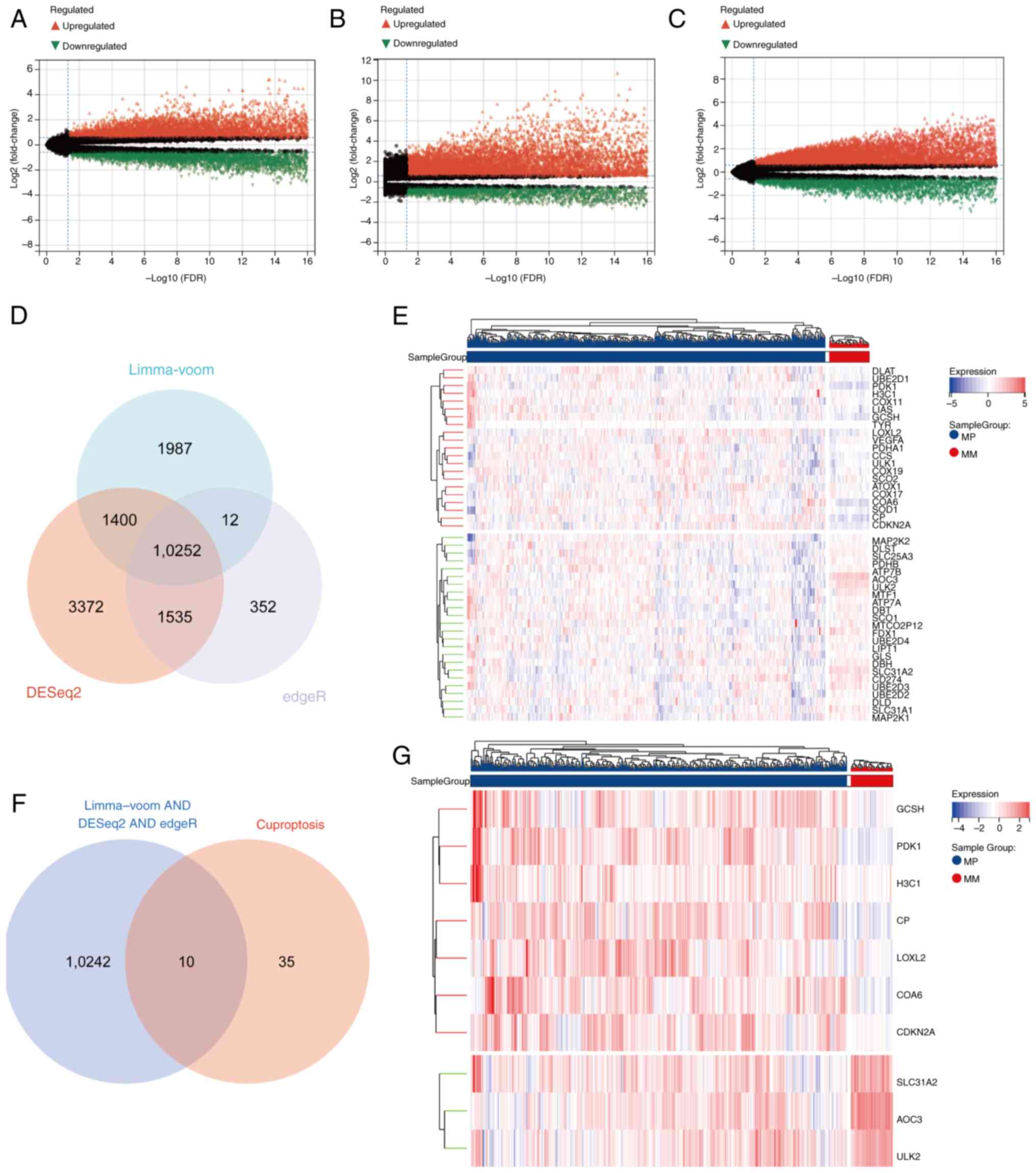
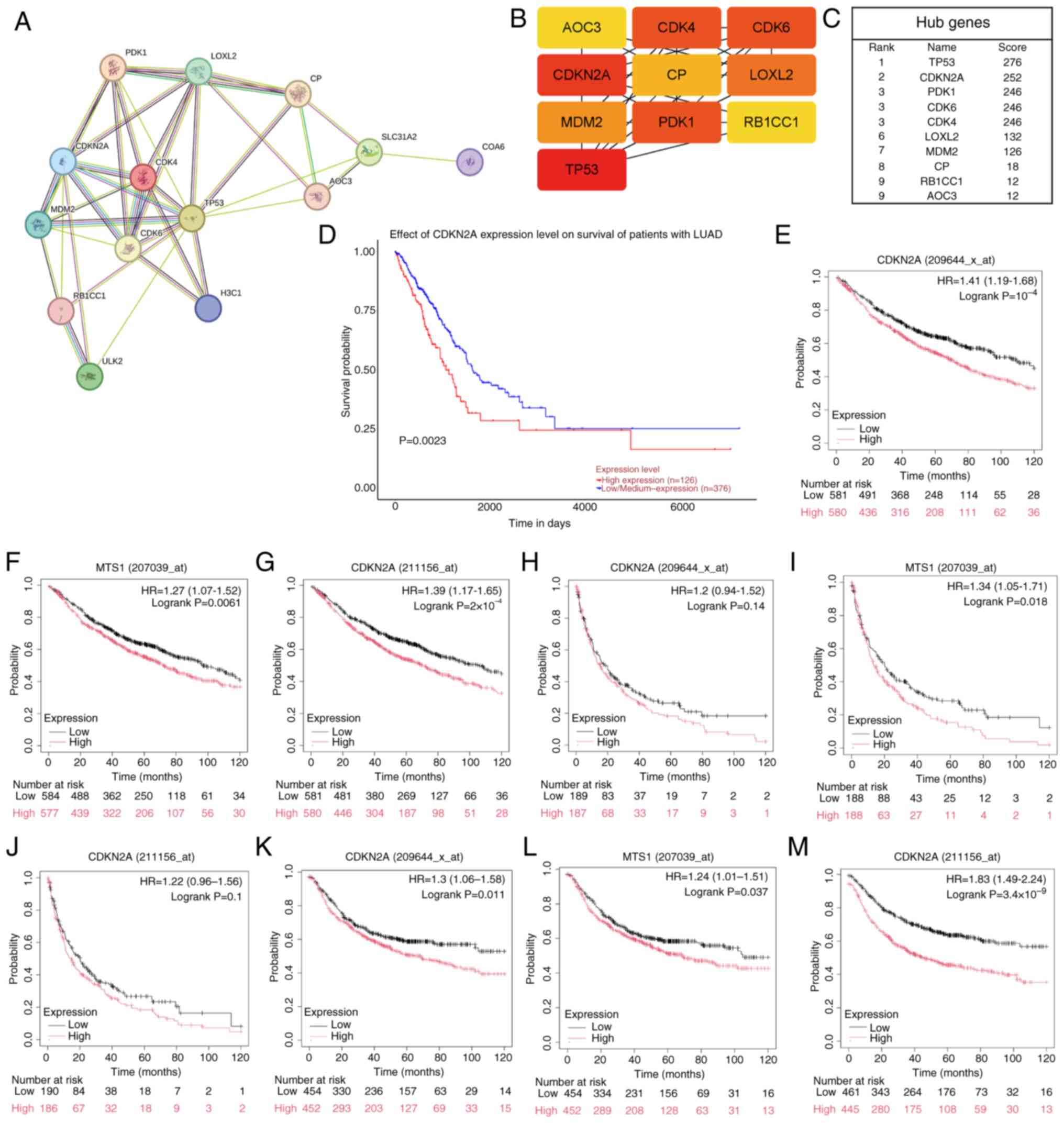
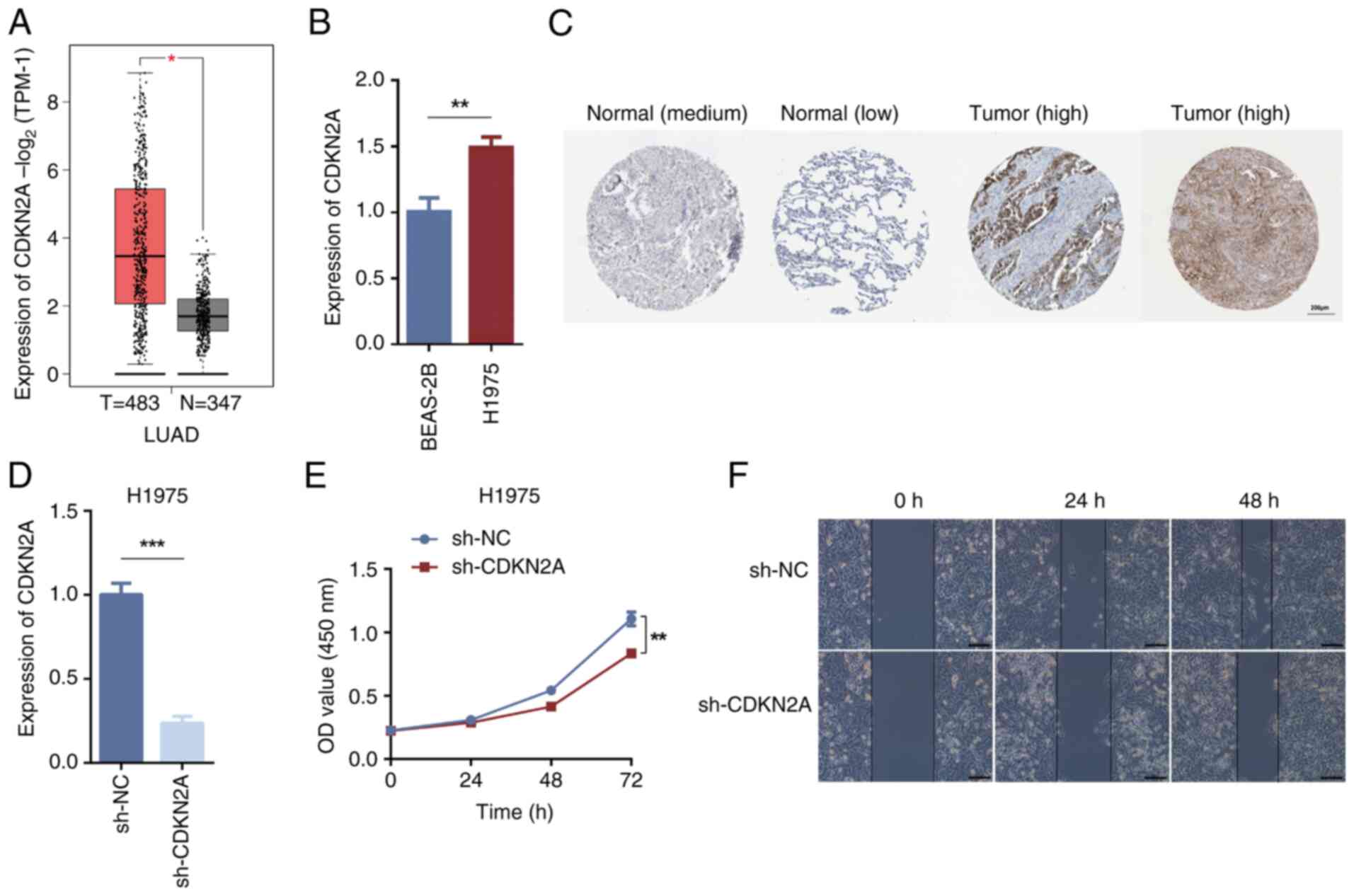
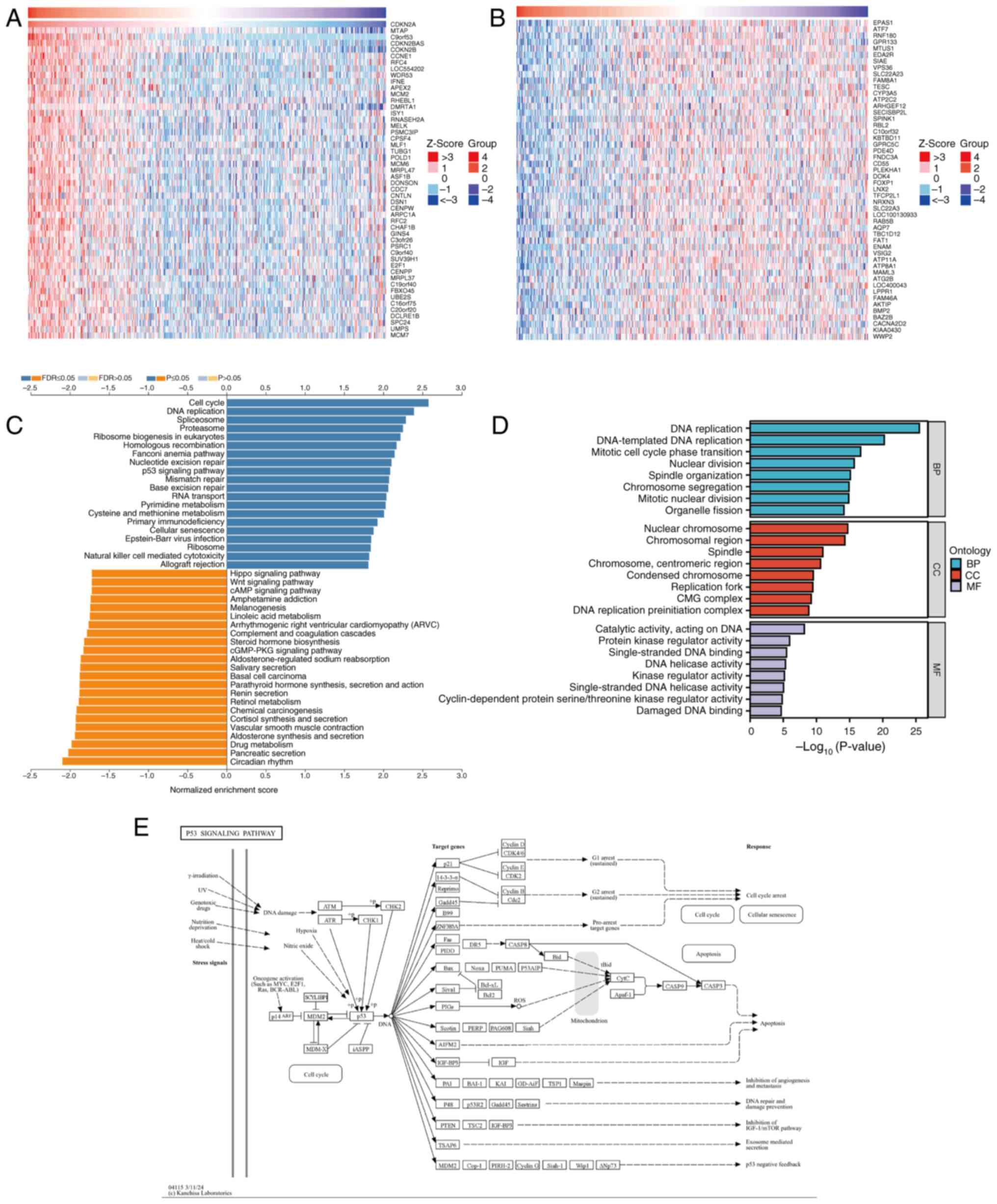
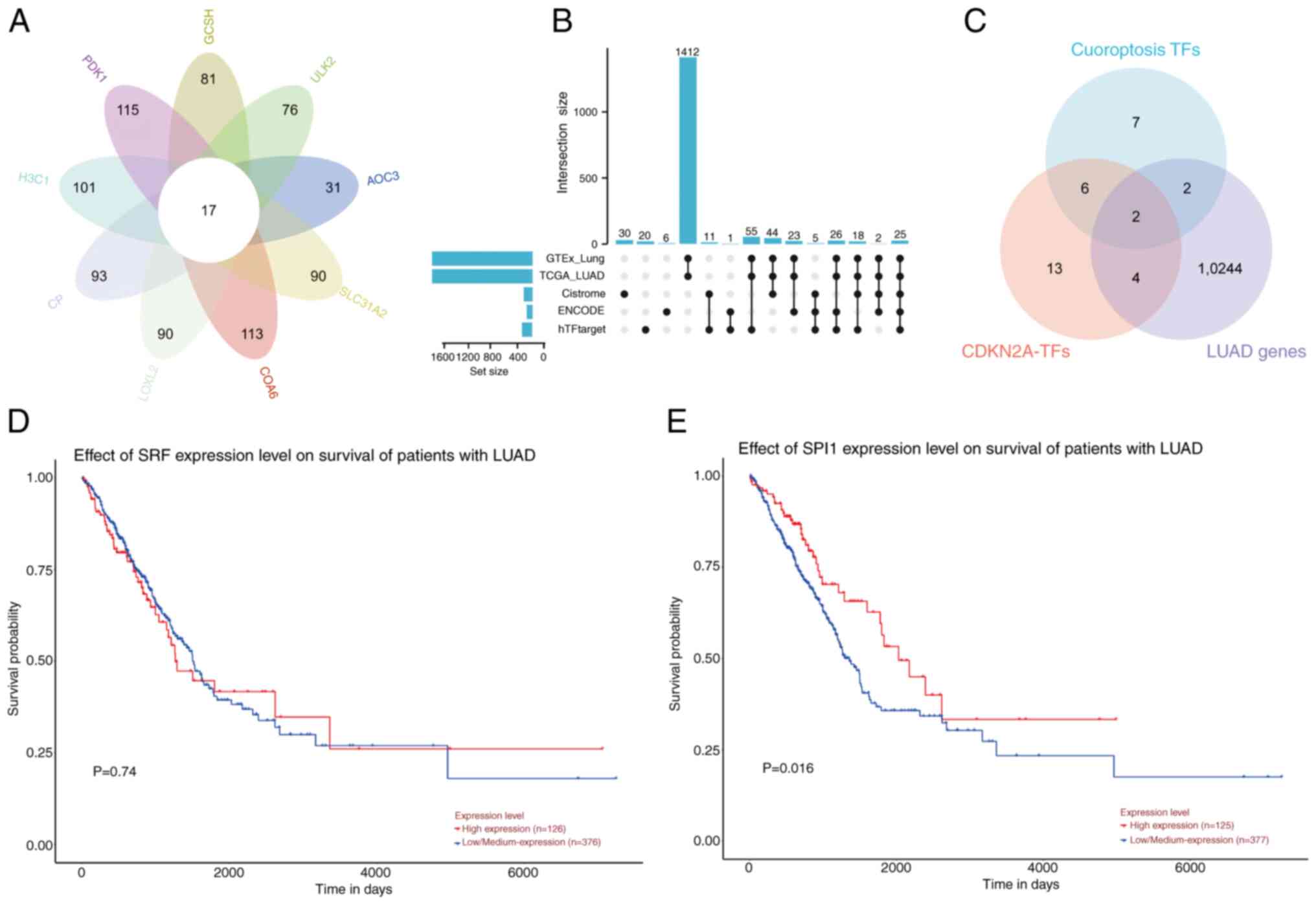
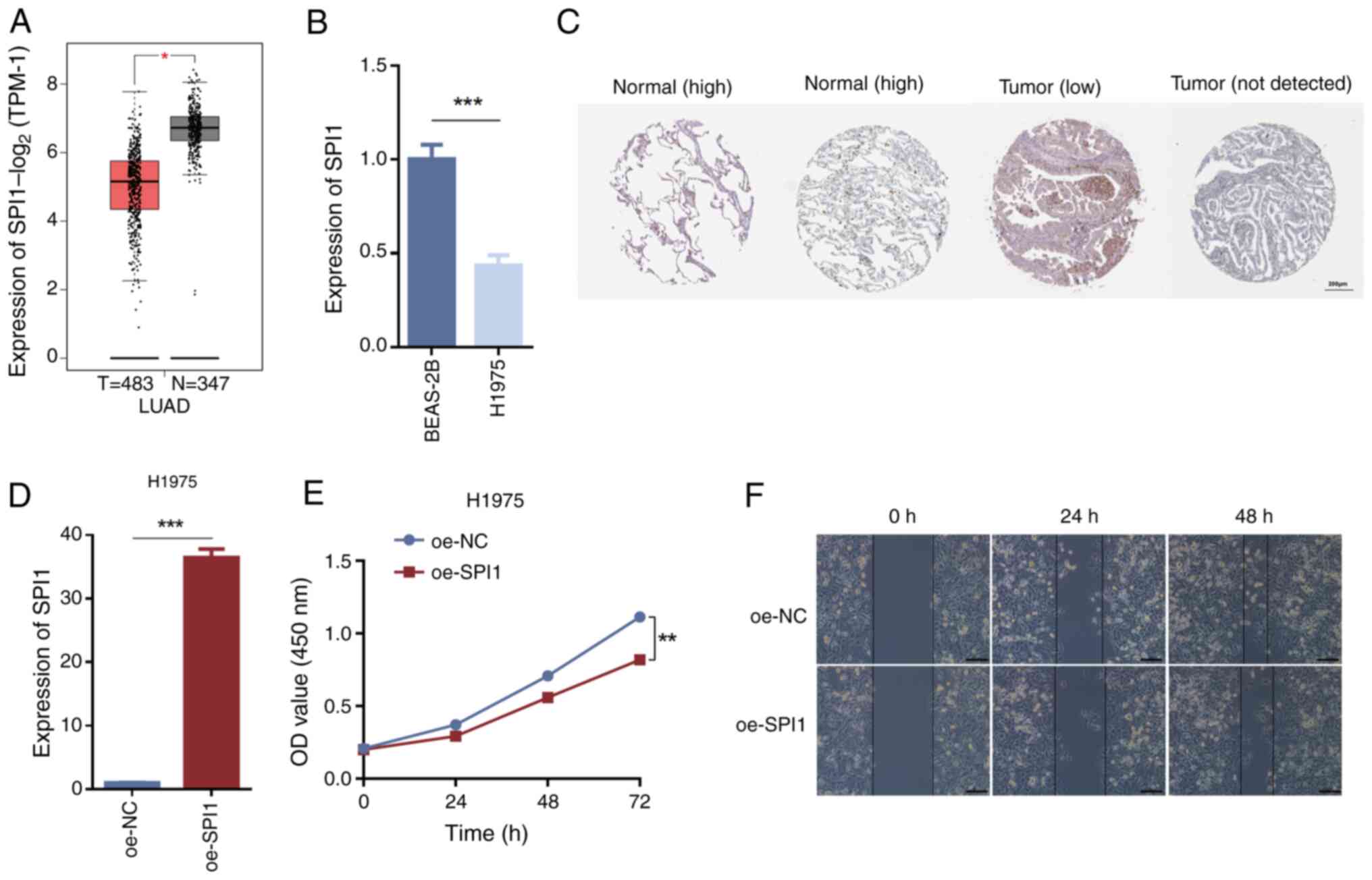
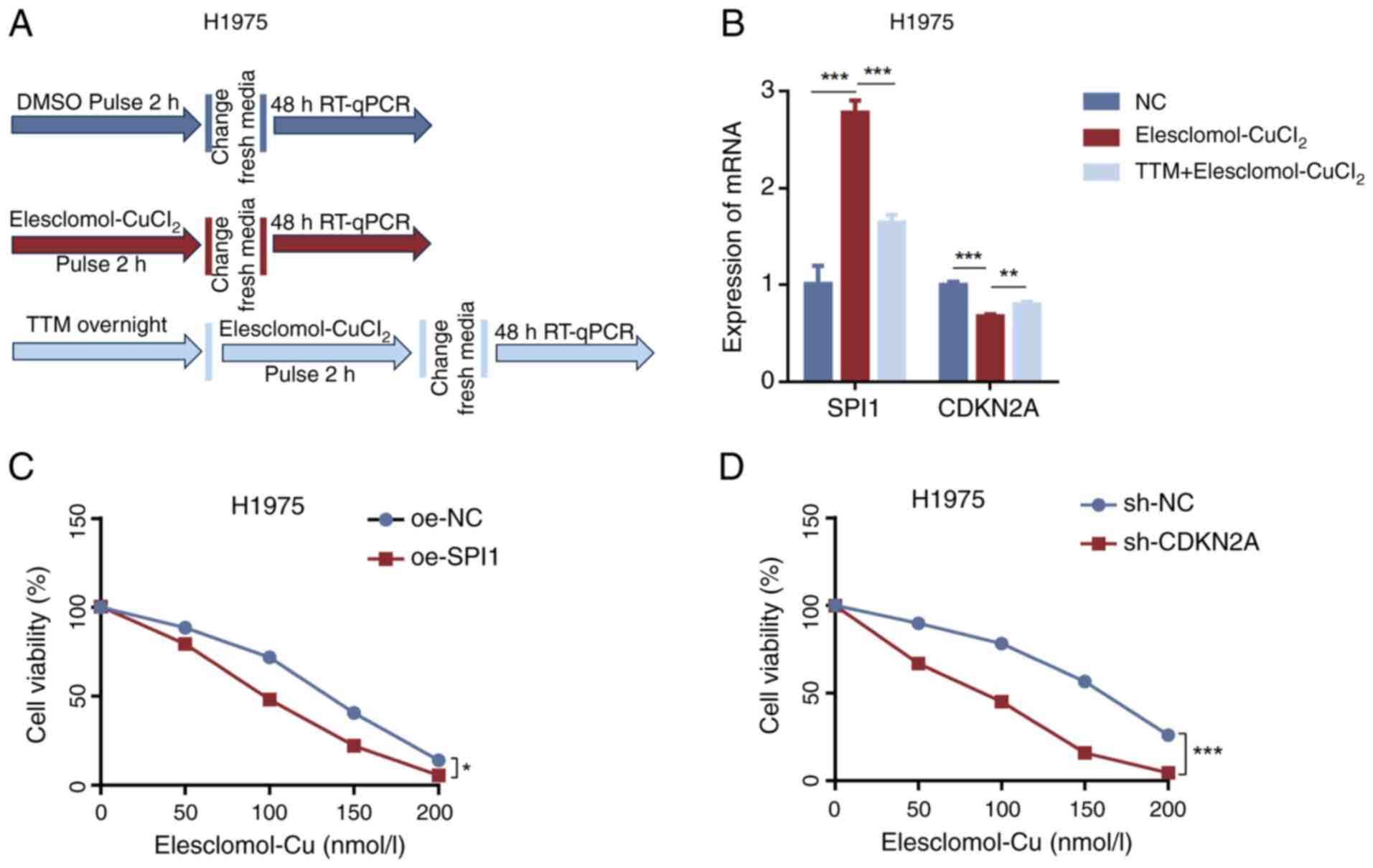
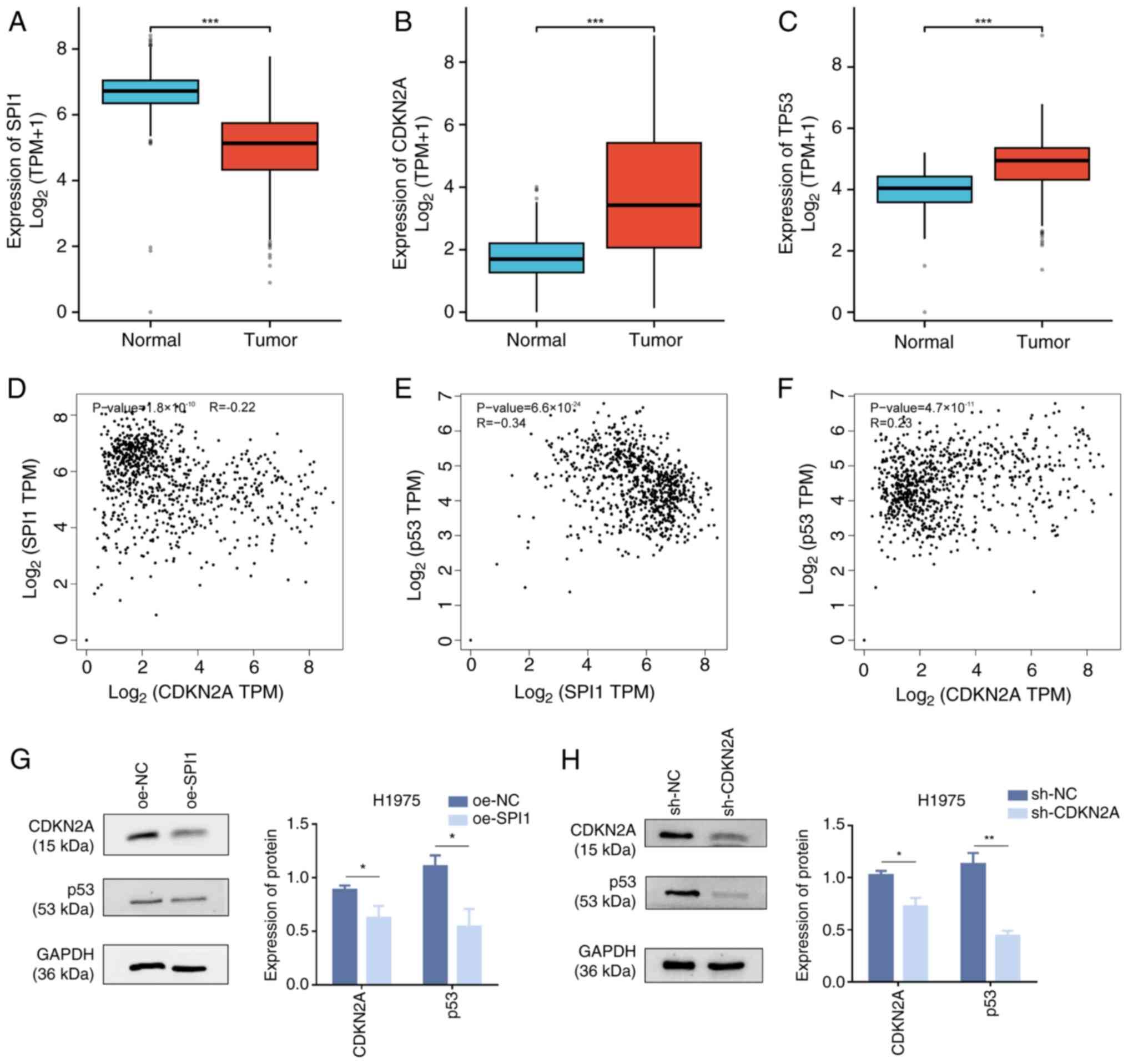
|
1
|
Siegel RL, Giaquinto AN and Jemal A:
Cancer statistics, 2024. CA Cancer J Clin. 74:12–49. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen P, Liu Y, Wen Y and Zhou C: Non-small
cell lung cancer in China. Cancer Commun (Lond). 42:937–970. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jurisic V, Vukovic V, Obradovic J,
Gulyaeva LF, Kushlinskii NE and Djordjević N: EGFR polymorphism and
survival of NSCLC patients treated with TKIs: A systematic review
and meta-analysis. J Oncol. 2020:19732412020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jiao G and Wang B: NK cell subtypes as
regulators of autoimmune liver disease. Gastroenterol Res Pract.
2016:69034962016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brand A, Singer K, Koehl GE, Kolitzus M,
Schoenhammer G, Thiel A, Matos C, Bruss C, Klobuch S, Peter K, et
al: LDHA-associated lactic acid production blunts tumor
immunosurveillance by T and NK cells. Cell Metab. 24:657–671. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma Q, Jiang H, Ma L, Zhao G, Xu Q, Guo D,
He N, Liu H, Meng Z, Liu J, et al: The moonlighting function of
glycolytic enzyme enolase-1 promotes choline phospholipid
metabolism and tumor cell proliferation. Proc Natl Acad Sci USA.
120:e22094351202023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen Y, Tang L, Huang W, Zhang Y, Abisola
FH and Li L: Identification and validation of a novel
cuproptosis-related signature as a prognostic model for lung
adenocarcinoma. Front Endocrinol (Lausanne). 13:9632202022.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tsvetkov P, Coy S, Petrova B, Dreishpoon
M, Verma A, Abdusamad M, Rossen J, Joesch-Cohen L, Humeidi R,
Spangler RD, et al: Copper induces cell death by targeting
lipoylated TCA cycle proteins. Science. 375:1254–1261. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mou Y, Wang J, Wu J, He D, Zhang C, Duan C
and Li B: Ferroptosis, a new form of cell death: Opportunities and
challenges in cancer. J Hematol Oncol. 12:342019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
D'Arcy MS: Cell death: A review of the
major forms of apoptosis, necrosis and autophagy. Cell Biol Int.
43:582–592. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fang Y, Tian S, Pan Y, Li W, Wang Q, Tang
Y, Yu T, Wu X, Shi Y, Ma P and Shu Y: Pyroptosis: A new frontier in
cancer. Biomed Pharmacother. 121:1095952020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Su Z, Yang Z, Xu Y, Chen Y and Yu Q:
Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol
Cancer. 14:482015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tsvetkov P, Detappe A, Cai K, Keys HR,
Brune Z, Ying W, Thiru P, Reidy M, Kugener G, Rossen J, et al:
Mitochondrial metabolism promotes adaptation to proteotoxic stress.
Nat Chem Biol. 15:681–689. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang Y, Yin D and Wu L: Identification of
cuproptosis-related subtypes and development of a prognostic
signature in colorectal cancer. Sci Rep. 12:173482022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu H and Tang T: Pan-cancer genetic
analysis of cuproptosis and copper metabolism-related gene set.
Front Oncol. 12:9522902022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Da Silva DA, De Luca A, Squitti R,
Rongioletti M, Rossi L, Machado CML and Cerchiaro G: Copper in
tumors and the use of copper-based compounds in cancer treatment. J
Inorg Biochem. 226:1116342022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salem A, Ahmed S, Khalfallah M, Hamadan N,
ElShikh W and Alfaki M: A comprehensive pan-cancer analysis reveals
cyclin-dependent kinase inhibitor 2A gene as a potential diagnostic
and prognostic biomarker in colon adenocarcinoma. Cureus.
16:e605862024.PubMed/NCBI
|
|
18
|
Stott FJ, Bates S, James MC, McConnell BB,
Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden KH and
Peters G: The alternative product from the human CDKN2A locus,
p14(ARF), participates in a regulatory feedback loop with p53 and
MDM2. EMBO J. 17:5001–5014. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu MT, Liu JY and Su J: CDKN2A gene in
melanoma. Zhonghua Bing Li Xue Za Zhi. 48:909–912. 2019.(In
Chinese). PubMed/NCBI
|
|
20
|
Chen Y: Identification and validation of
cuproptosis-related prognostic signature and associated regulatory
axis in uterine corpus endometrial carcinoma. Front Genet.
13:9120372022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou Z, Zhou Y, Liu D, Yang Q, Tang M and
Liu W: Prognostic and immune correlation evaluation of a novel
cuproptosis-related genes signature in hepatocellular carcinoma.
Front Pharmacol. 13:10741232022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi WK, Li YH, Bai XS and Lin GL: The cell
cycle-associated protein CDKN2A may promotes colorectal cancer cell
metastasis by inducing epithelial-mesenchymal transition. Front
Oncol. 12:8342352022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang D, Wang T, Zhou Y and Zhang X:
Comprehensive analyses of cuproptosis-related gene CDKN2A on
prognosis and immunologic therapy in human tumors. Medicine
(Baltimore). 102:e334682023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu C, Tan J, Wang X, Qin C, Long W, Pan Y,
Li Y and Liu Q: Pan-cancer analyses reveal molecular and clinical
characteristics of cuproptosis regulators. Imeta. 2:e682022.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The cancer genome atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–77. 2015.PubMed/NCBI
|
|
26
|
RStudio Team, . RStudio: Integrated
Development for R. RStudio, PBC; Boston, MA: 2020, http://www.rstudio.com/October 15–2023
|
|
27
|
Shen W, Song Z, Zhong X, Huang M, Shen D,
Gao P, Qian X, Wang M, He X, Wang T, et al: Sangerbox: A
comprehensive, interaction-friendly clinical bioinformatics
analysis platform. Imeta. 1:e362022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Law CW, Chen Y, Shi W and Smyth GK: voom:
Precision weights unlock linear model analysis tools for RNA-seq
read counts. Genome Biol. 15:R292014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Szklarczyk D, Gable AL, Nastou KC, Lyon D,
Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al:
The STRING database in 2021: Customizable protein-protein networks,
and functional characterization of user-uploaded gene/measurement
sets. Nucleic Acids Res. 49(D1): D605–D612. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Doncheva NT, Morris JH, Gorodkin J and
Jensen LJ: Cytoscape StringApp: Network analysis and visualization
of proteomics data. J Proteome Res. 18:623–632. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47((W1)): W556–W560.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nagy Á, Munkácsy G and Győrffy B:
Pancancer survival analysis of cancer hallmark genes. Sci Rep.
11:60472021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46(D1): D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kanehisa M, Araki M, Goto S, Hattori M,
Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T
and Yamanishi Y: KEGG for linking genomes to life and the
environment. Nucleic Acids Res. 36((Database Issue)): D480–D484.
2008.PubMed/NCBI
|
|
38
|
Wang S and Liu X: The UCSCXenaTools R
package: A toolkit for accessing genomics data from UCSC Xena
platform, from cancer multi-omics to single-cell RNA-seq. J Open
Source Softw. 4:16272019. View Article : Google Scholar
|
|
39
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Linder MC and Hazegh-Azam M: Copper
biochemistry and molecular biology. Am J Clin Nutr. 63:797S–811S.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li Y and Trush MA: DNA damage resulting
from the oxidation of hydroquinone by copper: Role for a
Cu(II)/Cu(I) redox cycle and reactive oxygen generation.
Carcinogenesis. 14:1303–1311. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Duan WJ and He RR: Cuproptosis:
Copper-induced regulated cell death. Sci China Life Sci.
65:1680–1682. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang H, Liu J, Zhu S, Miao K, Li Z, Qi X,
Huang L, Guo L, Wang Y, Cai Y and Lin Y: Comprehensive analyses of
genomic features and mutational signatures in adenosquamous
carcinoma of the lung. Front Oncol. 12:9458432022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ichikawa K, Imura J, Kawamata H, Takeda J
and Fujimori T: Down-regulated p16 expression predicts poor
prognosis in patients with extrahepatic biliary tract carcinomas.
Int J Oncol. 20:453–461. 2002.PubMed/NCBI
|
|
45
|
Li M, Tan Y, Li Z and Min L: Biological
characterization and clinical significance of cuproptosis-related
genes in lung adenocarcinoma. BMC Pulm Med. 25:132025. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yu YH, Li HJ, Yang XY, Yu LY and Tong XM:
Elesclomol-Cu induces cuproptosis in human acute myeloid leukemia
cells. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 32:389–394.
2024.PubMed/NCBI
|
|
47
|
Chen Y, Tang L, Huang W, Abisola FH, Zhang
Y, Zhang G and Yao L: Identification of a prognostic
cuproptosis-related signature in hepatocellular carcinoma. Biol
Direct. 18:42023. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Huo S, Wang Q, Shi W, Peng L, Jiang Y, Zhu
M, Guo J, Peng D, Wang M, Men L, et al: ATF3/SPI1/SLC31A1 signaling
promotes cuproptosis induced by advanced glycosylation end products
in diabetic myocardial injury. Int J Mol Sci. 24:16672023.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Moreau-Gachelin F, Tavitian A and
Tambourin P: Spi-1 is a putative oncogene in virally induced murine
erythroleukaemias. Nature. 331:277–280. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yuki H, Ueno S, Tatetsu H, Niiro H, Iino
T, Endo S, Kawano Y, Komohara Y, Takeya M, Hata H, et al: PU.1 is a
potent tumor suppressor in classical Hodgkin lymphoma cells. Blood.
121:962–970. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Endo S, Nishimura N, Toyoda K, Komohara Y,
Carreras J, Yuki H, Shichijo T, Ueno S, Ueno N, Hirata S, et al:
Decreased PU.1 expression in mature B cells induces
lymphomagenesis. Cancer Sci. 115:3890–3901. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jiang S, Willox B, Zhou H, Holthaus AM,
Wang A, Shi TT, Maruo S, Kharchenko PV, Johannsen EC, Kieff E and
Zhao B: Epstein-Barr virus nuclear antigen 3C binds to BATF/IRF4 or
SPI1/IRF4 composite sites and recruits Sin3A to repress CDKN2A.
Proc Natl Acad Sci USA. 111:421–426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xiong C, Ling H, Hao Q and Zhou X:
Cuproptosis: p53-regulated metabolic cell death? Cell Death Differ.
30:876–884. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xue Q, Kang R, Klionsky DJ, Tang D, Liu J
and Chen X: Copper metabolism in cell death and autophagy.
Autophagy. 19:2175–2195. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tschan MP, Reddy VA, Ress A, Arvidsson G,
Fey MF and Torbett BE: PU.1 binding to the p53 family of tumor
suppressors impairs their transcriptional activity. Oncogene.
27:3489–3493. 2008. View Article : Google Scholar : PubMed/NCBI
|