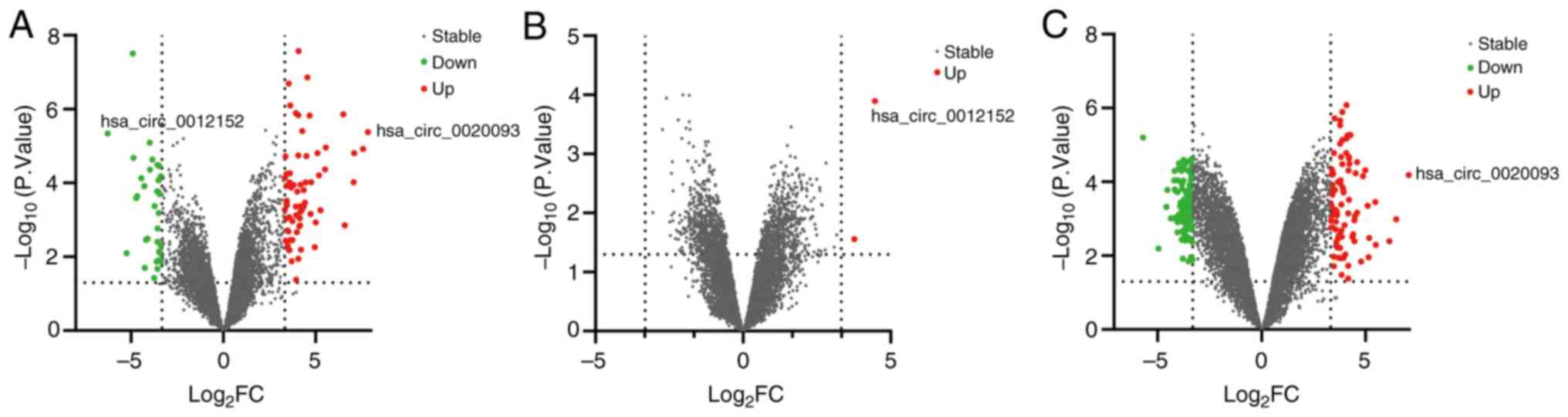
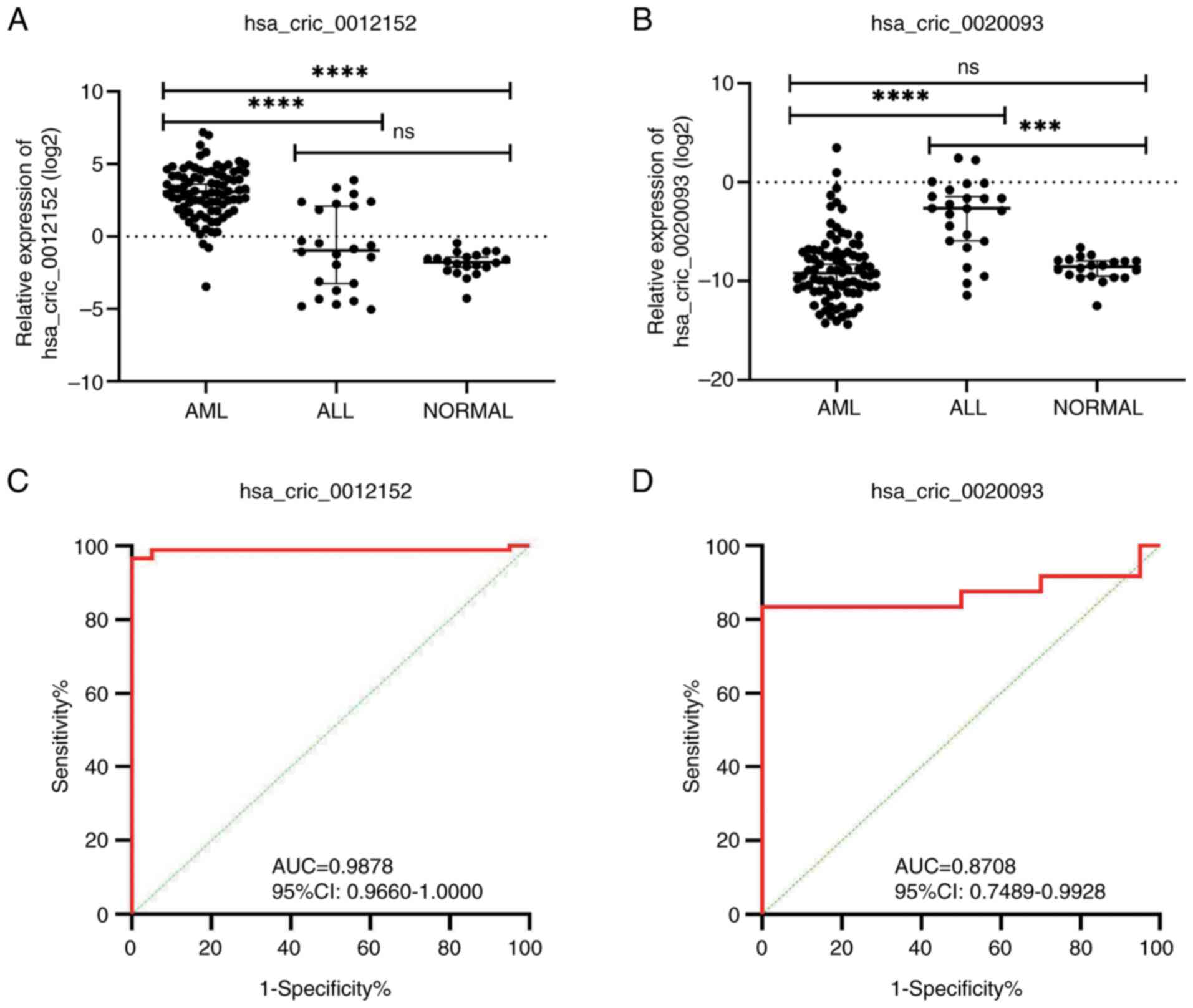
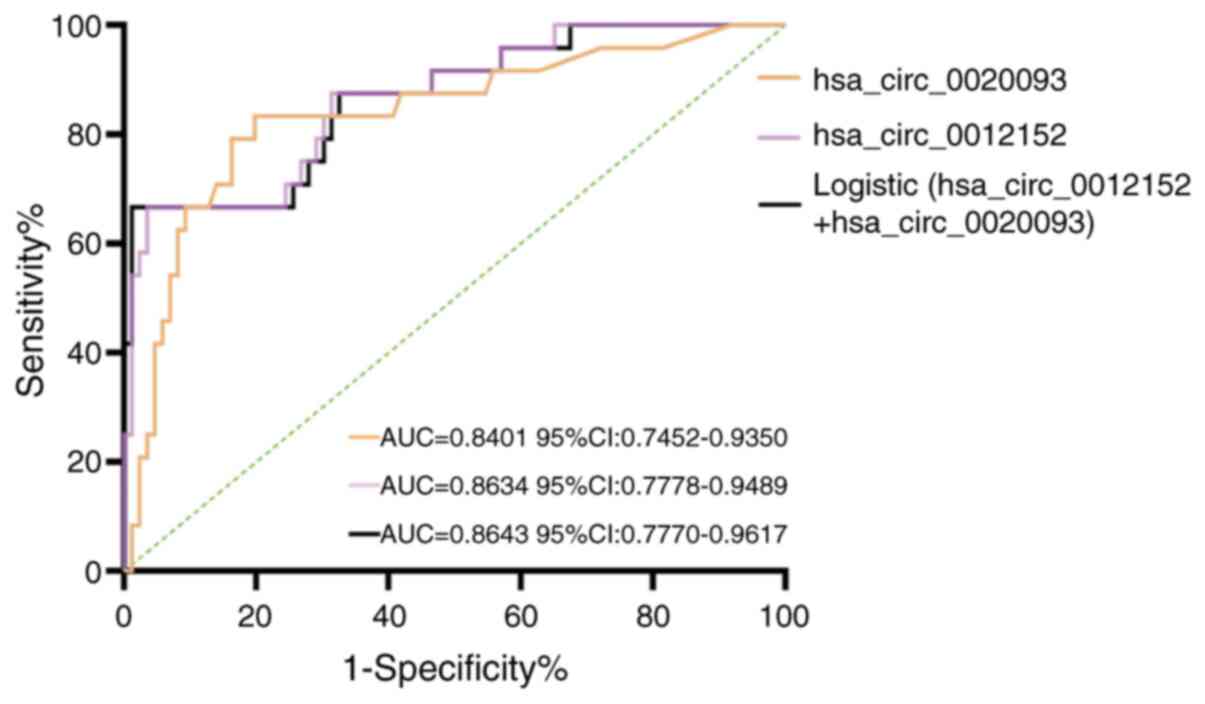
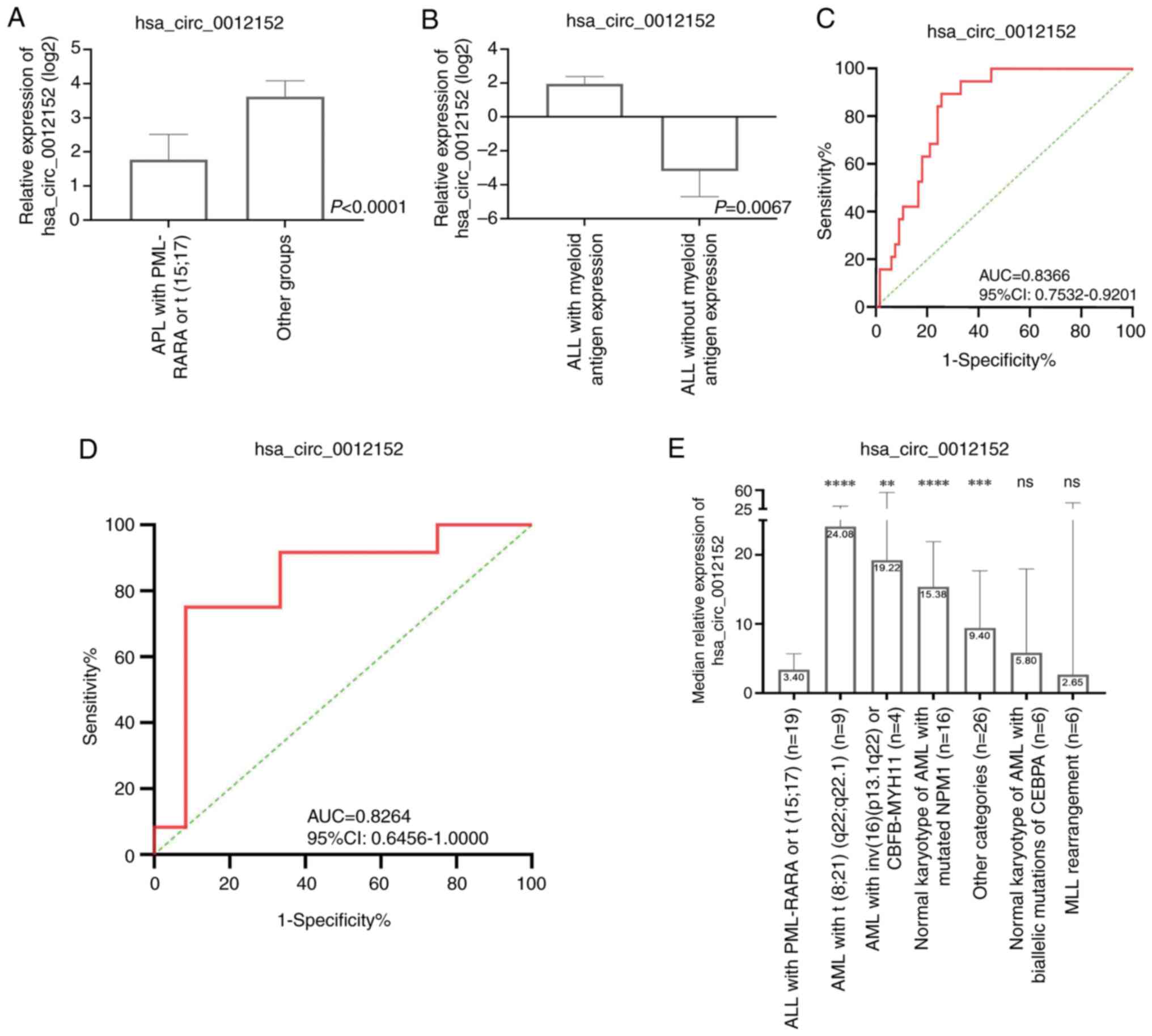
|
1
|
Devine SM and Larson RA: Acute leukemia in
adults: Recent developments in diagnosis and treatment. CA Cancer J
Clin. 44:326–352. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bennett JM, Catovsky D, Daniel MT,
Flandrin G, Galton DA, Gralnick HR and Sultan C: Proposals for the
classification of the acute leukaemias. French-American-British
(FAB) co-operative group. Br J Haematol. 33:451–458. 1976.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cheng H, Huang CM, Wang Y, Hu XX, Xu XQ,
Song XM, Tang GS, Chen L and Yang JM: Microarray profiling and
co-expression network analysis of the lncRNAs and mRNAs associated
with acute leukemia in adults. Mol Biosyst. 13:1102–1108. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pui CH, Schrappe M, Ribeiro RC and
Niemeyer CM: Childhood and adolescent lymphoid and myeloid
leukemia. Hematology Am Soc Hematol Educ Program. 118–145. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Miranda-Filho A, Piñeros M, Ferlay J,
Soerjomataram I, Monnereau A and Bray F: Epidemiological patterns
of leukaemia in 184 countries: A population-based study. Lancet
Haematol. 5:e14–e24. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Oliveira PD: Leukaemia prevalence
worldwide: Raising aetiology questions. Lancet Haematol. 5:e2–e3.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Porwit A and Béné MC: Acute leukemias of
ambiguous origin. Am J Clin Pathol. 144:361–376. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu XQ, Wang JM, Lu SQ, Chen L, Yang JM,
Zhang WP, Song XM, Hou J, Ni X and Qiu HY: Clinical and biological
characteristics of adult biphenotypic acute leukemia in comparison
with that of acute myeloid leukemia and acute lymphoblastic
leukemia: A case series of a Chinese population. Haematologica.
94:919–927. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rong D, Sun H, Li Z, Liu S, Dong C, Fu K,
Tang W and Cao H: An emerging function of circRNA-miRNAs-mRNA axis
in human diseases. Oncotarget. 8:73271–73281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lei K, Bai H, Wei Z, Xie C, Wang J, Li J
and Chen Q: The mechanism and function of circular RNAs in human
diseases. Exp Cell Res. 368:147–158. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo SS, Li BX, Zou DB, Yang SJ, Sheng LX,
Ouyang GF, Mu QT and Huang H: Tip of the iceberg: Roles of circRNAs
in hematological malignancies. Am J Cancer Res. 10:367–382.
2020.PubMed/NCBI
|
|
13
|
Yi YY, Yi J, Zhu X, Zhang J, Zhou J, Tang
X, Lin J, Wang P and Deng ZQ: Circular RNA of vimentin expression
as a valuable predictor for acute myeloid leukemia development and
prognosis. J Cell Physiol. 234:3711–3719. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li W, Zhong C, Jiao J, Li P, Cui B, Ji C
and Ma D: Characterization of hsa_circ_0004277 as a new biomarker
for acute myeloid leukemia via circular RNA profile and
bioinformatics analysis. Int J Mol Sci. 18:5972017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu J, Han Q, Gu Y, McGrath M, Qiao F, Chen
B, Song C and Ge Z: Circular RNA PVT1 expression and its roles in
acute lymphoblastic leukemia. Epigenomics. 10:723–732. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo S, Li B, Chen Y, Zou D, Yang S, Zhang
Y, Wu N, Sheng L, Huang H, Ouyang G and Mu Q: Hsa_circ_0012152 and
Hsa_circ_0001857 accurately discriminate acute lymphoblastic
leukemia from acute myeloid leukemia. Front Oncol. 10:16552020.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin CQ, Yang Y, Mao H, Zhao Q, Wu JQ, Li
DP and Huang YH: Clinical characteristics, prognostic factors and
misdiagnosis of bone marrow necrosis patients. Zhongguo Shi Yan Xue
Ye Xue Za Zhi. 29:1637–1644. 2021.(In Chinese). PubMed/NCBI
|
|
18
|
Ridgeway JA, Tinsley S and Kurtin SE:
Practical guide to bone marrow sampling for suspected
myelodysplastic syndromes. J Adv Pract Oncol. 8:29–39.
2017.PubMed/NCBI
|
|
19
|
Butler JT, Yashar WM and Swords R:
Breaking the bone marrow barrier: Peripheral blood as a gateway to
measurable residual disease detection in acute myelogenous
leukemia. Am J Hematol. 100:638–651. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hematology Oncology Committee, Chinese
Anti-Cancer Association, Leukemia & Lymphoma Group, Chinese
Society of Hematology and Chinese Medical Association, . Chinese
guideline for diagnosis and treatment of adult acute lymphoblastic
leukemia (2024). Zhonghua Xue Ye Xue Za Zhi. 45:417–429. 2024.(In
Chinese). PubMed/NCBI
|
|
21
|
Khoury JD, Solary E, Abla O, Akkari Y,
Alaggio R, Apperley JF, Bejar R, Berti E, Busque L, Chan JKC, et
al: The 5th edition of the World Health Organization classification
of haematolymphoid tumours: myeloid and histiocytic/dendritic
neoplasms. Leukemia. 36:1703–1719. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Alaggio R, Amador C, Anagnostopoulos I,
Attygalle AD, Araujo IBO, Berti E, Bhagat G, Borges AM, Boyer D,
Calaminici M, et al: The 5th edition of the World Health
Organization classification of haematolymphoid tumours: Lymphoid
neoplasms. Leukemia. 36:1720–1748. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Staples TL: Expansion and evolution of the
R programming language. R Soc Open Sci. 10:2215502023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Doncheva NT, Morris JH, Gorodkin J and
Jensen LJ: Cytoscape StringApp: Network analysis and visualization
of proteomics data. J Proteome Res. 18:623–632. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tan Y, Tan Y, Li J, Hu P, Guan P, Kuang H,
Liang Q, Yu Y, Chen Z, Wang Q, et al: Combined IFN-γ and IL-2
release assay for detect active pulmonary tuberculosis: A
prospective multicentre diagnostic study in China. J Transl Med.
19:2892021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Takahashi K, Uchiyama H, Yanagisawa S and
Kamae I: The logistic regression and ROC analysis of group-based
screening for predicting diabetes incidence in four years. Kobe J
Med Sci. 52:171–180. 2006.PubMed/NCBI
|
|
28
|
Löwenberg B, Downing JR and Burnett A:
Acute myeloid leukemia. N Engl J Med. 341:1051–1062. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Weir EG and Borowitz MJ: Flow cytometry in
the diagnosis of acute leukemia. Semin Hematol. 38:124–138. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
McKenna RW: Multifaceted approach to the
diagnosis and classification of acute leukemias. Clin Chem.
46:1252–1259. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang WT, Chen TQ, Zeng ZC, Pan Q, Huang W,
Han C, Fang K, Sun LY, Yang QQ, Wang D, et al: The lncRNA LAMP5-AS1
drives leukemia cell stemness by directly modulating DOT1L
methyltransferase activity in MLL leukemia. J Hematol Oncol.
13:782020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bárcenas-López DA, Núñez-Enríquez JC,
Hidalgo-Miranda A, Beltrán-Anaya FO, May-Hau DI, Jiménez-Hernández
E, Bekker-Méndez VC, Flores-Lujano J, Medina-Sansón A, Tamez-Gómez
EL, et al: Transcriptome analysis identifies LINC00152 as a
biomarker of early relapse and mortality in acute lymphoblastic
leukemia. Genes (Basel). 11:3022020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gentner B, Pochert N, Rouhi A, Boccalatte
F, Plati T, Berg T, Sun SM, Mah SM, Mirkovic-Hösle M, Ruschmann J,
et al: MicroRNA-223 dose levels fine tune proliferation and
differentiation in human cord blood progenitors and acute myeloid
leukemia. Exp Hematol. 43:858–868.e7. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hornick NI, Huan J, Doron B, Goloviznina
NA, Lapidus J, Chang BH and Kurre P: Serum exosome MicroRNA as a
minimally-invasive early biomarker of AML. Sci Rep. 5:112952015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Avigad S, Verly IRN, Lebel A, Kordi O,
Shichrur K, Ohali A, Hameiri-Grossman M, Kaspers GJL, Cloos J,
Fronkova E, et al: miR expression profiling at diagnosis predicts
relapse in pediatric precursor B-cell acute lymphoblastic leukemia.
Genes Chromosomes Cancer. 55:328–339. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang W, Fang K, Chen TQ, Zeng ZC, Sun YM,
Han C, Sun LY, Chen ZH, Yang QQ, Pan Q, et al: circRNA circAF4
functions as an oncogene to regulate MLL-AF4 fusion protein
expression and inhibit MLL leukemia progression. J Hematol Oncol.
12:1032019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhou J, Zhou LY, Tang X, Zhang J, Zhai LL,
Yi YY, Yi J, Lin J, Qian J and Deng ZQ: Circ-Foxo3 is positively
associated with the Foxo3 gene and leads to better prognosis of
acute myeloid leukemia patients. BMC Cancer. 19:9302019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Han F, Zhong C, Li W, Wang R, Zhang C,
Yang X, Ji C and Ma D: hsa_circ_0001947 suppresses acute myeloid
leukemia progression via targeting hsa-miR-329-5p/CREBRF axis.
Epigenomics. 12:935–953. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jamal M, Song T, Chen B, Faisal M, Hong Z,
Xie T, Wu Y, Pan S, Yin Q, Shao L and Zhang Q: Recent progress on
circular RNA research in acute myeloid leukemia. Front Oncol.
9:11082019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yuan DM, Ma J and Fang WB: Identification
of non-coding RNA regulatory networks in pediatric acute myeloid
leukemia reveals circ-0004136 could promote cell proliferation by
sponging miR-142. Eur Rev Med Pharmacol Sci. 23:9251–9258.
2019.PubMed/NCBI
|
|
41
|
Puil L, Liu J, Gish G, Mbamalu G, Bowtell
D, Pelicci PG, Arlinghaus R and Pawson T: Bcr-Abl oncoproteins bind
directly to activators of the Ras signalling pathway. EMBO J.
13:764–773. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ohanian M, Tari Ashizawa A, Garcia-Manero
G, Pemmaraju N, Kadia T, Jabbour E, Ravandi F, Borthakur G,
Andreeff M, Konopleva M, et al: Liposomal Grb2 antisense
oligodeoxynucleotide (BP1001) in patients with refractory or
relapsed haematological malignancies: A single-centre, open-label,
dose-escalation, phase 1/1b trial. Lancet Haematol. 5:e136–e146.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yan J, Jiang N, Huang G, Tay JL, Lin B, Bi
C, Koh GS, Li Z, Tan J, Chung TH, et al: Deregulated MIR335 that
targets MAPK1 is implicated in poor outcome of paediatric acute
lymphoblastic leukaemia. Br J Haematol. 163:93–103. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tomiyasu H, Watanabe M, Sugita K,
Goto-Koshino Y, Fujino Y, Ohno K, Sugano S and Tsujimoto H:
Regulations of ABCB1 and ABCG2 expression through MAPK pathways in
acute lymphoblastic leukemia cell lines. Anticancer Res.
33:5317–5323. 2013.PubMed/NCBI
|
|
45
|
You H, Wang D, Wei L, Chen J, Li H and Liu
Y: Deferoxamine inhibits acute lymphoblastic leukemia progression
through repression of ROS/HIF-1 α, Wnt/β-catenin, and p38MAPK/ERK
pathways. J Oncol. 2022:82812672022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Simioni C, Martelli AM, Zauli G, Melloni E
and Neri LM: Targeting mTOR in acute lymphoblastic leukemia. Cells.
8:1902019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Evangelisti C, Chiarini F, Cappellini A,
Paganelli F, Fini M, Santi S, Martelli AM, Neri LM and Evangelisti
C: Targeting Wnt/β-catenin and PI3K/Akt/mTOR pathways in T-cell
acute lymphoblastic leukemia. J Cell Physiol. 235:5413–5428. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ge Z, Song C, Ding Y, Tan BH, Desai D,
Sharma A, Gowda R, Yue F, Huang S, Spiegelman V, et al: Dual
targeting of MTOR as a novel therapeutic approach for high-risk
B-cell acute lymphoblastic leukemia. Leukemia. 35:1267–1278. 2021.
View Article : Google Scholar : PubMed/NCBI
|