Introduction
Cancer has long been a paramount concern in global
public health, with the complexity of its pathogenesis extending
far beyond initial understanding. At present, clinical treatments
for cancer include radiotherapy, chemotherapy and some targeted
drugs. However, these treatments still cannot effectively relieve
pain or prolong the quality of life of the patient (1,2).
Therefore, identifying effective therapeutic targets for precision
medicine is of great significance.
The occurrence and development of cancer are
inextricably linked to epigenetic modifications, which are
influenced by environmental factors (3). Epigenetics has therefore emerged as a
research focus. DNA methylation, a key epigenetic modification,
refers to the addition of methyl groups to cytosine bases within
CpG islands by DNA methyltransferases (DNMTs). CpG islands act as
‘clusters of switch buttons’ in gene promoter regions, dynamically
regulating methylation similar to a light switch for gene
expression. Methylation acts as an ‘off button,’ silencing tumor
suppressor genes (4,5). Under normal physiological conditions,
DNA methylation accurately regulates gene activation and
repression, governing human organ development and contributing to
cancer initiation. DNA methylation also plays an important role in
cancer development and is often closely associated with the
silencing of tumor suppressor genes. When the CpG islands in tumor
suppressor genes are highly methylated, the expression of these
genes decreases, allowing tumor cells to escape regulation and
undergo rampant proliferation and metastasis, which seriously
threatens human life and health (6,7).
Therefore, in-depth study of the DNA methylation changes in various
target genes in cancer not only helps to understand the mechanisms
of cancer occurrence and development but is also expected to
provide new biomarkers and therapeutic targets for early diagnosis,
prognosis assessment and the targeted therapy of cancer (8).
The 14-3-3 gene family has been extensively studied
in cancer research (9). The 14-3-3
protein comprises seven isoforms (β, ε, γ, η, σ, τ and ζ), each
with distinct functions. This family regulates the activity,
stability, subcellular localization and interactions of client
proteins by mediating direct binding or promoting interactions with
other signaling molecules. These regulatory processes govern key
cellular pathways, including cell cycle control, apoptosis,
proliferation and programmed necrosis, often involving proteins
such as p53, cell division cycle 25C (CDC25C) and Bad (9,10).
14-3-3σ (SFN) is one of the numerous proteins closely associated
with cancer. SFN is a dimeric small molecule protein present in all
eukaryotes, particularly in epithelial cells (11). First identified in bovine brain
tissue, SFN has since been detected in various human organs,
including breasts, pancreas, stomach, colorectum, liver and kidneys
(12). The diversity of SFN
functions makes it a strong candidate as a therapeutic target for a
variety of diseases. Drawing from the existing literature on SFN in
the fields of epigenetics and cancer, the present review
synthesizes the role of the DNA methylation of SFN in different
malignancies, dissects its underlying mechanisms and evaluates its
potential as a diagnostic or prognostic biomarker and therapeutic
target.
Structure and biological function of
SFN
The SFN protein is composed of 245 amino acids, with
a molecular weight of ~28 kDa. Similar to other family members, SFN
forms a typical ‘U’-shaped dimer structure via an N- and C-terminal
α-helix. Each monomer contains nine α-helices, among which helices
α3, α4 and α8 form a conserved phosphopeptide-binding channel
similar to a molecular locking structure, which recognize
phosphorylated proteins through charge matching (13). Through its unique structure and
multifunctional regulatory network, SFN modulates key biological
processes, including the cell cycle, DNA damage repair, apoptosis
and tumor suppression (14). In
recent years, the loss of SFN expression in cancer due to
epigenetic modifications (such as DNA methylation) has become a hot
topic of research, providing important clues for understanding the
tumorigenesis mechanism. The main functional regulatory networks of
the SFN protein include: i) Cell cycle arrest and DNA damage
response: SFN induces G2/M arrest by sequestering cell cycle
regulators (such as CDK2/cyclin E complexes) upon DNA damage,
preventing their nuclear translocation. For instance, in
p53-dependent responses, SFN maintains G2 arrest by binding
phosphorylated CDC25C and inhibiting CDK1 activation (15). ii) Apoptosis regulation and
pro-survival function: SFN inhibits apoptosis by binding to the
phosphorylated forms of pro-apoptotic proteins such as Bad and Bax,
preventing their mitochondrial localization and subsequent
cytochrome C release under unstressed conditions (16). However, under conditions of
sustained DNA damage or oncogene activation, SFN protein
degradation can release the inhibition of apoptotic signals and
promote cell clearance (17). iii)
Tumor suppressor function: SFN expression is silenced in a variety
of epithelial tumors due to hypermethylation of promoter CpG
islands. Silencing SFN leads to cell cycle checkpoint defects,
genomic instability and apoptotic resistance, thereby promoting
tumor progression (18).
Restoration of SFN expression has been shown to significantly
reduce tumor cell proliferation and metastasis by inhibiting CDK
activity and suppressing epithelial-mesenchymal transition (EMT)
pathways. The silencing of SFN expression is closely related to
tumorigenesis, with promoter methylation being the main regulatory
mechanism (19). SFN acts as a
molecular scaffold integrating multiple signaling pathways. Studies
of its structure-function relationship provide a new direction for
the development of targeted cancer therapies, such as demethylation
drugs or phosphatase inhibitors. In the future, it will be
necessary to further analyze its interaction network in different
tissue microenvironments and explore its clinical application
potential as a biomarker or therapeutic target.
Relationship between DNA methylation and
cancer
Epigenetic modifications (such as DNA methylation,
histone modification and non-coding RNA regulation) play a pivotal
role in cancer development, with abnormal DNA methylation being one
of the most common epigenetic changes in tumor cells. DNA
methylation refers to the covalent addition of methyl groups to the
5′ position of cytosine in CpG dinucleotides (forming
5-methylcytosine), catalyzed by DNMTs (20). A number of studies have shown that
DNA methylation alters chromatin structure, DNA conformation,
stability and protein-DNA interactions, thereby regulating gene
expression (21–23). In normal cells, DNA methylation
regulates gene function through mechanisms such as promoter CpG
island hypermethylation and global genome hypomethylation. In
cancer, DNA methylation patterns are significantly disrupted, often
manifested as: i) Global hypomethylation, which promotes chromosome
instability and proto-oncogene activation; and ii) local
hypermethylation, which is concentrated in the promoter regions of
tumor suppressor genes, resulting in the silencing of their
expression (24,25). Different detection methods employ
distinct thresholds for defining hypermethylation. For instance, in
methylation-specific PCR (MSP), hypermethylation is identified when
the intensity of the methylated band exceeds 50% of the
unmethylated band. Pyrosequencing defines hypermethylation as
either a single CpG site methylation rate >30% or a regional
average >50%. Exceeding these hypermethylation thresholds
represents the molecular mechanism for the transcriptional
silencing of SFN, which correlates with malignancy in specific
cancer types (26). DNA methylation
affects genome stability and signaling pathway activity by
regulating gene expression silencing, making it an important
research direction in cancer diagnosis and treatment. Promoter CpG
island hypermethylation acts as an epigenetic switch for tumor
suppressor genes. DNA methylation forms dense chromatin structures
by recruiting methyl-binding proteins (such as methyl-CpG binding
protein 2) and histone deacetylases (HDACs), resulting in the
transcriptional silencing of genes. For example, in breast and lung
cancer, hypermethylation of the SFN promoter leads to defects in
G2/M phase arrest and apoptosis resistance, significantly promoting
tumor progression (27).
O6-methylguanine-DNA methyltransferase is a DNA repair enzyme that
protects genomic stability by clearing DNA damage induced by
alkylating agents. Hypermethylation of its promoter region leads to
gene silencing and loss of repair function, thereby increasing
mutation accumulation and promoting tumorigenesis (28). In tumor cells, signal transducers
and activators of transcription 5A, involved in cell proliferation
and immune regulation, exhibits promoter hypermethylation that
suppresses antitumor immune responses (such as T cell activation)
and facilitates immune escape (29). The clinical value of DNA methylation
is emerging in early diagnosis, prognosis, therapeutic targeting,
drug resistance modulation and immunotherapy synergy. Future
challenges in cancer research include identifying actionable
methylation targets, developing dynamic methylation regulation
strategies, and constructing methylation-driven gene networks.
Abnormal methylation of SFN in cancer
Breast cancer (BC)
BC is one of the most common malignant tumors, with
an increasing incidence among tumors afflicting women. In 2018
alone, 268,670 new cases of BC were reported in the United States
(30). As a heterogeneous disease
influenced by both genetic and environmental factors, BC remains a
significant health burden despite recent therapeutic advances.
Current research efforts in personalized treatment (previously
guided by disease severity) now focus on underlying biological
mechanisms. The search for new targets and treatment strategies is
important for patient survival and quality of life.
In recent years, DNA methylation is a major
epigenetic modification, and SFN (a tumor suppressor gene) is
frequently silenced by this modification in BC (31). Studies have shown that SFN DNA
methylation frequency exceeds 90% in BC. Additionally, SFN
hypermethylation has been observed in breast hyperplasia samples,
while normal tissues remain unmethylated (as determined by MSP
assay) (32,33). The downregulation of SFN has been
detected in BC samples by serial analysis of gene expression, but
no specific gene locus at the SFN site that could account for its
downregulation was identified. After the treatment of BC cells with
the DNMT inhibitor (DNMTI), 5-Azacytidine, SFN expression was
re-detected and found to be activated, indicating that SFN
underwent DNA methylation modification mediated by DNMTs. This
suggests that DNA hypermethylation of SFN is closely related to the
occurrence and development of BC (34). Another study grouped 77 patients
with BC (no symptoms of disease) and 34 patients with metastatic BC
(symptoms of disease), then DNA methylation in the serum samples
from these two groups was detected. The DNA methylation level was
higher in the BC group, indicating that SFN may serve as a
predictor of BC progression (35).
Therefore, SFN can be considered as a biomarker for screening
metastatic BC, both in early detection and for monitoring
subsequent treatment responses.
The specific mechanism by which SFN affects BC
progression is as follows. A study has shown that SFN is a
p53-dependent negative regulator of the cell cycle, blocking the
G2/M phase by inhibiting the formation of the CDC2-cyclin B1
complex. However, under conditions of hypermethylation, the
inactivation of SFN fails to block the cell cycle, thereby
promoting cancer progression (36).
The hypermethylation of the SFN gene promoter not only facilitates
BC diagnosis but also drives the development of new treatments.
Inhibiting DNA methylation of SFN is expected to become a potential
therapeutic target for BC, and drug design targeting this mechanism
could provide a valuable reference for the further clinical
treatment of BC.
Lung cancer (LC)
LC is the leading cause of cancer-related death
worldwide and includes two main subtypes: Small cell LC and
non-small cell LC (NSCLC) (37).
NSCLC accounts for ~75% of all LC cases (38). Most patients with NSCLC present with
advanced disease at diagnosis, as obvious symptoms often emerge in
the mid-to-late stages. Current clinical treatments for LC involve
a combination of surgery, radiotherapy and chemotherapy (39), yet these approaches fail to
sufficiently improve the quality of life for patients with advanced
disease. The development of early diagnostic markers and targeted
therapeutics for LC remains a critical need. The upregulation of
oncogenes and the silencing of tumor suppressor genes are the main
causes of LC (40). A study has
shown that the loss of SFN expression caused by DNA methylation is
correlated with the carcinogenesis and prognosis of LC. SFN
methylation has been detected in the serum of 167 patients with
NSCLC, with methylation rates ranging from 64.6 to 100% (as
determined by MSP assay). Thus, SFN could be used as a marker for
the serological testing of NSCLC (41).
As for the specific mechanism of the involvement of
SFN in LC progression, current studies mainly focus on cell cycle
regulation and drug resistance. For instance, it has been shown
that when SFN is hypermethylated and its expression is absent in
LC, failure of cell cycle arrest leads to tumor cells escaping
constraints on abnormal proliferation, thereby promoting LC
progression (34). The therapeutic
effect of chemotherapy drugs is often affected by drug resistance.
In a cisplatin-treated NSCLC cell line (A549 cells), tripartite
motif containing 25-mediated cisplatin resistance is primarily due
to the downregulation of SFN, indicating that targeting SFN may be
a potential strategy to reverse cisplatin resistance (42). However, rare contradictory reports
exist regarding SFN expression in NSCLC. For instance, one study
documented that high SFN expression, as a result of SFN
hypomethylation, in orthotopic lung adenocarcinoma models is a key
driver of early-stage malignant progression (43). The conflicting observations
regarding SFN methylation in NSCLC fundamentally reflect the
complexity of epigenetic regulation. The methylation status of the
same gene across distinct genomic regions may drive tumorigenesis
through divergent mechanisms, while sample heterogeneity, technical
limitations and co-mutation backgrounds further exacerbate
interpretational discrepancies. Future research should integrate
spatial methylation profiling, single-cell multi-omics and clinical
prospective cohorts to establish dynamic SFN methylation models,
potentially uncovering novel epigenetic targets for precision
stratified therapy in NSCLC.
Kidney cancer (KC)
Worldwide, KC occurs more frequently in men than in
women. Although there have been advances in the treatment and
diagnosis of KC in recent years, KC remains one of the deadliest
malignancies of the urinary system; its incidence is increasing,
with an estimated 400,000 new cases per year, according to the most
recent data from the World Health Organization. The global
mortality rate is close to 175,000 deaths per year (44,45).
Risk factors for KC include smoking, obesity, hypertension, poor
diet and chronic kidney disease; acute/chronic kidney injury are
often precursors of KC (46,47).
At present, the main selected treatment for KC is based on the
location of the tumor and the stage of the disease. These options
can be divided into surgical and non-surgical treatments (48). While improved screening has
increased patient survival, rising KC prevalence and stagnant cure
rates highlight the need for early diagnostic markers and targeted
small-molecule inhibitors to prevent disease progression. Very
little is known about the role of SFN in KC, especially regarding
the effects of the DNA methylation of SFN. A study has shown
abnormal expression of SFN in 15 (38%) canine KC cases, with a
positive association between SFN expression and a significantly
reduced survival time observed (11). In human KC, SFN is typically
silenced by hypermethylation of its promoter. In a study of 31
patients with KC, methylation levels were found to increase
progressively with the malignancy of renal cancer, from normal to
cancerous kidney tissue. Among the 16 samples of renal cancer
tissues, 87.5% had complete hypermethylation of SFN (as determined
by MSP assay) (49). Additionally,
SFN can also influence cell cycle regulation, enabling tumor cells
to escape DNA damage caused by cisplatin and further promote tumor
cell proliferation. These findings indicate that SFN expression,
DNA methylation and the treatment of KC are closely related
(50). Whether SFN can unlock the
mystery of KC development or emerge as a key target for developing
KC treatment strategies remains to be further explored by
researchers.
Ovarian cancer (OC)
OC is the second most common cause of cancer-related
death in women worldwide, accounting for ~140,000 deaths annually.
This high mortality rate is largely due to a lack of clear early
diagnostic markers and delayed treatment (51). Most OC cases are definitively
diagnosed at an advanced stage, with a high recurrence rate. The
standard treatment for OC includes surgery and combination
chemotherapy. In the early stages, the disease can be successfully
treated with surgery alone. However, advanced stages require
complex chemotherapy regimens, with drugs such as bevacizumab
representing newer therapeutic options (52). While these targeted drugs have shown
progress in improving the prognosis of OC, it remains the deadliest
gynecological cancer in women. OC is generally divided into four
main types: Epithelial OC, germ cell tumors, sex cord-stromal
tumors and metastatic cancer (53).
DNA methylation, histone modification and gene silencing mediated
by non-coding RNAs are key processes in the development of OC and
represent new targets for cancer detection and treatment (54). In OC, SFN is primarily inactivated
by DNA methylation. A study evaluating SFN methylation as a
prognostic marker found that it is associated with OC but unable to
predict patient outcomes (MSP assay) (55). Hypermethylation of SFN promotes OC
progression.
Nasopharyngeal cancer (NPC)
NPC exhibits a significant geographical clustering,
with ~70% of the world's new cases concentrated in Southeast Asia
and East Asia. Among these regions, China has the highest incidence
of NPC, accounting for 47% of all cases (56). The main risk factors for NPC include
Epstein-Barr virus infection, genetic susceptibility, environmental
factors and diet (57). Current
prevention and control strategies focus on the following areas:
Screening of high-risk groups, risk factor intervention and
precision epidemiological studies. At present, only one study has
shown that SFN promoter methylation occurs in primary NPC tumors,
leading to decreased SFN expression; however, promoter
hypermethylation has not been detected in normal nasopharyngeal
epithelial cells (MSP-detected methylation rate: 63%). This
suggests that SFN expression may also serve as a prognostic marker
for NPC (58). SFN is a downstream
target gene of miR-597 and miR-675-5p. The downregulation of SFN by
miR-597 and miR-675-5p can drive EMT and promote the migration and
invasion of NPC (59,60). Additionally, SFN functions as a
tumor suppressor in vitro, inhibiting NPC cell invasion
through the SFN/EGFR/keratin-8 signaling axis (61). Collectively, these findings
highlight the research value of SFN in NPC progression.
Squamous cell carcinoma (SCC)
SCC can occur in multiple organs, including vulvar
SCC (VSCC), esophageal SCC (ESCC) and oral SCC (OSCC), among
others. SFN exhibits strong immune reactivity in SCCs from
different parts of the body (62).
VSCC is commonly found in young women, and the main treatment
method is radical surgery. However, the postoperative recurrence
rate is relatively high (63).
Targeted therapy has attracted attention, driving researchers to
identify tumor-associated factors and novel therapeutic markers. In
a previous study, SFN CpG methylation was found in ~53% of cases
(as determined by MSP assay) (64).
In another study of 302 cases of VSCC, it was confirmed that the
expression of SFN protein was downregulated, contributing to the
development of the disease (63).
This indicates that in VSCC, SFN is downregulated as a tumor
suppressor through DNA methylation, thereby promoting the
progression of the disease.
ESCC is one of the major types of esophageal cancer
in Asia and one of the most aggressive gastrointestinal cancer
types (65). The absence of
symptoms in early ESCC leads to late detection of the disease and
poor prognosis. SFN is downregulated in the early stage of ESCC,
and this downregulation is associated with a shortened survival
period, suggesting that SFN may serve as a biomarker for the early
detection of ESCC (66). Abnormal
expression of SFN affects the sensitivity of patients with ESCC to
chemotherapy drugs, which is the main pathway through which SFN is
involved in ESCC (67).
OSCC is a common cancer type and although it
accounts for only 1.5% of malignant tumors, its incidence rate is
increasing, which deserves attention. Existing studies have shown
that SFN is strongly expressed in OSCC, and the survival time of
patients with OSCC with high SFN expression is lower than that of
patients with low SFN expression, which is different from the
expression of SFN in other SCC types (68,69).
Inhibition of CpG methylation of SFN may inhibit the progression of
SCC.
Gallbladder cancer (GBC) and
cholangiocarcinoma
GBC is one of the few cancer types in developing
countries with a high mortality rate. The disease presents with
almost no obvious symptoms in its early stage, leading to most
patients being diagnosed in the middle or late stages, with a poor
prognosis (70). Given the
epithelial continuity between the gallbladder and bile duct, GBC
and cholangiocarcinoma share similar oncogenic and metastatic
mechanisms. Current effective treatments for GBC and
cholangiocarcinoma include complete tumor resection and adjuvant
therapy. However, the prognosis following surgical resection
remains poor and incidence has not declined significantly over
decades (71). Moreover, there has
been little progress in treatment (72). Therefore, it is urgent to identify
therapeutic targets and develop targeted drug therapies. At
present, there are few studies on the role and therapeutic
significance of SFN dysregulation in cholangiocarcinoma.
Epigenetics plays a key role in cancer development; however, there
is still no clear understanding of SFN DNA methylation in
cholangiocarcinoma and GBC (73,74).
As a result, research into new biomarkers for these two cancer
types has attracted significant interest from researchers. Only a
small number of studies have focused on GBC. In a study, SFN DNA
methylation was detected in 45 out of 50 GBC cases, and SFN
expression was downregulated in patients with advanced GBC (as
determined by MSP assay). These results suggest that low SFN
expression, driven by DNA methylation, may promote the progression
of GBC (75). Another study
confirmed that SFN gene upregulation is associated with an improved
prognosis, lower early cancer recurrence rates and reduced distant
metastasis after resection (72).
This suggests that detecting SFN DNA methylation or SFN expression
may represent a new approach in the treatment of GBC. Inhibiting
SFN DNA methylation may be a potential therapeutic strategy for
GBC; however, the role of SFN in the treatment of
cholangiocarcinoma remains unclear.
Pancreatic cancer (PC)
PC ranks among the highest in terms of cancer
mortality; >90% of patients with PC die due to a non-response to
treatment, which is attributed to the lack of effective diagnostic
and therapeutic strategies (76).
Therefore, it is urgent to identify the pathogenic targets in PC.
Whole-genome mapping has offered hope for improving PC management,
enabling the development of histological/serological markers and
immunotherapies. Through such efforts, SFN has emerged as a key
player in PC diagnosis and treatment (77). By analyzing the methylation patterns
of SFN, a high incidence of hypomethylation was observed in PC cell
lines and primary PC tissues, suggesting that SFN hypomethylation
is a common epigenetic event in PC. This study further showed that
SFN protein expression is significantly increased in PC (MSP
assay), and this elevated expression almost inversely correlated
with patients' survival rates (76). This suggests a potential link to PC
treatment failure. The mechanism of action of SFN in PC may involve
resistance to treatment-induced apoptosis and G2/M cell cycle
blockade, thereby causing resistance to anticancer drugs and
obstructing their therapeutic effects. For example, gemcitabine is
an important anticancer drug used in the treatment of various
cancer types, including PC. SFN methylation is regulated by DNMT1,
which is involved in the acquisition of gemcitabine resistance
(78). Elevated SFN expression also
results in resistance to mitoxantrone and doxorubicin, resistance
to drug-induced apoptosis and resistance to the G2/M cell cycle
blockade (79,80). Therefore, it is considered that SFN
may serve as a prognostic indicator to predict the survival of
patients with PC, as a biomarker to detect PC in its early stages
and as a potential target for therapeutic drugs to guide the
clinical treatment of patients. This approach is expected to bring
hope in improving the lifespan of patients with PC.
Gastric cancer (GC)
GC is a leading cause of cancer-related death
worldwide, ranking as the fifth most common cancer and third
deadliest globally (81). In China,
GC is the second most prevalent cancer and second leading cause of
cancer mortality, with a dismal prognosis (82). Despite recent progress in surgical
and pharmacological treatments, the survival rate for patients with
GC in China remains very low (~40%) (83). Therefore, it is urgent to identify
reliable therapeutic targets or develop small-molecule drugs to
treat GC or alleviate the pain it causes. Genetic predisposition
may play a key role in the development of GC (84). GC is a disease influenced by
environmental epigenetic modifications and often presents with
tumors that have a high frequency of abnormal CpG island
methylation (85). A study found
that abnormally elevated SFN expression is a biomarker of poor
prognosis in GC and is associated with shortened survival time in
patients with advanced GC (86).
Additionally, an analysis of SFN expression in tumor samples from
157 patients who underwent GC resection showed that SFN expression
was elevated in 48% of cases and was correlated with p53 expression
(86). A study has shown that after
SFN knockout, the proliferation and in vitro invasion
abilities of GC cells are reduced. Additionally, serum levels of
SFN are related to the therapeutic status of locally advanced
cancer. These findings suggest that SFN plays an important role in
the proliferation and metastasis of GC cells and may be a new
target for the detection and prevention of GC (87). Another study has shown that SFN
expression is increased in the gastric tissue of mice infected with
Helicobacter pylori. Since H. pylori is a recognized
factor in the development of GC, these results also indicate that
high expression of SFN is closely related to GC (88). Regarding the role of SFN in GC, its
effects vary across different stages of the disease. In some human
cancer types, SFN is often inactivated due to methylation of the
CpG islands, thereby losing its tumor suppressor function. This
mechanism may play an important role in undifferentiated GC. The
incidence of SFN hypermethylation was found to be 43% in primary GC
and higher in poorly differentiated adenocarcinomas, suggesting
that SFN hypermethylation is more prevalent in undifferentiated GC
than in differentiated GC (as determined by
bisulfite-bisulfite-single-strand conformation polymorphism assay
(89). In differentiated GC, SFN
interacts with p53 to stabilize and enhance its transcriptional
activity (87). In another study of
60 GC samples, SFN expression was positive in 64% of cases, with
positive staining observed in low, medium and highly differentiated
GC cells. This suggests that SFN is involved in the proliferation
of GC cells (90). Therefore, these
findings indicate that SFN may be a promising target for the
treatment of GC.
Colorectal cancer (CRC)
Compared with normal colorectal tissues, SFN
expression is significantly decreased in CRC tissues. Low
expression of SFN is also significantly correlated with low
survival rates in patients with CRC (91). SFN influences CRC progression
through transcriptional programs regulated by interacting factors
(such as Snail, c-JUN, Yes-associated protein 1 and Foxo1), which
promote tumorigenesis and growth (92,93).
Additionally, LIM and SH3 protein 1 (Lasp1) drives SFN-mediated CRC
progression via PI3K/AKT pathway activation. A combination of low
SFN and high Lasp1 expression is associated with a poorer overall
survival in patients with CRC (94). Notably, there are different
viewpoints. The latest research indicates that SFN functions as an
oncogene in CRC and is associated with treatment resistance
(9,95). This suggests that SFN may have
different effects in different regions of CRC, with upregulation of
SFN in the invasive areas promoting tumor progression. Epigenetic
silencing of the SFN gene through CpG hypermethylation has been
reported in a variety of cancer types; however, this mechanism is
not thought to apply to CRC, where SFN hypermethylation and
inactivation are considered rare events (96). In invasive CRC zones, the SFN gene
regulates cell cycle progression and tumor cell migration,
promoting metastasis (9). The
contradictory expression patterns of SFN may be attributed to
spatial heterogeneity, immune cell exhaustion and dynamic
epigenetic regulation. Similar discrepancies between gene
expression and function have been documented in other organs and
tissues. Cells in distinct anatomical regions not only exhibit
divergent gene expression profiles but also display distinct
epigenetic signatures and biological functions (29,97–99).
Other cancer types
SFN is also potentially linked to other cancer
types, including glioma, endometrial cancer (EC) and prostate
cancer (PCa) (34,100,101). In a study of 186 tumor samples
from patients with different grades of glioma, higher SFN
expression was associated with higher survival rates. SFN
inactivation in glioma was also due to its DNA methylation (as
determined by MSP assay). Compared with normal tissues, SFN
expression in glioma was downregulated, suggesting that targeting
SFN DNA methylation may offer a new opportunity to improve outcomes
in patients with this disease (100,102).
SFN is expressed in the normal epithelial cells of
most organs, and its epigenetic regulation is involved in
controlling specific expression in normal cells and gene silencing
in cancer cells (34). SFN is often
absent in EC and PCa. A study has shown that CpG island methylation
of SFN is closely related to its low expression (as determined by
MSP assay) (55). Most EC cases are
detected early due to vaginal bleeding and the recurrence rate is
low after treatment (103).
However, a study of 86 cases of EC and 46 cases of normal
endometrial tissue found that SFN hypermethylation, low expression
and inactivation in EC may be associated with recurrence (as
determined by MSP assay) (104).
The role of SFN in PCa has not been extensively studied. An early
study has shown that SFN is highly expressed in normal prostate
tissue, but its expression is significantly reduced in PCa
(105). As a cell cycle regulator
and tumor suppressor, SFN downregulation in PCa may promote
tumorigenesis by bypassing DNA damage checkpoints-possibly
independent of p53 (55,106). Detection of SFN CpG methylation
could potentially be used for diagnostic and prognostic purposes in
the future.
Tumor drug development targeting SFN and its
epigenetic modifications
Over the past decade, significant progress has been
made in the field of epigenetics research in cancer (107). Numerous studies have shown that
epigenetic modifications play a crucial regulatory role in the
dysregulation of tumor cells. The reversibility of epigenetic
modifications offers a chance to rectify their abnormal
alterations. Thus, employing epigenome-targeting agents to
facilitate the normalization of aberrant methylation in cancer
cells will serve as a key strategy for treating cancer (20). While research in this area has
advanced significantly in recent years, new challenges have arisen,
including low drug specificity and toxic side effects. Hence,
developing gene-specific targeted approaches holds notable
scientific and clinical value.
SFN methylation causes chromatin structure to become
more compact by recruiting methyl-binding proteins (such as
methyl-CpG binding domain protein 2) and HDACs, ultimately
inhibiting gene expression and blocking the binding of
transcription factors (such as p53) (108,109). The DNA methylation of SFN has the
dual potential of both being a therapeutic target and diagnostic
marker. As a diagnostic and prognostic biomarker, the SFN
methylation status is detectable in body fluids (such as plasma and
urine), which provides the possibility for non-invasive cancer
screening. For example, SFN methylation in the serum of patients
with BC is positively correlated with tumor stage (110). In CRC, the SFN methylation level
predicts resistance to 5-FU chemotherapy (111). This 5-FU predictive value not only
contributes to the formulation of individualized treatment plans
but also provides biomarkers for the efficacy monitoring of
epigenetic targeted drugs. Additionally, SFN methylation is also an
indicator of the response to demethylation therapy. SFN can work
synergistically with other epigenetic drugs, such as HDAC
inhibitors. For example, AR42 (OSU-HDAC42), an HDAC inhibitor,
shows a negative correlation with SFN expression and collaborates
to inhibit tumor growth (112).
The current literature reports that DNMTIs (such as 5-Aza-CdR) can
reverse SFN methylation and restore its expression, thereby
delaying tumor progression. For instance, 5-Aza-CdR restores SFN
expression, induces G2/M phase arrest and promotes apoptosis,
enhancing radiosensitivity in osteosarcoma and BC cells (113,114). Additionally, 5-Aza-CdR-induced SFN
upregulation triggers senescence in melanoma cells, inhibiting
tumor progression (115). In EC,
SFN-mediated G2/M arrest via 5-Aza-CdR exerts direct antitumor
effects. Notably, immunotherapy is a cornerstone of cancer
treatment. Recent studies have shown that DNMTIs can potentiate
immunotherapy [such as programmed death-ligand 1 (PD-L1) blockade]
(116,117). We therefore propose exploring
novel molecules integrating methylation reversal and
immunomodulation to reactivate SFN and enhance antitumor immunity,
with promising translational potential. SFN and microenvironment
therapy may also be related to tumor progression. For example, a
hypoxic microenvironment contributes to tumor progression, while
SFN inhibits metastasis and angiogenesis in CRC induced by tumor
hypoxia through regulating hypoxia-inducible factor-1α (HIF-1α)
(118). Hypoxic regions in gliomas
can induce HIF-1α activation, potentially influencing SFN
expression (119). Therefore, SFN
may also play a role in mediating responses to
microenvironment-targeted therapies. Although DNMTIs can reverse
SFN methylation, preclinical and clinical studies have revealed
notable limitations: Systemic toxicity, risk of proto-oncogene
activation, suboptimal targeting in solid tumors and off-target
effects. Most notably, robust clinical evidence supporting their
efficacy remains limited (120,121). To date, to the best of our
knowledge, there have been no clinical trials directly targeting
SFN methylation. In the existing studies, abnormal methylation of
SFN has been observed in the majority of cancer types (Table I). Therefore, developing antitumor
drugs targeting SFN methylation is also a promising strategy. In a
very small number of cancer types such as CRC, SFN exhibits a dual
role. However, the underlying reasons for these differences are not
clear, so further research is needed before conducting in-depth
studies on specific targeted treatment strategies.
 | Table I.Relationship between DNA methylation
state and the expression level of 14-3-3σ in different cancer
types. |
Table I.
Relationship between DNA methylation
state and the expression level of 14-3-3σ in different cancer
types.
| Cancer type | DNA methylation
state | Expression of
14-3-3σ | (Refs.) |
|---|
| Breast cancer | High | Low | (31) |
| Kidney cancer | High | Low | (49) |
| Ovarian cancer | High | Low | (55) |
| Nasopharyngeal
cancer | High | Low | (58) |
| Vulvar squamous
cell carcinoma | High | Low | (64) |
| Gallbladder
cancer | High | Low | (75) |
| Glioma | High | Low | (100) |
| Gastric cancer
(primary) | High | Low | (89) |
| Endometrial
cancer | High | Low | (55) |
| Lung cancer | High/Low | Low/High | (41,43) |
| Pancreatic
cancer | Low | High | (76) |
| Oral squamous cell
carcinoma | / | High | (68) |
| Prostatic
cancer | / | Low | (105) |
| Esophageal squamous
cell carcinoma | / | Low | (66) |
| Colorectal
cancer | / | Low | (91) |
Conclusions and perspectives
The SFN protein is a core molecule in cell cycle
regulation and the DNA damage response, and its functional
inactivation is closely related to cancer initiation and
progression. In the present review, the epigenetic regulation
mechanisms and clinical significance of this protein in various
tumor types were systematically summarized (Fig. 1 and Table SI). Silencing of SFN expression is
not only caused by gene mutation or deletion but also by abnormal
hypermethylation of promoter CpG islands. This epigenetic silencing
shows tissue-specific patterns in BC, PCa and LC and is positively
associated with tumor aggressiveness. Although the degree of DNA
methylation varies among different cancer types, SFN methylation
leads to G2/M checkpoint failure and apoptotic resistance, thereby
promoting genomic instability. There are also some specific
mechanisms. These findings suggest the pivotal role of SFN in
precision therapy.
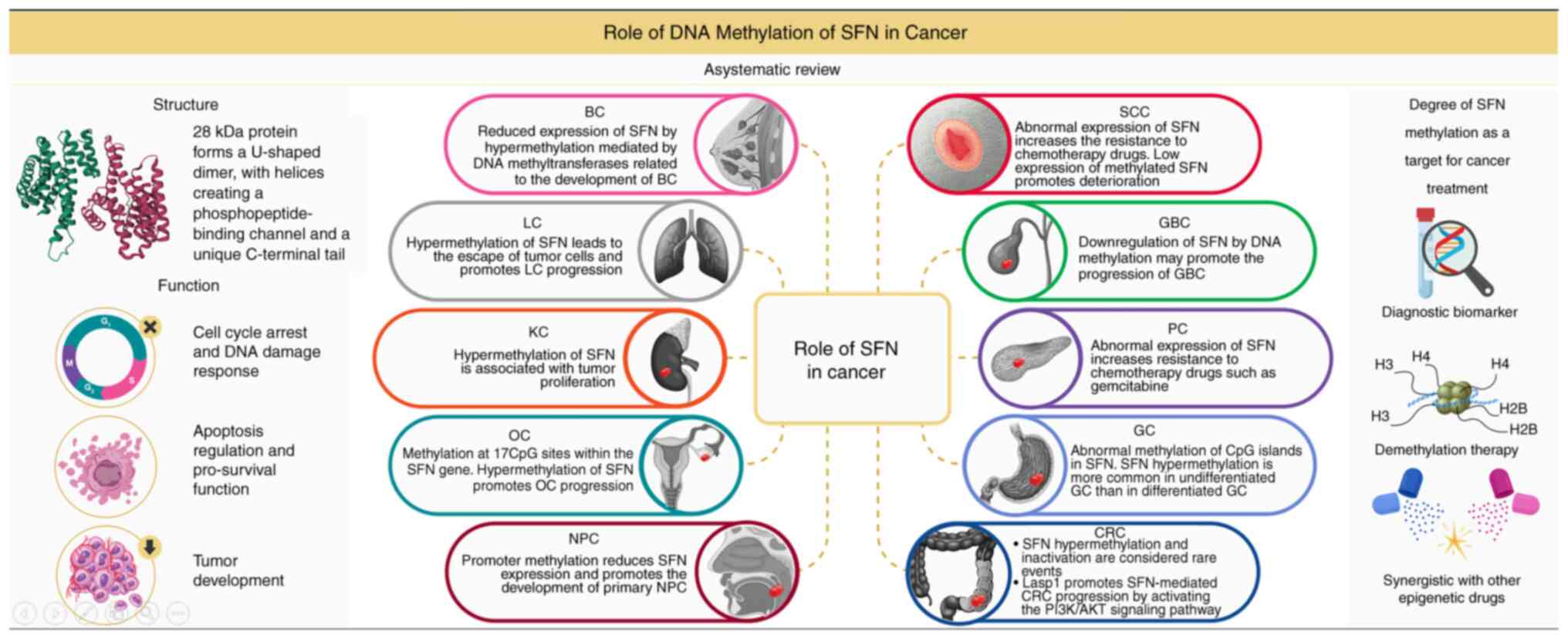 | Figure 1.Role of DNA methylation of SFN in
cancer. SFN, 14-3-3σ; BC, breast cancer; LC, lung cancer; KC,
kidney cancer; OC, ovarian cancer; NPC, nasopharyngeal cancer; SCC,
squamous cell carcinoma; GBC, gall blader cancer; PC, pancreatic
cancer; GC, gastric cancer; CRC, colorectal cancer. |
At present, to the best of our knowledge, there are
no clinical guidelines incorporating SFN methylation detection.
Through a literature review, we conclude that SFN methylation, as a
prognostic marker, holds certain clinical significance. SFN
methylation precedes abnormal protein expression, in contrast to
prostate-specific antigen/CA-125 that often serve as late-stage
indicators. SFN methylation is prognostically relevant across
multiple cancer types (such breast and lung cancer), supporting the
development of pan-cancer screening strategies. Additionally, SFN
methylation directly mediates resistance to cisplatin/gemcitabine,
a mechanism unaddressed by traditional markers. Thus, we consider
that SFN methylation profiling holds significant clinical utility
as a biomarker.
The existing challenges and breakthroughs highlight
the need for in-depth mechanistic research. At present, to the best
of our knowledge, the upstream driving factors of SFN methylation
have not been fully analyzed. The synergistic mechanisms between
methylation and other epigenetic modifications need to be clarified
through multi-omics integration analyses. The technical bottleneck
in clinical transformation lies in the insufficient sensitivity of
existing detection technologies for low-abundance methylated
circulating tumor DNA. At present, there is a lack of specific SFN
methylation inhibitors, and methyltransferase inhibitors also have
some unresolved clinical challenges. Systemic off-target effects of
DNMTIs, such as decitabine, may result from the activation of
proto-oncogenes. Therefore, further exploration of tumor-targeted
delivery strategies is needed. There is room for innovation in
therapeutic strategies. ‘Pulsed administration’ based on
methylation dynamics may balance demethylation efficacy and
toxicity. Combining epigenetic editing with immune checkpoint
blocking may break tumor immune tolerance. With the development of
single-cell epigenomics and spatial multi-omics techniques, it will
be possible to analyze the role of SFN methylation in the evolution
of tumor heterogeneity. We therefore propose that the development
of bi-functional molecules with both methylation reversal and
immune regulation functions, such as DNMT-PD-L1 dual-target
inhibitors, may provide a new paradigm for overcoming therapeutic
resistance. SFN is expected to become a signature target in the
field of cancer epigenetic therapy in these new modalities. Other
epigenetic modifications also need the attention of researchers,
such as RNA modifications and histone modifications, which may
drive cancer progression through synergistic or independent
effects. In the future, research should focus on multi-modification
interactions, dynamic regulation and precise intervention
strategies, combined with technological innovation and clinical
validation, to provide new targets and paradigms for cancer
treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
DH, YH wrote the main manuscript text and DN was
involved in drafting the manuscript and revising it critically for
important intellectual content. All authors read and approved the
final version of the manuscript. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
de Visser KE and Joyce JA: The evolving
tumor microenvironment: From cancer initiation to metastatic
outgrowth. Cancer Cell. 41:374–403. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arafeh R, Shibue T, Dempster JM, Hahn WC
and Vazquez F: The present and future of the cancer dependency map.
Nat Rev Cancer. 25:59–73. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mathers JC, Strathdee G and Relton CL:
Induction of epigenetic alterations by dietary and other
environmental factors. Adv Genet. 71:3–39. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gupta MK, Peng H, Li Y and Xu CJ: The role
of DNA methylation in personalized medicine for immune-related
diseases. Pharmacol Ther. 250:1085082023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bergstedt J, Azzou SAK, Tsuo K,
Jaquaniello A, Urrutia A, Rotival M, Lin DTS, MacIsaac JL, Kobor
MS, Albert ML, et al: The immune factors driving DNA methylation
variation in human blood. Nat Commun. 13:58952022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zeng Y and Chen T: DNA methylation
reprogramming during mammalian development. Genes (Basel).
10:2572019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tolmacheva EN, Vasilyev SA and Lebedev IN:
Aneuploidy and DNA methylation as mirrored features of early human
embryo development. Genes (Basel). 11:10842020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Klutstein M, Nejman D, Greenfield R and
Cedar H: DNA methylation in cancer and aging. Cancer Res.
76:3446–3450. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang Y, Yang M and Huang W: 14-3-3 σ: A
potential biomolecule for cancer therapy. Clin Chim Acta.
511:50–58. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iwahori S, Umaña AC, Kalejta RF and Murata
T: Serine 13 of the human cytomegalovirus viral cyclin-dependent
kinase UL97 is required for regulatory protein 14-3-3 binding and
UL97 stability. J Biol Chem. 298:1025132022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Suárez-Bonnet A, Lara-Garcia A, Stoll AL,
Carvalho S and Priestnall SL: 14-3-3σ protein expression in canine
renal cell carcinomas. Vet Pathol. 55:233–240. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gu Q, Cuevas E, Raymick J, Kanungo J and
Sarkar S: Downregulation of 14-3-3 proteins in Alzheimer's disease.
Mol Neurobiol. 57:32–40. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Obsil T, Ghirlando R, Klein DC, Ganguly S
and Dyda F: Crystal structure of the 14-3-3zeta:serotonin
N-acetyltransferase complex. A role for scaffolding in enzyme
regulation. Cell. 105:257–267. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mhawech P: 14-3-3 proteins-an update. Cell
Res. 15:228–236. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hermeking H and Benzinger A: 14-3-3
Proteins in cell cycle regulation. Semin Cancer Biol. 16:183–192.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Henry RE, Andrysik Z, Paris R, Galbraith
MD and Espinosa JM: A DR4:tBID axis drives the p53 apoptotic
response by promoting oligomerization of poised BAX. EMBO J.
31:1266–1278. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pozuelo-Rubio M: 14-3-3 Proteins are
regulators of autophagy. Cells. 1:754–773. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lodygin D and Hermeking H: The role of
epigenetic inactivation of 14-3-3sigma in human cancer. Cell Res.
15:237–246. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Raychaudhuri K, Chaudhary N, Gurjar M,
D'Souza R, Limzerwala J, Maddika S and Dalal SN: 14-3-3σ gene loss
leads to activation of the epithelial to mesenchymal transition due
to the stabilization of c-Jun protein. J Biol Chem.
291:16068–16081. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun L, Zhang H and Gao P: Metabolic
reprogramming and epigenetic modifications on the path to cancer.
Protein Cell. 13:877–919. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Meng H, Cao Y, Qin J, Song X, Zhang Q, Shi
Y and Cao L: DNA methylation, its mediators and genome integrity.
Int J Biol Sci. 11:604–617. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dai X, Ren T, Zhang Y and Nan N:
Methylation multiplicity and its clinical values in cancer. Expert
Rev Mol Med. 23:e22021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Suo XG, Wang JN, Zhu Q, Zhang MM, Ge QL,
Peng LJ, Wang YY, Ji ML, Ou YM, Yu JT, et al: METTL3 mediated m6A
modification of HKDC1 promotes renal injury and inflammation in
lead nephropathy. Int J Biol Sci. 21:3755–3775. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nishiyama A and Nakanishi M: Navigating
the DNA methylation landscape of cancer. Trends Genet.
37:1012–1027. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kulis M and Esteller M: DNA methylation
and cancer. Adv Genet. 70:27–56. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ku JL, Jeon YK and Park JG:
Methylation-specific PCR. Methods Mol Biol. 791:23–32. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Horie-Inoue K and Inoue S: Epigenetic and
proteolytic inactivation of 14-3-3sigma in breast and prostate
cancers. Semin Cancer Biol. 16:235–239. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jayaprakash C, Radhakrishnan R, Ray S and
Satyamoorthy K: Promoter methylation of MGMT in oral carcinoma: A
population-based study and meta-analysis. Arch Oral Biol.
80:197–208. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liang WW, Lu RJ, Jayasinghe RG, Foltz SM,
Porta-Pardo E, Geffen Y, Wendl MC, Lazcano R, Kolodziejczak I, Song
Y, et al: Integrative multi-omic cancer profiling reveals DNA
methylation patterns associated with therapeutic vulnerability and
cell-of-origin. Cancer Cell. 41:1567–1585.e7. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Barzaman K, Karami J, Zarei Z,
Hosseinzadeh A, Kazemi MH, Moradi-Kalbolandi S, Safari E and
Farahmand L: Breast cancer: Biology, biomarkers, and treatments.
Int Immunopharmacol. 84:1065352020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gheibi A, Kazemi M, Baradaran A, Akbari M
and Salehi M: Study of promoter methylation pattern of 14-3-3 sigma
gene in normal and cancerous tissue of breast: A potential
biomarker for detection of breast cancer in patients. Adv Biomed
Res. 1:802012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Luo J, Feng J, Lu J, Wang Y, Tang X, Xie F
and Li W: Aberrant methylation profile of 14-3-3 sigma and its
reduced transcription/expression levels in Chinese sporadic female
breast carcinogenesis. Med Oncol. 27:791–797. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Umbricht CB, Evron E, Gabrielson E,
Ferguson A, Marks J and Sukumar S: Hypermethylation of 14-3-3 sigma
(stratifin) is an early event in breast cancer. Oncogene.
20:3348–3353. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lodygin D and Hermeking H: Epigenetic
silencing of 14-3-3sigma in cancer. Semin Cancer Biol. 16:214–224.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zurita M, Lara PC, del Moral R, Torres B,
Linares-Fernández JL, Arrabal SR, Martínez-Galán J, Oliver FJ and
Ruiz de Almodóvar JM: Hypermethylated 14-3-3-sigma and ESR1 gene
promoters in serum as candidate biomarkers for the diagnosis and
treatment efficacy of breast cancer metastasis. BMC Cancer.
10:2172010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ko S, Kim JY, Jeong J, Lee JE, Yang WI and
Jung WH: The role and regulatory mechanism of 14-3-3 sigma in human
breast cancer. J Breast Cancer. 17:207–218. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Thai AA, Solomon BJ, Sequist LV, Gainor JF
and Heist RS: Lung cancer. Lancet. 398:535–554. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Petrella F, Rizzo S, Attili I, Passaro A,
Zilli T, Martucci F, Bonomo L, Del Grande F, Casiraghi M, De
Marinis F and Spaggiari L: Stage III non-small-cell lung cancer: An
overview of treatment options. Curr Oncol. 30:3160–3175. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Giaccone G and He Y: Current knowledge of
small cell lung cancer transformation from non-small cell lung
cancer. Semin Cancer Biol. 94:1–10. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ansari J, Shackelford RE and El-Osta H:
Epigenetics in non-small cell lung cancer: From basics to
therapeutics. Transl Lung Cancer Res. 5:155–171. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Raungrut P, Petjaroen P, Geater SL,
Keeratichananont W, Phukaoloun M, Suwiwat S and Thongsuksai P:
Methylation of 14-3-3σ gene and prognostic significance of 14-3-3σ
expression in non-small cell lung cancer. Oncol Lett. 14:5257–5264.
2017.PubMed/NCBI
|
|
42
|
Qin X, Qiu F and Zou Z: TRIM25 is
associated with cisplatin resistance in non-small-cell lung
carcinoma A549 cell line via downregulation of 14-3-3σ. Biochem
Biophys Res Commun. 493:568–572. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Radhakrishnan VM, Jensen TJ, Cui H,
Futscher BW and Martinez JD: Hypomethylation of the 14-3-3σ
promoter leads to increased expression in non-small cell lung
cancer. Genes Chromosomes Cancer. 50:830–836. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bahadoram S, Davoodi M, Hassanzadeh S,
Bahadoram M, Barahman M and Mafakher L: Renal cell carcinoma: An
overview of the epidemiology, diagnosis, and treatment. G Ital
Nefrol. 39:2022–vol3. 2022.PubMed/NCBI
|
|
45
|
Cirillo L, Innocenti S and Becherucci F:
Global epidemiology of kidney cancer. Nephrol Dial Transplant.
39:920–928. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bukavina L, Bensalah K, Bray F, Carlo M,
Challacombe B, Karam JA, Kassouf W, Mitchell T, Montironi R,
O'Brien T, et al: Epidemiology of renal cell carcinoma: 2022
Update. Eur Urol. 82:529–542. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chow WH, Dong LM and Devesa SS:
Epidemiology and risk factors for kidney cancer. Nat Rev Urol.
7:245–257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hancock SB and Georgiades CS: Kidney
cancer. Cancer J. 22:387–392. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liang S, Xu Y, Shen G, Zhao X, Zhou J, Li
X, Gong F, Ling B, Fang L, Huang C and Wei Y: Gene expression and
methylation status of 14-3-3sigma in human renal carcinoma tissues.
IUBMB Life. 60:534–540. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
50
|
Vasko R, Mueller GA, von Jaschke AK, Asif
AR and Dihazi H: Impact of cisplatin administration on protein
expression levels in renal cell carcinoma: A proteomic analysis.
Eur J Pharmacol. 670:50–57. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Penny SM: Ovarian cancer: An overview.
Radiol Technol. 91:561–575. 2020.PubMed/NCBI
|
|
52
|
Konstantinopoulos PA and Matulonis UA:
Clinical and translational advances in ovarian cancer therapy. Nat
Cancer. 4:1239–1257. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
O'Shea AS: Clinical staging of ovarian
cancer. Methods Mol Biol. 2424:3–10. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Thi HV, Ngo AD and Chu DT: Epigenetic
regulation in ovarian cancer. Int Rev Cell Mol Biol. 387:77–98.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Mhawech P, Benz A, Cerato C, Greloz V,
Assaly M, Desmond JC, Koeffler HP, Lodygin D, Hermeking H, Herrmann
F and Schwaller J: Downregulation of 14-3-3sigma in ovary, prostate
and endometrial carcinomas is associated with CpG island
methylation. Mod Pathol. 18:340–348. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Dee EC, Wang S, Ho FDV, Patel RR, Lapen K,
Wu Y, Yang F, Patel TA, Feliciano EJG, McBride SM and Lee NY:
Nasopharynx cancer in the United States: Racial and ethnic
disparities in stage at presentation. Laryngoscope. 135:1113–1119.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Su ZY, Siak PY, Lwin YY and Cheah SC:
Epidemiology of nasopharyngeal carcinoma: Current insights and
future outlook. Cancer Metastasis Rev. 43:919–939. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chan SY, To KF, Leung SF, Yip WW, Mak MK,
Chung GT and Lo KW: 14-3-3 sigma expression as a prognostic marker
in undifferentiated nasopharyngeal carcinoma. Oncol Rep.
24:949–955. 2010.PubMed/NCBI
|
|
59
|
Xie L, Jiang T, Cheng A, Zhang T, Huang P,
Li P, Wen G, Lei F, Huang Y, Tang X, et al: MiR-597 targeting
14-3-3σ enhances cellular invasion and EMT in nasopharyngeal
carcinoma cells. Curr Mol Pharmacol. 12:105–114. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang T, Lei F, Jiang T, Xie L, Huang P,
Li P, Huang Y, Tang X, Gong J, Lin Y, et al: H19/miR-675-5p
targeting SFN enhances the invasion and metastasis of
nasalpharyngeal cancer cells. Curr Mol Pharmacol. 12:324–333. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Huang WG, Cheng AL, Chen ZC, Peng F, Zhang
PF, Li MY, Li F, Li JL, Li C, Yi H, et al: Targeted proteomic
analysis of 14-3-3sigma in nasopharyngeal carcinoma. Int J Biochem
Cell Biol. 42:137–147. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Nakajima T, Shimooka H, Weixa P, Segawa A,
Motegi A, Jian Z, Masuda N, Ide M, Sano T, Oyama T, et al:
Immunohistochemical demonstration of 14-3-3 sigma protein in normal
human tissues and lung cancers, and the preponderance of its strong
expression in epithelial cells of squamous cell lineage. Pathol
Int. 53:353–360. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang Z, Tropè CG, Suo Z, Trøen G, Yang G,
Nesland JM and Holm R: The clinicopathological and prognostic
impact of 14-3-3 sigma expression on vulvar squamous cell
carcinomas. BMC Cancer. 8:3082008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Gasco M, Sullivan A, Repellin C, Brooks L,
Farrell PJ, Tidy JA, Dunne B, Gusterson B, Evans DJ and Crook T:
Coincident inactivation of 14-3-3sigma and p16INK4a is an early
event in vulval squamous neoplasia. Oncogene. 21:1876–1881. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Okumura H, Kita Y, Yokomakura N, Uchikado
Y, Setoyama T, Sakurai H, Omoto I, Matsumoto M, Owaki T, Ishigami S
and Natsugoe S: Nuclear expression of 14-3-3 sigma is related to
prognosis in patients with esophageal squamous cell carcinoma.
Anticancer Res. 30:5175–5179. 2010.PubMed/NCBI
|
|
66
|
Qi YJ, Wang M, Liu RM, Wei H, Chao WX,
Zhang T, Lou Q, Li XM, Ma J, Zhu H, et al: Downregulation of
14-3-3σ correlates with multistage carcinogenesis and poor
prognosis of esophageal squamous cell carcinoma. PLoS One.
9:e953862014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lai KKY, Chan KT, Choi MY, Wang HK, Fung
EYM, Lam HY, Tan W, Tung LN, Tong DKH, Sun RWY, et al: 14-3-3σ
confers cisplatin resistance in esophageal squamous cell carcinoma
cells via regulating DNA repair molecules. Tumour Biol.
37:2127–2136. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hayashi E, Kuramitsu Y, Fujimoto M, Zhang
X, Tanaka T, Uchida K, Fukuda T, Furumoto H, Ueyama Y and Nakamura
K: Proteomic profiling of differential display analysis for human
oral squamous cell carcinoma: 14-3-3 σ protein is upregulated in
human oral squamous cell carcinoma and dependent on the
differentiation level. Proteomics Clin Appl. 3:1338–1347. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Laimer K, Blassnig N, Spizzo G, Kloss F,
Rasse M, Obrist P, Schäfer G, Perathoner A, Margreiter R and
Amberger A: Prognostic significance of 14-3-3sigma expression in
oral squamous cell carcinoma (OSCC). Oral Oncol. 45:127–134. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wang S, Zheng R, Li J, Zeng H, Li L, Chen
R, Sun K, Han B, Bray F, Wei W and He J: Global, regional, and
national lifetime risks of developing and dying from
gastrointestinal cancers in 185 countries: A population-based
systematic analysis of GLOBOCAN. Lancet Gastroenterol Hepatol.
9:229–237. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Hu Z, Wang X, Zhang X, Sun W and Mao J: An
analysis of the global burden of gallbladder and biliary tract
cancer attributable to high BMI in 204 countries and territories:
1990-2021. Front Nutr. 11:15217702024. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Sirivatanauksorn V, Dumronggittigule W,
Dulnee B, Srisawat C, Sirivatanauksorn Y, Pongpaibul A, Masaratana
P, Somboonyosdech C, Sripinitchai S, Kositamongkol P, et al: Role
of stratifin (14-3-3 sigma) in adenocarcinoma of gallbladder: A
novel prognostic biomarker. Surg Oncol. 32:57–62. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Sharma A, Sharma KL, Gupta A, Yadav A and
Kumar A: Gallbladder cancer epidemiology, pathogenesis and
molecular genetics: Recent update. World J Gastroenterol.
23:3978–3998. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Nakaoka T, Saito Y and Saito H: Aberrant
DNA methylation as a biomarker and a therapeutic target of
cholangiocarcinoma. Int J Mol Sci. 18:11112017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Singh TD, Gupta S, Shrivastav BR and
Tiwari PK: Epigenetic profiling of gallbladder cancer and gall
stone diseases: Evaluation of role of tumour associated genes.
Gene. 576:743–752. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Li Z, Dong Z, Myer D, Yip-Schneider M, Liu
J, Cui P, Schmidt CM and Zhang JT: Role of 14-3-3σ in poor
prognosis and in radiation and drug resistance of human pancreatic
cancers. BMC Cancer. 10:5982010. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Rodriguez JA, Li M, Yao Q, Chen C and
Fisher WE: Gene overexpression in pancreatic adenocarcinoma:
Diagnostic and therapeutic implications. World J Surg. 29:297–305.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Qin L, Dong Z and Zhang JT: Reversible
epigenetic regulation of 14-3-3σ expression in acquired gemcitabine
resistance by uhrf1 and DNA methyltransferase 1. Mol Pharmacol.
86:561–569. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Dim DC, Jiang F, Qiu Q, Li T, Darwin P,
Rodgers WH and Peng HQ: The usefulness of S100P, mesothelin,
fascin, prostate stem cell antigen, and 14-3-3 sigma in diagnosing
pancreatic adenocarcinoma in cytological specimens obtained by
endoscopic ultrasound guided fine-needle aspiration. Diagn
Cytopathol. 42:193–199. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Qin L, Dong Z and Zhang JT: 14-3-3σ
regulation of and interaction with YAP1 in acquired gemcitabine
resistance via promoting ribonucleotide reductase expression.
Oncotarget. 7:17726–17736. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Venneman K, Huybrechts I, Gunter MJ,
Vandendaele L, Herrero R and Van Herck K: The epidemiology of
Helicobacter pylori infection in Europe and the impact of lifestyle
on its natural evolution toward stomach cancer after infection: A
systematic review. Helicobacter. 23:e124832018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhu Y, Jeong S, Wu M, Zhou JY, Jin ZY, Han
RQ, Yang J, Zhang XF, Wang XS, Liu AM, et al: Index-based dietary
patterns and stomach cancer in a Chinese population. Eur J Cancer
Prev. 30:448–456. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhang Y, Li Y, Lin C, Ding J, Liao G and
Tang B: Aberrant upregulation of 14-3-3σ and EZH2 expression serves
as an inferior prognostic biomarker for hepatocellular carcinoma.
PLoS One. 9:e1072512014. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Kim W, Kidambi T, Lin J and Idos G:
Genetic syndromes associated with gastric cancer. Gastrointest
Endosc Clin N Am. 32:147–162. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Chen YZ, Guo F, Sun HW, Kong HR, Dai SJ,
Huang SH, Zhu WW, Yang WJ and Zhou MT: Association between XPG
polymorphisms and stomach cancer susceptibility in a Chinese
population. J Cell Mol Med. 20:903–908. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Muhlmann G, Ofner D, Zitt M, Müller HM,
Maier H, Moser P, Schmid KW, Zitt M and Amberger A: 14-3-3 Sigma
and p53 expression in gastric cancer and its clinical applications.
Dis Markers. 29:21–29. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Jung JY, Koh SA, Lee KH and Kim JR: 14-3-3
Sigma protein contributes to hepatocyte growth factor-mediated cell
proliferation and invasion via matrix metalloproteinase-1
regulation in human gastric cancer. Anticancer Res. 42:519–530.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Nagappan A, Park HS, Park KI, Hong GE,
Yumnam S, Lee HJ, Kim MK, Kim EH, Lee WS, Lee WJ, et al:
Helicobacter pylori infection combined with DENA revealed altered
expression of p53 and 14-3-3 isoforms in Gulo-/- mice. Chem Biol
Interact. 206:143–152. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Suzuki H, Itoh F, Toyota M, Kikuchi T,
Kakiuchi H and Imai K: Inactivation of the 14-3-3 sigma gene is
associated with 5′ CpG island hypermethylation in human cancers.
Cancer Res. 60:4353–4357. 2000.PubMed/NCBI
|
|
90
|
Li YL, Liu L, Xiao Y, Zeng T and Zeng C:
14-3-3σ is an independent prognostic biomarker for gastric cancer
and is associated with apoptosis and proliferation in gastric
cancer. Oncol Lett. 9:290–294. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Young GM, Radhakrishnan VM, Centuori SM,
Gomes CJ and Martinez JD: Comparative analysis of 14-3-3 isoform
expression and epigenetic alterations in colorectal cancer. BMC
Cancer. 15:8262015. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Winter M, Rokavec M and Hermeking H:
14-3-3σ functions as an intestinal tumor suppressor. Cancer Res.
81:3621–3634. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Shao Q, Duong TN, Park I, Orr LM and
Nomura DK: Targeted protein localization by covalent 14-3-3
recruitment. J Am Chem Soc. 146:24788–24799. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Shao Z, Cai Y, Xu L, Yao X, Shi J, Zhang
F, Luo Y, Zheng K, Liu J, Deng F, et al: Loss of the 14-3-3σ is
essential for LASP1-mediated colorectal cancer progression via
activating PI3K/AKT signaling pathway. Sci Rep. 6:256312016.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Lonare A, Raychaudhuri K, Shah S, Madhu G,
Sachdeva A, Basu S, Thorat R, Gupta S and Dalal SN: 14-3-3σ
restricts YY1 to the cytoplasm, promoting therapy resistance, and
tumor progression in colorectal cancer. Int J Cancer. 156:623–637.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Ide M, Nakajima T, Asao T and Kuwano H:
Inactivation of 14-3-3sigma by hypermethylation is a rare event in
colorectal cancers and its expression may correlate with cell cycle
maintenance at the invasion front. Cancer Lett. 207:241–249. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Ben-Moshe S and Itzkovitz S: Spatial
heterogeneity in the mammalian liver. Nat Rev Gastroenterol
Hepatol. 16:395–410. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Postwala H, Shah Y, Parekh PS and
Chorawala MR: Unveiling the genetic and epigenetic landscape of
colorectal cancer: New insights into pathogenic pathways. Med
Oncol. 40:3342023. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Klemm SL, Shipony Z and Greenleaf WJ:
Chromatin accessibility and the regulatory epigenome. Nat Rev
Genet. 20:207–220. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Deng J, Gao G, Wang L, Wang T, Yu J and
Zhao Z: Stratifin expression is a novel prognostic factor in human
gliomas. Pathol Res Pract. 207:674–679. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Ito K, Suzuki T, Akahira J, Sakuma M,
Saitou S, Okamoto S, Niikura H, Okamura K, Yaegashi N, Sasano H and
Inoue S: 14-3-3sigma in endometrial cancer-a possible prognostic
marker in early-stage cancer. Clin Cancer Res. 11:7384–7391. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Liang S, Shen G, Liu Q, Xu Y, Zhou L, Xiao
S, Xu Z, Gong F, You C and Wei Y: Isoform-specific expression and
characterization of 14-3-3 proteins in human glioma tissues
discovered by stable isotope labeling with amino acids in cell
culture-based proteomic analysis. Proteomics Clin Appl. 3:743–753.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Iavazzo C, Gkegkes ID and Vrachnis N:
Early recurrence of early stage endometrioid endometrial carcinoma:
Possible etiologic pathways and management options. Maturitas.
78:155–159. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Nakayama H, Sano T, Motegi A, Oyama T and
Nakajima T: Increasing 14-3-3 sigma expression with declining
estrogen receptor alpha and estrogen-responsive finger protein
expression defines malignant progression of endometrial carcinoma.
Pathol Int. 55:707–715. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Evren S, Dermen A, Lockwood G, Fleshner N
and Sweet J: mTOR-RAPTOR and 14-3-3σ immunohistochemical expression
in high grade prostatic intraepithelial neoplasia and prostatic
adenocarcinomas: A tissue microarray study. J Clin Pathol.
64:683–688. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Lodygin D, Diebold J and Hermeking H:
Prostate cancer is characterized by epigenetic silencing of
14-3-3sigma expression. Oncogene. 23:9034–9041. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Chu DT, Ngo AD and Wu CC: Epigenetics in
cancer development, diagnosis and therapy. Prog Mol Biol Transl
Sci. 198:73–92. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Devailly G, Grandin M, Perriaud L, Mathot
P, Delcros JG, Bidet Y, Morel AP, Bignon JY, Puisieux A, Mehlen P
and Dante R: Dynamics of MBD2 deposition across methylated DNA
regions during malignant transformation of human mammary epithelial
cells. Nucleic Acids Res. 43:5838–5854. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Feng L, Pan M, Sun J, Lu H, Shen Q, Zhang
S, Jiang T, Liu L, Jin W, Chen Y, et al: Histone deacetylase 3
inhibits expression of PUMA in gastric cancer cells. J Mol Med
(Berl). 91:49–58. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Ye M, Huang T, Ying Y, Li J, Yang P, Ni C,
Zhou C and Chen S: Detection of 14-3-3 sigma (σ) promoter
methylation as a noninvasive biomarker using blood samples for
breast cancer diagnosis. Oncotarget. 8:9230–9242. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Sakai A, Otani M, Miyamoto A, Yoshida H,
Furuya E and Tanigawa N: Identification of phosphorylated serine-15
and −82 residues of HSPB1 in 5-fluorouracil-resistant colorectal
cancer cells by proteomics. J Proteomics. 75:806–818. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Balch C, Naegeli K, Nam S, Ballard B,
Hyslop A, Melki C, Reilly E, Hur MW and Nephew KP: A unique histone
deacetylase inhibitor alters microRNA expression and signal
transduction in chemoresistant ovarian cancer cells. Cancer Biol
Ther. 13:681–693. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Li Y, Geng P, Jiang W, Wang Y, Yao J, Lin
X, Liu J, Huang L, Su B and Chen H: Enhancement of radiosensitivity
by 5-Aza-CdR through activation of G2/M checkpoint response and
apoptosis in osteosarcoma cells. Tumour Biol. 35:4831–4839. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Wang L, Zhang Y, Li R, Chen Y, Pan X, Li
G, Dai F and Yang J: 5-aza-2′-Deoxycytidine enhances the
radiosensitivity of breast cancer cells. Cancer Biother Radiopharm.
28:34–44. 2013.PubMed/NCBI
|
|
115
|
Schultz J, Ibrahim SM, Vera J and Kunz M:
14-3-3sigma gene silencing during melanoma progression and its role
in cell cycle control and cellular senescence. Mol Cancer.
8:532009. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Yang Z, Chu B, Tu Y, Li L, Chen D, Huang
S, Huang W, Fan W, Li Q, Zhang C, et al: Dual inhibitors of DNMT
and HDAC remodels the immune microenvironment of colorectal cancer
and enhances the efficacy of anti-PD-L1 therapy. Pharmacol Res.
206:1072712024. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Huang W, Zhu Q, Shi Z, Tu Y, Li Q, Zheng
W, Yuan Z, Li L, Zu X, Hao Y, et al: Dual inhibitors of DNMT and
HDAC induce viral mimicry to induce antitumour immunity in breast
cancer. Cell Death Discov. 10:1432024. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Liu S, Guo R, Xu H, Yang J, Luo H, Yeung
SJ, Li K, Lee MH and Yang R: 14-3-3σ-NEDD4L axis promotes
ubiquitination and degradation of HIF-1α in colorectal cancer. Cell
Rep. 42:1128702023. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Bai J, Zhao Y, Shi K, Fan Y, Ha Y, Chen Y,
Luo B, Lu Y, Jie W and Shen Z: HIF-1α-mediated LAMC1 overexpression
is an unfavorable predictor of prognosis for glioma patients:
Evidence from pan-cancer analysis and validation experiments. J
Transl Med. 22:3912024. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Parker WB and Thottassery JV:
5-Aza-4′-thio-2′-deoxycytidine, a new orally bioavailable nontoxic
‘best-in-class’: DNA methyltransferase 1-depleting agent in
clinical development. J Pharmacol Exp Ther. 379:211–222. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Traube FR, Brás NF, Roos WP, Sommermann
CC, Diehl T, Mayer RJ, Ofial AR, Müller M, Zipse H and Carell T:
Epigenetic anti-cancer treatment with a stabilized carbocyclic
decitabine analogue. Chemistry. 28:e2022006402022. View Article : Google Scholar : PubMed/NCBI
|















