Acute myeloid leukemia (AML) is a hematological
malignancy characterized by the aberrant proliferation of
hematopoietic stem cells and is the most prevalent form of acute
leukemia in adults, accounting for ~80% of cases in this group
(1,2). Despite the continuous development of
medical technology, the 5-year survival rate of patients with AML
is 32% (with the rate as high as 50% in younger patients and lower
than 10% in those over 60 years old) and the prognosis remains poor
(3–5). Although traditional cytotoxic
chemotherapy has been the foundation of AML treatment for the past
50 years, researchers are investigating therapeutic approaches
aimed at enhancing patient survival (6,7).
Transcription is a complex biological event
involving the interaction of transcription factors, RNA polymerase
II and transcriptional cofactors with DNA regulatory elements. This
process is key for maintaining essential cell functions such as
cell proliferation, differentiation, and metabolic homeostasis
(8). It converts DNA sequences into
translatable mRNA, which regulates protein synthesis. Organisms
achieve precise gene expression through a sophisticated
transcriptional regulatory network that ensures internal
environmental stability and regulates normal cell development
(9–11). When transcriptional dysregulation
occurs, it can lead to abnormal cell proliferation and
differentiation, which ultimately results in tumors (12). Consequently, small-molecule targeted
therapies aimed at key proteins in transcriptional regulation have
become a focal point of current research (13). Menin protein inhibitors have
emerged, with several preclinical studies confirming their notable
potential as chromatin regulators (14–16).
The menin protein, encoded by the multiple endocrine
neoplasia type 1 gene, functions primarily as a scaffolding protein
that modulates gene transcription via interactions with various
gene regulators, such as histone methyltransferases and
transcription factors including JunD and SMAD (15,17,18).
In AML, menin serves a key role in epigenetic regulation by
interacting with genes such as lysine methyltransferase 2A (KMT2A)
and mutant nucleophosmin 1 (NPM1), thereby facilitating the
progression of AML (19,20). Inhibitors of the menin protein may
reverse aberrant transcription associated with tumors and restore
normal gene expression and cellular function by targeting this
essential epigenetic regulator (21). The present review aimed to provide a
comprehensive evaluation of the role of menin protein, assess the
data supporting its mechanisms in KMT2A/NPM1 mutations and discuss
the pharmacological characteristics and clinical challenges
associated with current inhibitors, such as functional disparities
of menin in various cellular contexts and optimization pathways for
combinatorial therapeutic strategies.
AML is categorized into subtypes based on genetic
and transcriptomic characteristics, one of which is defined by the
homeobox (HOX) gene upregulation and primarily includes NPM1-mutant
(NPM1-mut) and MLL-rearranged leukemia (22). MLL proteins (MLL1 and MLL2) belong
to the histone H3 position 4 lysine (H3K4) methyltransferase
family, which is essential for the maintenance of high HOX gene
expression, and activate myeloid ecotropic viral integration site 1
(MEIS1) through direct epigenetic regulation (16). Notably, MLL fusion proteins contain
specific Su(var)3-9, Enhancer-of-zeste, Trithorax) structural
domains, whereas the crystal structure of menin proteins exhibits a
rectangular conformation, which allows for the formation of deep
binding pockets that bind specifically to the N-terminal fragment
of MLL fusion proteins (23,24).
This interaction designates menin protein as an oncogenic cofactor
that facilitates histone H3 lysine trimethylation at position 4
(H3K4me3) via its engagement with MLL, subsequently enhancing
HOX/MEIS1 gene expression and precipitating leukemia (25). Utilizing a structure-based drug
design approach targeting the menin-MLL interaction interface,
Krivtsov et al (15)
developed a highly selective small molecule inhibitor, VTP50469,
which displaces menin from the fusion protein and prevents the
recruitment of MLL to target genes. Both cell and animal studies
(immunodeficient NSG mice xenotransplanted with human KMT2A-r
leukemia cells) have demonstrated notable anti-leukemic efficacy
(15,26). While preclinical data indicate that
the small molecule inhibitors exhibit potential anti-leukemic
efficacy, the extensive menin-MLL binding interface reveals that
the crystal structure of VTP50469 occupies a subregion of the
menin-MLL interface (15,27). This indicates the necessity for
potentially higher inhibitor concentrations to achieve complete
menin-MLL disruption, consistent with the typical dose-response
association of partial protein-protein interaction inhibitors
(28,29). This technical barrier may represent
a notable impediment in the translation of menin-MLL-targeted
therapies from laboratory research to clinical application.
The sirtuin family comprises seven highly conserved
members (SIRT1-SIRT7) that share evolutionary conservation, all of
which possess highly conserved catalytic structural domains,
differing markedly only at their N- and C-terminus (30,31).
This structure enables each member to exhibit
NAD+-dependent deacetylase and ADP-ribosyltransferase
activity (31). As components of
histone deacetylases, SIRTs serve a key role in cell proliferation,
survival, maintenance of genomic stability and metabolic regulation
by modulating gene expression and chromatin dynamics. A direct
interaction between the menin protein and SIRT1 has been identified
(30). Menin binds directly to the
deacetylase structural domain of SIRT1 through its C-terminal
domain, which forms a functional complex that regulates epigenetic
processes. In mouse hepatocytes, SIRT1 modulates CD36 gene
expression and intracellular triglyceride accumulation via histone
deacetylation, a process that is contingent upon the involvement of
the menin protein (32).
Furthermore, in hepatocellular carcinoma cells, menin enhances
NF-κB (p65) deacetylation by recruiting SIRT1, which underscores
its key role in the SIRT regulatory network (33). Numerous studies have demonstrated
that SIRT1 impacts apoptosis and inflammatory activation by
modulating the NF-κB pathway (34–36).
For example, in murine models, upregulation of SIRT1 promotes B
lymphocyte proliferation, inhibits apoptosis and advances
inflammatory responses by suppressing the NF-κB pathway (37,38).
Therefore, it is hypothesized that in acute leukemia, the
menin-SIRT interaction may also influence apoptosis and
inflammatory activation via the NF-κB pathway, potentially
inhibiting or promoting acute leukemia (39).
The human genome encodes a total of 11 PRMTs, which
regulate cell signaling networks by modifying arginine residues in
both histone and non-histone proteins (40). As important epigenetic regulators in
eukaryotes, all members of the PRMT family contain highly conserved
S-adenosylmethionine (SAM)-dependent methyltransferase structural
domains that facilitate the transfer of methyl groups from SAM to
the nitrogen atom of substrate arginine residues (41,42).
Menin protein can recruit PRMT5 to the growth arrest-specific
protein 1 (GAS1) gene locus, inhibiting GAS1 gene expression by
promoting the symmetric dimethylation of histone H4 at position 3
(H4R3me2) (43). However, in MLL,
the interaction between menin protein and PRMT5 leads to decreased
levels of H4R3me2 and fails to effectively inhibit GAS1 expression
(44,45). This suggests that the regulation of
PRMT by menin protein may be context-dependent (43). In MEN1 tumor syndrome and
mixed-lineage leukemia (MLL), the regulation of GAS1 by the
menin-PRMT5 complex exhibits opposite outcomes, suggesting an
oncogenic isoform of the menin-PRMT5 complex that reverses
epigenetic manifestations by altering the conformation of the
complex (43,44).
SUV39H1 is a histone H3 lysine 9 methyltransferase
(H3K9). Menin protein enhances trimethylation of H3K9 in the
promoter regions of target genes by interacting with SUV39H1,
thereby silencing transcription of associated genes (24,46).
In a previous study on IL-6 gene regulation, Song et al
(47) detected the enrichment of
menin protein and SUV39H1 around the IL-6 gene using chromatin
immunoprecipitation (ChIP). The aforementioned study reported that
menin protein and SUV39H1 are specifically recruited to the
auxiliary region of the IL-6 promoter and the recruitment of
SUV39H1 decreases with the depletion of menin protein, which
suggests menin protein may serve a key role in the regulation of
SUV39H1. Further analysis indicated that menin protein and SUV39H1
may regulate IL-6 gene expression at the protein level through H3K9
methylation (47). Numerous
preclinical and clinical studies have confirmed that IL-6 serves a
notable role in the development of acute leukemia (48–50).
Anti-IL-6 antibodies, such as cetuximab, have potential therapeutic
value in the treatment of various malignancies, including acute
leukemia, either alone or in combination with chemotherapy regimens
(51). However, the pathogenesis of
leukemia involves a complex regulatory network of multiple
signaling pathways - such as JAK/STAT, NF-κB and Wnt/β-catenin -
which limits the therapeutic efficacy of targeting IL-6 alone
(52–54). Following blocking IL-6 signaling,
leukemia cells sustain their survival and proliferative capacity by
activating alternative pathways (for example, Janus kinase/STAT),
which markedly reduces the clinical benefit of single-agent
therapy. These direct epigenetic regulatory interactions mediated
by menin are summarized in Table
I.
SMAD proteins are key downstream effector molecules
in the TGF-β signaling pathway and serve a direct role in TGF-β
signaling and gene transcriptional regulation (58). As transcriptional regulators, menin
protein binds SMAD proteins, thereby indirectly influencing the
activity of the TGF-β signaling pathway and regulating the
transcription of target genes. In pituitary secretory tumor cell
lines, menin protein markedly enhances the binding specificity of
SMAD3 to DNA sequences via interaction with SMAD3 (59,60).
More importantly, during osteoblast differentiation and maturation,
menin protein forms complexes with SMAD1, SMAD5 and the key
regulator of osteogenesis Runt-related transcription factor 2 to
promote the differentiation of mesenchymal stem cells into
osteoblasts (59,61). While the interaction of menin
protein with SMAD1, SMAD3 and SMAD5 has been demonstrated (59), the specific molecular mechanisms
underlying their synergistic activation of transcription warrant
further investigation.
As a key proto-oncogene, the dysregulated expression
of Myc is closely associated with >50% of malignant
tumorigenesis (62). Myc serves
primarily as a transcription factor through the classical
enhancer-box (E-box) region (63).
Wu et al (64) demonstrated
that Myc forms a regulatory complex with positive transcription
elongation factor b (P-TEEb) within the E-box region to activate
transcription, with the menin protein being a key factor in this
process. Menin regulates Myc-mediated transcriptional activity by
influencing the transcriptional elongation regulator P-TEEb. In the
KMT2A rearrangement (KMT2A-r) AML model, the expression of Myc and
its characteristic genes is markedly suppressed in leukemia cells
following treatment with a menin inhibitor (65). Zhou et al (65) found a notable positive correlation
between menin protein levels and Myc expression, which suggests Myc
may serve as a common target of menin protein. Based on these
findings, co-targeting menin protein and Myc may represent a novel
therapeutic strategy for leukemia treatment in future.
The FOX family is an evolutionarily conserved group
of transcription factors, all of which possess the distinctive
forkhead DNA-binding structural domain and are key for cell
proliferation and differentiation (66). Previous studies have indicated that
menin protein engages with several members of the FOX family, such
as FOXG1, FOXA1 and FOXO1 (67–69).
In FOXG1-associated encephalopathy, menin protein influences
α-thalassemia X-linked mental retardation protein-mediated FOXG1
transcription via modulation of the FOXG1 transcripts (67). Bonnavion et al (70) established that menin protein
interacts with FOXA2, which influences its trans-auto reactivation
ability and serves a role in the control of FOXA2 expression in
adult pancreatic α-cells.
The inaugural member of the Wnt family was
identified in 1982 and research has consistently validated the
essential function of the Wnt/β-catenin signaling pathway in
embryonic development and tissue regeneration (71–73),
Dysregulation of this system results in numerous illnesses, such as
colorectal and gastric cancer (74–77).
Menin protein serves as a reciprocal partner of β-catenin proteins,
the principal effector molecules of this signaling pathway, and can
facilitate their nuclear translocation. ChIP and chromosome
conformation capture (3C) studies demonstrated that the menin
protein augments the association of β-catenin proteins with the Myc
promoter (78–80). The mechanism of action of menin
protein inhibitors in the treatment of AML may partially arise from
their suppression of the Wnt/β-catenin protein signaling pathway
(81–83).
Nuclear receptors are a class of receptor proteins
located in the nucleus, which include the androgen receptor (AR),
estrogen, thyroid hormone, glucocorticoid, retinoid X, peroxisome
proliferator-activated, liver X and retinoic acid receptors
(84). These receptors not only
serve as biosensors to regulate key cellular activities such as
proliferation, differentiation, and apoptosis but also directly
bind to DNA to exert transcriptional regulation (85,86).
The interactions between nuclear receptors and tumor growth have
attracted notable research regarding their role in tumor
progression (87–89). Luo et al (90) identified that the menin protein
exhibits distinct pro-oncogenic actions in androgen
receptor-dependent prostate cancer cells by modulating AR
transcription and its target genes. Based on the presence of
nuclear receptor interaction sites in the amino acid sequence of
the menin protein, menin serves as an important coactivator of
nuclear receptor-mediated transcription (68,90).
The NF-κB family comprises five members: Rel or
c-Rel, RelA or p65, RelB, NF-κB1 or p50 and NF-κB2 or p52. Each
member exists in dimeric form and possesses a Rel homology domain
(91). The pro-carcinogenic role of
NF-κB is prevalent in hepatocellular carcinoma (92). Previous studies have demonstrated
that menin protein engages with NF-κB and suppresses p65-mediated
transcriptional activation via the recruitment of SIRT1 (33,93).
Another previous study revealed that the degree of menin-NF-κB
interaction changes the production of cell cycle protein D1, a
downstream signaling molecule of NF-κB that governs the G1/S phase
transition and affects cell proliferation (55). Irregularities in this mechanism may
result in tumorigenesis.
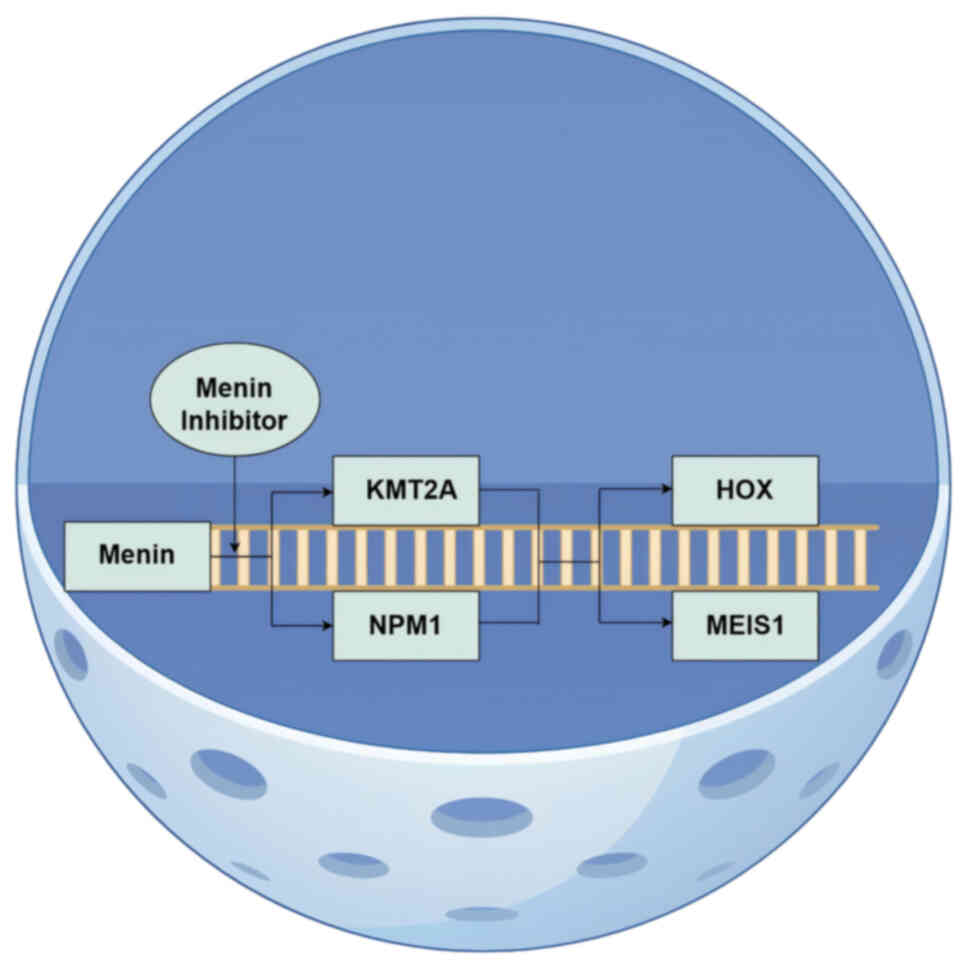
The menin-MLL complex activates the expression of
the HOX/MEIS1 gene cluster by mediating aberrant H3K4me3, a
well-established axis of epigenetic regulation implicated in
KMT2A-r and NPM1-mut leukemia (Fig.
1 (17,94). This mechanism was first elucidated
by Yokoyama et al (94), who
demonstrated that blocking the interaction between menin and the
methyltransferase KMT2A markedly decreases leukemia incidence in
mice (95). Further research has
confirmed the key role of menin protein in NPM1-mut leukemia
(96). Specifically, menin protein
contributes to KMT2A-r leukemia by binding to fusion proteins and
to NPM1-mut leukemias via aberrant nucleoplasmic transport pathways
(97). The role of the menin
protein in KMT2A-rearranged leukemia and NPM1-mutant leukemia cause
the dysregulated expression of the HOX and MEIS1 genes (98,99).
Menin inhibitors effectively suppress the abnormal proliferation
and differentiation of leukemia cells by specifically disrupting
the interaction between menin and KMT2A or NPM1, while
simultaneously downregulating the expression of HOX and MEIS1,
thereby demonstrating notable therapeutic potential (21).
Patients with acute leukemia characterized by
KMT2A-r, previously referred to as MLL, exhibit a long-term
survival rate <60% and poor prognosis across all age
demographics (100,101). This leukemia variant exhibits a
high prevalence among infants and children, accounting for >70%
of new acute lymphoblastic leukemia (ALL) diagnoses in infants, is
marked by notable aggressiveness, frequent relapse, substantial
medication resistance and presents considerable challenges for
therapeutic management as a high-risk genetic subtype (101).
KMT2A-r may lead to aberrant expression of the HOX
gene and its DNA-binding cofactor MEIS1, which may inhibit
hematological development and precipitate leukemia (21,102).
Although no specific medications have been approved for KMT2A-r
leukemia, preclinical studies have identified the chromatin
regulatory protein menin as a promising therapeutic target
(14,103,104). In KMT2A-driven leukemia, all KMT2A
fusion proteins contain menin-binding sequences, with menin protein
serving as a key cofactor that facilitates the interaction between
the KMT2A protein complex and the HOX gene promoter. In a study
using the KMT2A-mut leukemia model (105), it was demonstrated that the
inhibition of menin protein markedly reduces the transcript levels
of HOX and MEIS1, thereby reversing the leukemogenesis process.
Through the examination of gene expression in
pediatric and adult patients with primary AML, researchers
discovered that NPM1-mut leukemia exhibits notable similarities to
the KMT2A-r subtype and NPM1 is associated with the HOX/MEIS1 gene
cluster (106). NPM1-mut is among
the most prevalent genetic alterations in AML, affecting ~30% of
the total patient population (107). These mutations, primarily located
in the terminal exons of the NPM1 gene, enhance nuclear export
signaling activity and impair nucleolus localization signaling,
which results in the dysregulated expression of the HOXA/B and
MEIS1 genes (108). NPM1 may serve
as a transcriptional amplifier of gene expression, potentially
constituting a notable factor in the development of AML (109). Dillon et al (110) identified microscopic residual
lesions in patients with AML through pre-transplantation DNA
sequencing of hematopoietic stem cells, which revealed that
patients with NPM1-mut AML experience a significantly higher
recurrence rate and shorter survival compared with those without
NPM1-mut.
The persistence of NPM1-mut AML in an
undifferentiated state is attributed to the HOX-associated pathway,
which underscores the therapeutic potential of targeting this
pathway. Menin protein serves as a cofactor to promote H3K4me3
through interactions with MLL, thereby modulating the expression of
HOX and MEIS1 genes. This mechanism highlights the feasibility of
targeting menin protein for therapeutic interventions (96). In vivo experiments have
demonstrated that inhibitors of menin protein exhibit notable
anti-leukemic activity in NPM1-mut leukemia (20,21,111).
Previous studies conducted in NPM1-mutant leukemia models have
indicated that treatment with the menin inhibitor VTP50469, a
precursor drug to revumenib, leads to downregulation of oncogenic
cofactors such as MEIS1 and a marked reduction in the self-renewal
capacity of leukemic stem cells (15,104).
Understanding of the menin formation mechanism in
leukemia, alongside advancements in high-throughput screening
techniques and structural biology, facilitate creation of highly
selective small chemical inhibitors (112). Based on the efficacy of menin
inhibitors in KMT2A-r and NPM1-mut leukemia, a growing array of
menin inhibitors (such as revumenib and ziftomenib) exhibiting
enhanced pharmacological efficacy against these AML subtypes has
recently been introduced into clinical practice (21,113,114).
A total of seven menin inhibitors are undergoing
different phases of clinical development for acute myeloid
leukemia, with numerous candidates in development (Table II). The leading candidate is
revumenib, which demonstrated a promising safety and effectiveness
profile in a phase I open-label, dose-escalation and extension
study (AUGMENT-101) assessing the menin inhibitor revumenib for
KMT2A-r leukemia treatment (21,113).
The occurrence of grade ≤3 treatment-related side events was
minimal in treated patients, with asymptomatic QT interval
prolongation being the sole dose-limiting effect. Revumenib
achieved an overall remission rate of ≤53% and a complete remission
rate of ≤30% with partial hematological recovery (98,113).
In the subsequent phase II trial (AUGMENT-101), a total of 94
patients with KMT2A-r acute leukemia (comprising 78 patients with
AML, 14 with ALL and two with an indeterminate subtype) received
menin inhibitors, which achieved an overall remission rate of 63.2%
(of which, 68.2% exhibited no measurable residual disease;
unpublished data). In 57 patients with assessable efficacy, the
combined complete response and complete response with incomplete
hematological recovery rate reached 22.8% (113). Grade ≥3 adverse events included
neutropenia (37.2%), differentiation syndrome (16%) and QT interval
prolongation (13.8%). Most of these events were controllable and
transitory and menin inhibitors had a predictable safety
profile.
The combination therapy of menin inhibitors with
other targeted leukemia medications has potential due to the
promising efficacy and safety profile of menin inhibitor
monotherapy in the treatment of acute leukemia. Miao et al
(115) administered a menin
inhibitor and kinase inhibitor to NUP98-r leukemia samples, which
demonstrated that the combination therapy outperformed monotherapy,
with a combination index between 0.12 and 0.65, as determined by
the Chou-Talalay method, which indicated notable synergistic
effects. The combinatorial therapy induced a more pronounced
decrease in both the quantity and size of leukemic blasts in
NUP98-r leukemia specimens and was more effective in promoting cell
proliferation arrest and differentiation. Furthermore, the
combination of Brahma-related gene 1/Brahma inhibitors with menin
inhibitors for acute leukemia treatment has demonstrated notable
preclinical efficacy, resulting in a more substantial reduction in
leukemia burden and extended survival duration in mice compared
with monotherapy (116). A
clinical trial is currently examining menin inhibitors in
conjunction with azacitidine/vincristine for the treatment of acute
leukemia, specifically evaluating JNJ-75276617 with AML-targeted
treatments (trial no. NCT05453903) (117). The combined regimen inhibits the
immune evasion of acute leukemia cells following monotherapy and
diminishes relapse in treatment-resistant leukemia (117).
Common adverse effects of menin inhibitors include
gastrointestinal reactions, QT interval prolongation, cytopenia and
differentiation syndromes, however, these adverse effects are
within manageable limits and menin inhibitors are generally safe
(113,118). Furthermore, menin inhibitors are
not associated with notable off-target toxicity, which has enabled
the clinical scale-up of menin inhibitors and their emergence as a
potential option for long-term therapy (112). However, recent data have also
demonstrated that certain patients have menin mutations that
prevent binding of the inhibitor and thus mediate clinical
resistance, which leads to clinical relapse; therefore, resistance
to menin inhibitors remains a challenge (21,119).
Although menin inhibitors have demonstrated
significant efficacy in KMT2A-r and NPM1-mut leukemia, their
clinical application faces key challenges such as drug resistance
and optimization of combination strategies (21,98).
Future research should focus on overcoming resistance mediated by
menin protein mutations, including the development of allosteric
inhibitors and combination with epigenetic regulatory drugs to
block compensatory pathways. In terms of combination therapy, it is
necessary to optimize combination regimens with other drugs based
on synergy indices (such as the Chou-Talalay model) and explore the
potential advantages of sequential therapy. Furthermore, the
indication scope of menin inhibitors should be expanded to include
other subtypes dependent on the HOX/MEIS1 pathway, such as NUP98-r
leukemia and predictive biomarkers based on HOX gene expression
profiles or menin-MLL complex activity should be developed to
screen beneficiary populations. From a technical perspective,
structural biology and artificial intelligence-assisted design
should be leveraged to develop high-affinity inhibitors and
targeted delivery systems should be developed to enhance efficacy
and safety. In-depth studies of the menin protein regulatory
network may reveal its functional heterogeneity in different cell
environments and provide a theoretical basis for dual-targeting
strategies (such as menin-Myc co-inhibition). With the advancement
of multidisciplinary collaboration, menin inhibitors may become a
key therapy for specific leukemia subtypes and provide novel
directions for epigenetically targeted therapy.
Not applicable.
The present study was supported by the National Natural Science
Foundation of China (grant no. 81600129), Zhejiang Provincial
Natural Science Foundation of China (grant nos. LY21H080001 and
BY22H205675), the Medical and Health Research Project of Zhejiang
Province (grant nos. WKJ-ZJ-2444 and 2022KY944) and Zhejiang
Provincial Traditional Chinese Medicine Science and Technology
Project (grant no. 2022ZB276).
Not applicable.
HZ conceived, designed and supervised the study and
edited the manuscript. JB wrote the manuscript. Data authentication
is not applicable. Both authors have read and approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Pollyea DA, Bixby D, Perl A, Bhatt VR,
Altman JK, Appelbaum FR, de Lima M, Fathi AT, Foran JM, Gojo I, et
al: NCCN guidelines insights: Acute myeloid leukemia, version
2.2021. J Natl Compr Canc Netw. 19:16–27. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
De Kouchkovsky I and Abdul-Hay M: Acute
myeloid leukemia: A comprehensive review and 2016 update. Blood
Cancer J. 6:e4412016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shimony S, Stahl M and Stone RM: Acute
myeloid leukemia: 2025 update on diagnosis, risk-stratification,
and management. Am J Hematol. 100:860–891. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sasaki K, Ravandi F, Kadia TM, DiNardo CD,
Short NJ, Borthakur G, Jabbour E and Kantarjian HM: De novo acute
myeloid leukemia: A population-based study of outcome in the United
States based on the surveillance, epidemiology, and end results
(SEER) database, 1980 to 2017. Cancer. 127:2049–2061. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fruchtman H, Avigan ZM, Waksal JA, Brennan
N and Mascarenhas JO: Management of isocitrate dehydrogenase 1/2
mutated acute myeloid leukemia. Leukemia. 38:927–935. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Short NJ, Konopleva M, Kadia TM, Borthakur
G, Ravandi F, DiNardo CD and Daver N: Advances in the treatment of
acute myeloid leukemia: New drugs and new challenges. Cancer
Discov. 10:506–525. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Carter JL, Hege K, Yang J, Kalpage HA, Su
Y, Edwards H, Hüttemann M, Taub JW and Ge Y: Targeting multiple
signaling pathways: The new approach to acute myeloid leukemia
therapy. Signal Transduct Target Ther. 5:2882020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pei H, Guo W, Peng Y, Xiong H and Chen Y:
Targeting key proteins involved in transcriptional regulation for
cancer therapy: Current strategies and future prospective. Med Res
Rev. 42:1607–1660. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Milano L, Gautam A and Caldecott KW: DNA
damage and transcription stress. Mol Cell. 84:70–79. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Morgan MP, Finnegan E and Das S: The role
of transcription factors in the acquisition of the four latest
proposed hallmarks of cancer and corresponding enabling
characteristics. Semin Cancer Biol. 86:1203–1215. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vervoort SJ, Devlin JR, Kwiatkowski N,
Teng M, Gray NS and Johnstone RW: Targeting transcription cycles in
cancer. Nat Rev Cancer. 22:5–24. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Layden HM, Johnson AE and Hiebert SW:
Chemical-genetics refines transcription factor regulatory circuits.
Trends Cancer. 10:65–75. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kathman SG, Koo SJ, Lindsey GL, Her HL,
Blue SM, Li H, Jaensch S, Remsberg JR, Ahn K, Yeo GW, et al:
Remodeling oncogenic transcriptomes by small molecules targeting
NONO. Nat Chem Biol. 19:825–836. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Appel LM, Benedum J, Engl M, Platzer S,
Schleiffer A, Strobl X and Slade D: SPOC domain proteins in health
and disease. Genes Dev. 37:140–170. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Krivtsov AV, Evans K, Gadrey JY, Eschle
BK, Hatton C, Uckelmann HJ, Ross KN, Perner F, Olsen SN, Pritchard
T, et al: A Menin-MLL inhibitor induces specific chromatin changes
and eradicates disease in models of MLL-rearranged leukemia. Cancer
Cell. 36:660–673.e11. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kühn MW, Song E, Feng Z, Sinha A, Chen CW,
Deshpande AJ, Cusan M, Farnoud N, Mupo A, Grove C, et al: Targeting
chromatin regulators inhibits leukemogenic gene expression in NPM1
mutant leukemia. Cancer Discov. 6:1166–1181. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cuglievan B, Kantarjian H, Rubnitz JE,
Cooper TM, Zwaan CM, Pollard JA, DiNardo CD, Kadia TM, Guest E,
Short NJ, et al: Menin inhibitors in pediatric acute leukemia: A
comprehensive review and recommendations to accelerate progress in
collaboration with adult leukemia and the international community.
Leukemia. 38:2073–2084. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Agarwal SK, Guru SC, Heppner C, Erdos MR,
Collins RM, Park SY, Saggar S, Chandrasekharappa SC, Collins FS,
Spiegel AM, et al: Menin interacts with the AP1 transcription
factor JunD and represses JunD-activated transcription. Cell.
96:143–152. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang J, Gurung B, Wan B, Matkar S,
Veniaminova NA, Wan K, Merchant JL, Hua X and Lei M: The same
pocket in menin binds both MLL and JUND but has opposite effects on
transcription. Nature. 482:542–546. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fiskus W, Mill CP, Birdwell C, Davis JA,
Das K, Boettcher S, Kadia TM, DiNardo CD, Takahashi K, Loghavi S,
et al: Targeting of epigenetic co-dependencies enhances anti-AML
efficacy of Menin inhibitor in AML with MLL1-r or mutant NPM1.
Blood Cancer J. 13:532023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Issa GC, Aldoss I, DiPersio J, Cuglievan
B, Stone R, Arellano M, Thirman MJ, Patel MR, Dickens DS, Shenoy S,
et al: The menin inhibitor revumenib in KMT2A-rearranged or
NPM1-mutant leukaemia. Nature. 615:920–924. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gundry MC, Goodell MA and Brunetti L: It's
all about MEis: Menin-MLL inhibition eradicates NPM1-Mutated and
MLL-rearranged acute leukemias in mice. Cancer Cell. 37:267–269.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Borkin D, He S, Miao H, Kempinska K,
Pollock J, Chase J, Purohit T, Malik B, Zhao T, Wang J, et al:
Pharmacologic inhibition of the Menin-MLL interaction blocks
progression of MLL leukemia in vivo. Cancer Cell. 27:589–602. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Feng Z, Ma J and Hua X: Epigenetic
regulation by the menin pathway. Endocr Relat Cancer. 24:T147–T159.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yokoyama A and Cleary ML: Menin critically
links MLL proteins with LEDGF on cancer-associated target genes.
Cancer Cell. 14:36–46. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Adriaanse FRS, Schneider P,
Arentsen-Peters STCJM, Fonseca AMND, Stutterheim J, Pieters R,
Zwaan CM and Stam RW: Distinct responses to menin inhibition and
synergy with DOT1L inhibition in KMT2A-rearranged acute
lymphoblastic and myeloid leukemia. Int J Mol Sci. 25:60202024.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kurmasheva RT, Bandyopadhyay A, Favours E,
Pozo VD, Ghilu S, Phelps DA, McGeehan GM, Erickson SW, Smith MA and
Houghton PJ: Evaluation of VTP-50469, a menin-MLL1 inhibitor,
against Ewing sarcoma xenograft models by the pediatric preclinical
testing consortium. Pediatr Blood Cancer. 67:e282842020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nicholls SJ, Nissen SE, Fleming C, Urva S,
Suico J, Berg PH, Linnebjerg H, Ruotolo G, Turner PK and Michael
LF: Muvalaplin, an oral small molecule inhibitor of lipoprotein(a)
formation: A randomized clinical trial. JAMA. 330:1042–1053. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Groenland SL, Martínez-Chávez A, van
Dongen MGJ, Beijnen JH, Schinkel AH, Huitema ADR and Steeghs N:
Clinical pharmacokinetics and pharmacodynamics of the
cyclin-dependent kinase 4 and 6 inhibitors palbociclib, ribociclib,
and abemaciclib. Clin Pharmacokinet. 59:1501–1520. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ianni A, Kumari P, Tarighi S, Braun T and
Vaquero A: SIRT7: A novel molecular target for personalized cancer
treatment? Oncogene. 43:993–1006. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Goes JVC, Carvalho LG, de Oliveira RTG,
Melo MML, Novaes LAC, Moreno DA, Gonçalves PG, Montefusco-Pereira
CV, Pinheiro RF and Ribeiro Junior HL: Role of sirtuins in the
pathobiology of onco-hematological diseases: A PROSPERO-registered
study and in silico analysis. Cancers (Basel). 14:46112022.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cao Y, Xue Y, Xue L, Jiang X, Wang X,
Zhang Z, Yang J, Lu J, Zhang C, Wang W and Ning G: Hepatic menin
recruits SIRT1 to control liver steatosis through histone
deacetylation. J Hepatol. 59:1299–1306. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gang D, Hongwei H, Hedai L, Ming Z, Qian H
and Zhijun L: The tumor suppressor protein menin inhibits
NF-κB-mediated transactivation through recruitment of Sirt1 in
hepatocellular carcinoma. Mol Biol Rep. 40:2461–2466. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hernández-Jiménez M, Hurtado O, Cuartero
MI, Ballesteros I, Moraga A, Pradillo JM, McBurney MW, Lizasoain I
and Moro MA: Silent information regulator 1 protects the brain
against cerebral ischemic damage. Stroke. 44:2333–2337. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Teng Y, Huang Y, Yu H, Wu C, Yan Q, Wang
Y, Yang M, Xie H, Wu T, Yang H and Zou J: Nimbolide targeting SIRT1
mitigates intervertebral disc degeneration by reprogramming
cholesterol metabolism and inhibiting inflammatory signaling. Acta
Pharm Sin B. 13:2269–2280. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao X, Li M, Lu Y, Wang M, Xiao J, Xie Q,
He X and Shuai S: Sirt1 inhibits macrophage polarization and
inflammation in gouty arthritis by inhibiting the MAPK/NF-κB/AP-1
pathway and activating the Nrf2/HO-1 pathway. Inflamm Res.
73:1173–1184. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Q, Yan C, Xin M, Han L, Zhang Y and
Sun M: Sirtuin 1 (Sirt1) overexpression in BaF3 cells contributes
to cell proliferation promotion, apoptosis resistance and
pro-inflammatory cytokine production. Med Sci Monit. 23:1477–1482.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kotas ME, Gorecki MC and Gillum MP:
Sirtuin-1 is a nutrient-dependent modulator of inflammation.
Adipocyte. 2:113–118. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu QJ, Zhang TN, Chen HH, Yu XF, Lv JL,
Liu YY, Liu YS, Zheng G, Zhao JQ, Wei YF, et al: The sirtuin family
in health and disease. Signal Transduct Target Ther. 7:4022022.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Alinari L, Mahasenan KV, Yan F, Karkhanis
V, Chung JH, Smith EM, Quinion C, Smith PL, Kim L, Patton JT, et
al: Selective inhibition of protein arginine methyltransferase 5
blocks initiation and maintenance of B-cell transformation. Blood.
125:2530–2543. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Peng J, Ni B, Li D, Cheng B and Yang R:
Overview of the PRMT6 modulators in cancer treatment: Current
progress and emerged opportunity. Eur J Med Chem. 279:1168572024.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Qian K, Hu H, Xu H and Zheng YG: Detection
of PRMT1 inhibitors with stopped flow fluorescence. Signal
Transduct Target Ther. 3:62018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Abe Y and Tanaka N: Fine-Tuning of GLI
activity through arginine methylation: Its mechanisms and function.
Cells. 9:19732020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gurung B, Feng Z, Iwamoto DV, Thiel A, Jin
G, Fan CM, Ng JM, Curran T and Hua X: Menin epigenetically
represses Hedgehog signaling in MEN1 tumor syndrome. Cancer Res.
73:2650–2658. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kim H and Ronai ZA: PRMT5 function and
targeting in cancer. Cell Stress. 4:199–215. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Padeken J, Methot SP and Gasser SM:
Establishment of H3K9-methylated heterochromatin and its functions
in tissue differentiation and maintenance. Nat Rev Mol Cell Biol.
23:623–640. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Song TY, Lim J, Kim B, Han JW, Youn HD and
Cho EJ: The role of tumor suppressor menin in IL-6 regulation in
mouse islet tumor cells. Biochem Biophys Res Commun. 51:308–313.
2014. View Article : Google Scholar
|
|
48
|
Mei Y, Ren K, Liu Y, Ma A, Xia Z, Han X,
Li E, Tariq H, Bao H, Xie X, et al: Bone marrow-confined IL-6
signaling mediates the progression of myelodysplastic syndromes to
acute myeloid leukemia. J Clin Invest. 132:e1526732022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Burger R: Impact of interleukin-6 in
hematological malignancies. Transfus Med Hemother. 40:336–343.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kaser EC, Zhao L, D'Mello KP, Zhu Z, Xiao
H, Wakefield MR, Bai Q and Fang Y: The role of various interleukins
in acute myeloid leukemia. Med Oncol. 38:552021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yao X, Huang J, Zhong H, Shen N, Faggioni
R, Fung M and Yao Y: Targeting interleukin-6 in inflammatory
autoimmune diseases and cancers. Pharmacol Ther. 141:125–139. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Qin R, Wang T, He W, Wei W, Liu S, Gao M
and Huang Z: Jak2/STAT6/c-Myc pathway is vital to the pathogenicity
of Philadelphia-positive acute lymphoblastic leukemia caused by
P190(BCR-ABL). Cell Commun Signal. 21:272023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Di Francesco B, Verzella D, Capece D,
Vecchiotti D, Di Vito Nolfi M, Flati I, Cornice J, Di Padova M,
Angelucci A, Alesse E and Zazzeroni F: NF-κB: A druggable target in
acute myeloid leukemia. Cancers (Basel). 14:35572022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Láinez-González D, Alonso-Aguado AB and
Alonso-Dominguez JM: Understanding the Wnt signaling pathway in
acute myeloid leukemia stem cells: A feasible key against relapses.
Biology (Basel). 12:6832023.PubMed/NCBI
|
|
55
|
Liu P, Shi C, Qiu L, Shang D, Lu Z, Tu Z
and Liu H: Menin signaling and therapeutic targeting in breast
cancer. Curr Probl Cancer. 51:1011182024. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Paneni F, Osto E, Costantino S, Mateescu
B, Briand S, Coppolino G, Perna E, Mocharla P, Akhmedov A, Kubant
R, et al: Deletion of the activated protein-1 transcription factor
JunD induces oxidative stress and accelerates age-related
endothelial dysfunction. Circulation. 127:1229–1240. e1–e21. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gallo A, Cuozzo C, Esposito I, Maggiolini
M, Bonofiglio D, Vivacqua A, Garramone M, Weiss C, Bohmann D and
Musti AM: Menin uncouples Elk-1, JunD and c-Jun phosphorylation
from MAP kinase activation. Oncogene. 21:6434–6445. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dockray GJ: Keeping neuroendocrine cells
in check: Roles for TGFbeta, Smads, and menin? Gut. 52:1237–1239.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hendy GN, Kaji H, Sowa H, Lebrun JJ and
Canaff L: Menin and TGF-beta superfamily member signaling via the
Smad pathway in pituitary, parathyroid and osteoblast. Horm Metab
Res. 37:375–379. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Matkar S, Thiel A and Hua X: Menin: A
scaffold protein that controls gene expression and cell signaling.
Trends Biochem Sci. 38:394–402. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sowa H, Kaji H, Hendy GN, Canaff L, Komori
T, Sugimoto T and Chihara K: Menin is required for bone
morphogenetic protein 2- and transforming growth factor
beta-regulated osteoblastic differentiation through interaction
with Smads and Runx2. J Biol Chem. 279:40267–40275. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen H, Liu H and Qing G: Targeting
oncogenic Myc as a strategy for cancer treatment. Signal Transduct
Target Ther. 3:52018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chen X, Xu H, Yuan P, Fang F, Huss M, Vega
VB, Wong E, Orlov YL, Zhang W, Jiang J, et al: Integration of
external signaling pathways with the core transcriptional network
in embryonic stem cells. Cell. 133:1106–1117. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wu G, Yuan M, Shen S, Ma X, Fang J, Zhu L,
Sun L, Liu Z, He X, Huang D, et al: Menin enhances c-Myc-mediated
transcription to promote cancer progression. Nat Commun.
8:152782017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhou X, Zhang L, Aryal S, Veasey V, Tajik
A, Restelli C, Moreira S, Zhang P, Zhang Y, Hope KJ, et al:
Epigenetic regulation of noncanonical menin targets modulates menin
inhibitor response in acute myeloid leukemia. Blood. 144:2018–2032.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Tsai JW, Cejas P, Wang DK, Patel S, Wu DW,
Arounleut P, Wei X, Zhou N, Syamala S, Dubois FPB, et al: FOXR2 is
an epigenetically regulated pan-cancer oncogene that activates ETS
transcriptional circuits. Cancer Res. 82:2980–3001. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhuang K, Leng L, Su X, Wang S, Su Y, Chen
Y, Yuan Z, Zi L, Li J, Xie W, et al: Menin deficiency induces
autism-like behaviors by regulating foxg1 transcription and
participates in foxg1-related encephalopathy. Adv Sci (Weinh).
11:e23079532024. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Dreijerink KMA, Groner AC, Vos ESM,
Font-Tello A, Gu L, Chi D, Chi D, Reyes J, Cook J, Lim E, et al:
Enhancer-mediated oncogenic function of the menin tumor suppressor
in breast cancer. Cell Rep. 18:2359–2372. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Jiang Z, Shi D, Tu Y, Tian J, Zhang W,
Xing B, Wang J, Liu S, Lou J, Gustafsson JÅ, et al: Human proislet
peptide promotes pancreatic progenitor cells to ameliorate diabetes
through FOXO1/menin-mediated epigenetic regulation. Diabetes.
67:1345–1355. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Bonnavion R, Teinturier R, Gherardi S,
Leteurtre E, Yu R, Cordier-Bussat M, Du R, Pattou F, Vantyghem MC,
Bertolino P, et al: Foxa2, a novel protein partner of the tumour
suppressor menin, is deregulated in mouse and human MEN1
glucagonomas. J Pathol. 242:90–101. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Nusse R and Clevers H: Wnt/β-catenin
signaling, disease, and emerging therapeutic modalities. Cell.
169:985–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Bonnet C, Brahmbhatt A, Deng SX and Zheng
JJ: Wnt signaling activation: Targets and therapeutic opportunities
for stem cell therapy and regenerative medicine. RSC Chem Biol.
2:1144–1157. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Steinhart Z and Angers S: Wnt signaling in
development and tissue homeostasis. Development. 145:dev1465892018.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liu J, Xiao Q, Xiao J, Niu C, Li Y, Zhang
X, Zhou Z, Shu G and Yin G: Wnt/β-catenin signalling: function,
biological mechanisms, and therapeutic opportunities. Signal
Transduct Target Ther. 7:32022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Xiang Z, Wang Y, Ma X, Song S, He Y, Zhou
J, Feng L, Yang S, Wu Y, Yu B, et al: Targeting the
NOTCH2/ADAM10/TCF7L2 Axis-mediated transcriptional regulation of
Wnt pathway suppresses tumor growth and enhances chemosensitivity
in colorectal cancer. Adv Sci (Weinh). 12:e24057582025. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Hao J, Liu C, Gu Z, Yang X, Lan X and Guo
X: Dysregulation of Wnt/β-catenin signaling contributes to
intestinal inflammation through regulation of group 3 innate
lymphoid cells. Nat Commun. 15:28202024. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Feng Q, Nie F, Gan L, Wei X, Liu P, Liu H,
Zhang K, Fang Z, Wang H and Fang N: Tripartite motif 31 drives
gastric cancer cell proliferation and invasion through activating
the Wnt/β-catenin pathway by regulating Axin1 protein stability.
Sci Rep. 13:200992023. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Luo Y, Vlaeminck-Guillem V, Baron S,
Dallel S, Zhang CX and Le Romancer M: MEN1 silencing aggravates
tumorigenic potential of AR-independent prostate cancer cells
through nuclear translocation and activation of JunD and β-catenin.
J Exp Clin Cancer Res. 40:2702021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Hagège H, Klous P, Braem C, Splinter E,
Dekker J, Cathala G, de Laat W and Forné T: Quantitative analysis
of chromosome conformation capture assays (3C-qPCR). Nat Protoc.
2:1722–1733. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Sancho A, Li S, Paul T, Zhang F, Aguilo F,
Vashisht A, Balasubramaniyan N, Leleiko NS, Suchy FJ, Wohlschlegel
JA, et al: CHD6 regulates the topological arrangement of the CFTR
locus. Hum Mol Genet. 24:2724–2732. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wang Y, Krivtsov AV, Sinha AU, North TE,
Goessling W, Feng Z, Zon LI and Armstrong SA: The Wnt/beta-catenin
pathway is required for the development of leukemia stem cells in
AML. Science. 327:1650–1653. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wagstaff M, Coke B, Hodgkiss GR and Morgan
RG: Targeting β-catenin in acute myeloid leukaemia: Past present,
and future perspectives. Biosci Rep. 42:2022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Khan I, Eklund EE and Gartel AL:
Therapeutic vulnerabilities of transcription factors in AML. Mol
Cancer Ther. 20:229–237. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Font-Díaz J, Jiménez-Panizo A, Caelles C,
Vivanco MD, Pérez P, Aranda A, Estébanez-Perpiñá E, Castrillo A,
Ricote M and Valledor AF: Nuclear receptors: Lipid and hormone
sensors with essential roles in the control of cancer development.
Semin Cancer Biol. 73:58–75. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Yang Z, Gimple RC, Zhou N, Zhao L,
Gustafsson J and Zhou S: Targeting nuclear receptors for cancer
therapy: Premises, promises, and challenges. Trends Cancer.
7:541–556. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Lian F, Wang Y, Xiao Y, Wu X, Xu H, Liang
L and Yang X: Activated farnesoid X receptor attenuates apoptosis
and liver injury in autoimmune hepatitis. Mol Med Rep.
12:5821–5827. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Xu Y, Huangyang P, Wang Y, Xue L,
Devericks E, Nguyen HG, Yu X, Oses-Prieto JA, Burlingame AL,
Miglani S, et al: ERα is an RNA-binding protein sustaining tumor
cell survival and drug resistance. Cell. 184:5215–5229.e17. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Manickasamy MK, Jayaprakash S, Girisa S,
Kumar A, Lam HY, Okina E, Eng H, Alqahtani MS, Abbas M, Sethi G, et
al: Delineating the role of nuclear receptors in colorectal cancer,
a focused review. Discov Oncol. 15:412024. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Sun Y, Xie J, Cai S, Wang Q, Feng Z, Li Y,
Lu JJ, Chen W and Ye Z: Elevated expression of nuclear
receptor-binding SET domain 3 promotes pancreatic cancer cell
growth. Cell Death Dis. 12:9132021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Luo Y, Vlaeminck-Guillem V, Teinturier R,
Abou Ziki R, Bertolino P, Le Romancer M and Zhang CX: The scaffold
protein menin is essential for activating the MYC locus and
MYC-mediated androgen receptor transcription in androgen
receptor-dependent prostate cancer cells. Cancer Commun (Lond).
41:1427–1430. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhang T, Ma C, Zhang Z, Zhang H and Hu H:
NF-κB signaling in inflammation and cancer. MedComm (2020).
2:618–653. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
He G and Karin M: NF-κB and STAT3 - key
players in liver inflammation and cancer. Cell Res. 21:159–168.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Yeung F, Hoberg JE, Ramsey CS, Keller MD,
Jones DR, Frye RA and Mayo MW: Modulation of NF-kappaB-dependent
transcription and cell survival by the SIRT1 deacetylase. EMBO J.
23:2369–2380. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yokoyama A, Somervaille TC, Smith KS,
Rozenblatt-Rosen O, Meyerson M and Cleary ML: The menin tumor
suppressor protein is an essential oncogenic cofactor for
MLL-associated leukemogenesis. Cell. 123:207–218. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Mullard A: FDA approves first biparatopic
antibody therapy. Nat Rev Drug Discov. 24:72025. View Article : Google Scholar
|
|
96
|
Falini B, Brunetti L, Sportoletti P and
Martelli MP: NPM1-mutated acute myeloid leukemia: From bench to
bedside. Blood. 136:1707–1721. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Falini B, Gjertsen BT and Andresen V: The
acidic stretch and the C-terminal nuclear export signal motif of
NPM1 mutant: Are they druggable in AML? Leukemia. 37:2173–2175.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Perner F, Stein EM, Wenge DV, Singh S, Kim
J, Apazidis A, Rahnamoun H, Anand D, Marinaccio C, Hatton C, et al:
MEN1 mutations mediate clinical resistance to menin inhibition.
Nature. 615:913–919. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Brunetti L, Gundry MC, Sorcini D, Guzman
AG, Huang YH, Ramabadran R, Gionfriddo I, Mezzasoma F, Milano F,
Nabet B, et al: Mutant NPM1 maintains the leukemic state through
HOX expression. Cancer Cell. 34:499–512.e9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Krivtsov AV and Armstrong SA: MLL
translocations, histone modifications and leukaemia stem-cell
development. Nat Rev Cancer. 7:823–833. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Forgione MO, McClure BJ, Eadie LN, Yeung
DT and White DL: KMT2A rearranged acute lymphoblastic leukaemia:
Unravelling the genomic complexity and heterogeneity of this
high-risk disease. Cancer Lett. 469:410–418. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Thorsteinsdottir U, Kroon E, Jerome L,
Blasi F and Sauvageau G: Defining roles for HOX and MEIS1 genes in
induction of acute myeloid leukemia. Mol Cell Biol. 21:224–234.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Grembecka J, He S, Shi A, Purohit T,
Muntean AG, Sorenson RJ, Showalter HD, Murai MJ, Belcher AM,
Hartley T, et al: Menin-MLL inhibitors reverse oncogenic activity
of MLL fusion proteins in leukemia. Nat Chem Biol. 8:277–284. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Uckelmann HJ, Kim SM, Wong EM, Hatton C,
Giovinazzo H, Gadrey JY, Krivtsov AV, Rücker FG, Döhner K, McGeehan
GM, et al: Therapeutic targeting of preleukemia cells in a mouse
model of NPM1 mutant acute myeloid leukemia. Science. 367:586–590.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Di Fazio P: Targeting menin: A promising
therapeutic strategy for susceptible acute leukemia subtypes.
Signal Transduct Target Ther. 8:3842023. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Nadiminti KVG, Sahasrabudhe KD and Liu H:
Menin inhibitors for the treatment of acute myeloid leukemia:
Challenges and opportunities ahead. J Hematol Oncol. 17:1132024.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Wang R, Xu P, Chang LL, Zhang SZ and Zhu
HH: Targeted therapy in NPM1-mutated AML: Knowns and unknowns.
Front Oncol. 12:9726062022. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Uckelmann HJ, Haarer EL, Takeda R, Wong
EM, Hatton C, Marinaccio C, Perner F, Rajput M, Antonissen NJC, Wen
Y, et al: Mutant NPM1 directly regulates oncogenic transcription in
acute myeloid leukemia. Cancer Discov. 13:746–765. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Wang XQD, Fan D, Han Q, Liu Y, Miao H,
Wang X, Li Q, Chen D, Gore H, Himadewi P, et al: Mutant NPM1
hijacks transcriptional hubs to maintain pathogenic gene programs
in acute myeloid leukemia. Cancer Discov. 13:724–745. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Dillon LW, Gui G, Page KM, Ravindra N,
Wong ZC, Andrew G, Mukherjee D, Zeger SL, El Chaer F, Spellman S,
et al: DNA sequencing to detect residual disease in adults with
acute myeloid leukemia prior to hematopoietic cell transplant.
JAMA. 329:745–755. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Mill CP, Fiskus W, Das K, Davis JA,
Birdwell CE, Kadia TM, DiNardo CD, Daver N, Takahashi K, Sasaki K,
et al: Causal linkage of presence of mutant NPM1 to efficacy of
novel therapeutic agents against AML cells with mutant NPM1.
Leukemia. 37:1336–1348. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Huls G, Woolthuis CM and Schuringa JJ:
Menin inhibitors in the treatment of acute myeloid leukemia. Blood.
145:561–566. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Issa GC, Aldoss I, Thirman MJ, DiPersio J,
Arellano M, Blachly JS, Mannis GN, Perl A, Dickens DS, McMahon CM,
et al: Menin inhibition with revumenib for KMT2A-Rearranged
relapsed or refractory acute leukemia (AUGMENT-101). J Clin Oncol.
43:75–84. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Wang ES, Issa GC, Erba HP, Altman JK,
Montesinos P, DeBotton S, Walter RB, Pettit K, Savona MR, Shah MV,
et al: Ziftomenib in relapsed or refractory acute myeloid leukaemia
(KOMET-001): A multicentre, open-label, multi-cohort, phase 1
trial. Lancet Oncol. 25:1310–1324. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Miao H, Chen D, Ropa J, Purohit T, Kim E,
Sulis ML, Ferrando A, Cierpicki T and Grembecka J: Combination of
menin and kinase inhibitors as an effective treatment for leukemia
with NUP98 translocations. Leukemia. 38:1674–1687. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Fiskus W, Piel J, Collins M, Hentemann M,
Cuglievan B, Mill CP, Birdwell CE, Das K, Davis JA, Hou H, et al:
BRG1/BRM inhibitor targets AML stem cells and exerts superior
preclinical efficacy combined with BET or menin inhibitor. Blood.
143:2059–2072. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Kwon MC, Thuring JW, Querolle O, Dai X,
Verhulst T, Pande V, Marien A, Goffin D, Wenge DV, Yue H, et al:
Preclinical efficacy of the potent, selective menin-KMT2A inhibitor
JNJ-75276617 (bleximenib) in KMT2A- and NPM1-altered leukemias.
Blood. 144:1206–1220. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
An ZY and Zhang XH: Menin inhibitors for
acute myeloid leukemia: latest updates from the 2023 ASH Annual
Meeting. J Hematol Oncol. 17:522024. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Heikamp EB, Henrich JA, Perner F, Wong EM,
Hatton C, Wen Y, Barwe SP, Gopalakrishnapillai A, Xu H, Uckelmann
HJ, et al: The menin-MLL1 interaction is a molecular dependency in
NUP98-rearranged AML. Blood. 139:894–906. 2022. View Article : Google Scholar : PubMed/NCBI
|