Introduction
8p11 myeloproliferative syndrome (EMS), first
reported by Macdonald et al (1) in 1995, is a rare atypical
myeloproliferative disease, and is also termed stem cell
leukemia/lymphoma syndrome. The disease is characterized by a
significantly higher peripheral white blood cell count (WBC),
myeloid cell hyperplasia and eosinophilia in the bone marrow, lymph
node pathology for lymphoblastic lymphoma and involvement of the
short arm of chromosome 8 (8p11) for fibroblast growth factor-1
(FGFR1) gene translocation. Furthermore, EMS can progress to
acute leukemia in the short term (2–4). The
World Health Organization (WHO) designated EMS for myeloid/lymphoid
neoplasms with FGFR1 rearrangement (MLNAF) in 2008. The designation
was maintained in the fourth edition of the WHO classification of
hematopoietic and lymphoid tissue tumors revised in 2016 (5).
Globally, there have been only ~100 cases of MLNAF
reported to date. In 1992, Abruzzo et al (6), for the first time, reported the cases
of 3 patients with T lymphoblastic lymphoma and peripheral blood
eosinophilia. All patients subsequently developed myeloid
malignancy, and were gradually diagnosed with EMS. The occurrence
of MLNAF is closely associated with FGFR1 gene
abnormalities. FGFR1 encodes a receptor tyrosine kinase
transmembrane protein and belongs to the FGFR family. Under normal
conditions, FGFR1 exists in the form of an oligomer, which
binds to its ligand and induces homologous dimerization and
self-phosphorylation of FGFR1, thereby activating multiple
effectors and producing signals for proliferation and survival
(7). FGFR1 gene
abnormalities in patients with MLNAF often manifest as
translocation or insertion mutations, with gene translocation being
the most common. At present, 15 partner genes have been identified
for MLNAF, including ZNF198 (13p12), BCR (22q11), CEP110 (9q33),
FGFR1OP2 (12p11), FOP (6q27), TRIM24 (7q34), HERVK (19q13), MYO18A
(17q11), NUP98 (11p15), CPSF6 (12p11), CUX1 (7q22), RANBP2 (2q12),
TPR (1q25), SQSTM 1 (5q35) and LRRFIPI (2q37) (3,8). In
addition, two types of insertion changes, insertion
(13;8)(q12;p11p23) and insertion (12;8)(p11;p11;p22) (9,10) have
been observed. At present, no gene amplification or deletion
mutations have been reported. The products formed by balanced
translocation or insertion after FGR1 gene break exhibit
ligand independent FGFR1 tyrosine kinase activity, and
multiple downstream signaling pathways, including the
Ras/mitogen-activated protein kinase (Ras/MAPK),
phosphatidylinositol 3-kinase (PI3K), phospholipase C (PLC) 7 and
signal transducer and activator of transcription (STAT) pathways,
are continuously activated, leading to the development of MLNAF
(11).
Few cases concerning MLNAF with t(8;13)(p11;q12)
have been reported in the literature, and the outcome of most cases
of EMS is poor, even in patients that have been treated with
allogeneic stem cell transplantation. In the present study, the
case of a patient with MLNAF with t(8;13)(p11;q12) who was treated
with a tyrosine kinase inhibitor (TKI) combined with chemotherapy
was reported. The aim of the present study is to improve the
understanding of EMS.
Case report
A 62-year-old female with no history of
hematological disease was admitted to the Department of Hematology,
The Second Hospital of Hebei Medical University (Shijiazhuang,
China) in November 2019 due to a painless groin mass that had been
present for >2 months and a repeating recurrent rash lasting for
1 month. The patient had a history of an elevated peripheral
leukocyte level, first reported 11 months prior (range,
11.1–61.8×109/l; normal range,
3.5–9.5×109/l), which was left untreated. Physical
examination revealed extensive red papules and maculopapules on the
skin, as well as enlarged lymph nodes in the neck, armpits and
groin. The largest lymph node was located in the right groin,
measuring ~4×4 cm, with a hard texture and no tenderness. The liver
palpation indicated a protrusion of ~1 cm below the costal margin,
while the splenic palpation discerned an extension of ~6 cm below
the costal margin. The palpitations were homogeneous and had no
tenderness.
At admission, the initial peripheral blood analysis
revealed a WBC of 136.9×109/l (normal range,
3.5–9.5×109/l), with neutrophils accounting for 93.06%
(normal range, 40–75%) and eosinophils for 0.97% (normal range,
0.4–8%), a hemoglobin level of 151 g/l (normal range, 115–150 g/l)
and a platelet count of 208×109/l (normal range,
125–350×109/l). The eosinophil count, several serum
analysis results and eosinophil percentages in the bone marrow at
different stages of diagnosis, remission, relapse and AML
transformation are summarized in Table
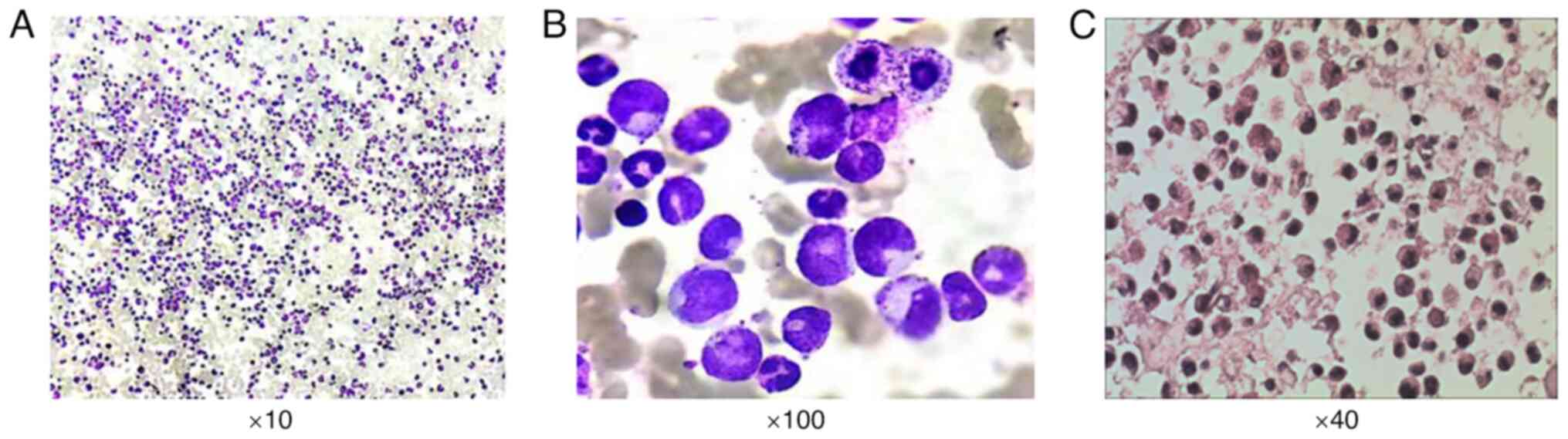
I. The bone marrow was hypercellular, with an increased
percentage of granules associated with eosinophilia, thus
indicating that the patient did not have chronic myeloid leukemia
(Fig. 1).
 | Table I.Summary of the eosinophil count,
serum analysis results and eosinophil percentages in bone marrow at
different stages. |
Table I.
Summary of the eosinophil count,
serum analysis results and eosinophil percentages in bone marrow at
different stages.
| Date | WBCs, n
(×109/l) | Es, % | Es, n
(×109/l) | Hb, g/l | PLTs, n
(×109/l) | LDH (U/l) | B2M (mg/l) | BM (E%) |
|---|
| November
2020-Diagnosis | 136.9 | 0.97 | 1.33 | 151 | 208 | 588 | 3.9 | 9.5 |
| April
2021-Remission | 9.6 | 3.8 | 0.4 | 100 | 200 | 253 | 2.9 | 2 |
| Early July
2021-Relapse | 29.1 | 3.1 | 0.9 | 106 | 276 | 634 | 2.9 | 11 |
| Late July 2021-AML
transformation | 36.3 | 0.8 | 0.3 | 96 | 45 | 1,219 | 4.3 | 8 |
Analyses of the breakpoint cluster region-abelson
leukemia virus (BCR-ABL) fusion gene, Janus kinase 2
(JAK2)-V617F, calreticulin (CALR) and
myeloproliferative leukemia virus oncogene (MPL) mutations,
and the next-generation sequencing of gene mutations related to
myeloid tumors all gave negative results. The BCR-ABL fusion
gene testing process was as follows: First, 0.8% ammonium chloride
red blood cell lysate was added to the bone marrow fluid and
centrifuged at 1,000 × g for 10 min at room temperature to obtain
white blood cells. RNA was extracted using the RNA pred Pure
Hi-Blood kit (Tiangen Biotech Co., Ltd.; cat. no. DP443), according
to the manufacturer's instructions. A total of 1 ml Trizol was
added to a 50-µl leukocyte suspension and mixed. The solution was
incubated at room temperature for 5 min, followed by the addition
of 0.2% chloroform. After shaking for 30 sec, it was left
undisturbed for 3 min and further centrifuged at 16,000 × g for 15
min at 4°C. The supernatant was transferred into a
diethylpyrocarbonatetreated EP tube and mixed with isopropyl
alcohol, and left at room temperature for 10 min. RNA was obtained
by centrifugation again at 16,000 × g for 5 min at 4°C. RNA
amplification was performed using the BCR-ABL fusion gene detection
kit (Bio-Rad Laboratories, Inc.; cat. no. 171V37145) in strict
accordance with the instructions provided. The LightCycler 480
fluorescence detector (Roche Diagnostics) was used, and the
amplification conditions included a temperature of 42°C for 30 min,
94°C for 5 min and 40 cycles. Fluorescence signals were collected
at 60°C during the second step of the PCR cycle, and data analysis
software Opticon Monitor realtime v2.02 (Bio-Rad Laboratories,
Inc.) was utilized. The JAK2V617F, MPL and CALR testing process was
as follows: First, 0.8% ammonium chloride red blood cell lysate was
added into the bone marrow fluid and centrifuged at 1,000 × g for
10 min at room temperature to obtain white blood cells. The Tianamp
genomic DNA kit (Tiangen Biotech Co., Ltd.; cat. no. DP304) was
used to extract intracellular DNA in strict accordance with the
instructions provided by the kit. A total of ~20 µl white blood
cells and 18 µl protease were mixed well. After which, 200 µl
buffer solution was added and the mixture was heated at 70°C for 10
min. A total of 200 µl anhydrous ethanol was added to the mixture,
mixed until white flocculent appeared and then transferred to the
centrifugal column. The sample was centrifuged at 13,400 × g for 1
min at room temperature and the waste liquid was discarded from the
collection pipe. A total of 500 µl GD was added to the centrifugal
column, and the sample was centrifuged at room temperature for 1
min at 13,400 × g. The waste liquid was discarded again and 500 µl
bleach solution per wash was added to the centrifugal column, and
centrifuged at room temperature for 1 min at 13,400 × g. The waste
liquid was discarded once more and centrifuged at 13,400 × g for
two min at room temperature. The centrifugal column was placed into
a new EP tube, 200 µl TE solution was added and at room temperature
centrifuged for 2 min at 13,400 × g in order to obtain DNA. The
mutations of JAK2V617F (Exon12), CALR (Exon9) and MPL (Exon10) were
qualitatively detected using the ipsogen JAK2 RGQ PCR Kit, ipsogen
MPL W515L/K MutaScreen Kit and CALR RGQ PCR Kit (Qiagen AB; cat.
nos. 673633, 676413 and 674013, respectively) The instructions of
the kit were strictly followed, and ABI PRISM fluorescent PCR
detector was used for detection. Amplification conditions included
40 cycles at 42°C for 5 min and 94°C for 3 min, and
fluorescencemyoproliferative tumor-related gene mutation detection
kit signals were collected at 60°C in the second step of the PCR
cycle. The analysis software used was Opticon Monitor realtime
v2.02 (Bio-Rad Laboratories, Inc.). The extraction steps of DNA and
RNA in next-generation sequencing were as aforementioned. The DNA
and RNA were extracted by commercial kits (Tiangen Biotech, Co.,
Ltd.; cat. nos. is DP340, DP304 and DP431). The purities and
concentrations of DNA and RNA were confirmed by Nanodrop 2000
(Thermo Scientific Scientific, Inc.) and Qubit 3.0 Fluorometer
(Thermo Fisher Scientific, Inc.). The Qsep400 nucleic acid fragment
analyzer (Hangzhou Houze Bio-Technology Co., Ltd.,) was utilized to
evaluate the integrity of DNA and RNA. The DNA were transformed
into libraries using KAPA EvoPlus Kits (Kapa Biosystems; Roche
Diagnostics; cat. no. 9420053001). The libraries were analyzed on
the Illumina sequencing platform NextSeq550 using 150-bp paired-end
sequencing. Sequencing was performed using the NextSeq 500/550 High
output kit v2.5 (300 cycles; cat. no. 20024908; Illumina Inc). To
quantify the final library concentration, the Qubit 3.0 Fluorometer
was employed. The loading concentration of the final library was
~14 pM.
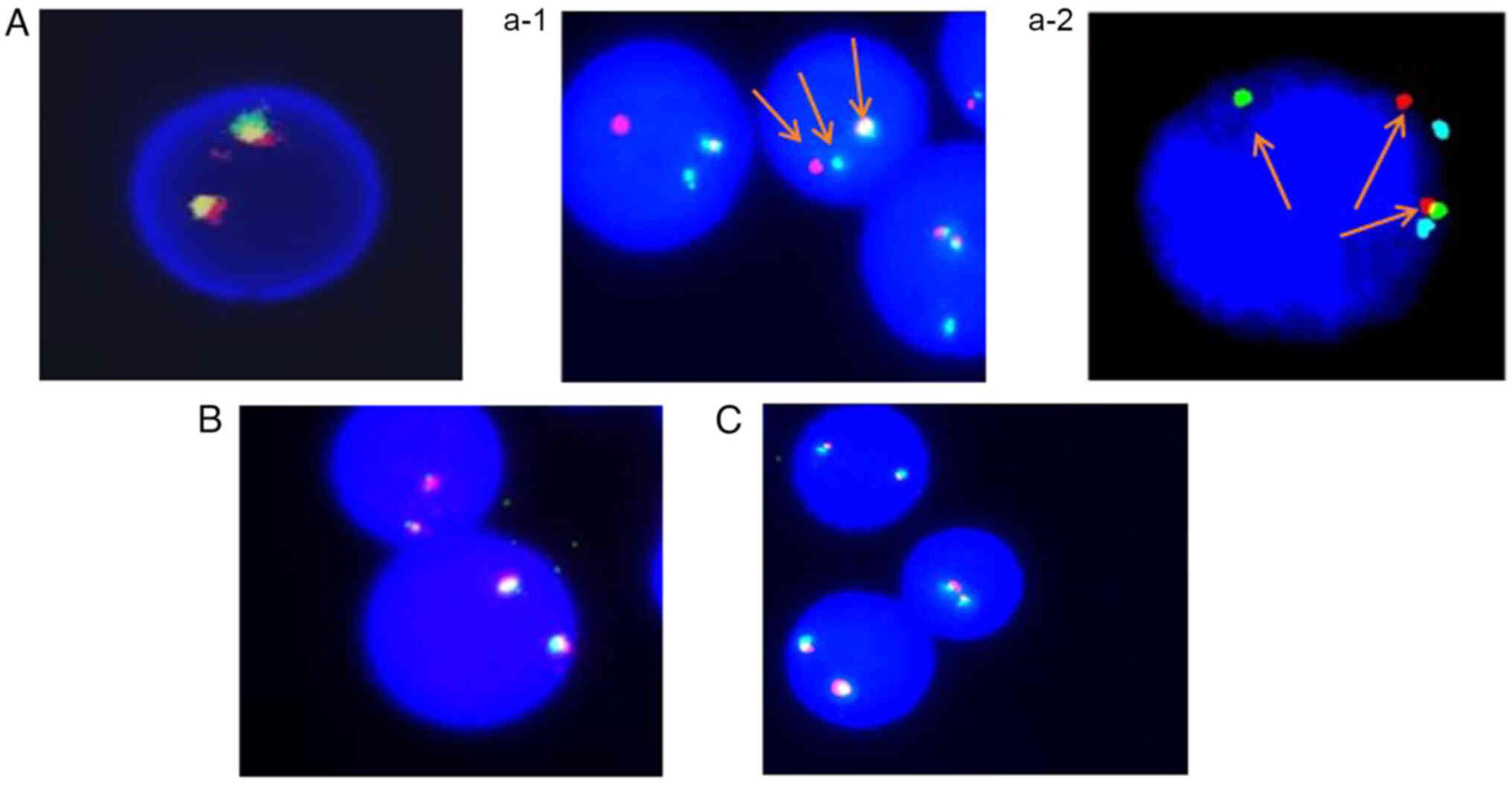
Fluorescence in situ hybridization (FISH)
analysis of the bone marrow showed positive FGFR1 gene
rearrangement (positive rate, 94%), with negative platelet-derived
growth factor receptor A (PDGFRA) and PDGFRB gene
rearrangements (Fig. 2). The
preparation of the peripheral bone marrow samples from patients for
FGFR1, PDGFRA, PDGFRB FISH detection was performed as follows:
Initially, the samples were fixed in a fixed solution consisting of
methanol to acetic acid (3:1), one pre-fixation step was performed
using a 10% fixed solution, and the sample was fixed three times.
Next, the fixed cells were placed onto a glass slide to create a
sample slide, allowed to air dry, the fragments were washed with 2X
saline sodium citrate buffer solution at 37°C for 30 min, and
dehydrated sequentially in 75, 85 and 100% alcohol for 1 min each.
The FISH probes FGFR1/D8Z2 (Anbiping; cat. no. F.01109–01), PDGFRA
(Anbiping; cat. no. F.01162–01) and PDGFRB (Anbiping; cat. no.
F.01033–01) were utilized with a hybridization instrument (Thermos;
cat. no. S500-24) for hybridization, denaturing at 78°C for 8 min,
and hybridizing at 42°C for 16 h. On the following day, the samples
were washed with 0.3% NP40 wash solution at 68°C for 2 min,
followed by washing with deionized water at 37°C for 1 min. DAPI
nuclear staining was performed for 20 min at room temperature, and
the results were observed using a fluorescence microscope (Olympus;
cat. no. BX63). Metasystem ISIS V5.8.11 (Metasystem Co., Ltd.) FISH
analysis software was used for photography and analysis purposes.
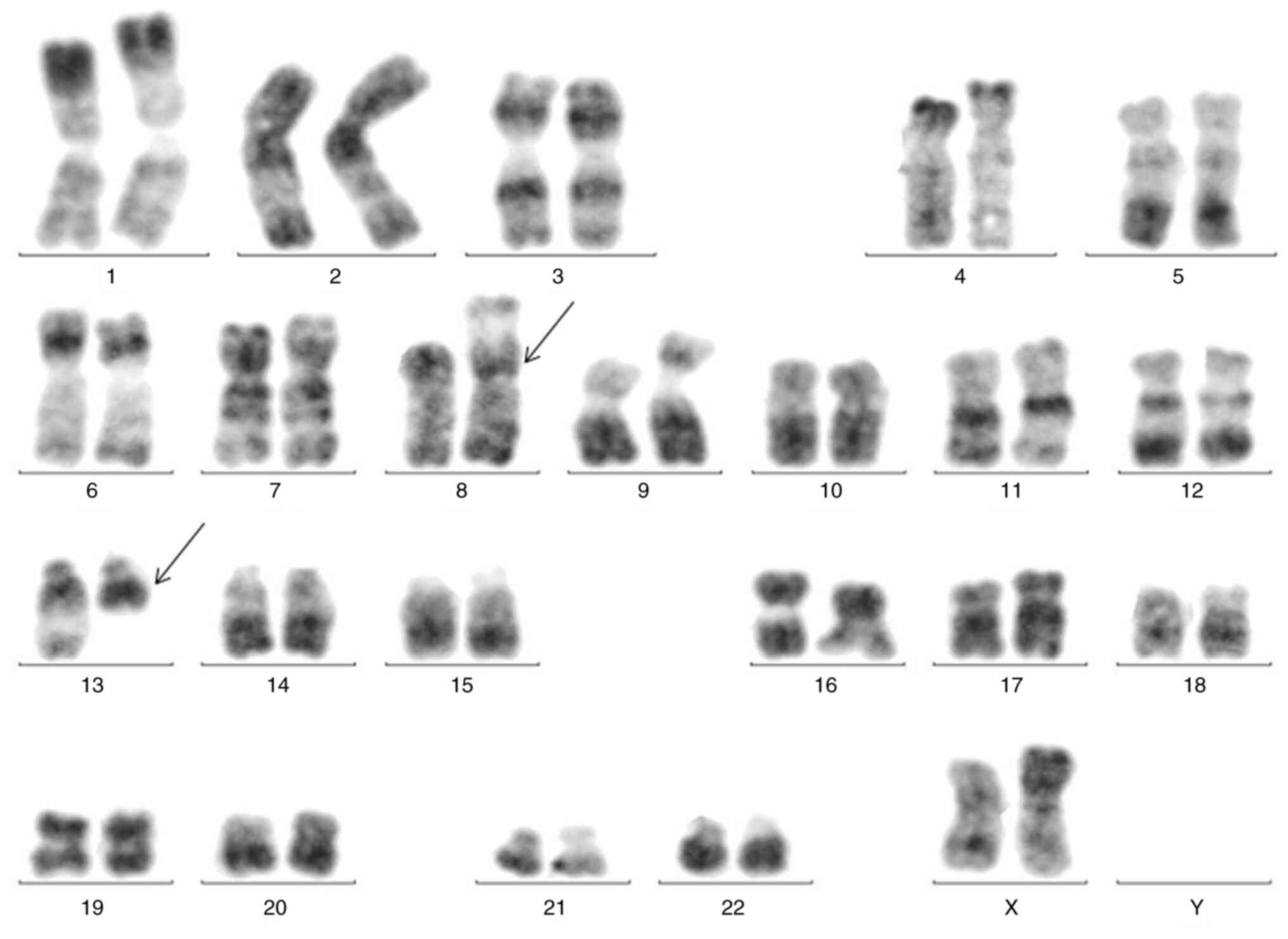
The karyotype showed a balanced translocation between chromosome 8
and chromosome 13: t(8;13)(p11;q12) (Fig. 3). Positron emission tomography
(PET)/computed tomography (CT) scans showed a diffuse increase in
bone marrow metabolic activity, multiple high metabolic lymph nodes
above and below the diaphragm, and splenomegaly with increased
metabolic activity, consistent with the manifestation of lymphoma
(Fig. S1).
A right inguinal lymph node biopsy revealed
disruption of the normal structure with diffuse infiltration by
lymphoblasts. The immunophenotype was as follows: CD3(+), CD5(+),
CD43(+), CD99(+), terminal deoxynucleotidyl transferase (partial
+), CD34(−), Ki-67(+ 70%), CD20(−), CD21(−), CD56(−),
myeloperoxidase(−), CD68 (loose +), BCL-2(−), BCL-6(−), CD10(−),
CD117(−), CD15(−), cyclinD1(−), mutated melanoma-associated antigen
1(−), paired box protein Pax-5(−) and Epstein-Barr virus-encoded
small RNA(−). In order to further investigate the expression of
immature markers in this patient with T lymphoblastic lymphoma, the
immunohistochemical staining of CD1A, CD4 and CD8 was also
performed. The results indicated that the samples were partially
positive for CD1A, positive for CD4 and negative for CD8 (Fig. 4). The lymph nodes were immersed in a
4% formaldehyde solution and fixed overnight at 4 °C. The following
day the tissue was dehydrated and embedded in paraffin wax. The
embedded material was cut into 4-µm slices, dewaxed and rehydrated,
and the antigens were extracted. The slices were incubated with 3%
hydrogen peroxide at room temperature for 4 min. After washing with
PBS, they were blocked with 5–10% normal goat serum (cat. no.
ab7481; Abcam) in PBS, incubated at room temperature for 10 min,
and the serum was removed. The following primary antibodies were
added: Anti-CD1A (cat. no. ab313875; 0.1 µg/ml; Abcam), anti-CD4
(cat. no. ab133616; 0.2 µg/ml; Abcam), anti-CD8 (cat. no. ab245118;
0.4 µg/ml; Abcam), anti-CD3 (cat. no. ab243874; 0.2 µg/ml; Abcam),
anti-CD43 (cat. no. ab101533; 0.5 µg/ml; Abcam), anti-CD5 (cat. no.
ab75877; 0.2 µg/ml; Abcam), anti-CD68 (cat. no. ab213363; 0.08
µg/ml; Abcam), anti-CD99 (cat. no. ab108297; 0.5 µg/ml; Abcam),
anti-terminal deucleotide transferase (cat. no. ab183341; 1 ug/ml;
Abcam), anti-Ki-67 (cat. no. ab92742; 1 µg/ml; Abcam), anti-Bcl-2
(cat. no. ab32124; 1 µg/ml; Abcam), anti-Bcl-6 (cat. no. ab172610;
0.5 µg/ml; Abcam), anti-CD10 (cat. no. ab208778; 0.1 µg/ml; Abcam),
anti-CD117 (cat. no. ab32363; 1 µg/ml; Abcam), anti-CD15 (cat. no.
ab218403; 5 µg/ml; Abcam), anti-CD20 (cat. no. ab219329; 1 µg/ml;
Abcam), anti-CD21 (cat. no. ab315160; 2 µg/ml; Abcam), anti-CD34
(cat. no. ab315820; 0.1 µg/ml; Abcam), anti-CD56 (cat. no.
ab313779; 0.2 µg/ml; Abcam), anti-cyclind1 (cat. no. ab273608; 1
µg/ml; Abcam), anti-myeloperoxidase (cat. no. ab134142; 0.1 µg/ml;
Abcam), anti-mutated melanoma-associated antigen 1 (cat. no.
ab247079; 0.5 µg/ml; Abcam), anti-pin-box protein Pax-5 (cat. no.
ab234402; 0.5 µg/ml; Abcam) and incubated at 3°C for 1–2 h or 4°C
overnight. After washing with PBS for three times, biotin-labeled
secondary antibody (HRP marker; cat. no. ab6721; 1 µg/ml; Abcam)
was added to the working solution and incubated at 37°C for 20–40
min. The sample was washed with PBS three times, streptavidin
coupled alkaline phosphatase was added and incubated at 37°C for
15–25 min. The sample was rinsed again with PBS and
3,3′-diaminobenzidine was added at room temperature for 3–15 min
for color development. Section cleaning, hematoxylin reverse
staining, dehydration, cleaning and mounting was performed. In
situ hybridization was used to detect encoded small RNA (EBER)
of Epstein-Barr virus. EBER in situ hybridization kit (cat.
no. ISH 7001) from ZSBG-Bio was used. The dewaxed and hydrated
lymph node sections were added with 1X protease K working solution
and incubated at room temperature for 5 min. After which, EBER
hybrid solution was added and incubated at 55°C for 60 min, and
finally transferred to 37°C for overnight incubation. After washing
with PBS, HRP-marked digoxin antibody was added. After incubation
at 37°C for 30 min, DAB color developing solution was added, and
hematoxylin was re-stained at room temperature for 1–2 min,
dehydrated and sealed. Representative images are taken with a light
microscope (Japan). This was consistent with the immunophenotype of
typical T lymphoblastic lymphoma. Based on the aforementioned
immunohistochemical staining, the biopsy of the right inguinal
lymph node was consistent with T lymphoblastic lymphoma.
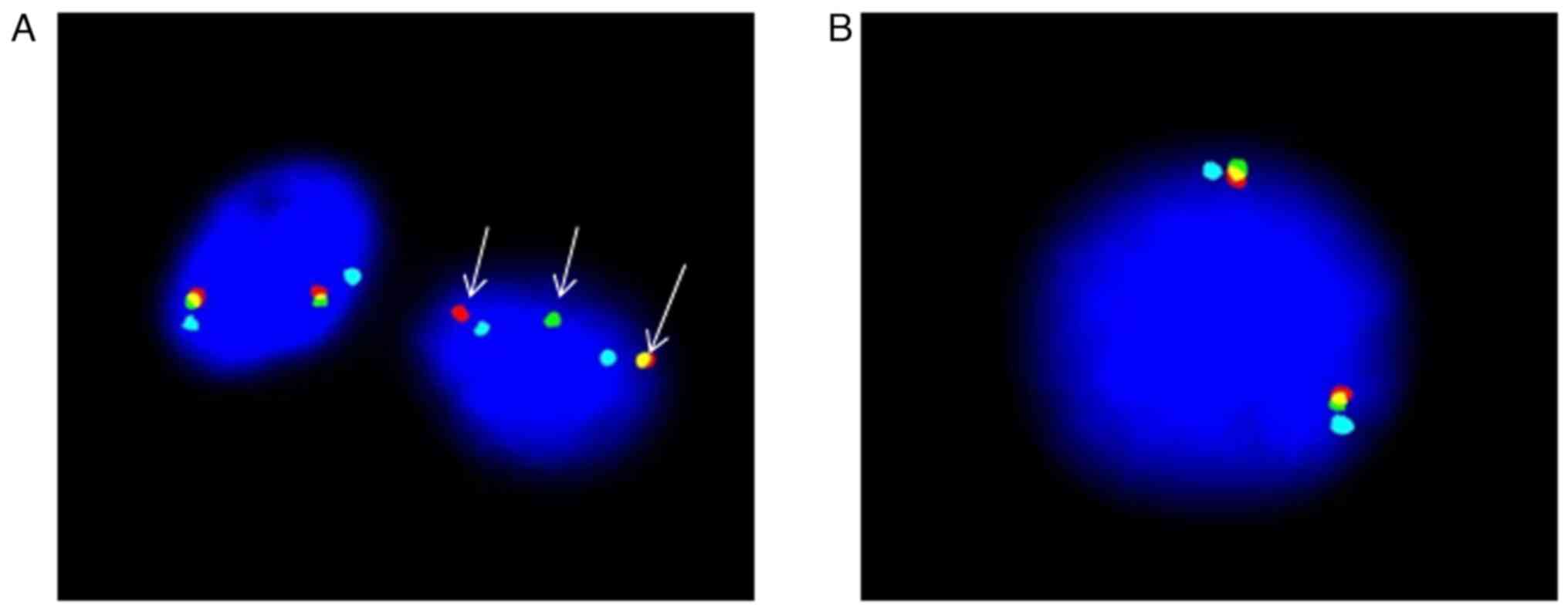
FGFR1 gene rearrangement was also positive in the right
inguinal lymph node tissue sections, as indicated by FISH (Fig. 5). Therefore, a diagnosis of MLNAF
with t(8;13)(p11;q12) was made. At that time, the FGFR1 partner
gene was not detected. Previously, paraffin-embedded samples of the
lymph nodes of patient were used to detect the presence of the
FGFR1-ZNF198 fusion gene by reverse transcription polymerase chain
reaction quantification (RT-qPCR. The lymph node paraffin-embedded
specimen was sliced and placed in a centrifuge tube. The tissue was
dewaxed using xylene. RNA was extracted using the RNA pred Pure
Hi-Blood kit (Tiangen Biotech Co., Ltd.; cat. no. DP443), in strict
accordance with the kit instructions. A total of 1 ml of Trizol
(Invitrogen; Thermo Fisher Scientific, Inc.) was added to
1×107 cell suspension and mixed. The mixture was
incubated at room temperature for 5 min, followed by the addition
of 0.2% chloroform. After shaking for 30 sec, it was left
undisturbed for 3 min and then centrifuged at 2–8°C at 12,000 × g
for 15 min. The supernatant was transferred into a DECP-treated EP
tube and mixed with isopropyl alcohol, which was left at room
temperature for 10 min. RNA was obtained by centrifugation again at
2–8°C at 12,000 × g for 5 min. cDNA was synthesized from 2 µg total
RNA using a first-strand cDNA synthesis kit (cat. no. K1622; Thermo
Fisher Scientific, Inc.). The PCR primers of the
FGFR1-ZNF198 fusion gene and normalization control β-actin
gene were designed based on the corresponding gene structure using
Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast), and
the sequences were follows: FGFR1-ZNF198, Forward,
5′-TCCCTGTGCCTGTGTATATCCC-3′ and reverse,
5′-CGGGAAGCTCATACTCAGAGAC-3′; and β-actin forward,
5′-AAGGCCAACCGCGAGAAGAT-3′ and reverse, 5′-TCGGTGAGGATCTTCATGAG-3′.
Using SYBR Green (Thermo Fisher Scientific, Inc.) as the
fluorophore, the amplification conditions included a temperature of
42°C for 30 min, 94°C for 5 min and 40 cycles. The method of
quantification was 2−ΔΔCq (Livak and Schmittgen 2001).
Due to the prolonged placement of the specimen, the mRNA was
severely degraded and could not be effectively reverse transcribed
into cDNA, thus the RT-qPCR could not be completed.
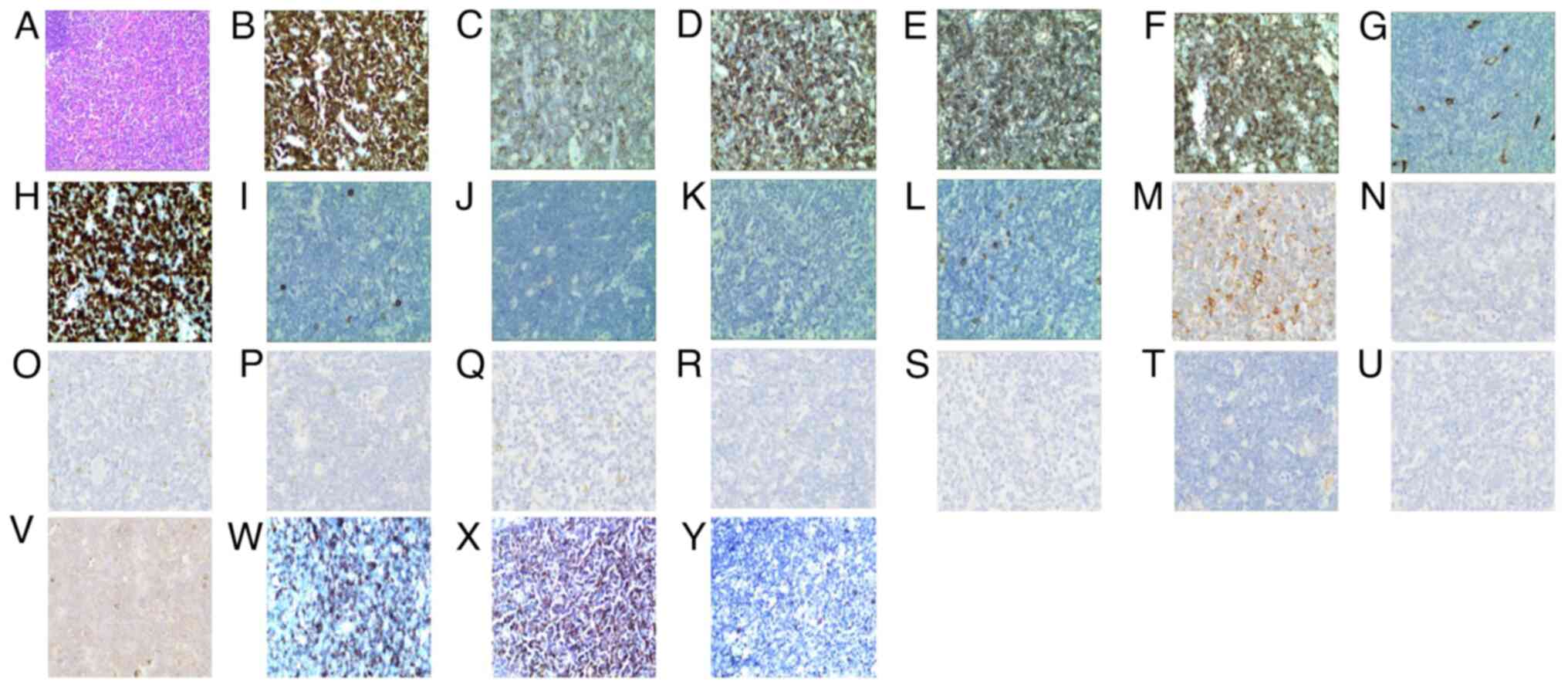 | Figure 4.Right inguinal lymph node biopsy with
an (A) hematoxylin and eosin-stained section (magnification, 100×)
showing partial disappearance of the normal structure. Tumor cells
(magnification, 400×) (B) CD3-diffusely bright positive, (C)
CD5-positive, (D) CD43-positive, (E) CD99-positive, (F) terminal
deoxynucleotidyl transferase partially positive, (G) CD34-negative,
(H) diffusely bright Ki-67-positive (+70%), (I) CD20-negative, (J)
CD21-negative, (K) CD56-negative, (L) myeloperoxidase-negative, (M)
CD68-(loose positive), (N) Bcl-2-negative, (O) Bcl-6-negative, (P)
CD10-negative, (Q) CD117-negative, (R) CD15-negative, (S)
cyclinD1-negative, (T) MUM-1-negative, (U) PAX-5-negative, (V)
EBER-negative, (W) CD1A-positive, (X) CD4-positive and (Y)
CD8-negative. |
The TKI dasatinib in combination with the CHOPE
chemotherapy (2 mg/m2 vindesine on day 1 + 750
mg/m2 cyclophosphamide on day 1 + 20 mg/m2
doxorubicin hydrochloride liposome on day 1 + 60 mg/m2
etoposide on days 1–3 + 1 mg/kg prednisone tablet on days 1–5) was
initiated for 1 cycle, every 21 days. After 4 cycles, the enlarged
lymph nodes of the patient disappeared, the liver returned to
normal and the spleen shrank to 1 cm below the left costal margin.
Furthermore, the peripheral blood count returned to normal, and the
proportion of FGFR1 gene rearrangement detected by bone
marrow FISH analysis decreased to 36% (Fig. 2). Finally, PET/CT scans showed
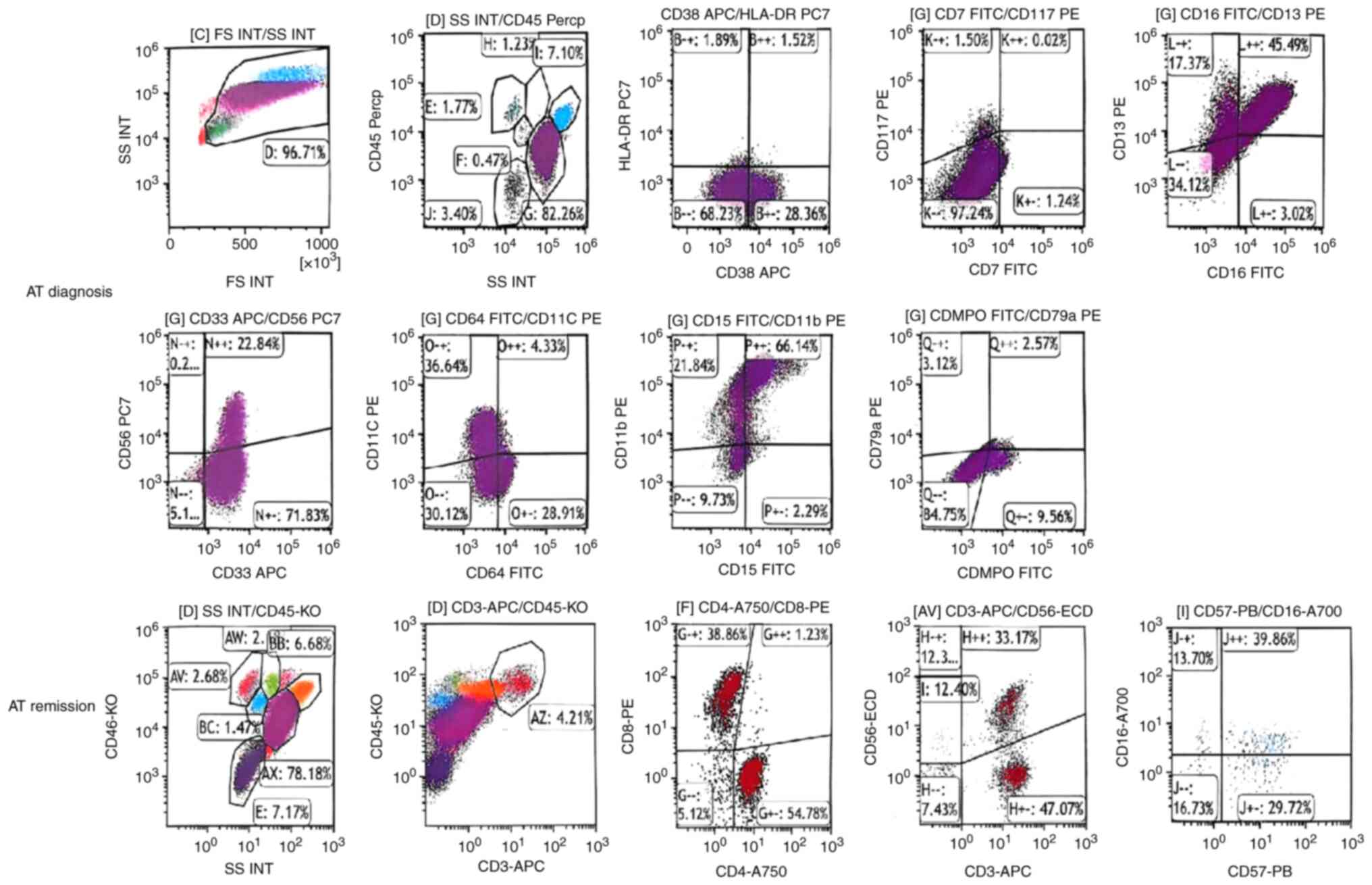
partial metabolic remission in April 2021 (Fig. 1). The flow cytometry results of the
patient's bone marrow at diagnosis and remission are presented in
Fig. 6. EDTA anticoagulant marrow
blood and the following antibodies were added: CD45 (cat. no.
ab40763; Abcam), CD38 (cat. no. ab108403; Abcam), CD117 (cat. no.
ab317843; Abcam), CD7 (cat. no. ab109296; Abcam), HLA-DR (cat. no.
ab92511; Abcam), CD16 (cat. no. ab223200; Abcam), CD13 (cat. no.
ab317440; Abcam), CD33 (cat. no. ab134115; Abcam), CD56 (cat. no.
ab220360; Abcam), CD64 (cat. no. ab109449; Abcam), CD11c (cat. no.
ab254183; Abcam), CD15 (cat. no. ab241552; Abcam), CD11b (cat. no.
ab224805; Abcam), MPO (cat. no. ab208670; Abcam) and CD79a (cat.
no. ab133483; Abcam) at 100 µl/each into the tube, shaken, and kept
in darkness at room temperature for 20 min. A total of 3%
paraformaldehyde was added and incubated in darkness 10 min at room
temperature. A total of 1 ml purified water was added and the
solution was kept in darkness at room temperature for 10 min after
shaking. The solution was centrifuged at 160 × g for 5 min at room
temperature and the supernatant was discarded. A total of 2 ml of
PBS solution was added, shaken and centrifuged at 160 × g for 5 min
at room temperature. The supernatant was discarded and 500 µl PBS
was added. Specimens were examined using Flow cytometer (Beckman
Coulter, Inc.) after mixing. The detected data was analyzed by
Kaluza 2.1.1 software(Beckman Coulter, Inc). The fluorescent dyes
were all obtained from Beckman Coulter, Inc, including fluorescein
isothiocyanate (FITC), phycoerythrin (PE), peridinin chlorophy II
protion (PreCP) and allophycocyanin (APC). The aforementioned
reagents and labeling methods were used to detect known positive
specimens as positive controls and the negative cell population of
the specimen was used as the negative control. However, upon
continuation of the regimen for a further two cycles, the disease
relapsed and new enlarged lymph nodes were found in the neck, and
the spleen notably increased in size. An adjusted treatment
regimen, including BCL2 inhibitor (venetoclax, 100 mg on day 1, 200
mg on day 2 and 400 mg on days 3–14) + demethylated agent (75
mg/m2 azacitidine on days 1–7) + histone deacetylase
inhibitors (20 mg chidamide twice a week for 2 weeks), was
administered. During the treatment process, the WBC count of the
patient gradually increased to 60×109/l, and as
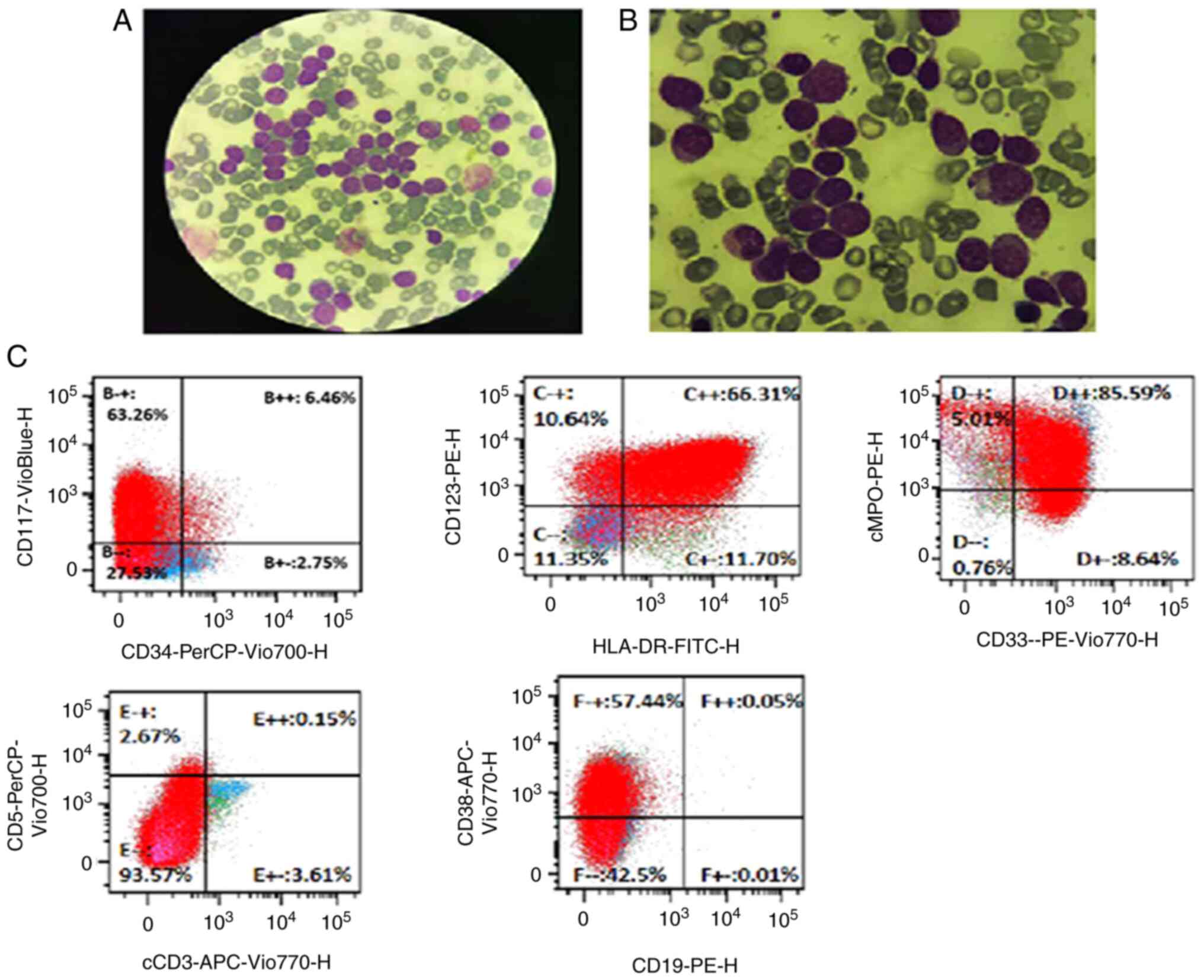
confirmed by bone marrow cell morphology and flow cytometry, the
patient was diagnosed with AML (Fig.
7). The proportion of FGFR1 gene rearrangement detected
by peripheral blood FISH analysis increased to 90%. After
diagnosis, the patient was changed to the IA chemotherapy regimen
(10 mg/m2 idarubicin on days 1–3 + 150 mg/m2
arabinoside on days 1–7) and entered the myelosuppression phase 1
week after chemotherapy. The patient subsequently experienced a
severe lung infection and died in August 2021.
Discussion
MLNAF is a malignant tumor originating from
hematopoietic stem cells, with various clinical manifestations and
aggressive progression; it is slightly more common in men than
women, with a median age of 32 years (range, 3–84 years). MLNAF
often presents with systemic symptoms such as fatigue, night
sweating, emaciation or fever. At initial diagnosis, local or
systemic lymph node enlargement and hepatic/splenomegaly are
commonly found, and some patients may have extranodal organ
involvement, such as that of the tonsils, lungs and mammary glands
(3). Approximately one-half of
patients with MLNAF possess the t(8;13)(p11;q12) mutation, forming
the ZNF198-FGFR1 fusion gene (3). These patients often show a significant
increase in WBC count, and increased eosinophils in the peripheral
blood and bone marrow, presenting the coexistence of
myeloproliferative neoplasm and T-cell lymphoma. Most of the
patients rapidly progress to acute leukemia, commonly AML, within
1–2 years (4). The molecular
pathogenesis of MLNAF is characterized by FGFR1
rearrangement, which forms a fusion gene through translocation,
insertion, inversion or deletion (12). This genetic variation impacts
FGFR1 mRNA transcription, thereby promoting the oncogenicity
and genetic diversity of the FGFR1 protein. FGFR1 fusion
genes can be divided into two types: Type I and type II. Type I
refers to the FGFR1 gene located at the 3′ end of the fusion
gene, and the FGFR1 tyrosine kinase domain is fused to the
N-terminal oligomerization domain of the partner protein. This
fusion type protein that cannot bind to the FGF ligand and causes a
conformational change in the FGFR1 tyrosine kinase domain.
This stimulates the function of FGFR1 oncogene and
constitutively activates its tyrosine kinase function, changes its
localization, and subsequently activates PI3K-AKT, RAS/MAPK, STAT
and PLC/PKC in the downstream cell pathways to transmit abnormal
signals, which ultimately leads to the transition from MLNAF to AML
(4,13). The fusion genes of FGFR1 and
its partners in MLNAF belong to type I. Conversely, type II fusion
proteins retain the extracellular domain of FGFR1, allowing them to
bind to FGF ligands, a characteristic commonly observed in solid
tumors. Even though the domains in the fusion proteins retained by
FGFR rearrangement are different, in all cases the protein retains
a complete kinase domain, suggesting that the kinase domain plays a
vital role in the function of the fusion protein. The expression of
ZNF198-FGFR1 is related to specific plasminogen activator
inhibitor-2-mediated anti-apoptosis, which is possibly one of the
reasons for the high malignancy of leukemia cells (14). The numbers and common phenotypes of
reported cases for MLNAF, and the reported response for
chemotherapy and TKIs have been described in the literature
(Table II) (15–36). A
review of this literature showed that the cohort of patients with
MNLF harboring ZNF198-FGFR1, representing the most prevalent
cases, predominantly exhibit resistance to chemotherapy (15). While a subset of patients may
experience transient responses to TKIs such as imatinib and
midostaurin, sustained efficacy remains elusive (16,17).
Notably, allogeneic hematopoietic stem cell transplantation was
pursued by 9 patients, resulting in remission for 7 patients;
however, 2 still experienced disease relapse (4). These findings underscore the
insufficiency of traditional chemotherapy and TKIs in addressing
this condition.
 | Table II.Numbers and common phenotypes of
reported cases for myeloid/lymphoid neoplasms with eosinophilia and
FGFR1 rearrangement, and the reported response for
chemotherapy and tyrosine kinase inhibitorsa. |
Table II.
Numbers and common phenotypes of
reported cases for myeloid/lymphoid neoplasms with eosinophilia and
FGFR1 rearrangement, and the reported response for
chemotherapy and tyrosine kinase inhibitorsa.
| Fusion genes | Number of
cases | Common
phenotypes | Physical and
laboratory examination | Sensitivity to
chemotherapy | Numbers and results
of allo-SCT | Sensitivity to
TKIs | (Refs.) |
|---|
|
ZNF198-FGFR1 | >30 |
T-LBL/T-lymphoma | Lymphadenopathy,
hepatosplenomegaly, eosinophilia or mono-cytosis or both | Insensitive | 7 remission; 2
recurrence | Sensitive
(imatinib, midostaurin) | (15–17) |
|
FOP1-FGFR1 | 5 | MPN, AML,
B-ALL | Polycythemia
without eosinophilia | Sensitive | No | Not tested | (18,19) |
|
CEP110-FGFR1 | >20 | AML, T-LBL | Lymphadenopathy,
purpura, skin lesions, eosinophilia and monocytosis | Insensitive | 7 remission; 1
recurrence | Sensitive
(imatinib, dasatinib, pemigatinib) | (20,21) |
|
HERVK-FGFR1 | 2 | AML, SM-AHNMD | Polycythemia,
poikilocyte, granulocytosis, abnormal megakaryocytes | Insensitive | 1 remission | Not tested | (22) |
|
BCR-FGFR1 | >30 | CML, aCML, AML,
B-ALL | Splenomegaly,
eosinophilia | Insensitive | 4 remission; 3
recurrence | Insensitive
(imatinib, dasatinib), Sensitive (ponatinib, pemigatinib) | (23,24) |
|
NUP98-FGFR1 | 2 | Therapeutic
AML | Granulocyte
hyperplasia with mononucleosis | Not tests | No | Not tested | (25) |
|
FOP2-FGFR1 | 2 | T-LBL, AML | Lymphadenopathy,
eosinophilia | Sensitive | No | Not tested | (26) |
|
TIF1-FGFR1 | 5 | CEL, AML | Eosinophilia | Resistant | No | Not tested | (27) |
|
MYO18A-FGFR1 | 2 | CML | Thrombocytopenia,
monocyte, eosinophilic and basophil increased | Resistant | No | Not tested | (28) |
|
CPSF6-FGFR1 | 1 | Not reported | Lymphadenopathy and
splenomegaly, neutrophils without eosinophilia | Resistant | No | Not tested | (29) |
|
LRRFIP1-FGFR1 | 1 | MDS, AML | Pancytopenia,
eosinophilia | Not tests | No | Not tested | (30) |
|
CUX1-FGFR1 | 1 | Pre-T-LBL | Neutrophils,
lymphocytes and monocytes increased without eosinophils | Resistant | No | Not tested | (31) |
|
TPR-FGFR1 | 4 | AML | Lymphadenopathy,
increasing monocytes | Insensitive | 1 remission | Not tested | (32) |
|
NUP358-FGFR1 | 2 | MDS | Splenomegaly, a
little eosinophilia | Sensitive | No | Not tested | (33) |
|
SQSTM1-FGFR1 | 1 | AML | Neutrophils and
monocytes increased, megakaryocytes | Not tests | No | Not tested | (34) |
|
TFG-FGFR1 | 1 | AML | Skin ecchymosis and
splenomegaly, eosinophilia | Insensitive | No | Resistant
(ponatinib) | (35) |
|
HOOK3-FGFR1 | 1 | MDS | Leukocytosis and
thrombocytopenia. | Insensitive | No | Resistant
(ponatinib) | (36) |
The patient described in the present study was an
elderly woman. The combination of chemotherapy and TKI treatment
achieved temporary complete remission; however, the condition
rapidly progressed to AML 6 months after the diagnosis, which is
consistent with previous literature reports (4,9). At
present, there is no unified standard for MLNAF treatment, and
conventional cytotoxic drugs have poor therapeutic effects, with
the overall survival rate in the literature reportedly <20%
(3). FGFR1, as a tyrosine kinase
inhibitor receptor, is hypothesized to be a therapeutic target, but
neither imatinib nor dasatinib have achieved ideal efficacy. New
TKIs, such as pemigatinib and futibatinib, which selectively
inhibit FGFR1 tyrosine kinase activity, have shown some efficacy in
in vitro and animal experiments (37,38),
but the exact therapeutic effect still needs to be verified by
further clinical trials. Hu et al (39) found that the activation of FGFR1 is
related to the upregulation of MYC, and there is a synergistic
effect between FGFR1 inhibitors and MYC-targeted inhibitors,
suggesting a new approach for MLNAF treatment. Currently, these new
targeted drugs are still in the experimental stage, and at present,
allogeneic hematopoietic stem cell transplantation (allo-HSCT) is
still considered to be the best option for the treatment of MLNAF.
A previous case study reported that patients with MLNAF achieved
disease-free survival times of up to 16 years when treated with
allo-HSCT (40). In addition,
another patient with refractory MLNAF received dual umbilical cord
blood transplantation and achieved 5 years of disease-free survival
(41). In the present study, the
patient obtained CR after combination chemotherapy + TKI, and was
recommended to receive allo-HSCT. However, due to economic
constraints, the patient did not receive the treatment and the
disease progressed to AML. Finally, the patient died of a pulmonary
infection after induction of remission treatment. The present study
further confirms that conventional TKIs combined with chemotherapy
have limited efficacy in the treatment of MLNAF, and that allo-HSCT
should be performed as early as possible after diagnosis.
In conclusion, MLNAF is a rare malignancy
originating from hematopoietic stem cells, which has diverse
clinical manifestations and may develop into multiple lineage
hematopoietic system tumors. The condition is easily misdiagnosed
in the early stage, and diagnostic gold-standard genetic
examination reveals FGFR1 gene breakage and chromosome 8p11
translocation. Conventional TKI chemotherapy has poor efficacy, and
new targeted drugs are still under research, which may bring hope
for the long-term survival of patients. However, at present,
allo-HSCT remains the first choice for MLNAF treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The sequencing results and raw data generated in the
present study may be found in the BioProject database under
accession number PRJNA1120252 or at the following URL: https://www.ncbi.nlm.nih.gov/sra/PRJNA1120252.
Authors' contributions
YG was responsible for clinical data collection,
interpretation of the results and drafting the manuscript. SQ
participated in the design of the study and provided general
support. JZ and XG assisted with the analysis. MM and TT collected
important background information, prepared the preliminary work of
the manuscript and assisted in preliminary data collection. All
authors have read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Written informed consent for publication of the case
report, including clinical details and images, was provided by the
patient's spouse.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Macdonald D, Aguiar RC, Mason PJ, Goldman
JM and Cross NC: A new myeloproliferative disorder associated with
chromosomal translocations involving 8p11: A review. Leukemia.
9:1628–1630. 1995.PubMed/NCBI
|
|
2
|
Fioretos T, Panagopoulos I, Lassen C,
Swedin A, Billström R, Isaksson M, Strömbeck B, Olofsson T,
Mitelman F and Johansson B: Fusion of the BCR and the fibroblast
growth factor receptor-1 (FGFR1) genes as a result of
t(8;22)(p11;q11) in a myeloproliferative disorder: The first fusion
gene involving BCR but not ABL. Genes Chromosomes Cancer.
32:302–310. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jackson CC, Medeiros LJ and Miranda RN:
8p11 myeloproliferative syndrome: A review. Hum Pathol. 41:461–476.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li T, Zhang G, Zhang X, Lin H and Liu Q:
The 8p11 myeloproliferative syndrome: Genotypic and phenotypic
classification and targeted therapy. Front Oncol. 12:10157922022.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arber DA, Orazi A, Hasserjian R, Thiele J,
Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M and Vardiman JW:
The 2016 revision to the World Health Organization classification
of myeloid neoplasms and acute leukemia. Blood. 127:2391–2405.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abruzzo LV, Jaffe ES, Cotelingam JD,
Whang-Peng J, Del Duca V Jr and Medeiros LJ: T-cell lymphoblastic
lymphoma with eosinophilia associated with subsequent myeloid
malignancy. Am J Surg Pathol. 16:236–245. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Katoh M and Nakagama H: FGF receptors:
Cancer biology and therapeutics. Med Res Rev. 34:280–300. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang CL, Tang GS, Guo MQ, Cheng H, Liu
MD, Yang JM and Gong SL: Clinical significance of FGFR1 gene
abnormalities in blood tumors. Zhongguo Shi Yan Xue Ye Xue Za Zhi.
28:983–988. 2020.(In Chinese). PubMed/NCBI
|
|
9
|
Grand EK, Grand FH, Chase AJ, Ross FM,
Corcoran MM, Oscier DG and Cross NC: Identification of a novel
gene, FGFR1OP2, fused to FGFR1 in 8p11 myeloproliferative syndrome.
Genes Chromosomes Cancer. 40:78–83. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou F, Chen S, Chao H, Zhang R, Zhou M
and Pan J: Clinical and gene involved of one case of 8p11
myeloproliferative syndrome with ins(13;8)(q12;p11p23). Zhonghua
Xue Ye Xue Za Zhi. 36:291–296. 2015.(In Chinese). PubMed/NCBI
|
|
11
|
Tiong KH, Mah LY and Leong CO: Functional
roles of fibroblast growth factor receptors (FGFRs) signaling in
human cancers. Apoptosis. 8:1447–1468. 2013. View Article : Google Scholar
|
|
12
|
Gallo LH, Nelson KN, Meyer AN and Donoghue
DJ: Functions of fibroblast growth factor receptors in cancer
defined by novel translocations and mutations. Cytokine Growth
Factor Rev. 26:425–449. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Greulich H and Pollock PM: Targeting
mutant fibroblast growth factor receptors in cancer. Trends Mol
Med. 17:283–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kasyapa CS, Kunapuli P, Hawthorn L and
Cowell JK: Induction of the plasminogen activator inhibitor-2 in
cells expressing the ZNF198/FGFR1 fusion kinase that is involved in
atypical myeloproliferative disease. Blood. 107:3693–3699. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Urrea Pineda LY, Perilla O,
Santiago-Pacheco V and Trujillo Montoya S: Myeloproliferative
syndrome with eosinophilia associated with translocation t(8; 13)
and T-cell lymphoblastic lymphoma: A case report and review of the
literature. Cureus. 14:e227342022.PubMed/NCBI
|
|
16
|
Buijs A, van Wijnen M, van den Blink D,
van Gijn M and Klein SK: A ZMYM2-FGFR1 8p11 myeloproliferative
neoplasm with a novel nonsense RUNX1 mutation and tumor lysis upon
imatinib treatment. Cancer Genet. 206:140–144. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen J, Deangelo DJ, Kutok JL, Williams
IR, Lee BH, Wadleigh M, Duclos N, Cohen S, Adelsperger J, Okabe R,
et al: PKC412 inhibits the zinc finger 198-fibroblast growth factor
receptor 1 fusion tyrosine kinase and is active in treatment of
stem cell myeloproliferative disorder. Proc Natl Acad Sci USA.
101:14479–14484. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vizmanos JL, Hernández R, Vidal MJ,
Larráyoz MJ, Odero MD, Marı́n J, Ardanaz MT, Calasanz MJ and Cross
NC: Clinical variability of patients with the t(6;8)(q27;p12) and
FGFR1OP-FGFR1 fusion: Two further cases. Hematol J. 5:534–537.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Popovici C, Zhang B, Gŕegoire MJ,
Jonveaux P, Lafage-Pochitaloff M, Birnbaum D and Pébusque MJ: The
t(6;8)(q27;p11) translocation in a stem cell myeloproliferative
disorder fuses a novel gene, FOP, to fibroblast growth factor
receptor 1. Blood. 93:1381–1389. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guasch G, Mack GJ, Popovici C, Dastugue N,
Birnbaum D, Rattner JB and Pébusque MJ: FGFR1 is fused to the
centrosome-associated protein CEP110 in the 8p12 stem cell
myeloproliferative disorder with t(8;9)(p12;q33). Blood.
95:1788–1796. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen MY, Shen HJ, Chao HY, Wang Q, Zhang
XW, He C, Cen JN, Chen SN, Zhang R and Zhu MQ: 8p11
myeloproliferative syndrome with CEP110-FGFR1 fusion in a child.
Zhonghua Er Ke Za Zhi. 57:297–300. 2019.(In Chinese). PubMed/NCBI
|
|
22
|
Mugneret F, Chaffanet M, Maynadié M,
Guasch G, Favre B, Casasnovas O, Birnbaum D and Pébusque MJ: The
8p12 myeloproliferative disorder. t(8;19)(p12;q13.3): A novel
translocation involving the FGFR1 gene. Br J Haematol. 111:647–649.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Manur R, Sung PJ, Loren AW, Ritchie EK,
Frank D, Bagg A, Geyer JT and Bogusz AM: Leukemic lineage switch in
a t(8;22)(p11.2;q11.2)/BCR-FGFR1-rearranged myeloid/lymphoid
neoplasm with RUNX1 mutation-diagnostic pitfalls and clinical
management including FGFR1 inhibitor pemigatinib. Leuk Lymphoma.
61:450–454. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Khodadoust MS, Luo B, Medeiros BC, Johnson
RC, Ewalt MD, Schalkwyk AS, Bangs CD, Cherry AM, Arai S, Arber DA,
et al: Clinical activity of ponatinib in a patient with
FGFR1-rearranged mixed-phenotype acute leukemia. Leukemia.
30:947–950. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Romana SP, Radford-Weiss I, Ben Abdelali
R, Schluth C, Petit A, Dastugue N, Talmant P, Bilhou-Nabera C,
Mugneret F, Lafage-Pochitaloff M, et al: NUP98 rearrangements in
hematopoietic malignancies: A study of the groupe francophone de
cytogénétique hématologique. Leukemia. 20:696–706. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Onozawa M, Ohmura K, Ibata M, Iwasaki J,
Okada K, Kasahara I, Yamaguchi K, Kubota K, Fujisawa S, Shigematsu
A, et al: The 8p11 myeloproliferative syndrome owing to rare
FGFR1OP2-FGFR1 fusion. Eur J Haematol. 86:347–349. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Belloni E, Trubia M, Gasparini P, Micucci
C, Tapinassi C, Confalonieri S, Nuciforo P, Martino B, Lo-Coco F,
Pelicci PG and Di Fiore PP: 8p11 myeloproliferative syndrome with a
novel t(7;8) translocation leading to fusion of the FGFR1 and TIF1
genes. Genes Chromosomes Cancer. 42:320–325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Walz C, Chase A, Schoch C, Weisser A,
Schlegel F, Hochhaus A, Fuchs R, Schmitt-Gräff A, Hehlmann R, Cross
NC and Reiter A: The t(8;17)(p11;q23) in the 8p11
myeloproliferative syndrome fuses MYO18A to FGFR1. Leukemia.
19:1005–1009. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hidalgo-Curtis C, Chase A, Drachenberg M,
Roberts MW, Finkelstein JZ, Mould S, Oscier D, Cross NC and Grand
FH: The t(1;9)(p34;q34) and t(8;12)(p11;q15) fuse pre-mRNA
processing proteins SFPQ (PSF) and CPSF6 to ABL and FGFR1. Genes
Chromosomes Cancer. 47:379–385. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Soler G, Nusbaum S, Varet B, Macintyre EA,
Vekemans M, Romana SP and Radford-Weiss I: LRRFIP1, a new FGFR1
partner gene associated with 8p11 myeloproliferative syndrome.
Leukemia. 23:1359–1361. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wasag B, Lierman E, Meeus P, Cools J and
Vandenberghe P: The kinase inhibitor TKI258 is active against the
novel CUX1-FGFR1 fusion detected in a patient with T-lymphoblastic
leukemia/lymphoma and t(7;8)(q22;p11). Haematologica. 96:922–926.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim SY, Kim JE, Park S and Kim HK:
Molecular identification of a TPR-FGFR1 fusion transcript in an
adult with myeloproliferative neoplasm, T-lymphoblastic lymphoma,
and a t(1;8)(q25;p11.2). Cancer Genet. 207:258–262. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gervais C, Dano L, Perrusson N, Hélias C,
Jeandidier E, Galoisy AC, Ittel A, Herbrecht R, Bilger K and
Mauvieux L: A translocation t(2;8)(q12;p11) fuses FGFR1 to a novel
partner gene, RANBP2/NUP358, in a
myeloproliferative/myelodysplastic neoplasm. Leukemia.
27:1186–1188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nakamura Y, Ito Y, Wakimoto N, Kakegawa E,
Uchida Y and Bessho M: A novel fusion of SQSTM1 and FGFR1 in a
patient with acute myelomonocytic leukemia with t(5;8)(q35;p11)
translocation. Blood Cancer J. 4:e2652014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang T, Wang Z, Zhang L, Wen L, Cai W,
Yang X, Pan J, Ruan C, Wu D, Sun A and Chen S: Identification of a
novel TFG-FGFR1 fusion gene in an acute myeloid leukaemia patient
with t(3;8)(q12;p11). Br J Haematol. 188:177–181. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang X, Wang F, Yan F, Huang D, Wang H,
Gao B, Gao Y, Hou Z, Lou J, Li W and Yan J: Identification of a
novel HOOK3-FGFR1 fusion gene involved in activation of the
NF-kappaB pathway. Cancer Cell Int. 22:402022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu PCC, Koblish H, Wu L, Bowman K,
Diamond S, DiMatteo D, Zhang Y, Hansbury M, Rupar M, Wen X, et al:
INCB054828 (pemigatinib), a potent and selective inhibitor of
fibroblast growth factor receptors 1, 2, and 3, displays activity
against genetically defined tumor models. PLoS One.
15:e02318772020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kalyukina M, Yosaatmadja Y, Middleditch
MJ, Patterson AV, Smaill JB and Squire CJ: TAS-120 cancer target
binding: Defining reactivity and revealing the first fibroblast
growth factor receptor 1 (FGFR1) irreversible structure.
ChemMedChem. 14:494–500. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hu T, Wu Q, Chong Y, Qin H, Poole CJ, van
Riggelen J, Ren M and Cowell JK: FGFR1 fusion kinase regulation of
MYC expression drives development of stem cell leukemia/lymphoma
syndrome. Leukemia. 32:2363–2373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu LP, Chen Y, Shi HX and Huang XJ: 8p11
myeloproliferative syndrome cured by allogeneic hematopoietic stem
cell transplantation: Two case reports and literature review.
Beijing Da Xue Xue Bao Yi Xue Ban. 45:993–996. 2013.(In Chinese).
PubMed/NCBI
|
|
41
|
Larosa F, Maddens S, Legrand F, Pouthier
F, Ferrand C, Saas P, Hayette S, Chabod J, Tiberghien P, Rohrlich
PS and Deconinck E: Early immune reconstitution and efficient graft
vs tumor effect after unrelated partially matched double cord blood
transplantation in refractory 8p11 syndrome. Bone Marrow
Transplant. 46:622–624. 2011. View Article : Google Scholar : PubMed/NCBI
|