Introduction
Gastric carcinoma (GC) is one of the most prevalent
cancers worldwide, ranking fifth in incidence and fourth in
cancer-related mortality globally (1). Risk factors for GC include
Helicobacter pylori infection, dietary habits, smoking and
genetic predispositions (2).
Despite advancements in surgical techniques, chemotherapy and
targeted therapies, early diagnosis of GC remains challenging due
to non-specific symptoms, often resulting in a poor prognosis at
the advanced stages (3). Current
challenges in GC management include the development of treatment
resistance and the need for personalized therapeutic approaches
(4). Phosphoinositide-3-kinase
regulatory subunit 1 (PIK3R1) is a key regulatory subunit of
phosphoinositide 3-kinases, acting as a tumor suppressor by
inhibiting the activity of p110α of class I PI3K (PIK3CA) (5). Mutations in PIK3R1 have been
implicated in tumorigenesis across multiple organs (6), and elevated levels of PIK3R1 are
associated with the progression of GC (7,8).
ATRX, a regulator of the chromatin state, gene
expression, cellular senescence and DNA damage repair, belongs to
the SWI/SNF protein family. Loss of ATRX expression is
associated with activation of the alternative lengthening of
telomeres pathway, contributing to the replicative immortality of
cancer cells (9,10). Loss-of-function (LOF) mutations in
ATRX are linked to aggressive traits such as tumour growth,
migration, invasion and metastasis in osteosarcoma (9). Although the role of ATRX in GC
is not well understood, female patients with GC harbouring
ATRX mutations exhibit microsatellite instability-high
subtypes, elevated tumour mutational burden and increased
programmed cell death ligand-1 (PD-L1) expression. Moreover, those
patients exhibit prolonged survival when treated with immune
checkpoint inhibitors (11).
RNA binding motif protein 10 (RBM10), an alternative
splicing factor protein, regulates RNA transcription and
expression, including pre-mRNA splicing, mRNA stabilisation and
mRNA transcription. RBM10 is recognised as a tumour
suppressor gene that inhibits proliferation, invasion and
metastasis while promoting apoptosis during tumorigenesis (12,13).
However, RBM10 has also been shown to promote cancer via a
negative feedback mechanism, driven by the upregulation of
RBM10 or its homolog (13).
As with ATRX, there have been few studies on RBM10 in
GC, but upregulation of RBM10 has been confirmed in The
Cancer Genome Atlas (TCGA) dataset (14).
Epstein-Barr virus (EBV) is an oncogenic virus that
can contribute to the malignant transformation of normal gastric
cells (15). The primary route by
which EBV infects epithelial cells is through direct cell-to-cell
contact mediated by B lymphocytes. Following persistent infection,
EBV enters latency, during which it does not integrate into the
host genome but replicates concurrently with the host cells
(16). EBV exhibits a
distinct gene expression profile depending on the latency type.
EBV-positive GC (EBVpGC) is closely associated with latency type 1
genes, including EBV-encoded small RNA 1 (EBER1), EBER2 and EBV
nuclear antigens 1 (16). EBVpGC is
distinguished from other types of GC by its characteristic genomic
aberrations, distinct clinicopathological features and generally
favourable prognosis (17). In GC,
PIK3R1 mutations are associated with EBVpGC cases, whereas
TP53 inactivation is typically identified in EBV-negative GC
(EBVnGC) cases (18).
In the present study, a GC case with a poor
prognosis, harbouring LOF mutations in the PIK3R1, ATRX and
RBM10 genes, with diverse histological features associated
with EBV infection and TP53 inactivation is reported.
Case report
In April 2023, a 62-year-old man presented to the
Emergency Department of Ewha Woman's University Mokdong Hospital
(Seoul, Republic of Korea) with hematemesis. The patient's initial
vital signs were stable. Although the patient had a history of
infarction in the left middle cerebral artery and was receiving
antithrombotic therapy (aspirin and clopidogrel), there was no
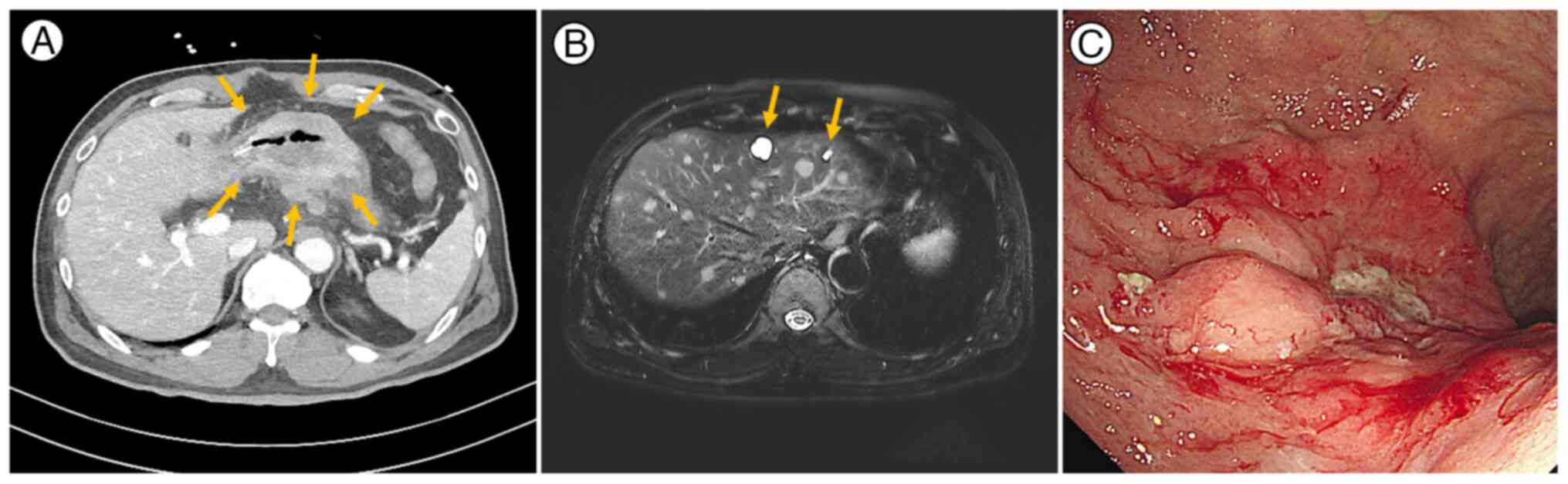
history of cancer. Abdominal computed tomography revealed an
encircling mass in the lower gastric body (Fig. 1A), multiple metastatic lymph nodes
near the celiac axis and gastroepiploic vessels as well as
peritoneal seeding, suggestive of unresectable advanced GC.
Following admission, numerous well-defined lesions of varying
sizes, highly suspected of representing hepatic metastasis, were
identified in both hepatic lobes via diffusion-weighted magnetic
resonance imaging (Fig. 1B).
Esophagogastroduodenoscopy confirmed active bleeding from exposed
vessels with detached blood clots (Fig.
1C). Given the active bleeding and the challenges of initiating
chemotherapy immediately, palliative subtotal gastrectomy and liver
excisional biopsy were selected as the initial intervention.
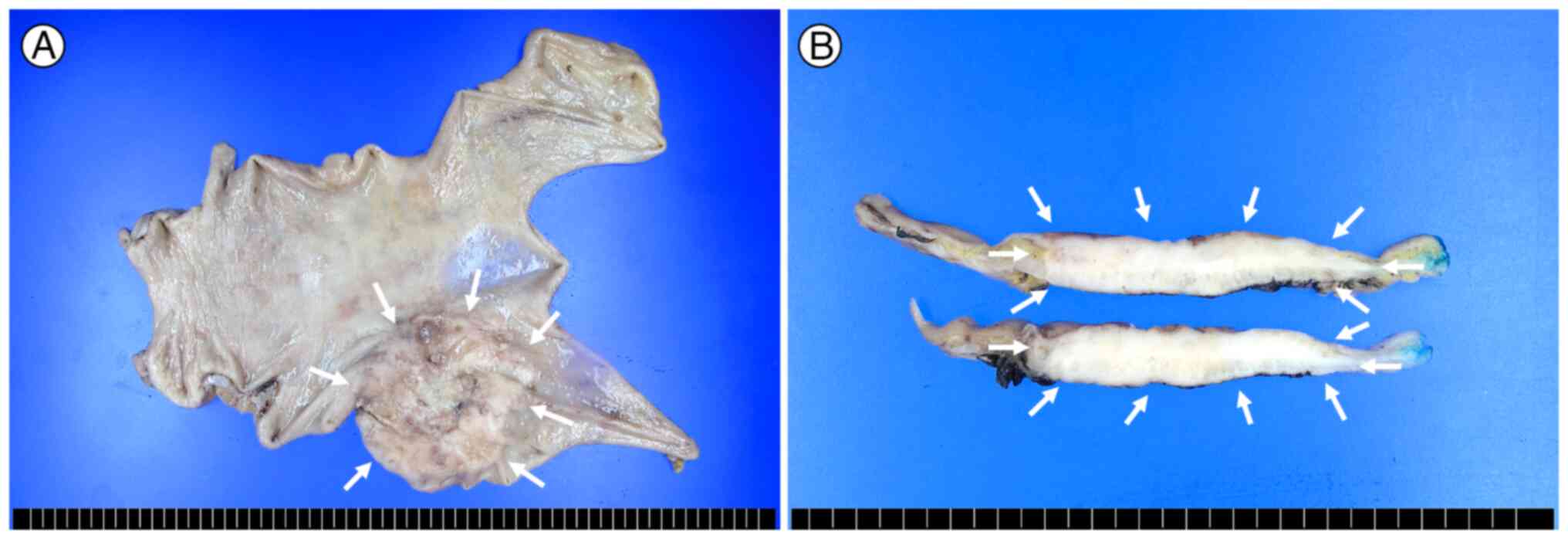
Macroscopically, a well-defined, ulceroinfiltrative mass measuring
7.0×6.0×1.2 cm was identified on the posterior wall of the gastric
body and antrum (Fig. 2A). The cut
surface was greyish-white, solid and necrosis-free, with direct
invasion into the serosa (Fig. 2B).
For histological examination, the tissues were paraffin-embedded
following fixation in 10% neutral-buffered formalin at room
temperature (~25°C) for 24 h. Sections were cut at a thickness of 4
µm and stained with haematoxylin for 5 min and eosin for 2 min at
room temperature. The prepared slides were examined using a light
microscope (Leica DM2000; Leica Microsystems, Inc.).
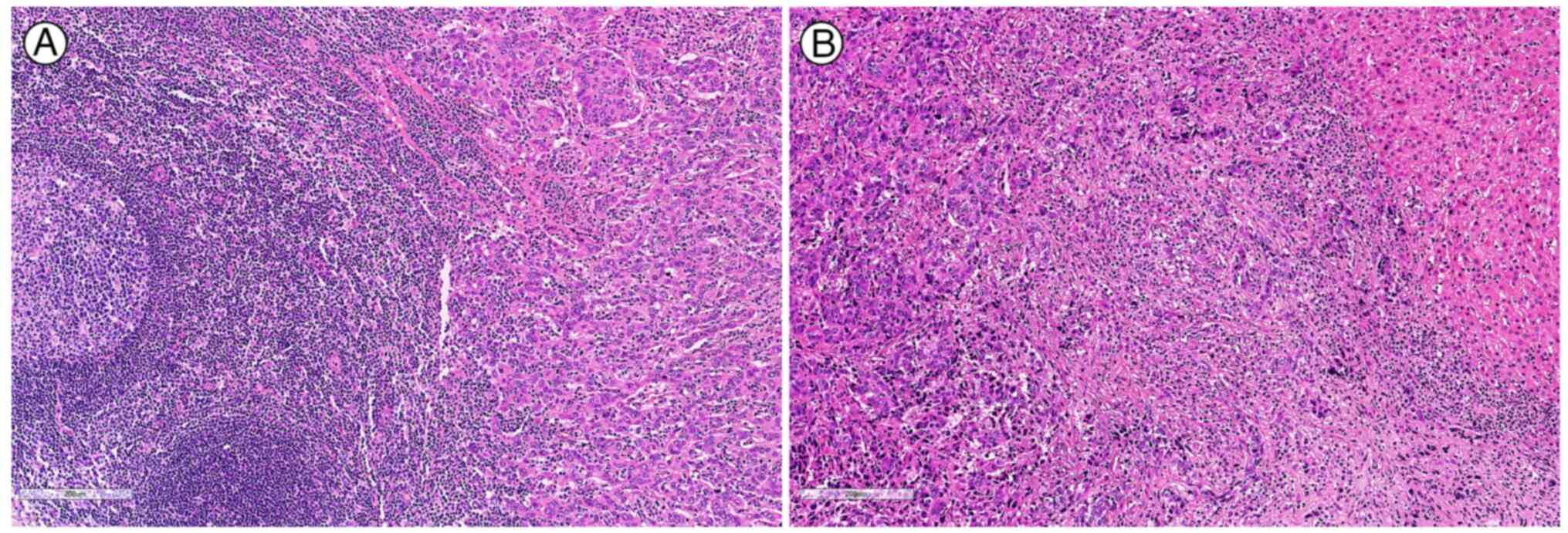
Microscopically, the tumour consisted of three distinct
histological components: Gastric adenosquamous carcinoma (GASC), GC
with lymphoid stroma (GCLS) and poorly differentiated
adenocarcinoma (PDAD), accounting for 30, 40 and 30% of the tumour
volume, respectively. The GASC component comprised of both
adenocarcinoma and squamous cell carcinoma, characterised by large
round-to-oval nuclei with abundant eosinophilic cytoplasm,
intercellular bridges and multifocal glandular formations (Fig. 3A). No keratin pearls were observed.
This component was primarily located in the mucosal and submucosal
layers. The GCLS component exhibited abundant lymphocytic
infiltration around undifferentiated carcinoma clusters with lacy
or tubular growth patterns, pleomorphic polygonal cells, prominent
nucleoli and vesicular chromatin (Fig.
3B). The PDAD component featured relatively small, monotonous,
hyperchromatic nuclei with coarse chromatin, prominent nucleoli and
brisk mitotic activity. This component had a mucinous stromal
background, with rare glandular formations and minimal lymphocytic
infiltration (Fig. 3C). The GCLS
and PDAD components were primarily located in the muscularis
propria and subserosa.
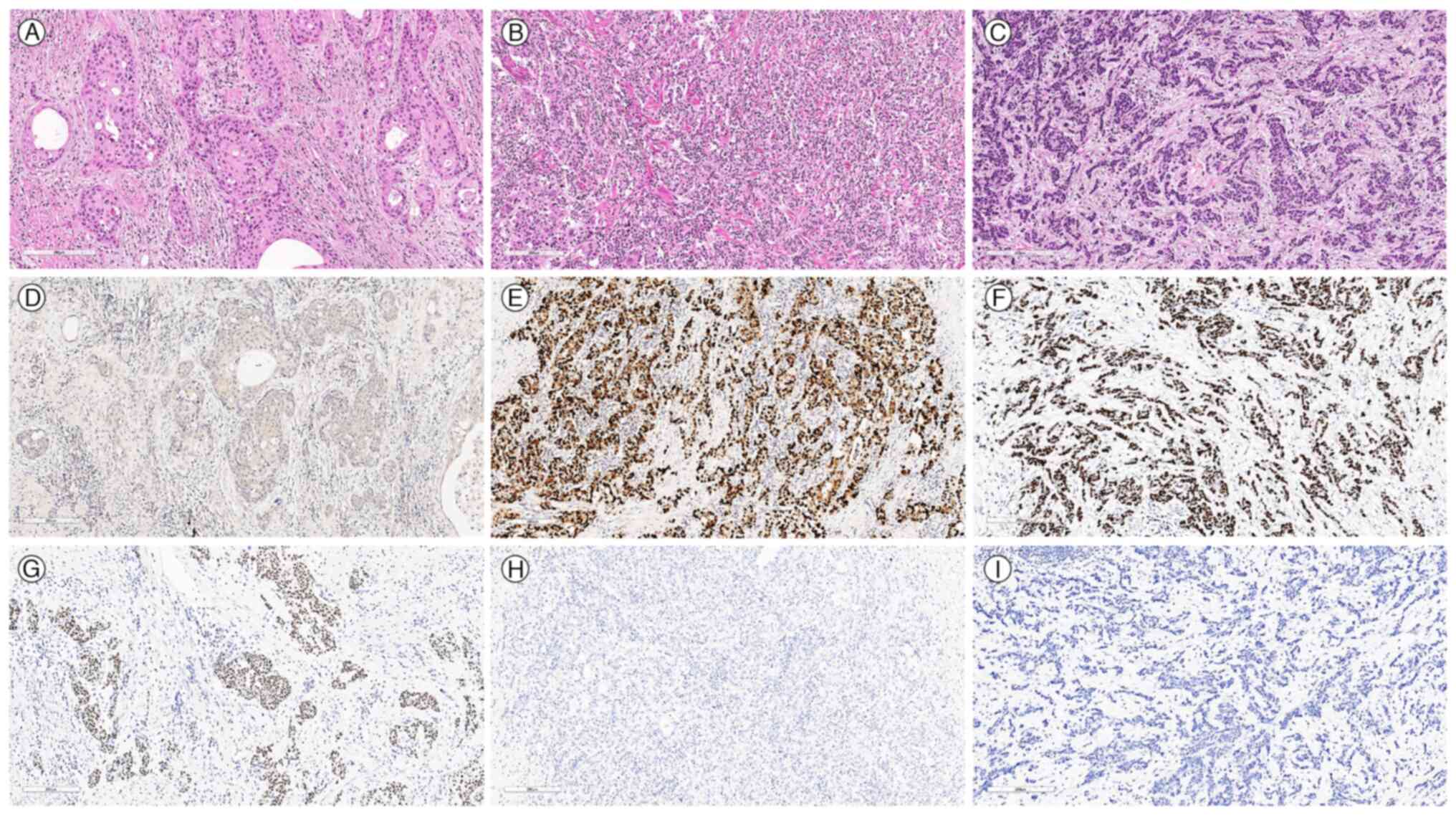 | Figure 3.Microscopic images depicting three
distinct histological components of the tumour alongside results
from EBV in situ hybridization and p40 staining. (A) GASC
component exhibiting large round to oval nuclei with abundant
eosinophilic cytoplasm, intercellular bridges and multifocal
glandular formation (H&E; scale bar, 200 µm). (B) GCLS
component exhibiting abundant lymphocytic infiltration around
undifferentiated carcinoma cell clusters (H&E; scale bar, 200
µm). (C) PDAD component exhibiting hyperchromatic nuclei with
mucinous stromal backgrounds with intermittent exhibition of
glandular formation (H&E; scale bar, 200 µm). EBV was not
detected in the (D) GASC component (scale bar, 200 µm), but was
detected in the (E) GCLS (scale bar, 200 µm) and (F) PDAD (scale
bar, 200 µm) components. Immunoreactivity for p40 was identified in
the (G) GASC component (scale bar, 200 µm), but was negative in the
(H) GCLS (scale bar, 200 µm) and (I) PDAD (scale bar, 200 µm)
components. GASC, gastric adenosquamous carcinoma; GCLS, gastric
carcinoma with lymphoid stroma; PDAD, poorly differentiated
adenocarcinoma; EBV, Epstein-Barr virus. |
To assess biomarker expression and the molecular
characteristics of the three histological components, mucicarmine
staining, immunohistochemical (IHC) analysis, EBV in situ
hybridisation and next-generation sequencing (NGS) were conducted.
For mucicarmine staining, the Artisan Mucicarmine Stain Kit
(Agilent Technologies; cat. no. AR168) was used. The protocol
involved deparaffinizing and hydrating sections, staining with
Weigert's Iron haematoxylin for 10 min at room temperature,
followed by incubation in Working Mucicarmine Solution for 30–60
min at room temperature. After washing, the sections were
counterstained with Tartrazine Solution for 30 sec to 1 min at room
temperature, then dehydrated, cleared and mounted. This method
stains mucin in pink, nuclei in black and the background in yellow.
For immunostaining, the tissues were paraffin-embedded and fixed in
10% neutral-buffered formalin at room temperature for 24 h.
Sections were cut at a thickness of 4 µm and dried at 55°C for 3 h.
Heat-induced epitope retrieval was performed at pH 9.0 for 20 min
using a Leica-Bond Autostainer and a Bond Polymer Refine Detection
Kit (cat. no. DS9800; Leica Biosystems). Endogenous peroxidase
activity was quenched using 3% hydrogen peroxide at room
temperature for 10 min. Primary antibodies were incubated at room
temperature for 1 h, with details of the antibodies and dilutions
provided in Table SI. Secondary
antibody incubation was performed using the aforementioned Bond
Polymer Refine Detection Kit, and staining was visualized with
diaminobenzidine (DAB) chromogen. The prepared slides were examined
using a light microscope (Leica DM2000; Leica Microsystems, Inc.).
p40 and p63 staining identified the squamous cell carcinoma
component, while synaptophysin and chromogranin staining detected
the neuroendocrine carcinoma component. To classify the molecular
subtype according to the Asian Cancer Research Group, mutL homolog
1 (MLH1), mutS homolog 2 (MSH2), p53 and E-cadherin staining were
performed (19). PD-L1 and
programmed cell death protein-1 (PD-1) are known to be frequently
upregulated, while human epidermal growth factor receptor 2 (HER2)
typically shows negative expression in EBVpGC (20). EBV in situ hybridisation was
performed to detect EBER RNA sequences in formalin-fixed,
paraffin-embedded tissue sections. The tissues were fixed in 10%
neutral-buffered formalin at room temperature for 24 h and embedded
in paraffin. Sections were cut at a thickness of 4 µm and
deparaffinized prior to hybridization, and permeabilized using
proteinase K (cat. no. S3020; Dako; Agilent Technologies, Inc.) at
37°C for 15 min. Hybridization was conducted using Bond
hybridization buffer (cat. no. PB0589; Leica Biosystems) with the
biotin-labelled Epstein-Barr Virus Early EBER RNA Probe
(Invitrogen; Thermo Fisher Scientific, Inc.; cat. no. 551019) at a
concentration of 0.5 µg/ml, incubated at 45°C for 20–60 min.
Post-hybridization washes were performed with saline-sodium citrate
buffer at 45°C to remove excess probes, followed by blocking
endogenous peroxidase activity using 3% hydrogen peroxide at room
temperature for 10 min. Detection was carried out using the Bond
Polymer Refine Detection Kit (Leica Biosystems, Ltd.), and
visualization was achieved with DAB chromogen. Counterstaining was
performed with haematoxylin for 5 sec. The slides were examined
under a light microscope (Leica DM2000; Leica Microsystems, Inc.).
For NGS, the 5-µm-thick formalin-fixed, paraffin-embedded tissue
sections were deparaffinized using alcohol and subsequently
hydrated. Sections containing tumour cell-rich regions were
microdissected with an ethanol-dipped scalpel. The tissue underwent
washing and digestion with phosphate-buffered saline and proteinase
K (Qiagen GmbH). DNA extraction was performed using a QIAamp DSP
DNA Kit (Qiagen GmbH). Nucleic acid quantification was carried out
using a Qubit 4 Fluorometer and fluorescence-based quantitation
assays (Thermo Fisher Scientific, Inc.). The quality and integrity
of the extracted DNA were assessed using agarose gel
electrophoresis and the purity was verified by measuring A260/A280
and A260/A230 ratios with a NanoDrop spectrophotometer (Thermo
Fisher Scientific, Inc.). Library preparation utilized the Ion
AmpliSeq Library Preparation kit with nucleic acid samples (Thermo
Fisher Scientific, Inc.; cat. no. 4475345). DNA sequencing was
conducted employing the IonTorrent S5 XL, alongside a control cell
line mixture (Horizon Discovery; Revvity Discovery Limited) and a
targeted gene panel, the Oncomine Comprehensive Assay Plus (Thermo
Fisher Scientific, Inc, cat. no. A48577). The sequencing utilized
200 bp single-end reads. The final library was loaded at a
concentration of 50 pM onto the Ion 550 Chip, optimized for the Ion
Torrent S5 XL system. This panel facilitates the identification of
single nucleotide variants and copy number variants from a pool of
500 gene mutations and 69 gene fusions. For genomic data analysis
and variant calling the Ion Reporter software v5.2 was employed
(Thermo Fisher Scientific, Inc.).
Representative histological features and images of
p40 staining and EBV in situ hybridisation for the three
tumour components are shown in Fig.
3. The GASC component revealed diffuse immunoreactivity for p40
(Fig. 3G) and p63 as well as focal
identification of mucin in the mucicarmine staining (Fig. S1) but was negative for EBV in
situ hybridisation (Fig. 3D).
The GCLS and PDAD components were diffusely positive for EBV in
situ hybridisation (Fig. 3E and
F, respectively) and negative for p40 (Fig. 3H and I, respectively) and p63
staining. All components showed negativity for chromogranin,
synaptophysin, PD-1 and HER2 staining, and were positive for MLH1,
MSH2 and E-cadherin staining (Fig.
S2). The p53 staining showed patchy positivity in all
components (Fig. S3). PD-L1
expression (SP142 and SP263) was <1% positive in immune cells
and negative in tumour cells in all components. In total, 29
somatic mutations and 14 deletions were identified using the NGS
panel; 2 missense mutations and 2 deletions were clinically
relevant, whereas the remaining 27 mutations and 12 deletions were
variants of uncertain significance. The tumour mutation burden
ranged from 5.68 to 6.63, with microsatellite stability in all
components. All three histologically distinct components shared
common a missense mutation in PIK3R1 (c.1690A>G, p.
Asn564Asp) and homozygous deletions of ATRX and RBM10
(both with copy number 0). The PDAD component exclusively harboured
a missense mutation in TP53: c.730G>A (p. Gly244Ser). No
PIK3CA and ARID1A mutations, which are prevalent in
EBVpGC (15), were identified. The
LOF or likely LOF events in PIK3R1, TP53, ATRX and
RBM10, which function as tumour suppressor genes, are
classified as oncogenic or likely oncogenic based on the OncoKB
database (https://www.oncokb.org/). OncoKB
compiles mutation effect information into a database based on
various experimental data, functional evidence and in silico
evidence. According to their standard operating procedure file, a
predictive algorithm was utilized, incorporating two programs: SIFT
and PolyPhen. The results of NGS analysis for each histologically
distinct component are presented in Table I.
 | Table I.Clinically relevant molecular
alterations identified by next-generation sequencing. |
Table I.
Clinically relevant molecular
alterations identified by next-generation sequencing.
| Gene | Type of genetic
alteration | Mutation effect
based on in silico analysis | Oncogenic
effect | HGVS
nomenclature | Associated
histology in the case |
|---|
| PIK3R1 | Missense
mutation | LOF | Oncogenic | c.1690A>G (p.
Asn564Asp) | GASC,
GCLSa and
PDADa |
| ATRX | Deletion | Likely LOF | Likely
oncogenic |
g.76763769_77041552del | GASC,
GCLSa and
PDADa |
| RBM10 | Deletion | Likely LOF | Likely
oncogenic |
g.47006798_47046088del | GASC,
GCLSa and
PDADa |
| TP53 | Missense
mutation | LOF | Oncogenic | c.730G>A (p.
Gly244Ser) | PDADa |
Lymph node metastasis was identified in 14 out of 19
lymph nodes (Fig. 4A) and hepatic
metastasis was identified via intraoperative liver biopsy (Fig. 4B). The histological features of the
metastasis were exclusively of the GCLS component. Accordingly, the
TNM stage was determined to be pT4aN3aM1 based on the 8th edition
of the American Joint Committee on Cancer Staging System (21). The patient was transferred to
another hospital immediately after surgery without chemotherapy or
radiotherapy and passed away 6 months later.
Discussion
In the present study a rare case of GC with three
distinct histological features, each displaying a different EBV
infection state and molecular alteration, yet all sharing common
mutations in PIK3R1, ATRX and RBM10 was reported.
Specifically, the tumour comprised a GASC component without EBV
infection, a GCLS component with EBV infection and active
TP53, and a PDAD component with concurrent EBV infection and
TP53 inactivation. Based on both the molecular and
histological findings, the pathogenesis of the present case may be
hypothesised as follows: A subset of the tumour, harbouring
mutations in PIK3R1, ATRX and RBM10, underwent
EBV-driven malignant transformation, manifesting histologically as
GCLS and PDAD. Subsequently, the PDAD component of this subset
acquired TP53 inactivation. Evidence from previous studies
supports this hypothesis. Knockdown of ATRX can induce EBV
reactivation (22). Additionally,
patients with activated PI3Kδ syndrome due to heterozygous
gain-of-function mutations in PIK3CD or PIK3R1 are
known to be susceptible to EBV, raising the possibility of EBV
susceptibility in patients with LOF mutations in PIK3R1
(23). Furthermore, RBM10
has antiviral properties, as upregulation in cells infected with
dengue virus inhibits viral replication, while RBM10
knockdown promotes replication (24). Thus, RBM10 loss of function
may have contributed to the development of EBVpGC.
The pathogenesis of GASC, which constitutes 0.25% of
all primary GC cases (20), remains
unclear due to its rarity. To date, no studies have investigated
the relationship between GASC and PIK3R1. EBVpGC is
characterised by distinctive genomic alterations, including
frequent DNA hypermethylation, PIK3CA and ARID1A
mutations, and an absence of TP53 mutations (15). EBVpGC is often associated with
abundant lymphocytic infiltration surrounding adenocarcinoma
clusters, a feature known as GCLS (20). Patients with EBVpGC generally
exhibit a longer median survival time compared with those with
EBVnGC, with survival times of 8.5 vs. 5.3 years, respectively
(17). Although EBVpGC accounts for
<10% of all GC cases, it is regarded as a unique entity,
distinct from typical gastric adenocarcinoma (15,17).
Mutations in PIK3R1 and TP53 in EBVpGC were
documented in a previous TCGA study. Of the 73 EBVpGC cases, 8
(11%) harboured PIK3R1 mutations and 11 (15.1%) had
TP53 mutations. By contrast, among the 75 EBVnGC cases, only
1 (1.3%) exhibited a PIK3R1 mutation, while TP53
mutations were found in 47 cases (62.7%) (18). These findings suggest that although
PIK3R1 mutations are rare in GC, they may be associated with
EBV infection. While most TP53 mutations are linked to
EBVnGC, they can also occur in EBVpGC.
In the present case, the GCLS component displayed
typical histological features of EBVpGC with EBV infection.
However, unlike prior studies suggesting a favourable prognosis for
EBVpGC, the GCLS component resulted in hepatic and lymph node
metastases, and the patient passed away within 6 months. This
outcome suggests that metastasis may have been driven by the
PIK3R1 mutation, consistent with reports of higher
PIK3R1 expression in advanced stages (III and IV) compared
with early stages (I and II) of GC (6).
To date, several cases of mixed EBVpGC with ASC have
been reported with histopathological and IHC analyses, but
molecular features have not been explored using NGS (25–27).
Although a previous study on GC with EBV heterogeneity exists, it
differs from the present case in that it did not demonstrate
histological heterogeneity or include alterations in PIK3R1,
ATRX and RBM10 (28).
The underlying cause of EBV infection heterogeneity remains
unknown. More comprehensive studies, including multicentre
research, are needed to further investigate the implications of EBV
infection heterogeneity across diverse histological features.
In conclusion, diverse molecular alterations and EBV
infection heterogeneity can be observed in mixed GC across distinct
histological components. As these alterations and heterogenous EBV
status may influence patient prognosis and treatment strategies, a
thorough evaluation of molecular features alongside histological
characteristics is crucial.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by a grant from Kyung Hee University in
2024 (grant no. KHU-20241076) and a grant from the Asan Institute
for Life Sciences, Asan Medical Center, Seoul, Korea (grant no.
2021IL0040).
Availability of data and materials
The NGS data generated in the present study may be
found in the NCBI Sequence Read Archive under accession no.
PRJNA1182944 or under the following URL: https://www.ncbi.nlm.nih.gov/sra/PRJNA1182944.
All other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
Conceptualisation and writing of the original draft
were conducted by SUJ. Case selection and investigation of the
clinical data was conducted by YK. Review of the H&E slides was
conducted by SUJ, EC and SK. NGS analysis and funding acquisition
was conducted by SK. Molecular pathology consultation was conducted
by JB. Immunohistochemical staining analysis and reviewing and
editing the manuscript was conducted by SUJ and SK. SUJ and SK
confirm the authenticity of all the raw data. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
This retrospective study involves experimental
analysis of human tissue specimens collected following surgical
procedures. The Institutional Review Board at Ewha Woman's
University Mokdong Hospital (Seoul, Republic of Korea) reviewed and
approved the study protocol (approval no. 2024-05-004), including a
waiver of consent due to the retrospective nature of the study and
the patient's death. The study was conducted in accordance with the
Declaration of Helsinki.
Patient consent for publication
The Institutional Review Board at Ewha Woman's
University Mokdong Hospital (Seoul, Republic of Korea) waived the
requirement of patient consent for publication due to the
retrospective nature of the study and the patient's death.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
Se Un Jeong ORCID, 0000-0001-8399-5792; Euno Choi
ORCD, 0000-0002-9284-9276; Yongil Kim ORCID, 0000-0002-4131-364X;
Jaeyoung Byeon ORCID, 0009-0002-7316-6549; So-Woon Kim ORCID,
0000-0002-9840-848X.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Plummer M, Franceschi S, Vignat J, Forman
D and de Martel C: Global burden of gastric cancer attributable to
helicobacter pylori. Int J Cancer. 136:487–490. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lordick F, Shitara K and Janjigian YY: New
agents on the horizon in gastric cancer. Ann Oncol. 28:1767–1775.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fattahi S, Amjadi-Moheb F, Tabaripour R,
Ashrafi GH and Akhavan-Niaki H: PI3K/AKT/mTOR signaling in gastric
cancer: Epigenetics and beyond. Life Sci. 262:1185132020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu Y, Wang D, Li Z, Li X, Jin M, Jia N,
Cui X, Hu G, Tang T and Yu Q: Pan-cancer analysis on the role of
PIK3R1 and PIK3R2 in human tumors. Sci Rep. 12:59242022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xia TF, Chen J, Wu K, Zhang J and Yan Q:
Long noncoding RNA NEAT1 promotes the growth of gastric cancer
cells by regulating miR-497-5p/PIK3R1 axis. Eur Rev Med Pharmacol
Sci. 23:6914–6926. 2019.PubMed/NCBI
|
|
8
|
Li Q, Tian Y, Liang Y and Li C:
CircHIPK3/miR-876-5p/PIK3R1 axis regulates regulation
proliferation, migration, invasion, and glutaminolysis in gastric
cancer cells. Cancer Cell Int. 20:3912020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartholf DeWitt S, Hoskinson Plumlee S,
Brighton HE, Sivaraj D, Martz EJ, Zand M, Kumar V, Sheth MU, Floyd
W, Spruance JV, et al: Loss of ATRX promotes aggressive features of
osteosarcoma with increased NF-κB signaling and integrin binding.
JCI Insight. 7:e1515832022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Aguilera P and López-Contreras AJ: ATRX, a
guardian of chromatin. Trends Genet. 39:505–519. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ge Y, Wei F, Du G, Fei G, Li W, Li X, Chu
J and Wei P: The association of sex-biased ATRX mutation in female
gastric cancer patients with enhanced immunotherapy-related
anticancer immunity. BMC Cancer. 21:2402021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cao Y, Geng J, Wang X, Meng Q, Xu S, Lang
Y, Zhou Y, Qi L, Wang Z and Wei Z: RNA-binding motif protein 10
represses tumor progression through the Wnt/β- catenin pathway in
lung adenocarcinoma. Int J Biol Sci. 18:124–139. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao Y, Di X, Zhang Q, Li R and Wang K:
RBM10 regulates tumor apoptosis, proliferation, and metastasis.
Front Oncol. 11:6039322021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun X, Jia D and Yu Y: Expression pattern,
immune signature, and prognostic value of RBM10 in human cancers.
Histol Histopathol. 187902024.
|
|
15
|
Saito M and Kono K: Landscape of
EBV-positive gastric cancer. Gastric Cancer. 24:983–989. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang J, Liu Z, Zeng B, Hu G and Gan R:
Epstein-barr virus-associated gastric cancer: A distinct subtype.
Cancer Lett. 495:191–199. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun K, Jia K, Lv H, Wang SQ, Wu Y, Lei H
and Chen X: EBV-positive gastric cancer: Current knowledge and
future perspectives. Front Oncol. 10:5834632020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
He CY, Qiu MZ, Yang XH, Zhou DL, Ma JJ,
Long YK, Ye ZL, Xu BH, Zhao Q, Jin Y, et al: Classification of
gastric cancer by EBV status combined with molecular profiling
predicts patient prognosis. Clin Transl Med. 10:353–362. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cristescu R, Lee J, Nebozhyn M, Kim KM,
Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al: Molecular
analysis of gastric cancer identifies subtypes associated with
distinct clinical outcomes. Nat Med. 21:449–456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Carneiro F: Gastric adenocarcinoma. WHO
Classification of Tumours: Digestive System Tumours. International
Agency for Research on Cancer; Lyon: 2019
|
|
21
|
Amin MB, Edge SB, Greene FL, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, et al: AJCC cancer staging manual. 8th edition.
Springer; New York, NY: 2017
|
|
22
|
Tsai K, Thikmyanova N, Wojcechowskyj JA,
Delecluse HJ and Lieberman PM: EBV tegument protein BNRF1 disrupts
DAXX-ATRX to activate viral early gene transcription. PLoS Pathog.
7:e10023762011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Carpier JM and Lucas CL: Epstein-barr
virus susceptibility in activated PI3Kδ syndrome (APDS)
immunodeficiency. Front Immunol. 8:20052018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pozzi B, Bragado L, Mammi P, Torti MF,
Gaioli N, Gebhard LG, García Solá ME, Vaz-Drago R, Iglesias NG and
García CC: Dengue virus targets RBM10 deregulating host cell
splicing and innate immune response. Nucleic Acids Res.
48:6824–6838. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cao F, Yan Y, Niu D, Huang X, Jia L, Diao
X and Li Z: Epstein-Barr virus-associated gastric adenosquamous
carcinoma with concurrent gastric carcinoma with lymphoid stroma: A
case report and review of the literature. BMC Gastroenterol.
22:3462022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miyake H, Miyasaka C, Ishida M, Miki H,
Inoue K and Tsuta K: Simultaneous gastric adenosquamous carcinoma
and gastric carcinoma with lymphoid stroma: A case report. Mol Clin
Oncol. 11:77–80. 2019.PubMed/NCBI
|
|
27
|
Kuroda N, Oonishi K, Inoue K, Ohara M,
Mizuno K and Lee GH: Lymphoepithelioma-like carcinoma of the
stomach associated with adenosquamous carcinoma. Med Mol Morphol.
43:170–173. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim HN, Ahn S and Kim KM: Gastric cancer
with epstein-barr virus heterogeneity: Evaluation of the frequency,
clinicopathologic features, and genomic profiles. Pathol Res Pract.
238:1541082022. View Article : Google Scholar : PubMed/NCBI
|