Introduction
Cancer remains a major global health burden and
continues to be a prominent cause of death, with incidence and
mortality rates rising sharply worldwide. Data from the
International Agency for Research on Cancer estimated that there
were 19.3 million new cancer cases and ~10 million cancer-related
deaths globally in 2020. Projections indicate that by 2040, the
global cancer burden will reach 28.4 million cases, an increase of
47% compared with 2020 levels (1,2).
Timely diagnosis and intervention are essential for enhancing both
curative outcomes and 5-year survival rates among patients
(3). However, the absence of
specific clinical manifestations often leads to delayed detection,
with a number of cases identified at advanced stages. Although
advancements in targeted therapy (4) and immunotherapy (5) have extended both the progression-free
survival (PFS) and overall survival (OS) of patients with advanced
malignancies, these modalities remain limited by narrow therapeutic
windows and do not consistently align with individual clinical
expectations. Immediate efforts are therefore warranted to identify
novel biomarkers capable of enabling early detection, precise
diagnosis and personalized therapeutic strategies, while also
offering predictive value for immunotherapy responsiveness
(6).
Damaged proteins pose a notable threat to cellular
function and viability. To preserve homeostasis, 80–90% of such
aberrant proteins are eliminated via the ubiquitin-proteasome
system (UPS) (7). This degradation
process depends on the sequential action of three enzymes: E1, E2
and E3. The E1 ubiquitin-activating enzyme utilizes ATP hydrolysis
to activate ubiquitin by forming a thioester bond between its
cysteine residue and the C-terminal glycine of ubiquitin. Activated
ubiquitin is subsequently transferred to the E2
ubiquitin-conjugating enzyme. In coordination with the E3 ubiquitin
ligase, E2 mediates the covalent attachment of ubiquitin to
specific substrate proteins, thereby marking them for recognition
and degradation by the 26S proteasome complex (8). Accumulating evidence indicates that
dysregulation or mutation of UPS components is strongly correlated
with tumorigenesis, positioning the UPS as a central target in
contemporary antitumor therapeutic strategies (9–12).
Ubiquitin-conjugating enzyme 2T (UBE2T), a key
element of the UPS, comprises a conserved UBC fold and a C-terminal
extension. UBE2T has been previously identified as a regulatory
factor in the DNA repair pathway associated with Fanconi anemia
(13). Elevated UBE2T mRNA
expression has been observed in multiple myeloma (14), breast cancer (15), renal cell carcinoma (16), ovarian cancer (9), cervical cancer (17) and retinoblastoma (18) compared with adjacent normal tissues.
Furthermore, increased UBE2T expression has been shown to be
correlated with reduced OS and PFS, indicating its potential role
in tumor progression.
Although accumulating evidence implicates
UBE2T in the pathogenesis of various malignancies,
comprehensive analyses across tumor types remain scarce. The
present study systematically evaluated UBE2T across diverse
cancer types, analyzing gene and protein expression profiles,
clinical phenotypes, survival outcomes, genetic alterations and
drug sensitivity. Additionally, correlations with immune checkpoint
genes, tumor-infiltrating immune cells and immune-related molecular
signatures were explored. Data integration was performed using R
software and datasets from The Cancer Genome Atlas (TCGA) (19), Genotype-Tissue Expression (GTEx),
UALCAN, GEPIA2, Tumor Immune Estimation Resource (TIMER)2.0, GSCA
and cBioPortal.
Materials and methods
Analysis of UBE2T expression
profiles
The ‘Gene DE’ module of the TIMER 2.0 database
(20) was employed to compare the
UBE2T expression levels between tumor tissues and adjacent
normal tissues across various cancer types. Gene expression
distributions were visualized using box plots, with statistical
significance assessed via the Wilcoxon test and denoted by asterisk
notation. To reinforce and expand these comparisons, the ‘Box
Plots’ feature in GEPIA2 (http://gepia2.cancer–pku.cn/#index) (21), integrating TCGA and GTEx datasets,
was utilized under the thresholds of P<0.01 and
log2FoldChange=1. Protein expression data for UBE2T in
pan-cancer contexts were retrieved from the UALCAN database
(http://ualcan.path.uab.edu/index.html), which supports
TCGA-based cancer data analysis. UBE2T mRNA profiles across
cancer cell lines were sourced from the Cancer Cell Line
Encyclopedia (https://sites.broadinstitute.org/ccle/tools) (22).
Expression levels of UBE2T mRNA and protein
in pancreatic cancer cell lines [PANC1, ASPC, BXPC3, MIA2
(Mia-paca-2), SW1990 and CAPAN1] and normal pancreatic epithelial
cells (HPDE) were assessed via reverse transcription-quantitative
PCR (RT-qPCR) and western blotting. These 7 cell lines were
purchased from American Type Culture Collection and underwent
cultivation in Dulbecco's modified Eagle's medium (cat. no.
C11995500BT; Gibco; Thermo Fisher Scientific, Inc.) with the
addition of 10% fetal bovine serum (cat. no. 10099-141 C; Gibco;
Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml
streptomycin (cat. no. 15140-122; Gibco; Thermo Fisher Scientific,
Inc.). These cell cultures were nurtured under humidified
conditions at 37°C with 5% CO2 utilizing the Thermo
Scientific HERACELL 240i CO2 Incubator (240i; Thermo
Fisher Scientific, Inc.). Western blotting was conducted using
established protocols. Briefly, cell lysates were prepared in Radio
Immunoprecipitation Assay buffer (cat. no. 87787; Thermo Fisher
Scientific, Inc.) supplemented with protease (cat. no. 04693124001;
Roche Diagnostics) and phosphatase inhibitors (cat. no. B15001-A;
Selleck Chemicals), followed by incubation on ice for 30 min. The
lysates underwent centrifugation at 13,580 × g for 15 min at 4°C,
and the supernatant was subsequently collected with care. A total
of 20 µg/lane of total protein was separated by 10%
SDS-polyacrylamide gel electrophoresis. Post-electrophoresis,
proteins were transferred onto polyvinylidene difluoride membranes
(0.45 µm; MilliporeSigma). Membranes were incubated in 5% bovine
serum albumin (cat. no. SLBN9354V; MilliporeSigma) to block
non-specific binding sites for 1 h at room temperature. Primary
antibodies specific to UBE2T (1:2,000; cat. no. A6853;
Abclonal Biotech Co., Ltd.) and β-actin (1:2,000; cat. no. 4967S;
Cell Signaling Technology, Inc.) were applied and maintained at 4°C
overnight. After primary incubation, membranes were exposed to
horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (H+L)
secondary antibodies (1:5,000; cat. no. 7074S; Cell Signaling
Technology, Inc.) for 1 h at room temperature. Signal detection was
achieved using Super ECL Detection Reagent (cat. no. 36208ES60;
Shanghai Yeasen Biotechnology Co., Ltd.) and imaged with the
Bio-Rad ChemiDoc MP Chemiluminescence Gel Imaging system (cat. no.
1708280; Bio-Rad Laboratories, Inc.). Quantification of western
blot bands was performed using Image Lab™ software (Version 6.1;
Bio-Rad Laboratories, Inc.).
For RT-qPCR, total RNA was extracted by lysing cells
in 1 ml RNAiso Plus reagent (cat. no. 9109; Takara Biotechnology
Co., Ltd.), ensuring complete homogenization. Following
centrifugation (12,000 × g; 4°C; 15 min), the supernatant was
removed and the RNA pellet was washed with 75% ethanol to improve
purities. RNA extraction and subsequent reverse transcription were
performed according to the protocols in the PrimeScript™ RT Master
Mix (cat. no. RR036A; Takara Biotechnology Co., Ltd.) and TB
Green® Premix Ex Taq™ II (cat. no. RR820A; Takara
Biotechnology Co., Ltd.) kits. qPCR was carried out on the 7500
Real-Time PCR System (cat. no. RR820A; Takara Biotechnology Co.,
Ltd.) using standard operational guidelines, and performed with the
SYBR® Premix TM kit (Invitrogen; Thermo Fisher
Scientific, Inc.). The conditions for RT-qPCR were as follows: 95°C
for 10 min, 40 cycles of 94°C for 30 sec, 60°C for 30 sec and 72°C
for 30 sec, and a final extension of 10 min at 72°C. Relative mRNA
expression levels were quantified using the 2−ΔΔCq
method (23) and normalized against
β-actin as an internal control. The primer sequences used as
follows: UBE2T forward (F), 5′-ATCCCTCAACATCGCAACTGT-3′ and
reverse (R), 5′-CAGCCTCTGGTAGATTATCAAGC-3′; β-actin F,
5′-CGTGCGTGACATTAAGGAGAA-3′ and R, 5′-AGGAAGGAAGGCTGGAAGAG-3′.
Statistical analysis was conducted using GraphPad
Prism version 9.01 (Dotmatics). Comparisons between each of the six
pancreatic cancer cells and the single control group were performed
using independent samples t-tests, with statistical significance
assessed at a Bonferroni-corrected alpha level of α=0.01 (0.05/5
comparisons). Data are presented as mean ± standard deviation and
were obtained from three independent repeats. All statistical
analyses were two-sided. P<0.05 was considered to indicate a
statistically significant difference.
Clinical phenotype analysis and
receiver operating characteristics (ROC) curves for diagnosis and
prognostic value
The association between UBE2T expression and
clinical phenotype was assessed using R software (version 4.2.1;
http://www.r-project.org/). The results
were visualized through violin plots and histograms. The diagnostic
and prognostic relevance of UBE2T in tumors was further
evaluated via Xiantao Academic (https://xiantaozi.com).
Survival analysis
Kaplan-Meier plots for OS and disease-free survival
(DFS) across 33 tumor types were generated using the ‘Survival
Analysis’ module in the GEPIA2 database, with ‘Median’ specified as
the group cut-off. Hazard ratios were estimated via the Cox
proportional hazards model, with 95% confidence intervals indicated
by dashed lines and statistical significance defined as
P<0.05. Associations between UBE2T expression and
OS, progression-free interval (PFI) and disease-specific survival
(DSS) in pan-cancer contexts were further examined through Cox
regression. The R packages ‘survival’ and ‘survminer’ were employed
for this analysis, and outcomes were visualized using forest
plots.
Genetic characterization and
alteration analysis
Genomic location, subcellular distribution, protein
tertiary structure and known protein interactions of UBE2T
were retrieved from the GeneCards database (https://www.genecards.org/). The cBioPortal platform
(https://www.cbioportal.org/) integrates
multi-dimensional genomic and clinical datasets (24), offering analyses of somatic
mutations, DNA copy number variations (CNVs) and associated
oncogenic alterations. Through modules such as ‘Cancer Types
Summary’, ‘Plots’, ‘Mutations’ and ‘Survival’, cBioPortal was used
to delineate the mutation prevalence, classification, prognostic
relevance and potential epitope modification sites of UBE2T
across various cancer types.
CNV, single nucleotide variant (SNV)
and drug sensitivity analyses of UBE2T in pan-cancer
A database known as GSCALite (https://guolab.wchscu.cn/GSCA/#/) integrates genomic
and pharmacological data including >10,000 tumor samples from 33
TCGA cancer types and >750 small-molecule agents from the
Genomics of Drug Sensitivity in Cancer (GDSC) and Cancer
Therapeutics Response Portal (CTRP) repositories (25). UBE2T was systematically
analyzed within this platform for its association with drug
sensitivity, CNV and SNV profiles.
Correlation between UBE2T expression
levels and immune checkpoint genes and immune markers
The ‘Gene_Corr’ module of the TIMER2.0 database was
employed to assess the association between UBE2T expression
and a spectrum of immune checkpoint genes, including TNFSF9, CD27,
IDO1, CD274, TNFRSF9, CD28, TIGIT, CD40, CD70, PDCD1, CD86, LAG3,
ICOS, HHLA2, ICOSLG, CTLA4, IDO2, CD80, CD276 and BTLA (26). Heatmap visualization was generated
under the ‘Purity Adjustment’ mode to reduce the impact of sample
composition variability. Key immunotherapy response predictors,
such as mismatch repair (MMR), microsatellite instability (MSI),
tumor mutation burden (TMB) and neoantigens (NEO), were further
examined. Expression profiles for MSI and 5 core MMR genes (EPCAM,
MSH6, PMS2, MSH2 and MLH1) were retrieved from TCGA. Pearson
correlation analysis was conducted to quantify associations between
UBE2T expression and MMR, MSI, TMB and NEO metrics.
Statistical significance was determined at P<0.05.
Analysis of immune infiltration
The ESTIMATE algorithm infers stromal and immune
cell infiltration in tumor tissues via single-sample gene set
enrichment analysis (ssGSEA) based on transcriptomic profiles
(27), yielding quantitative
metrics termed ‘stromalscore’ and ‘immunescore’. This computational
procedure, executed using the R package ‘estimate’, visualizes the
outcomes through scatter plots. UBE2T expression data were
retrieved from the normalized pan-cancer dataset available in the
UCSC database (http://genome.ucsc.edu/cgi-bin/hgGateway), with
expression levels log2-transformed as log2(x
+ 0.001). Immune cell infiltration associated with UBE2T
across multiple cancer types was further evaluated using the R
algorithms ‘Timer,’ ‘EPIC’ and ‘Cibersort’.
Gene Ontology (GO), Kyoto Encyclopedia
of Genes and Genomes (KEGG), GSEA and cellular functional state
analyses
Given the elevated expression of UBE2T and
its association with adverse prognosis across various malignancies,
functional characterization was pursued through GO/KEGG and GSEA
analyses. Additionally, KEGG analysis results for pancreatic cancer
were derived from the LinkedOmics database (https://linkedomics.org/login.php). The specific steps
were as follows: Firstly, ‘Pancreatic adenocarcinoma’ was selected
as the ‘cancer cohort’, the data type was set to RNAseq,
UBE2T was set as the dataset attribute, the data type in the
Target dataset was also set as RNAseq, and finally, the Pearson
correlation test was chosen as the statistical method. A total of
123 genes, identified as UBE2T-related or interacting
partners via GEPIA2 and STRING databases (https://cn.string-db.org/), were subjected to GO and
KEGG pathway enrichment using the R packages ‘clusterprofiler’ and
‘pathview’. To examine the biological relevance of UBE2T in
tumor subtypes such as breast invasive carcinoma (BRCA) and
pancreatic adenocarcinoma (PAAD), GSEA was conducted to assess
pathway-level enrichment between phenotypic states. Moreover, the
CancerSea platform (http://biocc.hrbmu.edu.cn/CancerSEA/), including
93,475 single cancer cells across 74 single cell RNA-sequencing
datasets from 27 cancer types and defining 14 cellular functional
states (28), was employed to
interrogate UBE2T-related cellular programs at single-cell
resolution.
Results
Expression profiles of UBE2T
Analysis using the TIMER2.0 database revealed
differential expression of UBE2T between pan-cancer and
normal tissues within TCGA dataset. Elevated UBE2T
expression was observed in bladder urothelial carcinoma (BLCA),
BRCA, cervical squamous cell carcinoma and endocervical
adenocarcinoma (CESC), cholangiocarcinoma (CHOL), colon
adenocarcinoma (COAD), esophageal carcinoma (ESCA), glioblastoma
multiforme (GBM), head and neck squamous cell carcinoma (HNSC),
kidney renal clear cell carcinoma (KIRC), kidney renal papillary
cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung
adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), PAAD,
pheochromocytoma and paraganglioma (PCPG), prostate adenocarcinoma
(PRAD), rectum adenocarcinoma (READ), stomach adenocarcinoma
(STAD), thyroid carcinoma (THCA) and uterine corpus endometrial
carcinoma (UCEC) compared with the matched normal tissues, whereas
expression in kidney chromophobe (KICH) was reduced (Fig. 1A). Data from adrenocortical
carcinoma (ACC), acute myeloid leukemia (LAML), mesothelioma
(MESO), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC),
brain lower grade glioma (LGG), ovarian serous cystadenocarcinoma
(OV), sarcoma (SARC), testicular germ cell tumor (TGCT), thymoma
(THYM), uterine carcinosarcoma (UCS) and uveal melanoma (UVM)
lacked sufficient normal tissue samples for reliable comparison. To
strengthen and extend these observations, UBE2T expression
across 33 tumor types and normal tissues was analyzed using the
GEPIA2 platform. Consistent with the initial findings, UBE2T
was generally upregulated in tumors, excluding MESO and UVM due to
the absence of normal controls. No significant expression
differences were identified for CHOL, KICH, KIRC, LGG, PCPG, PRAD,
SARC and THCA (Fig. 1B). Notably,
normal LAML tissues exhibited higher UBE2T expression
relative to tumor counterparts. Results from both databases
demonstrated notable concordance. The results shown in Fig. 1-E revealed increased UBE2T
mRNA and protein expression in pancreatic cancer cell lines
relative to normal pancreatic cells. Additionally, the results
shown in Fig. 1F indicated higher
UBE2T protein levels in LIHC, HNSC, LUSC, OV, LUAD, RCC and UCEC
compared with the respective normal tissues.
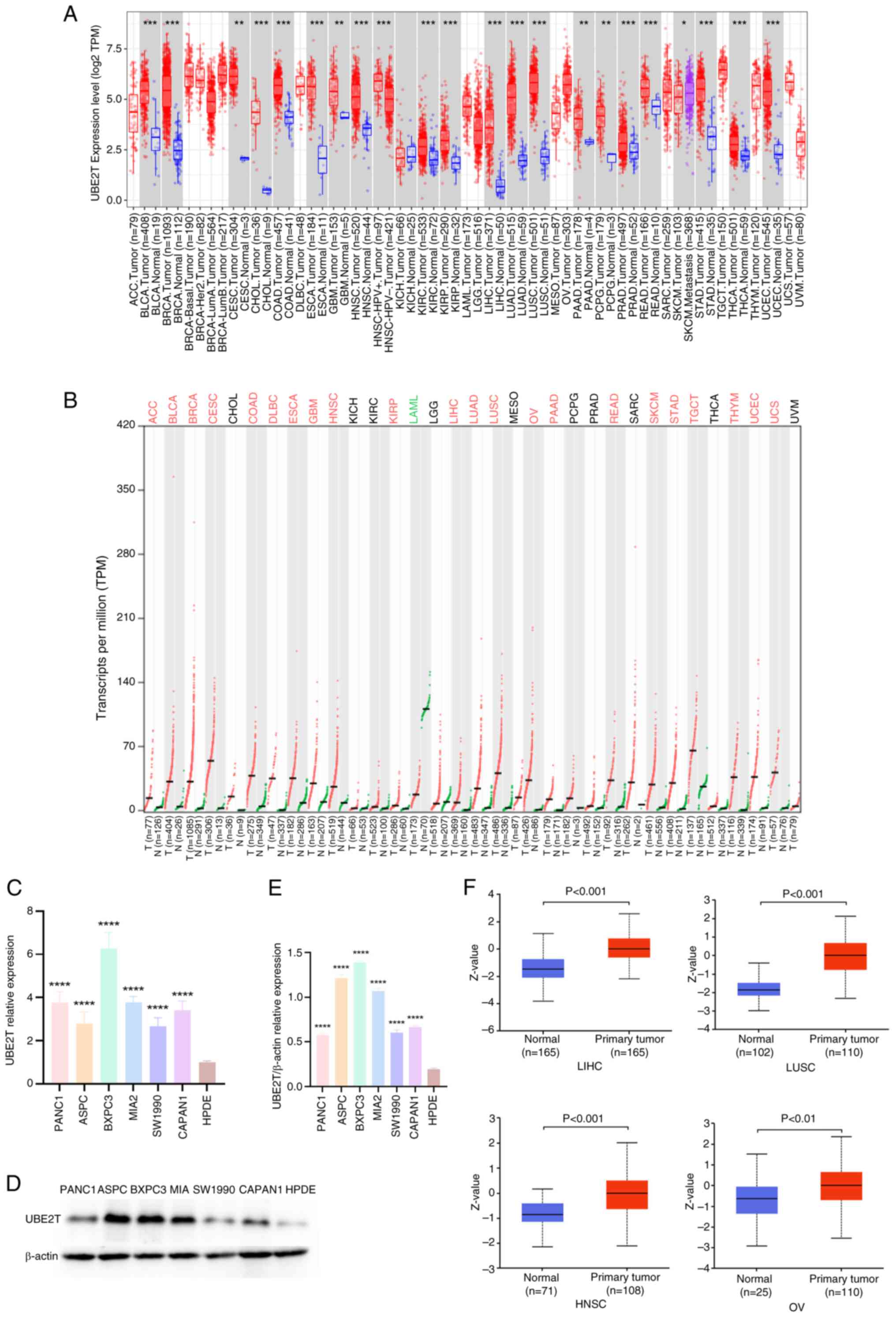 | Figure 1.UBE2T expression profiles in
pan-cancer. (A) Difference in UBE2T expression between
pan-cancer and normal tissues in TCGA; (B) difference in
UBE2T expression between pan-cancer and normal tissues in
TCGA + GTEx. Difference in UBE2T (C) mRNA and (D and E)
protein expression between pancreatic cancer cells including PANC1,
ASPC, BXPC3, MIA2, SW1990, CAPAN1 and normal pancreatic cells
(HPDE). (F) Difference in the protein level of UBE2T in LIHC, LUSC,
HNSC and OV compared with matched paracancerous tissues.
*P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. UBE2T,
ubiquitin-conjugating enzyme 2T; TCGA, The Cancer Genome Atlas;
TPM, transcripts per million. |
Association between UBE2T and clinical
phenotype
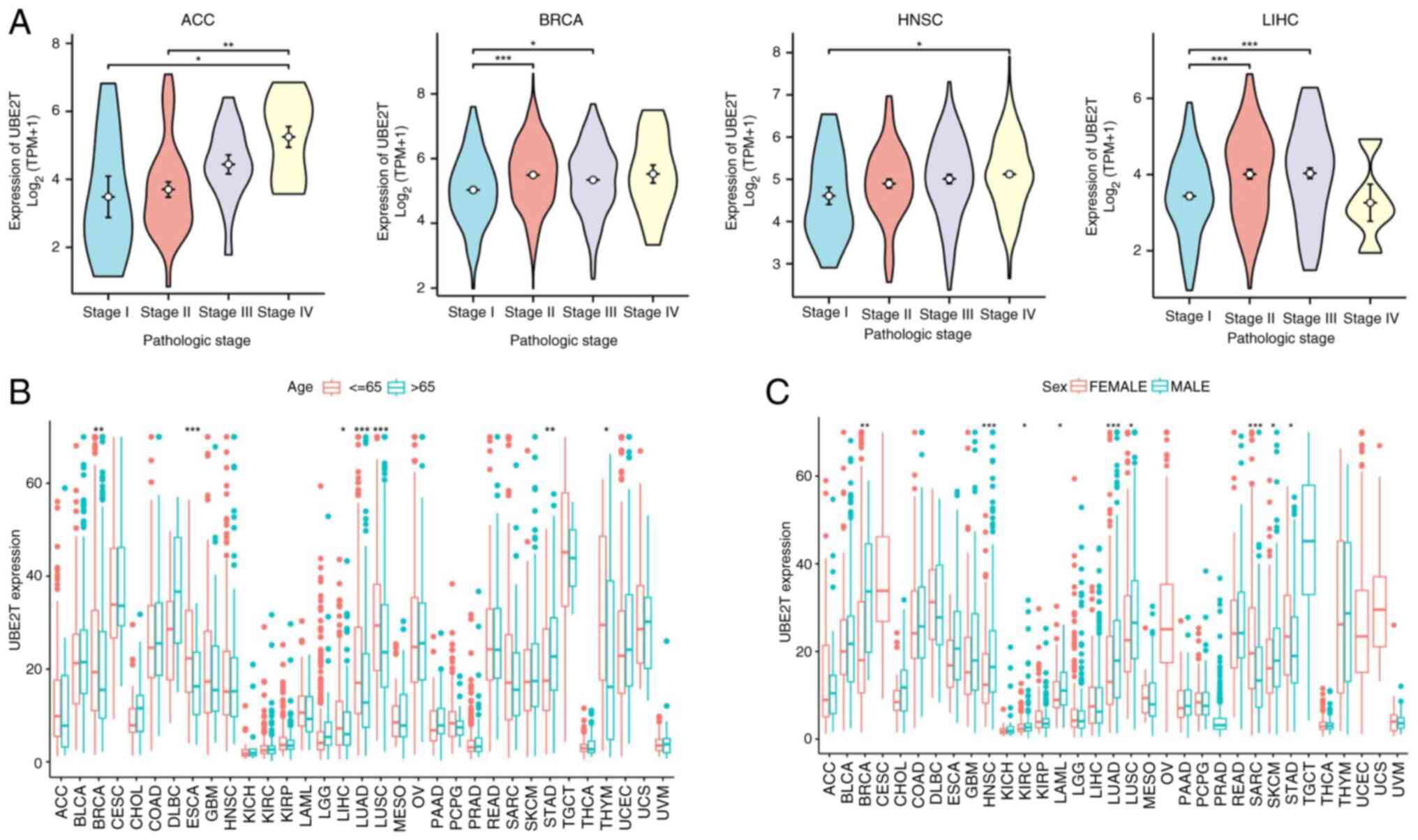
Analysis across 33 tumor types revealed significant
associations between UBE2T expression and clinical characteristics.
As depicted in Fig. 2A, elevated
UBE2T levels in ACC, BRCA, KIRP, KICH, LIHC, HNSC, KIRC and THCA
were associated with advanced pathological stages. Furthermore,
UBE2T expression was higher in individuals aged ≤65 in BRCA, ESCA,
LIHC, LUAD, LUSC and THYM, whereas STAD exhibited an inverse
pattern (Fig. 2B). Increased UBE2T
expression was observed in male patients with BRCA, HNSC, KIRC,
LAML, LUAD, LUSC and skin cutaneous melanoma (SKCM), in contrast to
lower expression levels in men with SARC and STAD (Fig. 2C).
ROC curves for diagnostic and
prognostic value of UBE2T in pan-cancer
The identification of reliable biomarkers for early
tumor detection and prognostication remains a primary objective in
oncology. ROC analysis demonstrated that UBE2T achieved area
under the curve (AUC) values >0.9 in BRCA, BLCA, CHOL, COADREAD,
CESC, HNSC and LIHC, supporting its diagnostic applicability
(Fig. 3A-C). Furthermore,
UBE2T displayed AUC values >0.7 in predicting 1-, 3- and
5-year survival in ACC, KICH, GBMLGG and MESO (Fig. 3D), indicating its potential utility
in prognostic stratification.
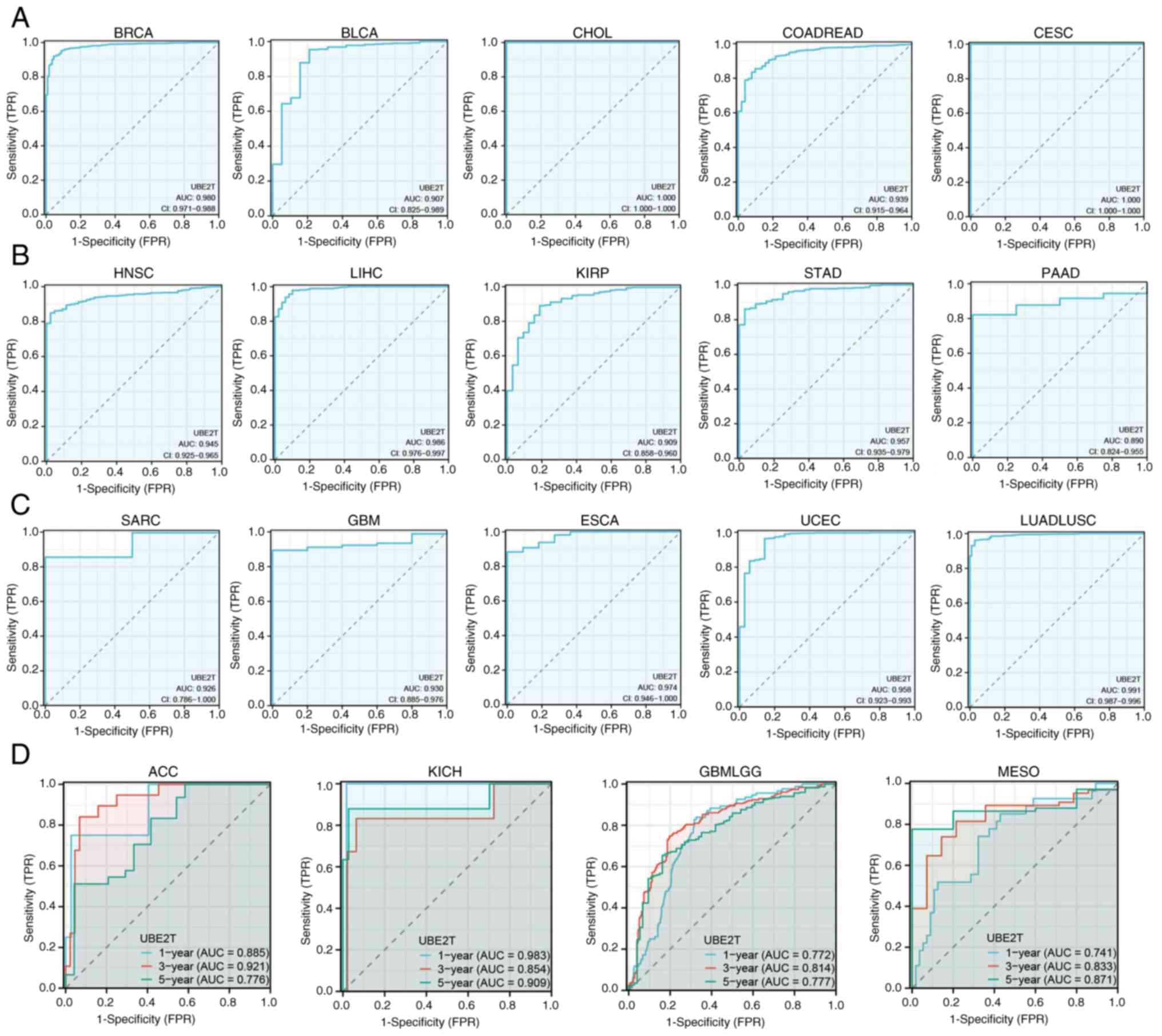 | Figure 3.ROC curves. ROC curves for the
diagnostic value of UBE2T in (A) BRCA, BLCA, CHOL, COADREAD
and CESC, (B) HNSC, LIHC, KIRP, STAD and PAAD and (C) SARC, GBM,
ESCA, UCEC and LUADLUSC. (D) ROC curves for the prognostic value of
UBE2T in ACC, KICH, GBMLGG and MESO. ROC, receiver operating
characteristics; UBE2T, ubiquitin-conjugating enzyme 2T; AUC, area
under the curve; CI, confidence interval; TPR, true positive rate;
FPR, false positive rate. |
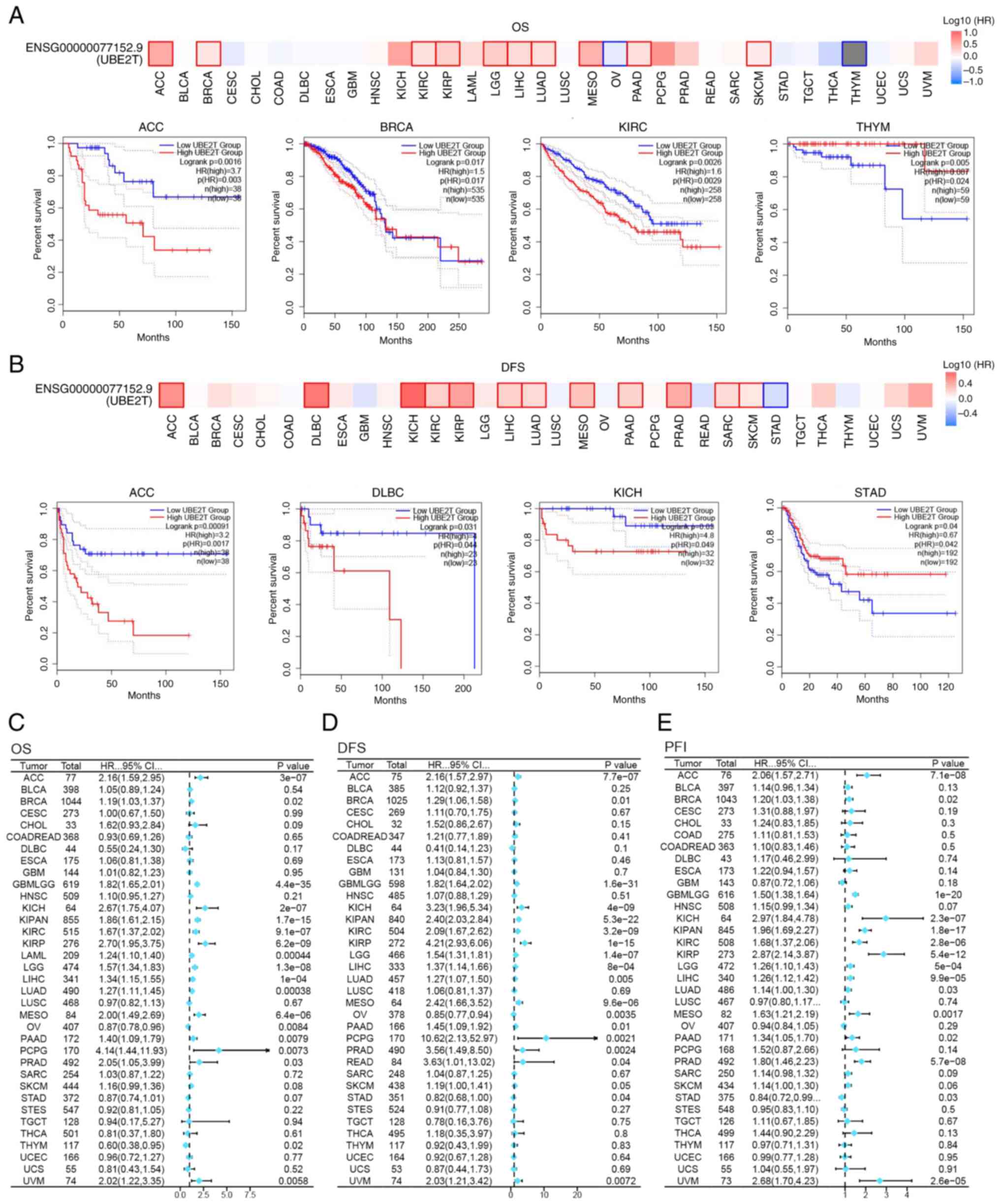
Survival analysis
Patients were stratified into high- and
low-expression cohorts according to the median UBE2T
expression level, followed by survival analysis to evaluate the
prognostic relevance of UBE2T. Analysis via GEPIA2 revealed
that elevated UBE2T expression was associated with reduced
DFS in ACC, DLBC, KICH, KIRC, KIRP, LIHC, LUAD, MESO, PAAD, PRAD,
SARC and SKCM, whereas a favorable correlation with DFS was
observed in STAD (Fig. 4B).
Similarly, increased UBE2T expression was linked to
decreased OS in ACC, BRCA, KIRC, KIRP, LGG, LIHC, LUAD, MESO, PAAD
and SKCM, while improved OS outcomes were observed in OV and THYM
(Fig. 4A). Further evaluation
through Cox regression analysis reinforced the prognostic value of
UBE2T across multiple cancer types. As illustrated in
Fig. 4C, elevated UBE2T
expression predicted unfavorable outcomes in ACC, BRCA, GBMLGG,
KICH, KIPAN, KIRC, KIRP, LAML, LGG, LIHC, LUAD, MESO, PCPG, PAAD
and UVM. Additionally, Fig. 4D
indicated a negative association between high UBE2T
expression and DFS in approximately half of the tumor types
analyzed. In parallel, a shortened PFI in ACC, BRCA, KICH, PAAD,
KIRP, LUAD and others was linked to high UBE2T expression
(Fig. 4E). Collectively, the data
suggest that UBE2T functions as a negative prognostic
indicator in a broad spectrum of tumors and may offer diagnostic
utility or therapeutic relevance in oncological contexts.
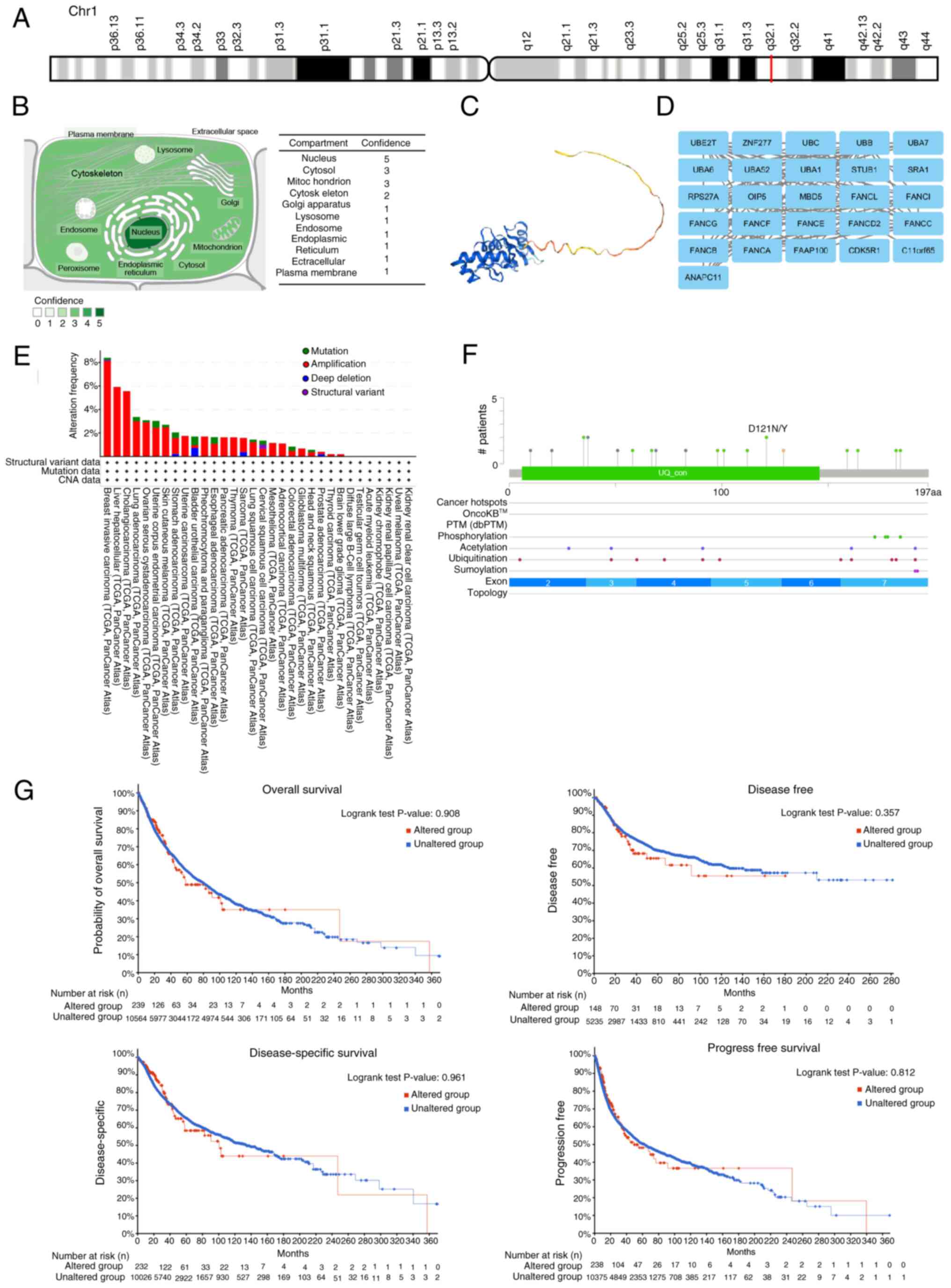
UBE2T gene characterization and
genetic variation analysis
The UBE2T gene was mapped to chromosome
1q32.1 (Fig. 5A), with
high-confidence localization to the nucleus, cytosol and
mitochondria (Fig. 5B). The
tertiary protein structure of UBE2T is illustrated in Fig. 5C. Protein-protein interaction
analysis via the STRING database identified 25
UBE2T-associated proteins, including ZNF277, UBC, UBB, UBA7,
UBA6, FANCF and CDK5R1 (Fig. 5D).
Genomic alterations of UBE2T across 10,967 tumor samples
from 33 cancer types were analyzed using cBioPortal. As presented
in Fig. 5E, gene amplification
represents the most frequent alteration, particularly in BRCA,
followed by LIHC, CHOL and LUAD. Mutations are most prevalent in
BLCA, while deep deletions were also observed in BLCA, SARC, PRAD
and STAD. Predicted post-translational modifications of
UBE2T included phosphorylation, acetylation, ubiquitination
and sumoylation (Fig. 5F). Survival
analysis revealed no statistically significant association between
UBE2T mutations and OS (P=0.908), DSS (P=0.961), DFS
(P=0.357) or PFI (P=0.812) for all individuals with cancer
(Fig. 5G).
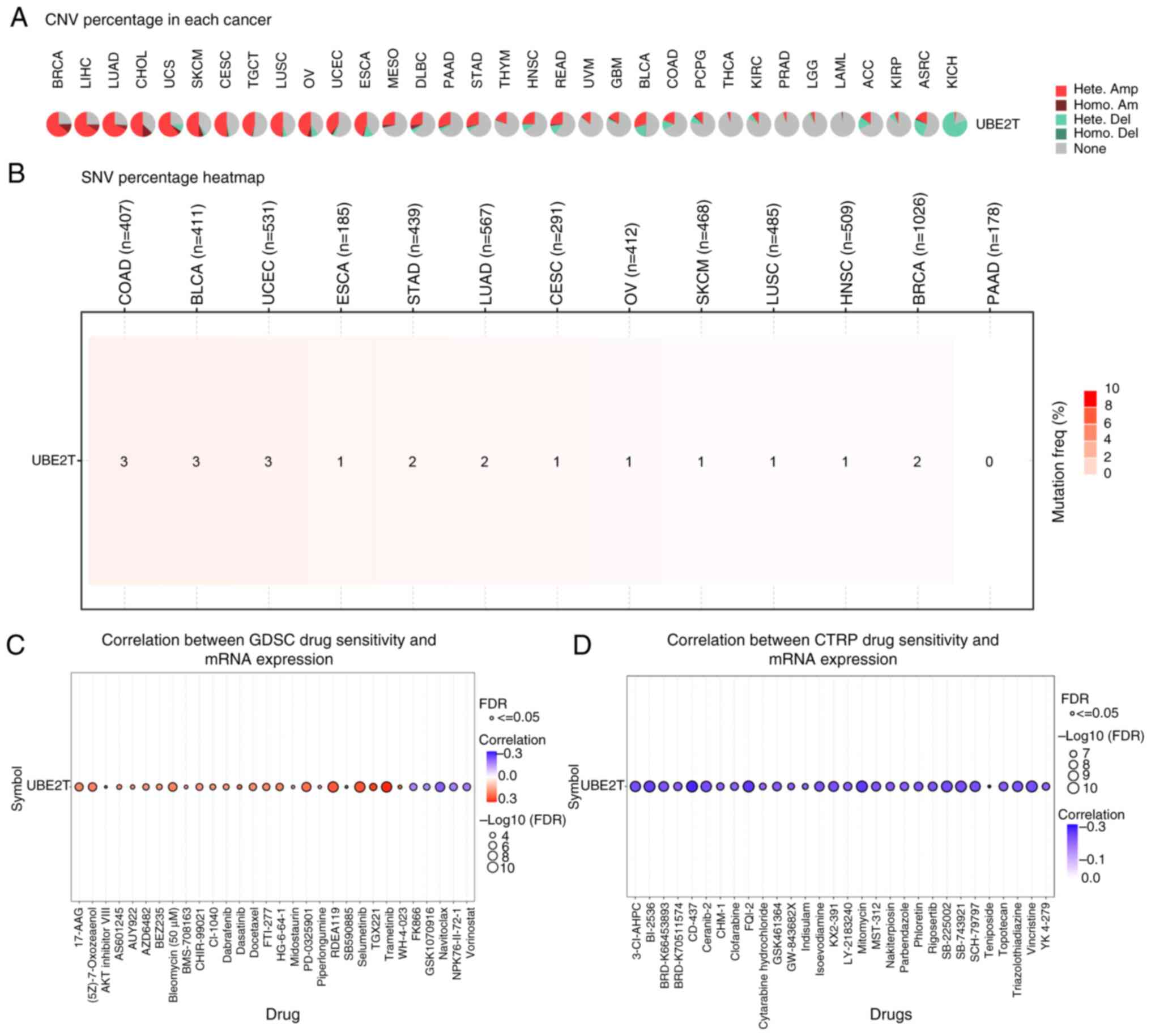
CNV, SNV and drug sensitivity analyses
of UBE2T in pan-cancer
CNV and SNV represent prevalent genetic alterations
in tumors. UBE2T was subjected to CNV, SNV and drug
sensitivity profiling via the GSCALite platform. As illustrated in
Fig. 6A, heterozygous amplification
emerged as the dominant CNV subtype of UBE2T, frequently
observed in malignancies including BRCA, LIHC, LUAD, CHOL, CESC,
UCS and SKCM. The results shown in Fig.
6B indicated that UBE2T SNVs were more frequently
detected in COAD, BLCA and UCEC, whereas no SNVs were identified in
PAAD. Drug response data were obtained from GDSC and CTRP. Analysis
based on the GDSC dataset revealed significant positive
correlations between UBE2T expression and sensitivity to
trametinib, selumetinib, RDEA119 and PD-0325901, while inverse
correlations were noted with navitoclax, vorinostat and FK866
(Fig. 6C). Additionally, CTRP-based
results demonstrated significant negative associations between
UBE2T expression and responses to CD-437, mitomycin,
SB-225002, vincristine and BI-2536 (Fig. 6D).
Association of UBE2T expression with
tumor immunomarkers and immune checkpoint genes
MMR, MSI, TMB and NEO serve as established
indicators for predicting responses to tumor immunotherapy
(29,30). The present analysis evaluated the
relationship between UBE2T expression and these markers. In
a pan-cancer context, UBE2T exhibited strong positive
associations with nearly all MMR-related genes (EPCAM, PMS2, MLH1,
MSH2 and MSH6) across diverse tumor types, with significant
correlations observed in BLCA, BRCA-Basal, CESC, COAD, ESCA, HNSC,
KIRC, KIRP, LGG, LIHC, LUSC, OV, PRAD and UVM (Fig. 7A). As shown in Fig. 7B, UBE2T expression was
positively associated with the majority of immune checkpoint genes
in BLCA, BRCA, HNSC, KIRC, LGG, LIHC, LUAD, PAAD, PRAD and TGCT,
whereas inverse associations were detected in THCA and THYM. These
patterns imply that UBE2T may modulate the tumor immune
microenvironment through immune checkpoint gene regulation in
select malignancies. Additionally, UBE2T demonstrated
significant positive correlations with MSI in BLCA, UCS, UCEC,
THCA, TGCT, STAD, SARC, PRAD, MESO, HNSC and ESCA (Fig. 7C). According to Fig. 7D, UBE2T expression showed positive
correlations across most tumor types, except for THYM, which
exhibited a negative association, and UVM, UCS, TGCT, LAML, KIRP,
GBM, ESCA, DLBC, CHOL and CESC, where no significant correlation
was noted. Fig. 7E indicated a
significant positive relationship between UBE2T and NEO in
GBM, GBMLGG, LUAD, COADREAD, BRCA and PRAD, while a negative
correlation was identified in TGCT. Collectively, these data
indicate that UBE2T expression is strongly associated with
tumor immunogenicity across multiple cancer types.
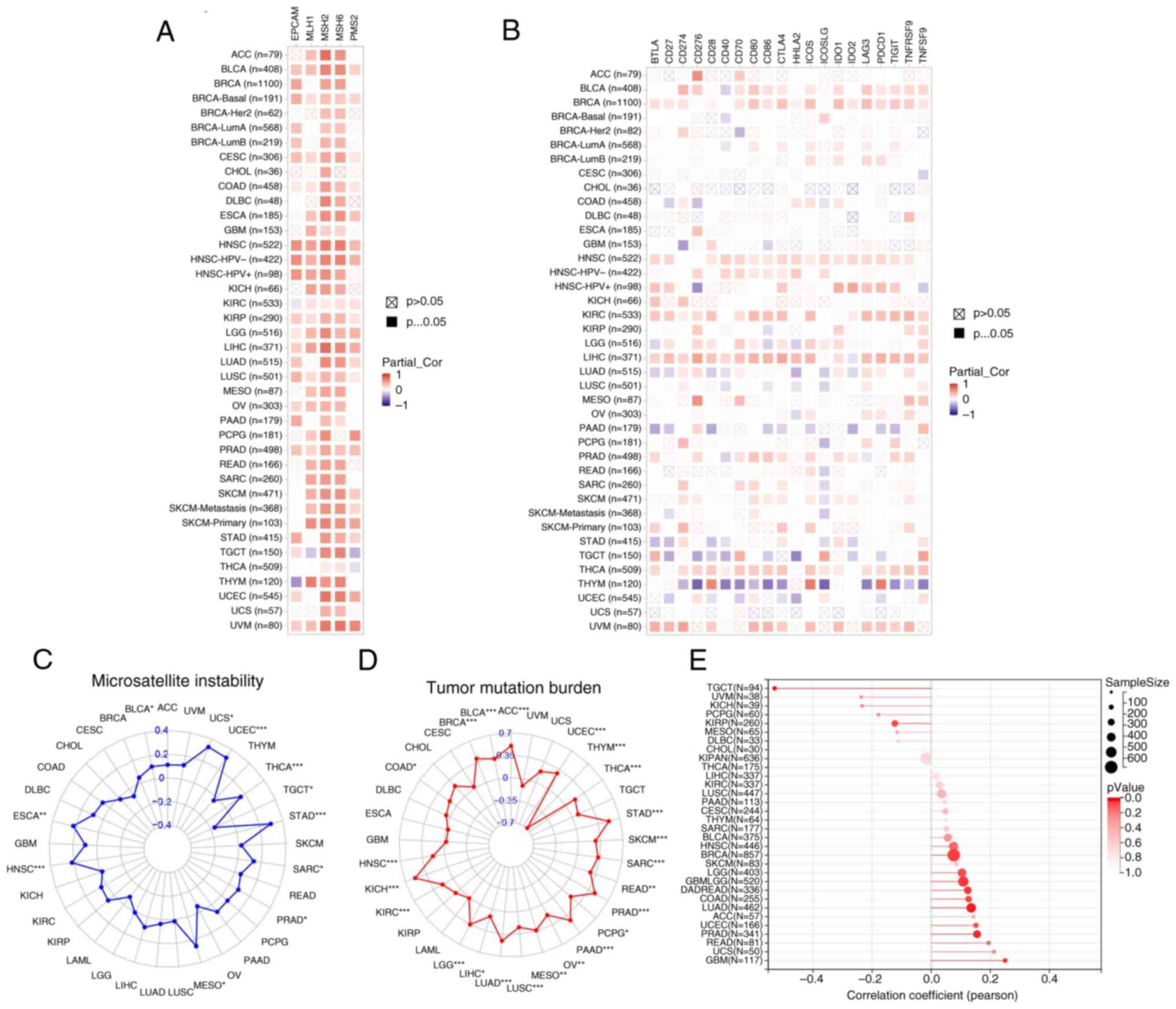 | Figure 7.Correlation between UBE2T and
immune checkpoint genes, as well as its association with
immunomarkers. (A) Correlation with MMR genes (including EPCAM,
MSH6, PMS2, MSH2 and MLH1). (B) Correlation with immune checkpoint
genes (including TNFSF9, CD27, IDO1, CD274, TNFRSF9, CD28, TIGIT,
CD40, CD70, PDCD1, CD86, LAG3, ICOS, HHLA2, ICOSLG, CTLA4, IDO2,
CD80, CD276 and BTLA). (C) Association with MSI, (D) TMB and (E)
NEO in pan-cancer. *P<0.05; **P<0.01; ***P<0.001. UBE2T,
ubiquitin-conjugating enzyme 2T; MMR, mismatch repair; MSI,
microsatellite instability; TMB, tumor mutational burden; NEO,
neoantigens. |
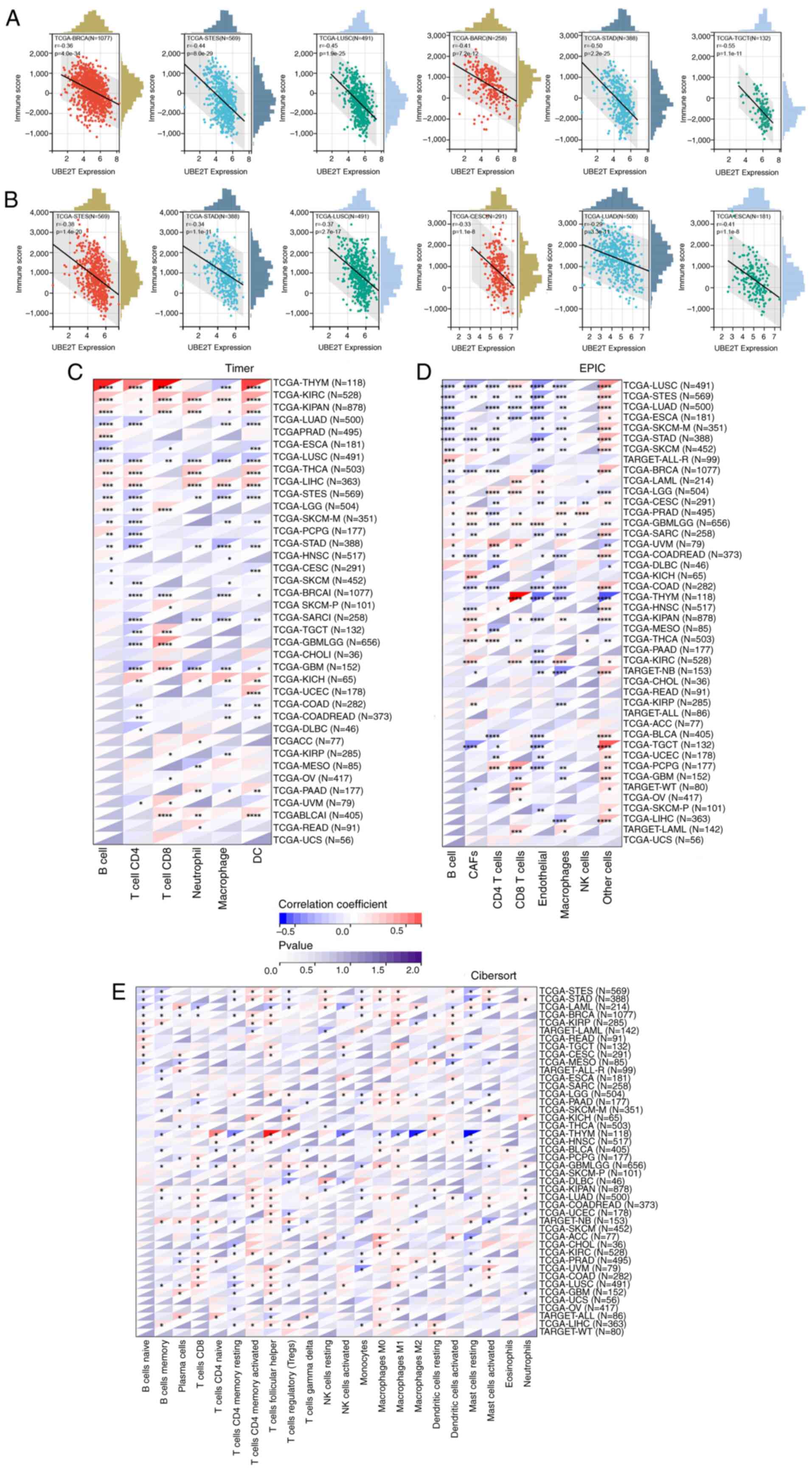
Immune infiltration analysis
Tumor-infiltrating immune cells represent a central
component of the tumor microenvironment, exerting substantial
influence on tumor progression and clinical outcomes (31–33).
The present study assessed the relationship between UBE2T
expression and both StromalScore and ImmuneScore. As shown in
Fig. 8A, UBE2T expression
exhibited a negative correlation with StromalScore in BRCA, stomach
and esophageal carcinoma (STES), LUSC, SARC, STAD and TGCT, among
others. A similar inverse association was observed between
UBE2T and ImmuneScore in STES, STAD, LUSC, CESC, LUAD and
ESCA (Fig. 8B).
To further characterize the immune landscape, TIMER,
EPIC and CIBERSORT algorithms were applied to evaluate associations
between UBE2T expression and immune cell infiltration.
According to Fig. 8C, UBE2T
expression showed positive correlations with B cells,
CD4+ T cells, CD8+ T cells and dendritic
cells (DCs) in THYM, KIRC, KIPAN, THCA and LIHC. Additionally, a
strong positive association with cancer-associated fibroblasts
(CAFs) was observed in PRAD, GBMLGG, KICH, KIPAN, MESO, THCA, KIRC
and KIRP, while most remaining immune cell types demonstrated
negative correlations. EPIC-based analysis (Fig. 8D) indicated that UBE2T
expression was predominantly negatively associated with immune cell
infiltration across pan-cancer, with the notable exception of a
positive correlation with CD8+ T cells in THYM.
Comparable patterns were identified using the CIBERSORT algorithm
(Fig. 8E). Collectively, these data
highlight the intricate and context-dependent role of UBE2T
in modulating the tumor immune microenvironment. UBE2T may
contribute to immune evasion through the expansion of
immunosuppressive populations in certain tumors, whereas in others,
it may enhance the recruitment or activity of cytotoxic immune
subsets, thereby supporting antitumor responses.
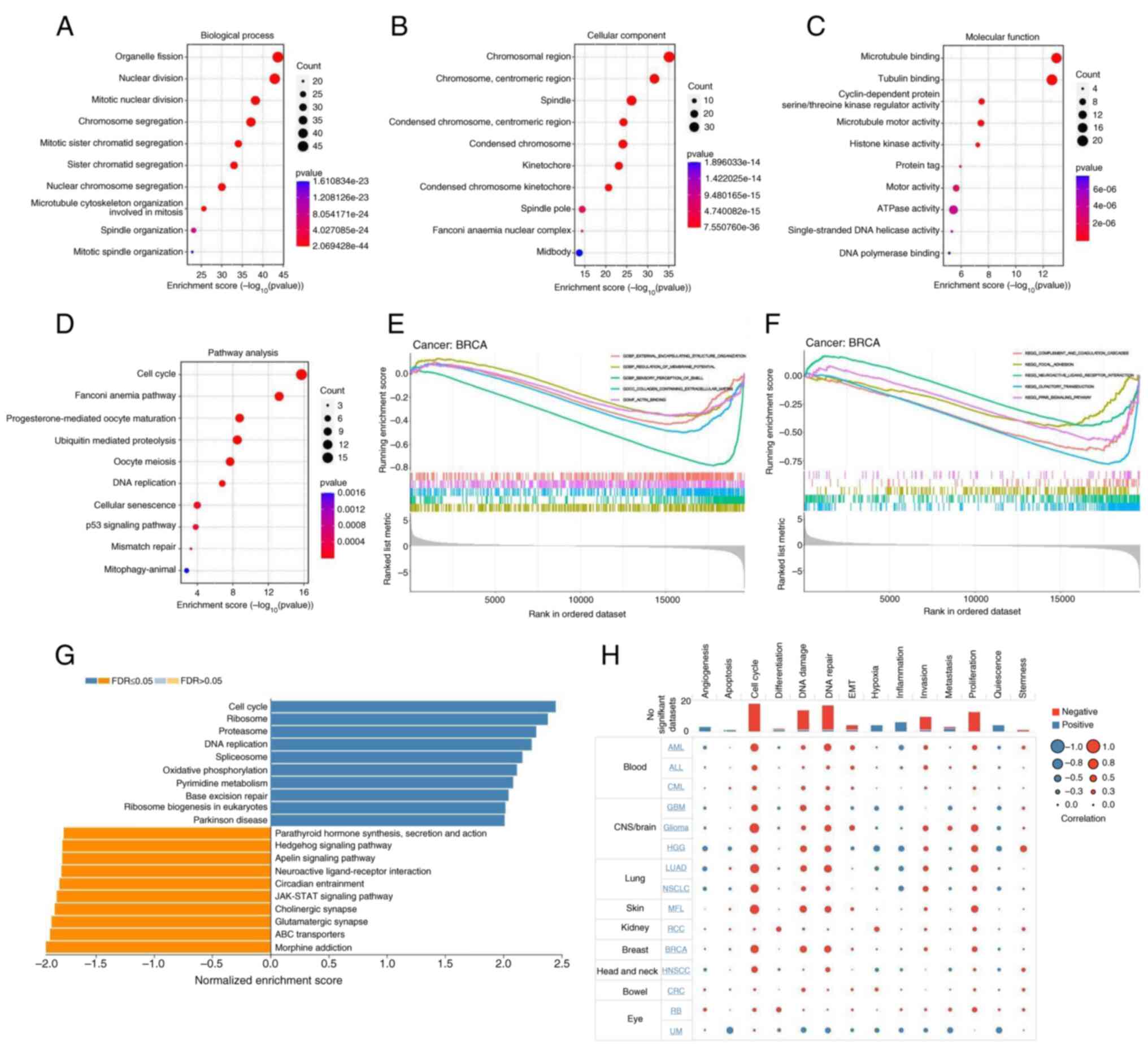
GO, KEGG, GSEA and cellular functional
state analyses
The GO database classifies gene functions into three
categories: Biological Process (BP), Cellular Component (CC) and
Molecular Function (MF). As shown in Fig. 9A, UBE2T and its associated
genes were predominantly involved in BPs such as ‘organelle
fission’, ‘nuclear division’, ‘mitotic nuclear division’ and
‘chromosome segregation’. Corresponding CC terms included
‘chromosomal region’, ‘chromosome, centromeric region’ and
‘spindle’ (Fig. 9B), while MF
annotations primarily included ‘microtubule binding’ and ‘tubulin
binding’ (Fig. 9C). KEGG pathway
enrichment (Fig. 9D) revealed
significant associations with ‘cell cycle’, ‘Fanconi anemia
pathway’, ‘ubiquitin mediated proteolysis’, ‘oocyte meiosis’, ‘DNA
replication’ and ‘p53 signaling pathway’. Notably, ‘mismatch
repair’ also appeared as a key pathway, indicating a strong link
between UBE2T and tumor immune contexture, as well as
potential responsiveness to immunotherapeutic strategies. The GSEA
results for BRCA (Fig. 9E)
identified BPs including ‘external encapsulating structure
organization’, ‘regulation of membrane potential’ and ‘sensory
perception of smell’. Enriched CCs involved ‘collagen-containing
extracellular matrix’ and MF terms included ‘actin binding’.
GSEA-based KEGG results for BRCA further implicated pathways such
as ‘complement and coagulation cascades’, ‘focal adhesion’ and
‘PPAR signaling pathway’ in UBE2T-related mechanisms
(Fig. 9F). Analysis of KEGG results
for pancreatic cancer revealed the mechanism by which UBE2T
promotes the occurrence and development of pancreatic cancer
(Fig. 9G). The results in Fig. 9H showed that UBE2T induced
different changes in cellular function in different tumor types,
but mainly focused on ‘Cell cycle’, ‘DNA damage’, ‘DNA repair’,
‘proliferation’, ‘invasion’, ‘EMT’, ‘inflammation’ and
‘angiogenesis’.
Discussion
The UPS governs the degradation of aberrant
proteins, with E2 ubiquitin-conjugating enzymes playing a central
role in the ubiquitination cascade. UBE2T encodes a member
of this enzyme class and has been implicated in tumorigenesis.
Accumulating evidence suggests that UBE2T contributes to
oncogenic processes across multiple malignancies. Specifically,
Huang et al (9) observed
elevated UBE2T expression in ovarian cancer, which was
associated with an unfavorable prognosis, and demonstrated that
UBE2T drove epithelial-mesenchymal transition (EMT) via the
AKT/mTOR axis, enhancing cellular proliferation and invasiveness.
Similarly, Hu et al (10)
reported increased UBE2T levels in nasopharyngeal carcinoma
tissues, with immunohistochemical analysis linking its upregulation
to adverse clinical outcomes. Functional assays in vitro and
in vivo confirmed that UBE2T enhanced proliferation,
invasion and metastasis through activation of the
AKT/GSK3β/β-catenin pathway. Cui et al (11) further validated
UBE2T-mediated EMT induction via the PI3K/AKT pathway in
ovarian cancer, reinforcing its role in tumor progression. In
breast cancer, Ueki et al (12) demonstrated that silencing
UBE2T upregulated BRCA1 expression, thereby attenuating
tumor cell development. In colorectal cancer, UBE2T was
found to be upregulated and implicated in tumor advancement by
accelerating p53 protein degradation (34). Based on this collective evidence,
the present study explored the relationship between UBE2T
expression and tumor prognosis, clinical characteristics,
immune-related biomarkers, immune checkpoint proteins, immune cell
infiltration and associated signaling pathways.
The results of the present study indicated that
UBE2T expression was elevated across a broad spectrum of
tumor types, including BLCA, BRCA, CESC, CHOL, COAD, KIRC, LUSC,
PCPG and UCEC. Protein-level upregulation in LIHC, HNSC, LUSC, OV,
LUAD, RCC and UCEC was corroborated by the UALCAN database,
aligning with previous studies (9,14–18,9–12,34).
Furthermore, diagnostic analyses in the present study indicated
that UBE2T exhibited high discriminative power in multiple
tumor types, with ROC values >0.9. Genomic instability, a
central feature of tumorigenesis (35,36),
was reflected in UBE2T gene alterations, predominantly
amplifications and mutations, consistent with observed CNV and SNV
profiles. These genetic aberrations likely contribute to
UBE2T upregulation and its tumor-promoting function.
Clinically, elevated UBE2T expression, more prevalent among
male patients aged ≤65, was linked to advanced disease stages in
ACC, BRCA, KIRP, KICH, LIHC, LUSC, HNSC, KIRC and THCA. This
pattern partially accounts for the association between high
UBE2T expression and poor survival outcomes in pan-cancer
cohorts, as confirmed in the present study. Similar associations
have been previously reported. For instance, Zhu et al
(37) observed UBE2T
upregulation in gallbladder cancer, correlating it with an advanced
clinical stage and an unfavorable prognosis. Increased UBE2T
expression was also documented in gastric cancer, where it's
correlated with a high T classification, low differentiation and
reduced survival (38). Parallel
trends were observed in colorectal (39), pancreatic (40), esophageal squamous cell carcinoma
(41), as well as in breast and
lung cancer (42). Cumulatively,
these data support the classification of UBE2T as an
oncogene and a viable therapeutic target. In the present study,
mechanistic analyses revealed UBE2T involvement in pathways
including ‘cell cycle’, ‘ubiquitin-mediated proteolysis’, ‘p53
signaling pathway’, ‘mismatch repair’, and ‘DNA replication’.
Functionally, UBE2T contributed to altered cellular states
characterized by enhanced proliferation, invasion and EMT,
consistent with earlier studies (9–11,42–44).
Immunotherapy represents a central strategy in
contemporary oncology, with immune-related biomarkers such as
StromalScore, ImmunoScore, TMB, MMR, MSI and NEO widely employed to
assess and predict treatment efficacy (45,46).
In the present study, UBE2T exhibited a negative correlation
with the StromalScore and ImmunoScore across most cancer types,
suggesting its involvement in shaping the tumor immune
microenvironment. Immune checkpoint genes modulate T-cell activity
to preserve immune equilibrium (47), and monoclonal antibodies targeting
these checkpoints have demonstrated sustained therapeutic benefit
in malignancies such as colorectal cancer (48). In the present study, UBE2T
showed widespread associations with immune checkpoint genes in
pan-cancer, implying that UBE2T may influence antitumor
immune responses by regulating these genes and altering immune cell
infiltration patterns (49). MMR
genes are essential for preserving genomic integrity during DNA
replication, and their dysfunction can result in MSI, contributing
to tumorigenesis through increased somatic mutation burden
(50–52), NEO generation (53) and the recruitment of immune effector
cells and inflammatory mediators (54,55).
The findings of the present study revealed significant associations
between UBE2T expression and both MMR gene expression and
MSI status in multiple tumor types. TMB, a widely recognized
biomarker in immunotherapy (56),
displayed a positive correlation with UBE2T in 23 cancer
types, further supporting a potential immunogenomic role. By
contrast, UBE2T expression was positively correlated with
NEO in only a limited subset of tumors and negatively in TGCT,
indicating heterogeneity in its relationship with NEO presentation.
Collectively, the evidence suggests that UBE2T may influence
immunotherapeutic outcomes through multifaceted interactions with
immune-related genomic features.
The tumor microenvironment comprises malignant
cells alongside surrounding immune and inflammatory components,
tumor-associated fibroblasts, microvasculature and a complex array
of chemokines and cytokines. Among immune constituents,
CD8+ T cells and Th1-polarized CD4+ T cells
exert potent cytotoxic effects, with elevated infiltration levels
inversely associated with tumor recurrence (27). An enhanced presence of DCs and
natural killer (NK) cells has also been linked to favorable
clinical outcomes (33,57). By contrast, immune evasion
mechanisms are supported by myeloid-derived suppressor cells, CAFs,
regulatory T cells, tumor-associated macrophages and M2
macrophages, which suppress T-cell-mediated antitumor responses
(58). In the present study,
analysis revealed a positive correlation between UBE2T
expression and immunocidal cell infiltration in THYM, KIRC, KIPAN,
THCA and LIHC. Conversely, UBE2T expression exhibited
negative correlations with macrophages, B cells, CD4+
and CD8+ T cells, neutrophils and DCs in LUSC, STES,
TGCT, STAD, SKCM, and COAD, with additional negative associations
observed in SKCM-M, PCPG, STAD, BRCA and SARC. EPIC algorithm-based
evaluation further indicated a positive correlation between
UBE2T and CD4+ T cells in LGG, UVM, THCA and
PCPG, as well as with CD8+ T cells in LUSC, LAML, LGG,
UVM, THYM and PCPG. Notably, UBE2T expression correlated
positively with NK cells only in PRAD and THCA. Complementary
analysis via ssGSEA demonstrated a positive correlation between Th2
cell infiltration and anaplastic grade in retinoblastoma, as
reported by Wang et al (18). Despite accumulating data, the
relationship between UBE2T and immune cell infiltration
remains unclear. Further experimental validation is warranted to
elucidate the underlying mechanisms and confirm these
associations.
Although the present study comprehensively analyzed
the UBE2T gene from various aspects, with an emphasis on
pancreatic cancer, it still has some shortcomings, mainly including
a lack of sufficient experimental verification and reliance on
publicly available databases. Therefore, further functional and
mechanistic experiments are needed to refine the data.
In summary, the present study demonstrated that
abnormal expression of UBE2T in tumor tissues, as identified
through analysis of public databases, is associated with
unfavorable clinical outcomes. Furthermore, the genetic profile of
UBE2T, its immunological correlations and underlying
regulatory mechanisms have been delineated. These insights
collectively characterize UBE2T as a proto-oncogene with
therapeutic relevance.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
PL made contributions to conception and design. ZZ
obtained the database data. ANX analyzed the data using R software.
YHC obtained the experimental data. Data analysis and
interpretation was completed by ZZ. ZZ, PL, ANX and YHC confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
UBE2T
|
ubiquitin-conjugating enzyme 2T
|
|
ACC
|
adrenocortical carcinoma
|
|
BLCA
|
bladder urothelial carcinoma
|
|
BRCA
|
breast invasive carcinoma
|
|
CESC
|
cervical squamous cell carcinoma and
endocervical adenocarcinoma
|
|
CHOL
|
cholangiocarcinoma
|
|
COAD
|
colon adenocarcinoma
|
|
DLBC
|
lymphoid neoplasm diffuse large
B-cell lymphoma
|
|
ESCA
|
esophageal carcinoma
|
|
GBM
|
glioblastoma multiforme
|
|
HNSC
|
head and neck squamous cell
carcinoma
|
|
KICH
|
kidney chromophobe
|
|
KIRC
|
kidney renal clear cell carcinoma
|
|
KIRP
|
kidney renal papillary cell
carcinoma
|
|
LAML
|
acute myeloid leukemia
|
|
LGG
|
brain lower grade glioma
|
|
LIHC
|
liver hepatocellular carcinoma
|
|
LUAD
|
lung adenocarcinoma
|
|
LUSC
|
lung squamous cell carcinoma
|
|
MESO
|
mesothelioma
|
|
OV
|
ovarian serous cystadenocarcinoma
|
|
PAAD
|
pancreatic adenocarcinoma
|
|
PCPG
|
pheochromocytoma and
paraganglioma
|
|
PRAD
|
prostate adenocarcinoma
|
|
READ
|
rectum adenocarcinoma
|
|
SARC
|
sarcoma
|
|
SKCM
|
skin cutaneous melanoma
|
|
STAD
|
stomach adenocarcinoma
|
|
STES
|
stomach and esophageal carcinoma
|
|
TGCT
|
testicular germ cell tumor
|
|
THCA
|
thyroid carcinoma
|
|
THYM
|
thymoma
|
|
UCEC
|
uterine corpus endometrial
carcinoma
|
|
UCS
|
uterine carcinosarcoma
|
|
UVM
|
uveal melanoma
|
|
TCGA
|
The Cancer Genome Atlas
|
|
GTEx
|
Genotype-Tissue Expression
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
ROC
|
receiver operating
characteristics
|
|
AUC
|
area under the curve
|
|
OS
|
overall survival
|
|
DFS
|
disease-free survival
|
|
DSS
|
disease-specific survival
|
|
PFS
|
progression-free survival
|
|
PFI
|
progression-free interval
|
|
TMB
|
tumor mutation burden
|
|
MSI
|
microsatellite instability
|
|
NEO
|
neoantigens
|
|
CNV
|
copy number variation
|
|
SNV
|
single nucleotide variant
|
|
BP
|
Biological Process
|
|
CC
|
Cellular Component
|
|
MF
|
Molecular Function
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
2
|
Miller KD, Ortiz AP, Pinheiro PS, Bandi P,
Minihan A, Fuchs HE, Tyson DM, Tortolero-Luna G, Fedewa SA, Jemal
AM and Siegel RL: Cancer statistics for the US Hispanic/Latino
population, 2021. CA Cancer J Clin. 71:466–487. 2021.PubMed/NCBI
|
|
3
|
Yang J, Xu R, Wang C, Qiu J, Ren B and You
L: Early screening and diagnosis strategies of pancreatic cancer: A
comprehensive review. Cancer Commun (Lond). 41:1257–1274. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun W, Shi Q, Zhang H, Yang K, Ke Y, Wang
Y and Qiao L: Advances in the techniques and methodologies of
cancer gene therapy. Discov Med. 27:45–55. 2019.PubMed/NCBI
|
|
5
|
Abbott M and Ustoyev Y: Cancer and the
immune system: The history and background of immunotherapy. Semin
Oncol Nurs. 35:1509232019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang Z, Wu L, Wang A, Tang W, Zhao Y, Zhao
H and Teschendorff AE: dbDEMC 2.0: Updated database of
differentially expressed miRNAs in human cancers. Nucleic Acids
Res. 45:D812–D818. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Park J, Cho J and Song EJ:
Ubiquitin-proteasome system (UPS) as a target for anticancer
treatment. Arch Pharm Res. 43:1144–1161. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Johnson DE: The ubiquitin-proteasome
system: Opportunities for therapeutic intervention in solid tumors.
Endocr Relat Cancer. 22:T1–T17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang W, Huang H, Xiao Y, Wang L, Zhang T,
Fang X and Xia X: UBE2T is upregulated, predicts poor prognosis,
and promotes cell proliferation and invasion by promoting
epithelial-mesenchymal transition via inhibiting autophagy in an
AKT/mTOR dependent manner in ovarian cancer. Cell Cycle.
21:780–791. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu W, Xiao L, Cao C, Hua S and Wu D: UBE2T
promotes nasopharyngeal carcinoma cell proliferation, invasion, and
metastasis by activating the AKT/GSK3β/β-catenin pathway.
Oncotarget. 7:15161–15172. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cui P, Li H, Wang C, Liu Y, Zhang M, Yin
Y, Sun Z, Wang Y and Chen X: UBE2T regulates epithelial-mesenchymal
transition through the PI3K-AKT pathway and plays a carcinogenic
role in ovarian cancer. J Ovarian Res. 15:1032022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ueki T, Park JH, Nishidate T, Kijima K,
Hirata K, Nakamura Y and Katagiri T: Ubiquitination and
downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T
overexpression in human breast cancer cells. Cancer Res.
69:8752–8760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Alpi AF, Chaugule V and Walden H:
Mechanism and disease association of E2-conjugating enzymes:
Lessons from UBE2T and UBE2L3. Biochem J. 473:3401–3419. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Alagpulinsa DA, Kumar S, Talluri S,
Nanjappa P, Buon L, Chakraborty C, Samur MK, Szalat R, Shammas MA
and Munshi NC: Amplification and overexpression of E2 ubiquitin
conjugase UBE2T promotes homologous recombination in multiple
myeloma. Blood Adv. 3:3968–3972. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dutta R, Guruvaiah P, Reddi KK, Bugide S,
Bandi DS, Edwards YJK, Singh K and Gupta R: UBE2T promotes breast
cancer tumor growth by suppressing DNA replication stress. NAR
Cancer. 4:zcac0352022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hao P, Kang B, Li Y, Hao W and Ma F: UBE2T
promotes proliferation and regulates PI3K/Akt signaling in renal
cell carcinoma. Mol Med Rep. 20:1212–1220. 2019.PubMed/NCBI
|
|
17
|
Liu Y, Ji W, Yue N and Zhou W:
Ubiquitin-conjugating enzyme E2T promotes tumor stem cell
characteristics and migration of cervical cancer cells by
regulating the GRP78/FAK pathway. Open Life Sci. 16:1082–1090.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Z, Chen N, Liu C, Cao G, Ji Y, Yang W
and Jiang Q: UBE2T is a prognostic biomarker and correlated with
Th2 cell infiltrates in retinoblastoma. Biochem Biophys Res Commun.
614:138–144. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hutter C and Zenklusen JC: The cancer
genome atlas: Creating lasting value beyond its data. Cell.
173:283–285. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48:W509–W514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ghandi M, Huang FW, Jané-Valbuena J,
Kryukov GV, Lo CC, McDonald ER III, Barretina J, Gelfand ET,
Bielski CM, Li H, et al: Next-generation characterization of the
cancer cell line encyclopedia. Nature. 569:503–508. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu CJ, Hu FF, Xia MX, Han L, Zhang Q and
Guo AY: GSCALite: A web server for gene set cancer analysis.
Bioinformatics. 34:3771–3772. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao Y, Zhang M, Pu H, Guo S, Zhang S and
Wang Y: Prognostic implications of pan-cancer CMTM6 expression and
its relationship with the immune microenvironment. Front Oncol.
10:5859612021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yuan H, Yan M, Zhang G, Liu W, Deng C,
Liao G, Xu L, Luo T, Yan H, Long Z, et al: CancerSEA: A cancer
single-cell state atlas. Nucleic Acids Res. 47:D900–D908. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schumacher TN and Schreiber RD:
Neoantigens in cancer immunotherapy. Science. 348:69–74. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yarchoan M, Hopkins A and Jaffee EM: Tumor
mutational burden and response rate to PD-1 inhibition. N Engl J
Med. 377:2500–2501. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tada K, Kitano S, Shoji H, Nishimura T,
Shimada Y, Nagashima K, Aoki K, Hiraoka N, Honma Y, Iwasa S, et al:
Pretreatment immune status correlates with progression-free
survival in chemotherapy-treated metastatic colorectal cancer
patients. Cancer Immunol Res. 4:592–599. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Angelova M, Charoentong P, Hackl H,
Fischer ML, Snajder R, Krogsdam AM, Waldner MJ, Bindea G, Mlecnik
B, Galon J and Trajanoski Z: Characterization of the
immunophenotypes and antigenomes of colorectal cancers reveals
distinct tumor escape mechanisms and novel targets for
immunotherapy. Genome Biol. 16:642015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sanz-Pamplona R, Melas M, Maoz A, Schmit
SL, Rennert H, Lejbkowicz F, Greenson JK, Sanjuan X, Lopez-Zambrano
M, Alonso MH, et al: Lymphocytic infiltration in stage II
microsatellite stable colorectal tumors: A retrospective prognosis
biomarker analysis. PLoS Med. 17:e10032922020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu M, Li X, Huang W, Chen Y, Wang B and
Liu X: Ubiquitin-conjugating enzyme E2T(UBE2T) promotes colorectal
cancer progression by facilitating ubiquitination and degradation
of p53. Clin Res Hepatol Gastroenterol. 45:1014932021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao L, Jiang L, Wang L, He J, Yu H, Sun
G, Chen J, Xiu Q and Li B: UbcH10 expression provides a useful tool
for the prognosis and treatment of non-small cell lung cancer. J
Cancer Res Clin Oncol. 138:1951–1961. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
37
|
Zhu X, Li T, Niu X, Chen L and Ge C:
Identification of UBE2T as an independent prognostic biomarker for
gallbladder cancer. Oncol Lett. 20:442020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yu H, Xiang P, Pan Q, Huang Y, Xie N and
Zhu W: Ubiquitin-Conjugating enzyme E2T is an independent
prognostic factor and promotes gastric cancer progression. Tumour
Biol. 37:11723–11732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Luo M and Zhou Y: Comprehensive analysis
of differentially expressed genes reveals the promotive effects of
UBE2T on colorectal cancer cell proliferation. Oncol Lett.
22:7142021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zheng YW, Gao PF, Ma MZ, Chen Y and Li CY:
Role of ubiquitin-conjugating enzyme E2T in the carcinogenesis and
progression of pancreatic cancer. Oncol Lett. 20:1462–1468. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang X, Liu Y, Leng X, Cao K, Sun W, Zhu J
and Ma J: UBE2T contributes to the prognosis of esophageal squamous
cell carcinoma. Pathol Oncol Res. 27:6325312021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Perez-Peña J, Corrales-Sánchez V, Amir E,
Pandiella A and Ocana A: Ubiquitin-conjugating enzyme E2T (UBE2T)
and denticleless protein homolog (DTL) are linked to poor outcome
in breast and lung cancers. Sci Rep. 7:175302017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang Y, Leng H, Chen H, Wang L, Jiang N,
Huo X and Yu B: Knockdown of UBE2T inhibits osteosarcoma cell
proliferation, migration, and invasion by suppressing the PI3K/Akt
signaling pathway. Oncol Res. 24:361–369. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gong YQ, Peng D, Ning XH, Yang XY, Li XS,
Zhou LQ and Guo YL: UBE2T silencing suppresses proliferation and
induces cell cycle arrest and apoptosis in bladder cancer cells.
Oncol Lett. 12:4485–4492. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang Y and Zhang Z: The history and
advances in cancer immunotherapy: Understanding the characteristics
of tumor-infiltrating immune cells and their therapeutic
implications. Cell Mol Immunol. 17:807–821. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Peng M, Mo Y, Wang Y, Wu P, Zhang Y, Xiong
F, Guo C, Wu X, Li Y, Li X, et al: Neoantigen vaccine: An emerging
tumor immunotherapy. Mol Cancer. 18:1282019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu Y, Zugazagoitia J, Ahmed FS, Henick
BS, Gettinger SN, Herbst RS, Schalper KA and Rimm DL: Immune cell
PD-L1 colocalizes with macrophages and is associated with outcome
in PD-1 pathway blockade therapy. Clin Cancer Res. 26:970–977.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ciardiello D, Vitiello PP, Cardone C,
Martini G, Troiani T, Martinelli E and Ciardiello F: Immunotherapy
of colorectal cancer: Challenges for therapeutic efficacy. Cancer
Treat Rev. 76:22–32. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Topalian SL, Drake CG and Pardoll DM:
Immune checkpoint blockade: A common denominator approach to cancer
therapy. Cancer Cell. 27:450–461. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Baretti M and Le DT: DNA mismatch repair
in cancer. Pharmacol Ther. 189:45–62. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sha D, Jin Z, Budczies J, Kluck K,
Stenzinger A and Sinicrope FA: Tumor mutational burden as a
predictive biomarker in solid tumors. Cancer Discov. 10:1808–1825.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ballhausen A, Przybilla MJ, Jendrusch M,
Haupt S, Pfaffendorf E, Seidler F, Witt J, Sanchez AH, Urban K,
Draxlbauer M, et al: The shared frameshift mutation landscape of
microsatellite-unstable cancers suggests immunoediting during tumor
evolution. Nat Commun. 11:47402020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li J, Wu C, Hu H, Qin G, Wu X, Bai F,
Zhang J, Cai Y, Huang Y, Wang C, et al: Remodeling of the immune
and stromal cell compartment by PD-1 blockade in mismatch
repair-deficient colorectal cancer. Cancer Cell. 41:1152–1169.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xie N, Shen G, Gao W, Huang Z, Huang C and
Fu L: Neoantigens: Promising targets for cancer therapy. Signal
Transduct Target Ther. 8:92023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Shin SJ, Kim SY, Choi YY, Son T, Cheong
JH, Hyung WJ, Noh SH, Park CG and Kim HI: Mismatch repair status of
gastric cancer and its association with the local and systemic
immune response. Oncologist. 24:e835–e844. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hou W, Yi C and Zhu H: Predictive
biomarkers of colon cancer immunotherapy: Present and future. Front
Immunol. 13:10323142022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gulubova MV, Ananiev JR, Vlaykova TI,
Yovchev Y, Tsoneva V and Manolova IM: Role of dendritic cells in
progression and clinical outcome of colon cancer. Int J Colorectal
Dis. 27:159–169. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Veglia F, Perego M and Gabrilovich D:
Myeloid-derived suppressor cells coming of age. Nat Immunol.
19:108–119. 2018. View Article : Google Scholar : PubMed/NCBI
|