Kirste S, Stegmaier P, McNulty M, Karachaliou N, Rosell R,
et al: SMARCA4/BRG1 is a novel prognostic biomarker predictive of
cisplatin-based chemotherapy outcomes in resected non-small cell
lung cancer. Clin Cancer Res. 22:2396–2404. 2016. View Article : Google Scholar : PubMed/NCBI
|