Introduction
Ovarian cancer (OC), one of the most lethal
malignancies of the female reproductive system, has a morbidity
rate of 3.7% and a mortality rate of 4.7%, threatening the lives
and health of patients (1). As the
ovary is located in the deep pelvis, early lesions often show
nonspecific symptoms, such as abdominal distension, abdominal pain
and indigestion, which can be easily neglected or misdiagnosed as
other diseases. Most patients are already in an advanced stage when
diagnosed with OC, and the 5-year survival rate is <45%, which
seriously affects patient quality of life (2). At present, the diagnosis of OC mainly
relies on comprehensive methods, such as imaging, serum tumor
marker detection and histopathology; however, these methods have
limitations regarding sensitivity and specificity for early
diagnosis (3). Therefore, exploring
new diagnostic and therapeutic targets is of great importance for
improving the early diagnosis rate and therapeutic effects of OC
(4).
Long non-coding (lnc)RNA are a class of non-coding
RNA molecules with a length of >200 nucleotides that do not
encode proteins but serve an important biological function in
tumorigenesis and development. They can interact with DNA and
transcription factors to regulate the transcription initiation,
elongation and termination of genes (5). They also serve as molecular scaffolds,
recruit relevant RNA-binding proteins, form ribonucleoprotein
complexes and participate in mRNA splicing, translocation and
stability regulation (6). lncRNAs
typically act as competitive endogenous RNAs and bind to microRNAs
(miRNAs/miRs), interfering with the normal regulation of their
target mRNAs by miRNAs, thus affecting the stability and
translation efficiency of mRNAs (7). Several studies have reported that
lncRNAs are associated with cancer development and generally
exhibit abnormal expression patterns (8–10). For
example, in hepatocellular carcinoma, aberrant expression of
lncRNAs, such as zinc finger E-box binding homeobox 1
(ZEB1)-antisense RNA 1 (AS1), hepatocellular
carcinoma upregulated lncRNA and metastasis associated lung
adenocarcinoma transcript 1 (MALAT1), directly promotes
tumor growth and metastasis, including abnormal cell proliferation,
invasion and metastasis (11,12).
The lncRNA integrin (ITG)B8-AS1 regulates colorectal cancer
cell proliferation by sequestering let-7c-5p/let-7d-5p and
miR-33b-5p, which in turn affects the expression of ITG family
members, such as ITGB3 and ITGA3, thereby modulating
focal adhesion signaling (13). In
OC, HOX transcript antisense RNA upregulation in OC cells
was reported to enhance cell growth and migration by interacting
with miR-222-3p (14).
MALAT1, a key cancer-associated lncRNA, is overexpressed in
OC and regulates cell proliferation and DNA synthesis via the
miR-506-iASPP axis (15). lncRNA
toll like receptor 8 (TLR8)-AS1 was reported to
enhance OC chemoresistance and metastasis by increasing the
stability of TLR8 mRNA (16). Conversely, overexpression of
maternally expressed 3 in OC was reported to be beneficial
for patient prognosis and modulate the miR-219a-5p/EGFR axis
(17). Homeobox D-AS1 can
enhance the invasion and epithelial-mesenchymal transition process
of OC cells by targeting miR-133a-3p and activating the
Wnt/β-catenin signaling pathway, making tumor cells more metastatic
and malignant (4).
Resveratrol (RES), a polyphenolic phytonutrient with
several biological properties, has shown promising anticancer
potential in preclinical studies, exerting cytotoxic effects and
inducing apoptosis in tumor cells (18). RES can inhibit cell proliferation by
targeting key signaling pathways that regulate cyclins or induce
apoptosis by activating apoptosis-related proteins. The anticancer
effects of RES have been reported in several cancer types, which
are associated with the regulation of lncRNAs (19). Research has revealed that RES exerts
its antitumor effects by downregulating the expression of
tumor-promoting lncRNAs, including MALAT1 and small
nucleolar RNA host gene (SNHG)16 (20). Our previous study further
demonstrated that RES markedly inhibited the proliferation of A2780
cells whilst regulating both circular RNA and miRNA expression.
Several important candidate non-coding RNAs were also screened
(21). However, the specific
molecular mechanisms of RES in the lncRNA regulatory network
affecting cell proliferation and apoptosis have not yet been fully
elucidated.
The present study aimed to assess the key biological
processes and signaling pathways regulated by RES in OC cells using
A2780 cell line as a model. Changes in lncRNA expression profiles
after RES treatment were analyzed using high-throughput sequencing
technology, screening five highly responsive RES hub genes, and
constructing lncRNA-mRNA interaction networks. The findings of the
present study could deepen the understanding of the molecular
mechanism of RES against OC, provide potential biomarkers for the
early diagnosis of OC, and lay a theoretical foundation for the
development of novel targeted therapeutic strategies based on
lncRNA regulation.
Materials and methods
Cell culture and RES treatment
The human A2780 OC cell line [cat. no. iCell-h004,
Mirror Qidian (Shanghai) Cell Technology Co., Ltd.] was cultivated
in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.),
supplemented with 10% FBS and 1% antibiotic-antimycotic solution
(all Gibco; Thermo Fisher Scientific, Inc.) at 37°C and 5%
CO2 atmospheric conditions. Cells were incubated with 75
µM RES (Beijing Solarbio; Beijing, China) at 37°C for 24 h, where
indicated, a dose previously reported to approximate an
IC50 dose (21). The
cells were confirmed to be free of Mycoplasma contamination
and subjected to short tandem repeat identification.
RNA sequencing and expression of
lncRNAs
A2780 cells were treated with or without 75 µM RES
for 24 h, and TRIzol™ (Invitrogen™; Thermo Fisher Scientific, Inc.)
were used to isolate total RNA from cells. RNA quality was
quantified using RNase-free agarose gel electrophoresis using an
Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). After
eliminating ribosomal RNAs from total RNA, the samples were diluted
in fragmentation buffer before reverse transcription of the
mRNA/non-coding RNA transcripts with random primers using the VAHTS
Universal V6 RNA-seq Library Prep Kit for Illumina (Vazyme Biotech
Co., Ltd., Nanjing, China, Cat no. NR604). First-strand cDNA
synthesis was performed at 25°C for 10 min, 42°C for 15 min, and
70°C for 15 min; second-strand cDNA synthesis was then performed at
16°C for 30 min, 65°C for 15 min. The cDNA fragments were purified
using the QiaQuick PCR extraction kit (cat no. 28104, Qiagen, Inc.)
and subsequently ligated to an Illumina sequencing adapter
(Illumina, Inc.), to construct cDNA libraries. After digestion, PCR
products were amplified and the library concentration was measured
using the DNA 1000 assay kit (Agilent Technologies, Inc.), with
quantification and pooling performed using the ABI StepOnePlus
Real-Time PCR System (Thermo Fisher Scientific, Inc.). The
libraries, with a loading concentration of 3 ng/µl, were then
sequenced by Guangzhou Gene Denovo Biotechnology Co., Ltd. using
the HiSeq 2500 Sequencing System (Illumina, Inc.) with a paired-end
150 bp sequencing strategy. Raw sequence reads were screened using
fastp (version 0.18.0) to eliminate low-quality bases and adapter
sequences, and high-quality clean reads were obtained (22). Once the reference genome index was
constructed, clean reads from RNA sequencing were mapped to the
reference genome using HISAT2 (version 2.1.0) (23). Transcript reconstruction was
performed using StringTie (version 1.3.4) in combination with
HISAT2 to identify the splice variants of known and novel genes
(24–26). The protein-coding ability of the
novel transcripts was evaluated using CNCI (version 2), CPC
(version 0.9-r2) and FEELNC (version v0.2) with default parameters
(27–29). The non-protein-coding ability
results were intersected as lncRNAs, which were then divided into
five types according to their comparison with protein-coding genes:
Bidirectional, intergenic, antisense, intronic and
sense-overlapping lncRNAs. For lncRNAs and mRNAs, the differential
expression of their transcripts was assessed separately using
DESeq2 (version 1.20.0) between groups and edgeR (version 3.20.9)
between samples (30,31). Differential expression of
genes/transcripts was determined using |fold change|≥2 and false
discovery rate of <0.05.
Bioinformatics analysis
RNAplex (version 0.2) and the Vienna RNA package
were used to predict the complementary relationships between
antisense lncRNAs and mRNA (32).
Optimal base pairing was predicted using minimal free-energy
prediction based on thermodynamics. lncRNAs can function to
cis-regulate the genes surrounding an identical allele
(33). Upstream lncRNAs that
interact with promoters or additional cis-elements can
modulate gene expression transcriptionally or
post-transcriptionally (33).
Typically, lncRNAs in the 3′untranslated region or downstream can
show additional regulatory effects. Therefore, lncRNAs annotated as
‘unknown region’ were reannotated as cis-regulators at
<10 kb up/downstream of one specific gene. lncRNAs can also
trans-regulate long-distance genes (33). Pearson's correlation coefficients
were calculated between lncRNA and protein-coding genes, with the
pairs satisfying |correlation|>0.999, and Gene Ontology (GO) and
Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were
performed. Moreover, the functions of the lncRNA parental genes
were evaluated using GO and KEGG analyses. GO annotation was
performed using GO resources (http://www.geneontology.org/).
lncRNA-mRNA association analysis
Based on the results of KEGG enrichment analysis,
combined with published evidence supporting the anticancer
mechanism pathways, including the p53 signaling pathway, oocyte
meiosis and cell cycle pathways, differentially expressed lncRNAs
(DELs) and mRNAs and in these pathways were screened for
co-expression analysis (34–36).
The lncRNA-mRNA co-expression network was visualized using
Cytoscape software (version 3.9.1, cytoscape.org/).
Validation of DELs and mRNAs using
RT-quantitative (qPCR)
RT-qPCR was performed to assess DELs and mRNA
expression. Total RNA was extracted using the Ultrapure RNA kit
(DNase I, Cat no. CW0597S, Jiangsu Cowin Biotech Co., Ltd). cDNA
was synthesized using Easyscript one-step gDNA removal and cDNA
synthesis supermix (Cat no. AE-311, TransGen Biotech Co., Ltd.)
with incubation at 42°C for 1 h. Specific primers were designed for
RT-qPCR assays in a 20 µl reaction (Table SI). Reactions were performed using
a Light-Cycler96 instrument (Roche Diagnostics) with the TransStart
green qPCR supermix containing SYBR GREEN (Cat no. AQ131-01,
TransGen Biotech Co., Ltd.). The thermocycling conditions were as
follows: pre-denaturation at 94°C for 30 sec, followed by 40 cycles
with denaturation at 94°C for 5 sec and annealing at 61°C for 35
sec. Post-amplification, melting curve analysis was performed at
95°C for 10 sec, 65°C for 60 sec, and 97°C for 1 sec. Relative RNA
expression subsequently calculated as 2−ΔΔCq values
based on GAPDH as the housekeeping gene (37). Each experiment was performed in
triplicate to ensure reproducibility.
Cell transfection
pcDNA3.1-SNHG15-203 overexpression plasmids
(Nanjing GenScript Co., Ltd.) and small interfering (si)RNAs
targeting SNHG15-203 (Table
SII, Shanghai GenePharma Co., Ltd.) were constructed and
transfected into A2780 cells. Specifically, 200 nM siRNA duplexes
or the overexpression plasmid, and 2.5 µl Lipofectamine™ 2000
(Thermo Fisher Scientific, Inc.) were diluted in 100 µl Opti-MEM,
mixed, and incubated at 25°C for 20 min before being added to each
well of a 12-well plate containing target cells. An additional 800
µl Opti-MEM was introduced, followed by gentle shaking.
Transfection was performed at 25°C. The transfection mixture was
replaced with fresh medium without antibiotics after 6–8 h, and the
cells were further cultured at 37°C for 48 h before harvesting.
RT-qPCR was performed to validate the overexpression and knockdown
efficiency, as aforementioned.
Cell Counting Kit-8 (CCK-8) assay
Following transfection, cell proliferation was
assessed using a CCK-8 kit (Biosharp Life Sciences). Briefly, the
CCK-8 reagent was added to the treated cells cultured in a 96-well
plate. After incubation for 48 h, the optical density of each well
was measured at 450 nm using a microplate reader (NanJing DeTie
Laboratory Equipment Co., Ltd.). Each experiment was performed in
triplicate.
Transwell assay
Cell migration was assessed using Transwell chambers
(Corning, Inc.) with a pore size of 8 µm. A total of
2×105 cells in 1 ml serum-free medium was added to the
upper Transwell chamber, and 600 µl complete medium containing 10%
FBS was added to the lower chamber. After 24 h incubation at 37°C,
cells were fixed in paraformaldehyde at room temperature for 15 min
and stained with crystal violet at room temperature for 10 min. The
number of migrating cells in random areas was counted under an
inverted microscope (IX51; Olympus Corporation).
Statistical analysis
GraphPad Prism statistical software (version 8.0;
Dotmatics) was used for experimental data processing. For
comparisons between two groups, an unpaired two-tailed Student's
t-test was used if the data were normally distributed and had equal
variances; otherwise, the Mann-Whitney U test was applied. For
comparisons among multiple groups, one-way analysis of variance was
used for parametric data, followed by Tukey's HSD test for multiple
comparisons; otherwise, the Kruskal-Wallis H-test was used,
followed by Dunn's test for post hoc pairwise comparisons.
Pearson's correlation coefficient was used for correlation
analysis. Data are expressed as mean ± standard error of the mean
of ≥3 independent experimental repeats. P<0.05 was considered to
indicate a statistically significant difference.
Results
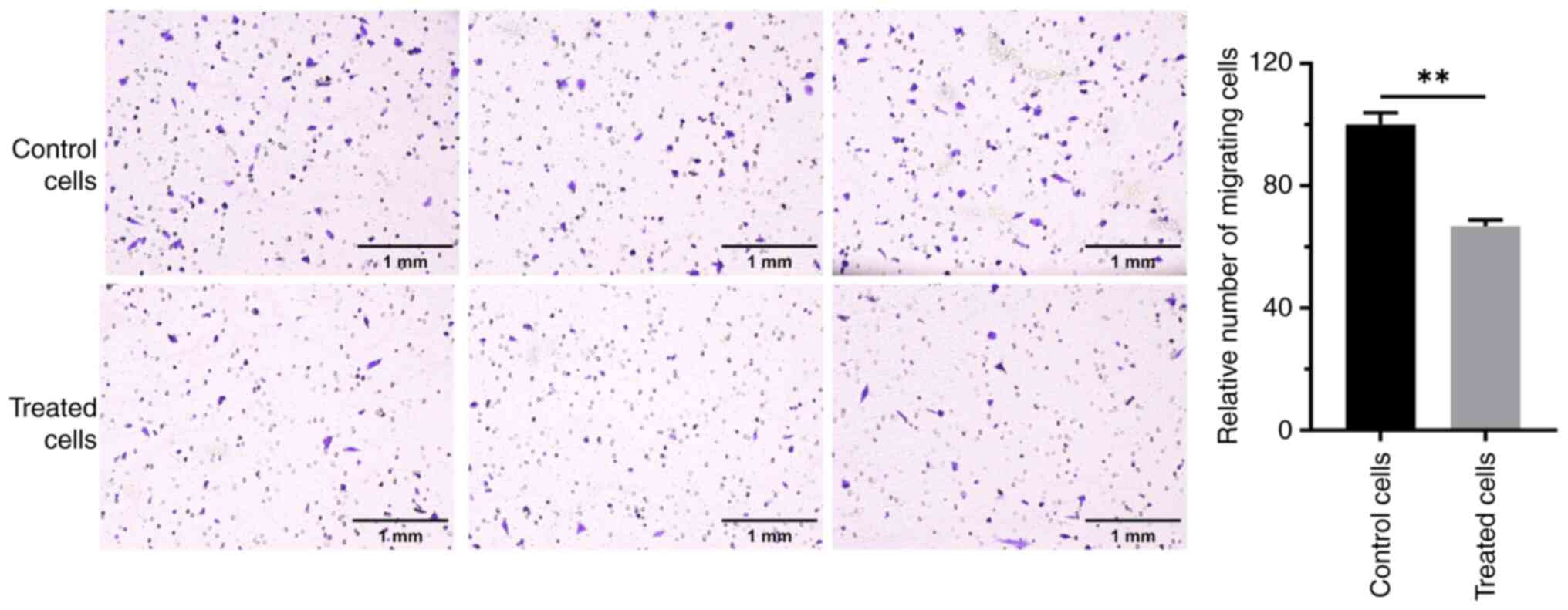
Effect of RES on the migration of OC
cells
The effects of RES treatment on the migration of the
OC cell line, A2780, were assessed. Transwell assays revealed that
cells in the control group had a strong migration ability. The
number of cells passing through the membrane was high, and numerous
cells were observed on the lower surface of the membrane in the
field of view (Fig. 1). By
contrast, the migration ability of cells treated with RES for 24 h
was notably altered, with the number of cells crossing the membrane
was greatly reduced. The difference was significant compared with
that in the control group (P<0.01). These results indicate that
RES significantly inhibited the migration of A2780 OC cells.
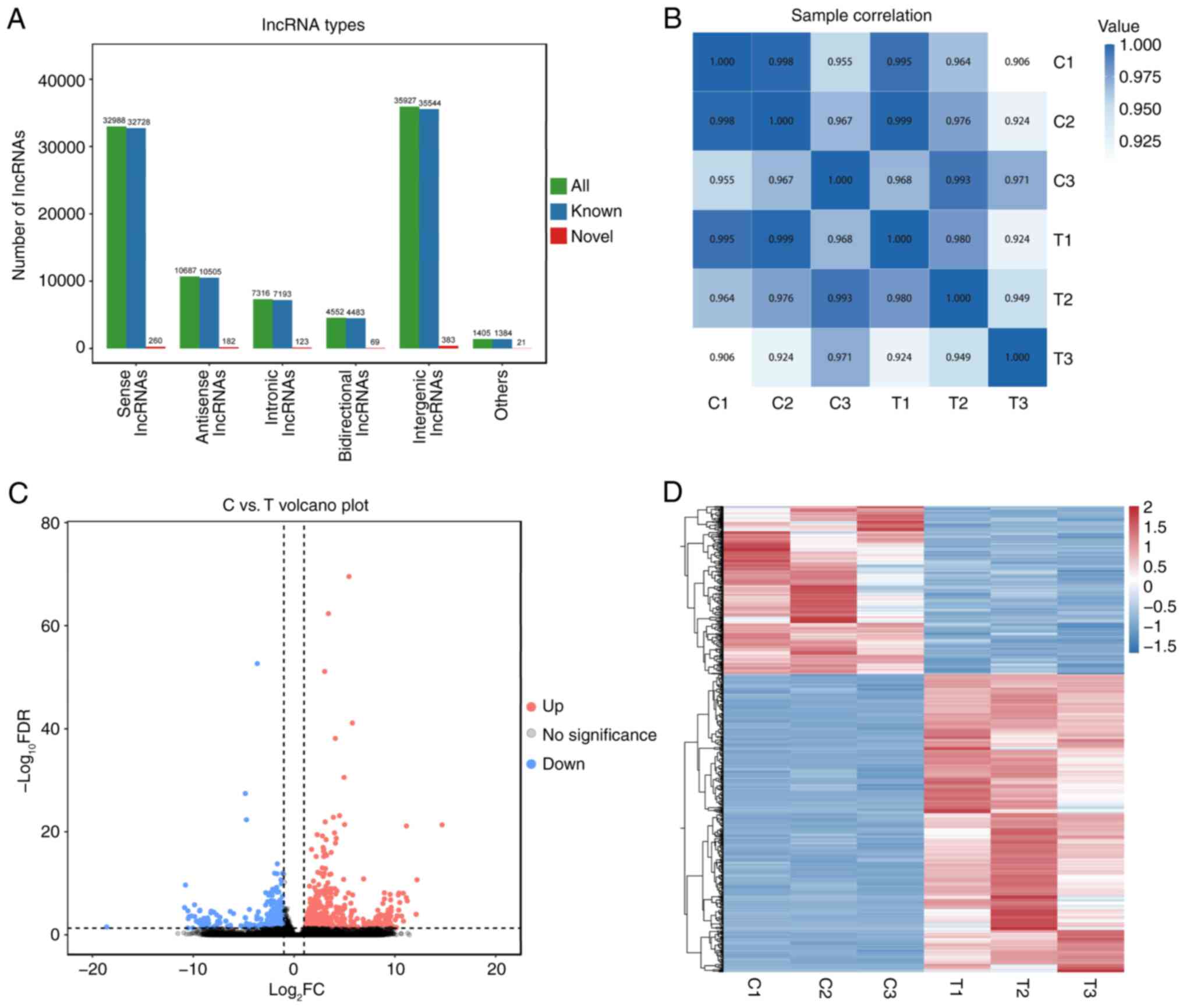
RES treatment alters the lncRNA
expression profile of OC cells
To evaluate the potential mechanism underlying the
inhibitory effect of RES on the proliferation of OC cells,
lncRNA-sequencing analysis was performed on the OC cell line A2780
in the control and RES-treated groups. Novel lncRNAs were first
identified and categorized into the following five groups based on
their positions in the genome: 32,988 sense lncRNAs, 10,687
antisense lncRNAs, 7,316 intronic lncRNAs, 4,552 bidirectional
lncRNAs and 35,927 intergenic lncRNAs (Fig. 2A). Pearson's correlation analysis
demonstrated that lncRNA expression among biological replicates
within groups showed a high degree of similarity, with significant
differences between groups (Fig.
2B). Further analysis revealed 721 DELs, of which 461 were
upregulated and 260 were downregulated (Fig. 2C). Hierarchical clustering analysis
demonstrated significant differences in the lncRNA expression
patterns between the two groups (Fig.
2D). Notably, lncRNAs H19-203 and vacuole membrane
protein 1 (VMP1)-217 were highly expressed in the
RES-treated group, whereas lncRNAs enolase 1 (ENO1)-210,
SNHG15-203 and SNHG1-266 were markedly downregulated
(Table SIII). Meanwhile, 24 SNHG
family lncRNAs were identified, including lncRNA
SNHG15-203.
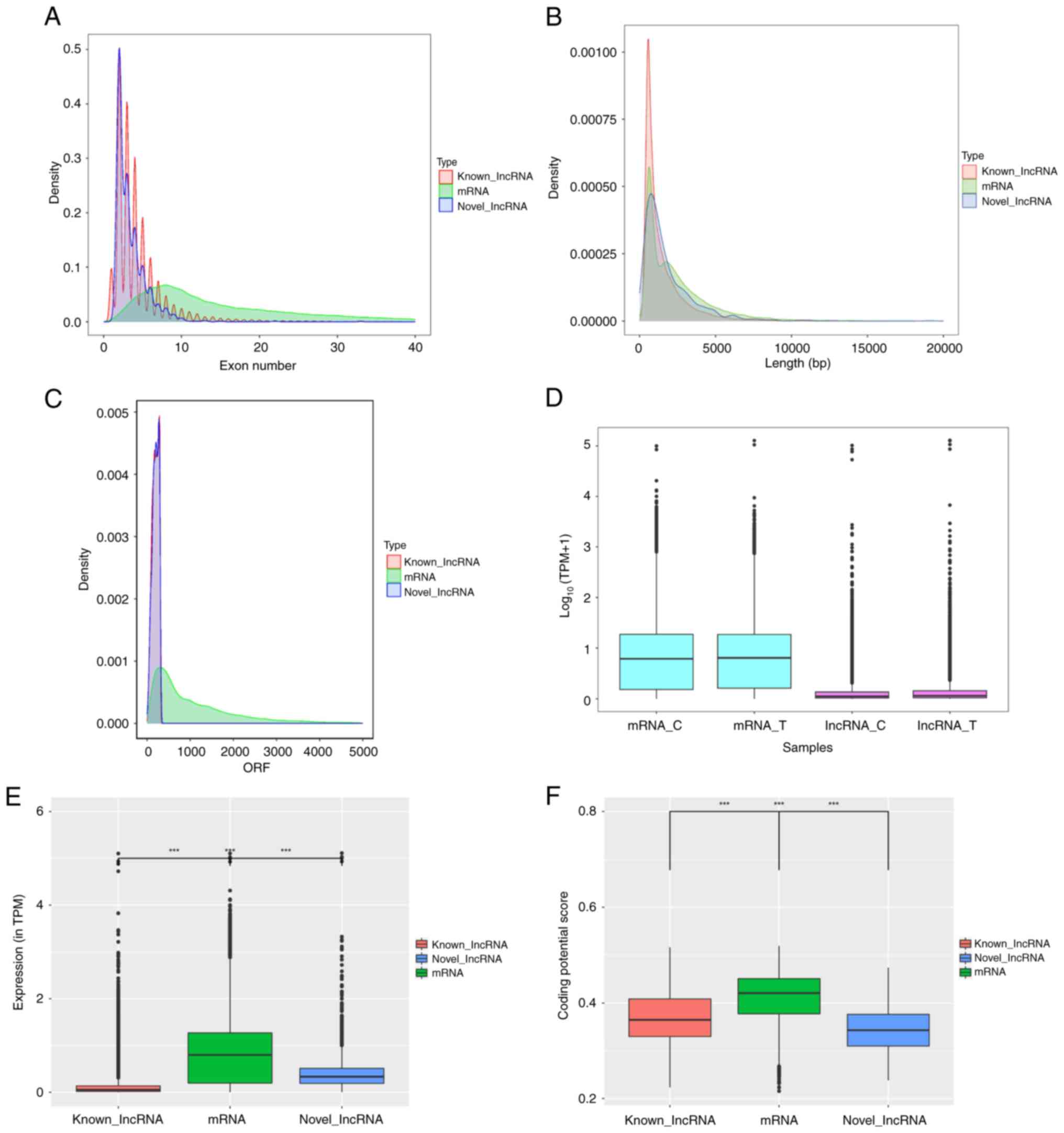
Feature distribution comparison of
lncRNAs and mRNAs
Previously, mRNA sequencing in the RES-treated OC
cell line A2780 revealed significant changes in mRNA expression in
RES-treated cells and clarified its key role in cell proliferation
and immunomodulation. Based on this, the potential roles of lncRNAs
and their synergistic regulation of mRNAs were further explored.
Compared with mRNAs, both annotated and novel lncRNAs exhibited
fewer exons, shorter lengths and lower coding potentials (Fig. 3A-C). Analysis of total lncRNA
expression data demonstrated markedly lower levels than those of
mRNA, and the abundances of lncRNAs and mRNAs were notably
increased by RES treatment (Fig.
3D). Differences in transcript abundance were also observed
between DELs and mRNAs (Fig. 3E).
In addition, the coding potential scores of differentially
expressed mRNAs were higher than those of the lncRNAs, and those of
novel lncRNAs were significantly lower than those of the annotated
lncRNAs (Fig. 3F).
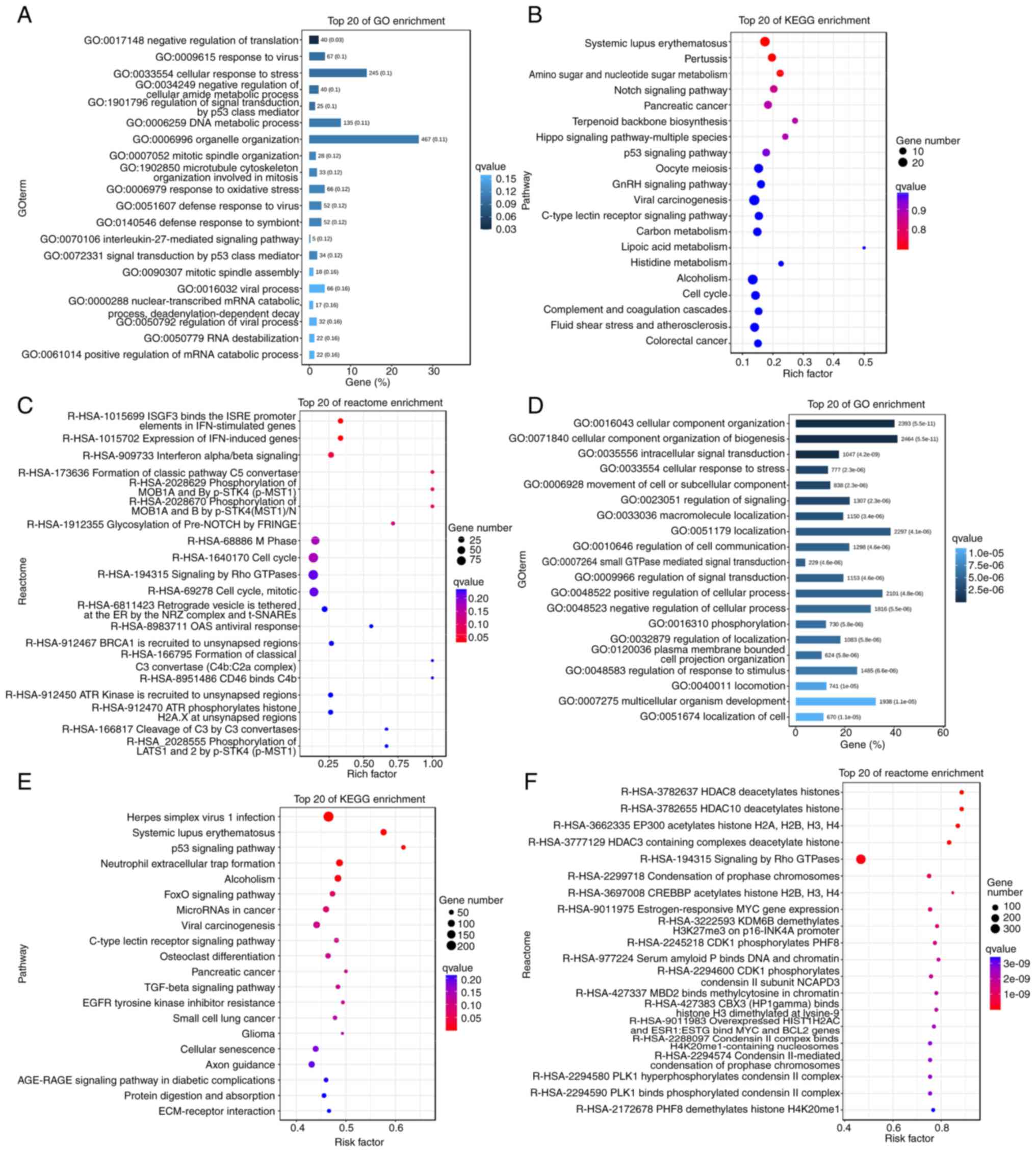
Target gene prediction and functional
enrichment analysis of lncRNAs
To further assess the potential function of lncRNAs
in the inhibition of OC cell proliferation by RES, the
complementary binding relationship between lncRNAs and mRNAs was
analyzed and the potential function of lncRNAs was inferred based
on the functions of known mRNAs. The DELs were first predicted for
cis and trans roles, and 5,503 and 1,780,009
lncRNA-mRNA target gene pairs were obtained, respectively.
Subsequently, GO functional enrichment analysis of the DELs was
performed. The results revealed that the target genes associated
with these differentially expressed cis-acting lncRNAs were
significantly enriched in several key biological processes, such as
response to viruses, regulation of signal transduction by p53 class
mediators, DNA metabolic processes and response to oxidative stress
(Fig. 4A). KEGG enrichment analysis
further indicated that these cis-acting lncRNAs were mainly
involved in carcinogenesis, Notch, Hippo, cell cycle and other
classical tumor-related signaling pathways (Fig. 4B). The Reactome pathways involved
also covered cell cycle progression and antiviral activity, which
corroborated the GO and KEGG analysis results (Fig. 4C). the GO function of the
differentially expressed trans-acting lncRNAs were analyzed,
which revealed that they were mainly enriched in signal
transduction and response to stress and stimuli (Fig. 4D). The results of KEGG enrichment
analysis demonstrated that the trans-acting lncRNAs were
mainly involved in p53, Forkhead Box (Fox)O signaling pathway,
cellular senescence, extracellular matrix-receptor interaction and
cancer-related signaling pathways, whilst the Reactome pathways
included deacetylation of histones, Signaling by Rho GTPases and
phosphorylation (Fig. 4E and
F).
Furthermore, certain genes were revealed to be
involved in multiple important biological functions (Fig. 4A-F). For example, cyclin
dependent kinase inhibitor 1A (CDKN1A) and sequestosome
(SQSTM) were enriched in cellular entries, anatomical
entities, binding, cellular processes and biological regulation,
and were associated with cellular senescence and cell cycle
signaling pathways. Reactome analysis revealed certain specific
pathways, such as estrogen-responsive MYC gene expression,
lysine demethylase 6B demethylation of H3K27me3 on the p16-INK4A
promoter, CDK1 phosphorylation of PHD finger protein 8,
binding of serum amyloid P to DNA and chromatin, and binding of
methyl-CpG binding domain protein 2 to methylcytosine in chromatin.
These specific pathways contain several members of the histone H2A
family, such as H2AC7, H2AC20 and H2AC18. Therefore,
we hypothesize that the regulatory network centered on CDKN1A,
SQSTM1 and histones may serve a crucial role in the inhibition
of A2780 cell proliferation by RES.
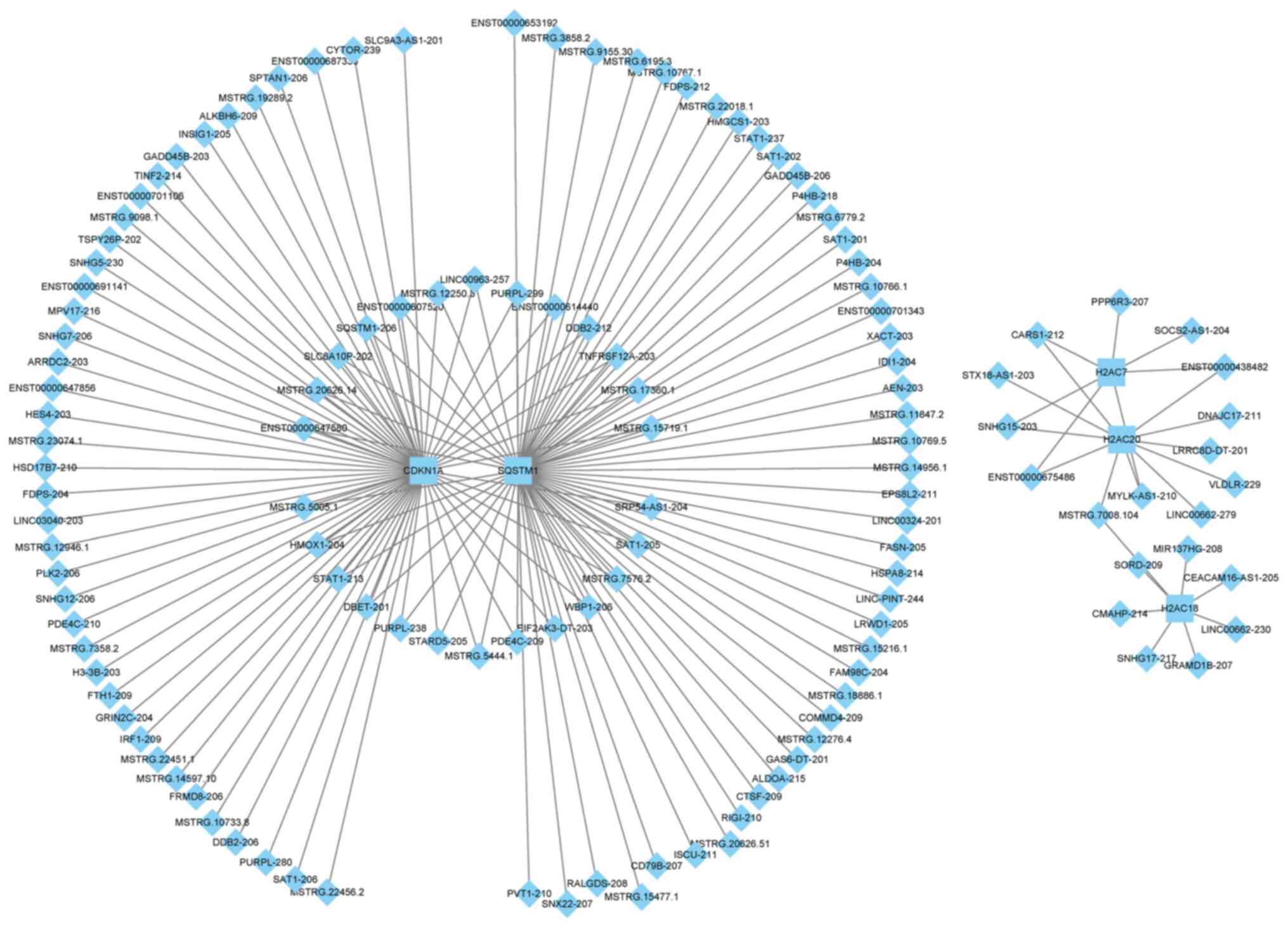
lncRNA-mRNA networks
A total of five mRNAs associated with the p53
signaling pathway, oocyte meiosis, cellular senescence, cell cycle
and their associated lncRNAs were selected and lncRNA-mRNA
regulatory networks were constructed using Cytoscape. The results
revealed that CDKN1A and SQSTM1 formed a large
regulatory network, whereas the other genes, H2AC7, H2AC20
and H2AC18, formed a small network. Moreover, there was no
overlap in the lncRNAs involved in the two networks (Fig. 5 and Table SIV). In the large network,
CDKN1A and SQSTM1 demonstrated extensive regulatory
associations, with shared regulatory links with 26 lncRNAs,
including lnc-SQSTM1-206 and MSTRG.5444.1. It also
formed unique regulatory relationships with MSTRG.22456.2
and MSTRG.3858.2. In the small network, H2AC7 and
H2AC20 were targeted by five lncRNAs, including
SNHG15-203 and cysteinyl-tRNA synthetase 1–212. A specific
linkage between H2AC18, H2AC2 and MSTRG.7008.104
allowed the integration of the three histone genes into the same
network.
Validation of expression levels of
lncRNAs and genes
To verify the accuracy of the data, five lncRNAs
(ENO1-210, SNHG1-266, SNHG15-203, VMP1-217 and
H19-203) were randomly selected and their expression levels
were assessed using RT-qPCR. The expression levels of ENO1-210,
SNHG1-266 and SNHG15-203 were significantly
downregulated, and the expression levels of VMP1-217 and
H19-203 were significantly upregulated in the RES-treated
group compared with in the control group (Fig. 6A). These results are consistent with
the RNA sequencing results (Table
SIII).
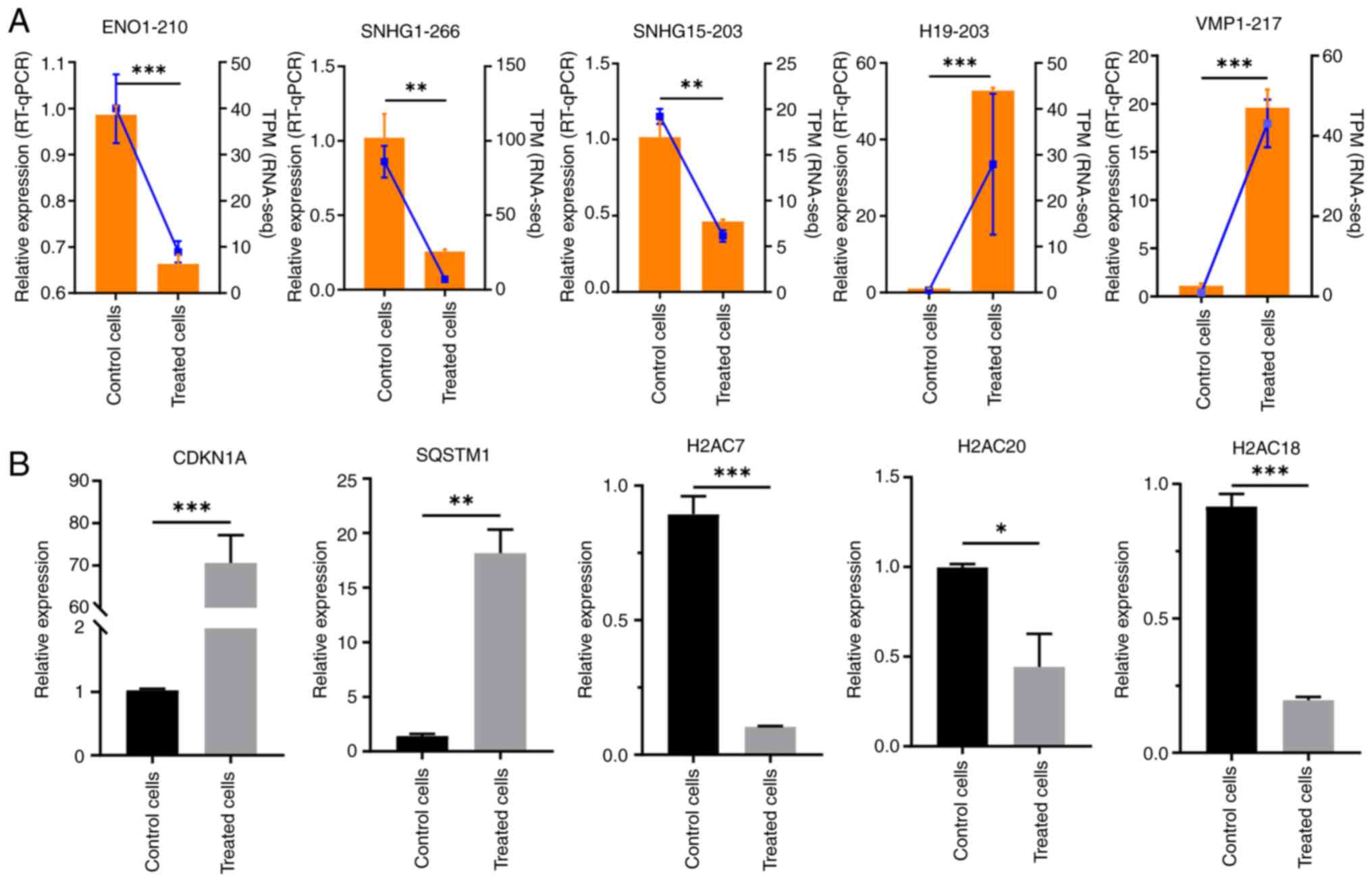 | Figure 6.RT-qPCR validation of long non-coding
RNA and mRNA expression in 75 µM resveratrol-treated ovarian cancer
cells. (A) RT-qPCR and RNA-seq quantification results of lncRNA.
Orange bars indicate RT-qPCR results, and blue lines indicate
RNA-seq results. (B) RT-qPCR results of mRNA expression.
*P<0.05; **P<0.01; ***P<0.001. RT-qPCR, reverse
transcription-quantitative PCR; RNA-seq, RNA-sequencing; TPM,
transcripts per million; ENO1, enolase 1; SNHG, small nucleolar RNA
host gene; VMP1, vacuole membrane protein 1; CDKN1A, cyclin
dependent kinase inhibitor 1A; SQSTM1, sequestosome 1; H2AC, H2A
clustered histone. |
The expression levels of key mRNAs associated with
the lncRNAs were further evaluated. A total of five key genes,
CDKN1A, SQSTM1, H2AC7, H2AC20 and H2AC18, were
evaluated for changes in expression levels, confirming alterations
in the mRNA levels of each gene after RES treatment. Among these,
CDKN1A and SQSTM1 were significantly upregulated,
whereas H2AC7, H2AC20 and H2AC18 were significantly
downregulated, compared with in the control group (Fig. 6B).
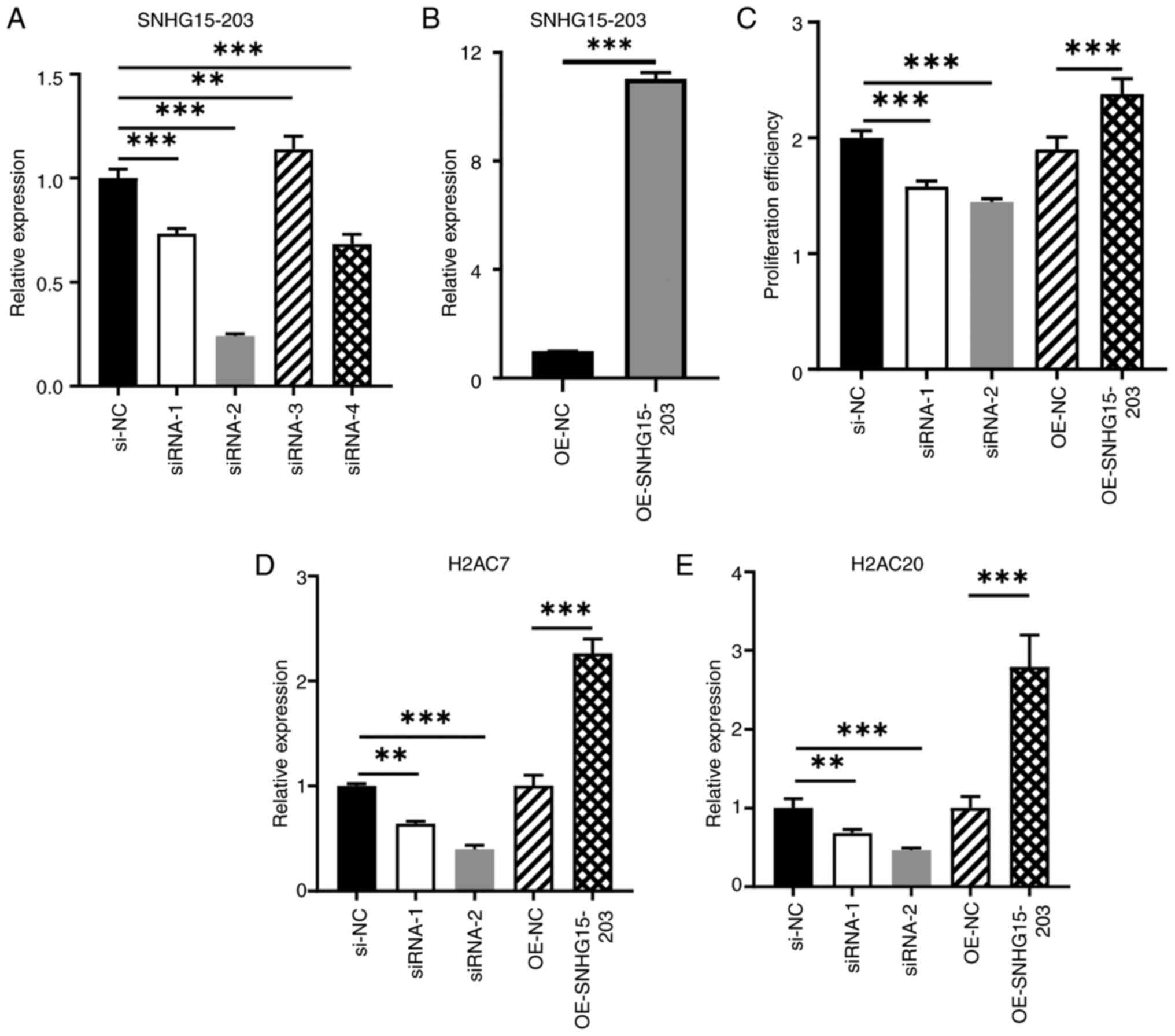
lnc-SNHG15-203 regulates OC cell
proliferation and target gene expression
To assess the predicted regulatory network,
lnc-SNHG15-203 was selected for an in-depth study, as it was
predicted to regulate both H2AC7 and H2AC20. First,
four siRNAs and plasmids were designed to knock-down and
overexpress lnc-SNHG15-203 in A2780 cells, respectively. The
knockdown efficiencies of different siRNAs varied, with the
knockdown efficiency of siRNA-2 at >70%, whereas that of siRNA-1
was ~30% (Fig. 7A). By contrast,
the overexpression group demonstrated significant upregulation,
with the expression level increasing by ~12-fold compared with that
in the control group (Fig. 7B). For
further validation, siRNA-1 and siRNA-2 were also used. The CCK-8
cell proliferation assay results demonstrated that both siRNA-1 and
siRNA-2 significantly inhibited the proliferation of A2780 cells
compared with the siRNA negative control, and lnc-SNHG15-203
overexpression notably promoted cell proliferation compared with
the empty vector control group (Fig.
7C). Compared with the negative control group, the expression
levels of its potential target genes, H2AC7 and
H2AC20, were notably downregulated after
lnc-SNHG15-203 knock-down (Fig.
7D and E). Conversely, compared with the empty vector control
group, lnc-SNHG15-203 overexpression significantly
upregulated H2AC7 and H2AC20, with their expression
levels increased by ~2- and 3-fold, respectively. These results
suggest that knockdown of lnc-SNHG15-203, a type of lncRNA
that is downregulated by RES, could inhibit the expression of its
target genes and cell proliferation, whereas its overexpression
exerts the opposite effects.
Discussion
OC is the leading cause of mortality in women with
gynecological cancers, with marked heterogeneity. Currently,
surgery combined with chemotherapy is the mainstream treatment for
OC. Although initial responses to therapy are often encouraging,
there is a high recurrence rate, leading to a poor prognosis.
Therefore, there is an urgent need to investigate the progression
mechanisms of OC and develop novel therapeutic strategies. The
natural product RES has been reported to have complex anticancer
activities (38). Therefore, the
present study analyzed the changes in lncRNA expression profiles
after RES treatment in A2780 cells, a prototypical model of OC, to
reveal the molecular mechanism by which RES inhibits the
proliferation of OC cells by regulating the lncRNA-mRNA network.
The analysis identified 1,038 novel lncRNA transcripts and 721
DELs. RES-regulated DELs were demonstrated to be mainly involved in
the p53, FoxO, cellular senescence and cell cycle signaling
pathways, which are biological processes associated with
tumorigenesis and development (34–36).
Reactome enrichment identified special pathways containing members
of the histone H2A family of genes. These genes serve important
regulatory roles in gene expression, DNA repair and structural
stability of chromatin (39,40).
By combining the functional pathways and effect of
RES on the mRNA expression profile from a previous study (21), the present study constructed a
lncRNA-mRNA regulatory network. This yielded two discrete networks:
A larger network formed by the upregulated CDKN1A and
SQSTM1 genes, and a smaller network involving the
downregulated H2AC7, H2AC20 and H2AC18 genes. In
particular, key lncRNAs in the CDKN1A-SQSTM1 regulatory
network serve important regulatory roles in several cancers. For
instance, detected as part of the CDKN1A-SQSTM1 network,
lnc-MAP3K13-7:1 suppresses OC proliferation in the context
of polycystic ovary syndrome by inducing hypomethylation of the
CDKN1A promoter by downregulating DNA methyltransferase 1
(41). lnc-cancer susceptibility 15
was reported to modulate GC cell proliferation, migration and
epithelial-mesenchymal transition via ZEB1 and CDKN1A
(42). Moreover, the lncRNA RUN
and SH3 domain containing 1-AS1 was reported to enhance breast
cancer cell growth through the epigenetic silencing of CDKN1A,
Kruppel-like factor transcription factor 2 and Misato homolog 2
pseudogene (MSTO2P) upregulation in colorectal cancer, which
promotes growth via enhancer of zeste homolog 2-mediated
epigenetic silencing of CDKN1A (43,44).
MSTO2P inhibition inhibited the growth and migration of HT29
and SW480 colorectal cancer cells, leading to cell cycle arrest and
apoptosis. MNX1-AS1 is also markedly upregulated in GC
compared with its matched counterpart tissues, affecting overall
patient survival. Furthermore, it is functionally implicated in GC
cell invasion and migration. MNX1-AS1 overexpression has
also been reported to downregulate CDKN1A expression
(45). Additionally, prostate
cancer-associated transcript 1 was reported to promote the
growth, invasion and migration of MGC-803 and AGS cells via
CDKN1A (46). High
expression of CDKN1A is considered an important prognostic
factor for improving the overall survival of OC, and SQSTM1
has a notable impact on the multidrug resistance of OC (47,48).
Nevertheless, this network involving multiple molecular
interactions and regulation, requires further in-depth studies to
elucidate the specific mechanisms underlying OC progression.
Although the roles of CDKN1A and
SQSTM1 in tumors have been widely reported, therapeutic
strategies may be limited due to drug resistance (49). Therefore, the present study focused
on H2AC7/H2AC20. Multiple studies have reported that
abnormal histone modifications serve a key role in the occurrence
and development of OC, and that H2A family members can participate
in malignant tumor phenotypes by regulating chromatin structure
(50–52). The associations between the
H2AC7-H2AC20-H2AC18 network and cancer have not been
well-defined, and their roles in OC have not been described.
However, these genes have also been studied in other contexts. For
example, H2AC7 was identified as one of the 14 placental
candidate genes in a screen for causal genes associated with
spontaneous abortion (53).
H2AC20 was suggested as a potential DNA methylation
candidate gene during infection with Mycobacterium avium
paratuberculosis, whereas H2AC18 was reported to be one
of the most altered genes in the peripheral white blood cells of
cows following embryo transfer (54,55).
Members of the histone H2A family may influence the occurrence and
progression of cancer by affecting the chromatin structure and
regulating gene expression. lncRNAs participate in the regulation
of biological processes by regulating target genes (56). The potential associations among
H2AC7, H2AC20, H2AC18 and lncRNAs in cancer warrant further
exploration to reveal their complex regulatory mechanisms in cancer
initiation and progression. Moreover, lncRNA SNHGs are host genes
for small nucleolar RNA. The SNHG family includes 22 members, most
of which exert inhibitory or promoting functions in the initiation
and progression of cancer, and their expression is abnormal in
multiple cancers (57). As a
lncRNA, SNHG is involved in tumorigenesis through several molecular
regulatory mechanisms. The present study identified 24 SNHG
lncRNAs, which may affect the expression of cell
proliferation-associated genes through different mechanisms of
action, serve a role in the treatment of OC and provide a possible
direction for the development of novel therapeutic strategies based
on the SNHG family lncRNAs. The network built in the present study
also revealed lnc-SNHG15-203 in the core. Furthermore, given
the lack of cancer research, the specific effects of
lnc-SNHG15-203 on A2780 cells in vitro were assessed.
In the network map, lnc-SNHG15-203 was evaluated, which is
predicted to regulate H2AC7 and H2AC20. Similar to
RES treatment, it was demonstrated that silencing
lnc-SNHG15-203 significantly reduced the expression of
H2AC7 and H2AC20 in A2780 cells. Moreover, this
treatment inhibited cell proliferation, indicating that
lncRNA-SNHG15-203 may normally promote cell proliferation by
upregulating H2AC7 and H2AC20, and that RES acts to
counter this signaling axis. The results infer that the SNHG family
lncRNAs may inhibit the development of OC by influencing processes
such as cell proliferation and may become potential targets.
Furthermore, the RNA-sequencing (RNA-seq) results
suggest that DELs may be important in the inhibition of A2780 cell
proliferation by RES by regulating its target genes, and
participating in cell proliferation, senescence and
apoptosis-related signaling pathways. By analyzing the expression
profiles of lncRNAs, specific lncRNAs that were upregulated or
downregulated in OC were identified. These lncRNAs, including
SNHG15-203, could serve as potential biomarkers. These
findings demonstrate the utility of lncRNA-mRNA regulatory networks
in revealing RES targets and their underlying regulatory
mechanisms, and emphasize the importance of considering lncRNAs and
related genes in the treatment process, suggesting that the data in
the present study and the construction of additional networks are
important for OC treatment.
However, the present study has certain limitations.
First, the in vitro experiments were performed using the
A2780 OC cell line only, which may not fully reflect the
heterogeneity of the different subtypes. Second, the lack of in
vivo validation prevents confirmation of whether the regulatory
effects observed in vitro are valid in vivo.
Additionally, due to the lack of immunoprecipitation-level
antibodies against the target proteins and database prediction of
no binding sites, the present study was unable to confirm the
specific mechanism of interaction between lnc-SNHG15-203 and
H2AC7/H2AC20. Future research should be expanded to
include multiple cell models and in vivo experiments, and
alternative approaches to elucidate the molecular interaction
mechanisms to enhance the generalizability of the conclusions.
In summary, the present study comprehensively
assessed the role of RES in inhibiting OC cell proliferation by
regulating lncRNAs using RNA-seq. The expression profiles of
lncRNAs involved in RES-mediated inhibition of OC cell
proliferation were systematically identified. These DELs were
associated with several important signaling pathways. Subsequently,
the lncRNA-mRNA regulatory networks were constructed. In addition,
the results demonstrated that lnc-SNHG15-203 regulates A2780
cell proliferation and H2AC7 and H2AC20 expression. These
findings provide a new perspective for the treatment of OC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present research was funded by the Scientific Research
Project of Colleges and Universities in Anhui Province (grant no.
2024AH050832), Natural Science Research Project of Clinical College
of Anhui Medical University (grant no. 2024XJKY001) and Research
Platform Project of Clinical College of Anhui Medical University:
Collaborative Innovation Platform for Tumor Translational Medicine
(grant no. 2025PT005).
Availability of data and materials
The original RNA-sequencing data generated and in
the present study may be found in the BioProject database under
accession number PRJNA1119553 or at the following URL: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1119553.
All other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
WZ conceived and designed the study, developed the
methodology and wrote the manuscript. JF and YZ performed formal
analysis. JF and QZ conducted the investigation. HH and YY
interpreted the results. JF edited the manuscript. WZ and JF
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Webb PM and Jordan SJ: Global epidemiology
of epithelial ovarian cancer. Nat Rev Clin Oncol. 21:389–400. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang A, Wang Y, Du C, Yang H, Wang Z, Jin
C and Hamblin MR: Pyroptosis and the tumor immune microenvironment:
A new battlefield in ovarian cancer treatment. Biochim Biophys Acta
Rev Cancer. 1879:1890582024. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sideris M, Menon U and Manchanda R:
Screening and prevention of ovarian cancer. Med J Aust.
220:264–274. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Y, Dun Y, Zhou S and Huang XH:
LncRNA HOXD-AS1 promotes epithelial ovarian cancer cells
proliferation and invasion by targeting miR-133a-3p and activating
Wnt/beta-catenin signaling pathway. Biomed Pharmacother.
96:1216–1221. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu J, Zhang Q, Yang D, Xie F and Wang Z:
The role of long non-coding RNAs in angiogenesis and
anti-angiogenic therapy resistance in cancer. Mol Ther Nucleic
Acids. 28:397–407. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu YS and Zhu J: Molecular and cellular
functions of long non-coding RNAs in prostate and breast cancer.
Adv Clin Chem. 106:91–179. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou Z, Leng C, Wang Z, Long L, Lv Y, Gao
Z, Wang Y, Wang S and Li P: The potential regulatory role of the
lncRNA-miRNA-mRNA axis in teleost fish. Front Immunol.
14:10653572023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao Y, Yuan D, Zhu D, Xu T, Huang A,
Jiang L, Liu C, Qian H and Bu X: LncRNA-MSC-AS1 inhibits the
ovarian cancer progression by targeting miR-425-5p. J Ovarian Res.
14:1092021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fang W and Xia Y: LncRNA HLA-F-AS1
attenuates the ovarian cancer development by targeting
miR-21-3p/PEG3 axis. Anticancer Drugs. 33:671–681. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dong Q, Qiu H, Piao C, Li Z and Cui X:
LncRNA SNHG4 promotes prostate cancer cell survival and resistance
to enzalutamide through a let-7a/RREB1 positive feedback loop and a
ceRNA network. J Exp Clin Cancer Res. 42:2092023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tan YT, Lin JF, Li T, Li JJ, Xu RH and Ju
HQ: LncRNA-mediated posttranslational modifications and
reprogramming of energy metabolism in cancer. Cancer Commun (Lond).
41:109–120. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou D, Wang Y, Hu H, Liu H, Deng J, Li L
and Zheng C: lncRNA MALAT1 promotes HCC metastasis through the
peripheral vascular infiltration via miRNA-613: A primary study
using contrast ultrasound. World J Surg Oncol. 20:2032022.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lin X, Zhuang S, Chen X, Du J, Zhong L,
Ding J, Wang L, Yi J, Hu G, Tang G, et al: lncRNA ITGB8-AS1
functions as a ceRNA to promote colorectal cancer growth and
migration through integrin-mediated focal adhesion signaling. Mol
Ther. 30:688–702. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fan L, Lei H, Lin Y, Zhou Z, Li J, Wu A,
Shu G, Roger S and Yin G: Hotair promotes the migration and
proliferation in ovarian cancer by miR-222-3p/CDK19 axis. Cell Mol
Life Sci. 79:2542022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lei R, Xue M, Zhang L and Lin Z: Long
noncoding RNA MALAT1-regulated microRNA 506 modulates ovarian
cancer growth by targeting iASPP. Onco Targets Ther. 10:35–46.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu Q, Lin YB, Li L and Liu J: LncRNA
TLR8-AS1 promotes metastasis and chemoresistance of ovarian cancer
through enhancing TLR8 mRNA stability. Biochem Biophys Res Commun.
526:857–864. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang L, Yu M and Zhao S: lncRNA MEG3
modified epithelial-mesenchymal transition of ovarian cancer cells
by sponging miR-219a-5p and regulating EGFR. J Cell Biochem.
120:17709–17722. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ren B, Kwah MX, Liu C, Ma Z, Shanmugam MK,
Ding L, Xiang X, Ho PC, Wang L, Ong PS and Goh BC: Resveratrol for
cancer therapy: Challenges and future perspectives. Cancer Lett.
515:63–72. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rauf A, Imran M, Butt MS, Nadeem M, Peters
DG and Mubarak MS: Resveratrol as an anti-cancer agent: A review.
Crit Rev Food Sci Nutr. 58:1428–1447. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Asemi R, Rajabpoor Nikoo N, Asemi Z,
Shafabakhsh R, Hajijafari M, Sharifi M, Homayoonfal M, Davoodvandi
A and Hakamifard A: Modulation of long non-coding RNAs by
resveratrol as a potential therapeutic approach in cancer: A
comprehensive review. Pathol Res Pract. 246:1545072023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu W, Zhang Y, Zhou Q, Zhen C, Huang H
and Liu X: Identification and Comprehensive analysis of
circRNA-miRNA-mRNA regulatory networks in A2780 cells treated with
resveratrol. Genes (Basel). 15:9652024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen S, Zhou Y, Chen Y and Gu J: Fastp: An
ultra-fast all-in-one FASTQ preprocessor. Bioinformatics.
34:i884–i890. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Trapnell C, Williams BA, Pertea G,
Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ and Pachter
L: Transcript assembly and quantification by RNA-Seq reveals
unannotated transcripts and isoform switching during cell
differentiation. Nat Biotechnol. 28:511–515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pertea M, Pertea GM, Antonescu CM, Chang
TC, Mendell JT and Salzberg SL: StringTie enables improved
reconstruction of a transcriptome from RNA-seq reads. Nat
Biotechnol. 33:290–295. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pertea M, Kim D, Pertea GM, Leek JT and
Salzberg SL: Transcript-level expression analysis of RNA-seq
experiments with HISAT, StringTie and Ballgown. Nat Protoc.
11:1650–1667. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C,
Liu Y, Chen R and Zhao Y: Utilizing sequence intrinsic composition
to classify protein-coding and long non-coding transcripts. Nucleic
Acids Res. 41:e1662013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ,
Wei L and Gao G: CPC: Assess the protein-coding potential of
transcripts using sequence features and support vector machine.
Nucleic Acids Res. 35:W345–W349. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wucher V, Legeai F, Hedan B, Rizk G,
Lagoutte L, Leeb T, Jagannathan V, Cadieu E, David A, Lohi H, et
al: FEELnc: A tool for long non-coding RNA annotation and its
application to the dog transcriptome. Nucleic Acids Res.
45:e572017.PubMed/NCBI
|
|
30
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tafer H and Hofacker IL: RNAplex: A fast
tool for RNA-RNA interaction search. Bioinformatics. 24:2657–2663.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nadile M, Retsidou MI, Gioti K, Beloukas A
and Tsiani E: Resveratrol against Cervical Cancer: Evidence from in
vitro and in vivo studies. Nutrients. 14:52732022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu X, Li P, Yan K, Du Y, Peng K, Li M,
Cui K, Zhang H, Yang X, Lu S and Liang X: Resveratrol ameliorates
the defects of meiotic maturation in lipopolysaccharide exposed
porcine oocytes. Reprod Toxicol. 115:85–93. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu H, Chen L, Zhu F, Han X, Sun L and Chen
K: The cytotoxicity effect of resveratrol: Cell cycle arrest and
induced apoptosis of breast cancer 4T1 cells. Toxins (Basel).
11:7312019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu XL, Deng SL, Lian ZX and Yu K:
Resveratrol targets a variety of oncogenic and oncosuppressive
signaling for ovarian cancer prevention and treatment. Antioxidants
(Basel). 10:17182021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Poyton MF, Feng XA, Ranjan A, Lei Q, Wang
F, Zarb JS, Louder RK, Park G, Jo MH, Ye J, et al: Coordinated DNA
and histone dynamics drive accurate histone H2A.Z exchange. Sci
Adv. 8:eabj55092022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Oberdoerffer P and Miller KM: Histone H2A
variants: Diversifying chromatin to ensure genome integrity. Semin
Cell Dev Biol. 135:59–72. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Geng X, Zhao J, Huang J, Li S, Chu W, Wang
WS, Chen ZJ and Du Y: lnc-MAP3K13-7:1 inhibits ovarian GC
proliferation in PCOS via DNMT1 Downregulation-mediated CDKN1A
promoter hypomethylation. Mol Ther. 29:1279–1293. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu Q, Xiang S, Ma J, Hui P, Wang T, Meng
W, Shi M and Wang Y: Long non-coding RNA CASC15 regulates gastric
cancer cell proliferation, migration and epithelial mesenchymal
transition by targeting CDKN1A and ZEB1. Mol Oncol. 12:799–813.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hu CC, Liang YW, Hu JL, Liu LF, Liang JW
and Wang R: LncRNA RUSC1-AS1 promotes the proliferation of breast
cancer cells by epigenetic silence of KLF2 and CDKN1A. Eur Rev Med
Pharmacol Sci. 23:6602–6611. 2019.PubMed/NCBI
|
|
44
|
Guo M and Zhang X: LncRNA MSTO2P promotes
colorectal cancer progression through epigenetically silencing
CDKN1A mediated by EZH2. World J Surg Oncol. 20:952022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ma JX, Yang YL, He XY, Pan XM, Wang Z and
Qian YW: Long noncoding RNA MNX1-AS1 overexpression promotes the
invasion and metastasis of gastric cancer through repressing
CDKN1A. Eur Rev Med Pharmacol Sci. 23:4756–4762. 2019.PubMed/NCBI
|
|
46
|
Bi M, Yu H, Huang B and Tang C: Long
non-coding RNA PCAT-1 over-expression promotes proliferation and
metastasis in gastric cancer cells through regulating CDKN1A. Gene.
626:337–343. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu C, Chen C, Xu Y, Li Z, Chen H and Wang
G: Prognostic significance of CDK1 in Ovarian and Cervical Cancers.
J Cancer. 16:1656–1667. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pan Y, Zhou J, Zhang W, Yan L, Lu M, Dai
Y, Zhou H, Zhang S and Yang J: The Sonic Hedgehog signaling pathway
regulates autophagy and migration in ovarian cancer. Cancer Med.
10:4510–4521. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Manousakis E, Miralles CM, Esquerda MG and
Wright RHG: CDKN1A/p21 in breast cancer: Part of the problem, or
part of the solution? Int J Mol Sci. 24:174882023. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhao S, Bellone S, Lopez S, Thakral D,
Schwab C, English DP, Black J, Cocco E, Choi J, Zammataro L, et al:
Mutational landscape of uterine and ovarian carcinosarcomas
implicates histone genes in epithelial-mesenchymal transition. Proc
Natl Acad Sci USA. 113:12238–12243. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Saravi S, Katsuta E, Jeyaneethi J, Amin
HA, Kaspar M, Takabe K, Pados G, Drenos F, Hall M and Karteris E:
H2A histone family member X (H2AX) is upregulated in ovarian cancer
and demonstrates utility as a prognostic biomarker in terms of
overall survival. J Clin Med. 9:28442020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yuan Y, Cao W, Zhou H, Qian H and Wang H:
H2A.Z acetylation by lincZNF337-AS1 via KAT5 implicated in the
transcriptional misregulation in cancer signaling pathway in
hepatocellular carcinoma. Cell Death Dis. 12:6092021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu A, Zhou L, Huang Y and Peng D:
Analysis of copy number variants detected by sequencing in
spontaneous abortion. Mol Cytogenet. 17:132024. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ibeagha-Awemu EM, Bissonnette N, Bhattarai
S, Wang M, Dudemaine PL, McKay S and Zhao X: Whole genome
methylation analysis reveals role of DNA methylation in Cow's ileal
and ileal lymph node responses to mycobacterium avium subsp.
paratuberculosis infection. Front Genet. 12:7974902021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
De Los Santos JA, Andrade JPN, Cangiano
LR, Iriarte A, Penagaricano F and Parrish JJ: Transcriptomic
analysis reveals gene expression changes in peripheral white blood
cells of cows after embryo transfer: Implications for pregnancy
tolerance. Reprod Domest Anim. 58:946–954. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lei K, Qu L, Liu F, Hao N, Chen J, Liu J
and Lin A: Long non-coding RNAs regulate fatty acid and cholesterol
metabolism. Genome Instability Dis. 3:70–82. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Braga EA, Filippova EA, Uroshlev LA,
Lukina SS, Pronina IV, Kazubskaya TP, Kushlinskiy DN, Loginov VI,
Fridman MV, Burdennyy AM and Kushlinskii NE: LncRNA genes of the
SNHG family: Co-methylation and common functions in ovarian cancer.
Biochemistry (Mosc). 89:2051–2068. 2024. View Article : Google Scholar : PubMed/NCBI
|