Introduction
Lung cancer is the leading cause of cancer-related
deaths in the developed world, with lung adenocarcinoma (LUAD)
being the most prevalent histological type. Various grading methods
based on a histological perspective have been proposed to predict
prognosis. The micropapillary subtype is a high-grade histological
element that serves as a marker of poor prognosis (1,2),
particularly among patients with EGFR-mutated LUAD (3). We previously suggested that the
micropapillary element may develop from a pure lepidic type
via intermediate forms exhibiting a low papillary structure
(4). The 5th edition of the World
Health Organization (WHO) classification defines this low papillary
structure as a filigree pattern, consisting of tumor cells growing
in delicate, lace-like, narrow stacks of three cells without
fibrovascular cores (5). As the
filigree and micropapillary patterns exhibit comparable malignant
potential, the filigree type should be classified as a variant of
the micropapillary element (6).
This supported our theory and has prompted us to investigate the
molecular basis of the lepidic-filigree micropapillary
(filigree)-conventional/overt micropapillary (mPAP) progression
pathway.
The Visium Spatial Gene Expression Profiling System
(10× Genomics, Pleasanton, CA, USA) has facilitated comparisons of
mRNA expression among various histologically distinct elements from
a single tissue section (7).
Conventional methods compare mRNA levels between tissue elements
within a single tumor by separately isolating RNA from each region
using microdissection. However, LUAD is histologically
heterogeneous within a single tumor, limiting the effectiveness of
conventional methods for analyzing spatial gene expression.
Notably, the Visium Spatial Gene Expression Profiling System
compares different histological elements while preserving
morphological information within a single tissue section (8). Recently, detailed analysis using the
Visium spatial transcriptomics platform has revealed the mechanism
by which lung tumors develop through interactions with their
surrounding microenvironment (9).
In the present study, we aimed to identify key
molecules promoting LUAD progression using the Visium Spatial Gene
Expression Solution, which offers better resolution than
microdissection. We examined gene expression profiles specific to
lepidic, filigree, and mPAP elements in identical histological
sections to reveal key molecules that promote the
lepidic-filigree-mPAP pathway.
Materials and methods
Patients
All LUAD tissues analyzed in this study were
surgically resected at the Kanagawa Prefectural Cardiovascular and
Respiratory Center between January 1997 and December 2022. The
proportion of smokers in lung adenocarcinoma in our study [50.5%
(104/207)] was comparable with the previous study investigating
Japanese cohort (56.2–61.9%) (10–12).
For the prognostic analysis, patients with lung cancer who
underwent surgery between 1997 and 2014 were included. All patients
provided written informed consent for the use of these samples for
research purposes. This study was approved by the ethics committee
at the Kanagawa Prefectural Cardiovascular and Respiratory Center
[Approval ID: KCRC-17-0016; October 1, 2018] and the materials
(patient tissues and data) were used based on a mixed retrospective
and prospective study design.
Histopathological analyses
Formalin-fixed paraffin- embedded (FFPE) tissue
sections were sliced and stained with hematoxylin and eosin (HE).
The proportions of the histological elements (lepidic, acinar,
papillary, mPAP, and solid elements) were recorded in 5% increments
according to the WHO classification (5). The filigree pattern was classified as
part of the micropapillary subtype (6,13).
Spatial gene expression profiling
Four LUAD samples were analyzed using the Visium
Spatial Gene Expression Profiling System (10× Genomics, Pleasanton,
CA, USA). Briefly, the quality of RNA was analyzed using FFPE
slides, and all samples that satisfied the DV200 requirement as a
standard of RNA integrity were then subjected to the adhesion test.
Eight tissue sections (n=2 each from the four LUAD samples) that
included lepidic, filigree, and mPAP elements were mounted onto
Visium Spatial Gene Expression Slides in oligo-barcoded capture
areas (6.5×6.5 mm). Sequencing libraries were generated using
Visium Spatial Gene Expression kits, followed by spatial transcript
analysis using Visium Spatial Gene Expression Solution (10×
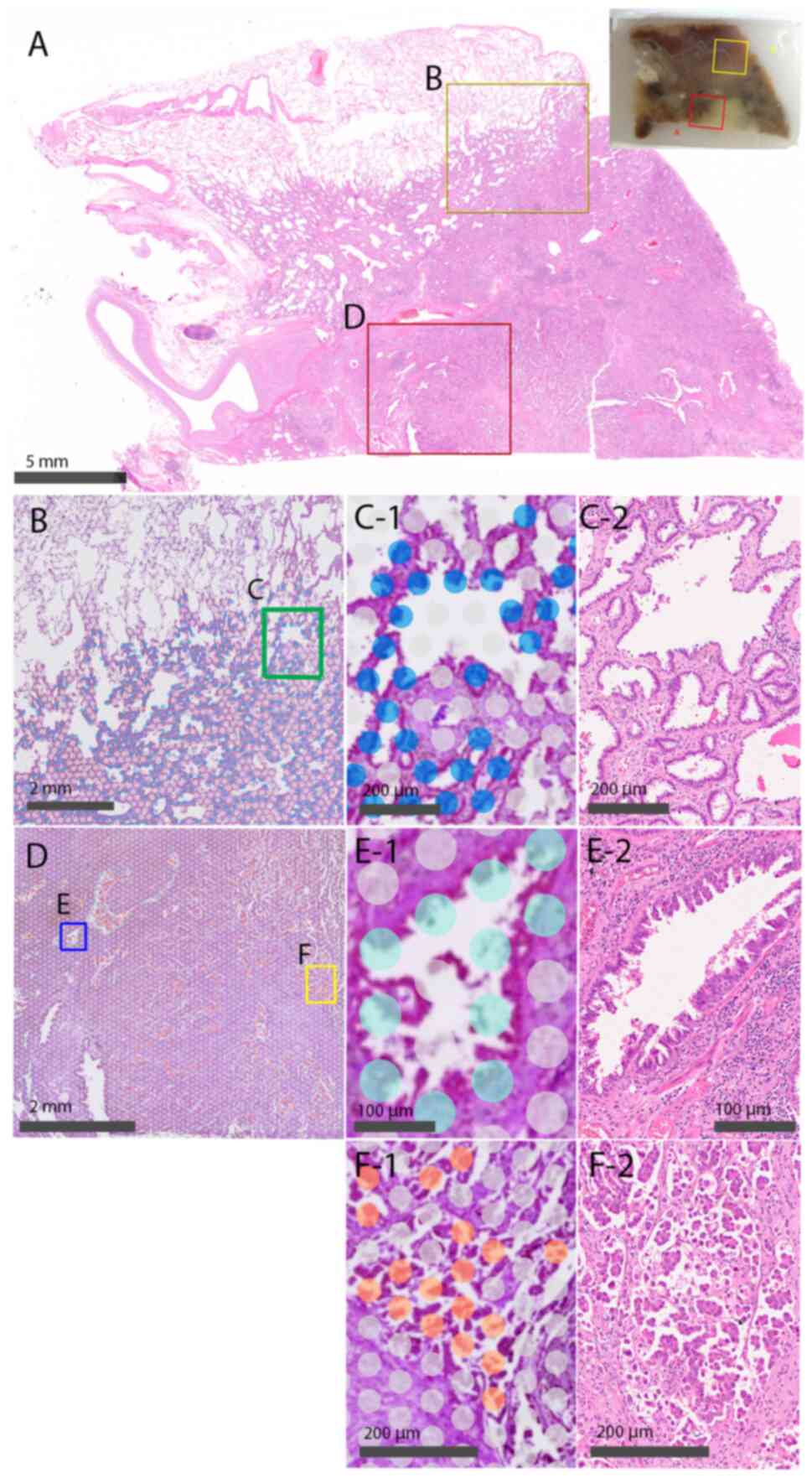
Genomics) (Fig. 1). The Visium
Spatial Gene Expression Profiling System contains approximately
5,000 circular spots with a diameter of 55 µm within a 6.5×6.5
mm2 analysis area on an FFPE slide, with each spot
containing 3–5 cells. RNA transcriptome data were obtained from
individual spots corresponding to each histological element
(Fig. 1C-1: lepidic element, D-1:
filigree element, E-1: mPAP element). Quantitative gene expression
data were normalized and mapped onto tissue sections using a Bio
Turing Lens (Bio Turing, San Diego, CA, USA). The mRNA levels in
the lepidic, filigree, and mPAP components were then compared and
analyzed. Differentially expressed genes among the three elements
were identified using t-tests with significance criteria set at
fold change (FC) >1.5 and a false discovery rate (FDR)
<0.05.
Immunohistochemical analyses
Immunohistochemical analysis was performed on 207
surgically resected LUAD samples. FFPE tissue sections were
incubated with primary antibodies against cellular retinoic acid
binding protein 2 (CRABP2; polyclonal; Sigma-Aldrich, St. Louis,
MO, USA), carcinoembryonic antigen-related cell adhesion molecule 5
(CEACAM5; clone CB30; Lifespan Biosciences, Washington, DC, USA),
and mucin 21 (MUC21; polyclonal; Novus Biologicals, Littleton, CO,
USA). The slides were autoclaved to retrieve antigens, and
immunoreactivity was then visualized using Simple Stain MAX-PO
(MULTI; Nichirei, Tokyo, Japan). The reactions were further
developed using 3,3′-diaminobenzidine (DAB) and counterstained with
hematoxylin. The intensity of CRABP2, CEACAM5, and MUC21 staining
in neoplastic cells was judged as negative (0), weak (1), or strong (2) and scored as: 1× (proportion of area
with weak intensity) + 2× (proportion of area with strong intensity
(%, in 5% increments).
Statistical analysis
Categorical and continuous variables were
respectively analyzed using Pearson chi-square tests or Fisher
exact tests, the Kruskal-Wallis test with Dunn's multiple
comparison test and the Cox proportional hazards model. Receiver
operating characteristic (ROC) curves of recurrence were generated
to establish a cut-off to distinguish ‘high’ from ‘low’
immunohistological scores. Kaplan-Meier curves of recurrence and
survival were plotted, and differences in recurrence-free survival
(RFS) and overall survival (OS) rates were analyzed using log-rank
tests. Values with P<0.05 were considered significant. All data
were statistically analyzed using JMP 9.0.2 (SAS Institute, Cary,
NC, USA).
Results
Changes in gene expression in each
LUAD subtype
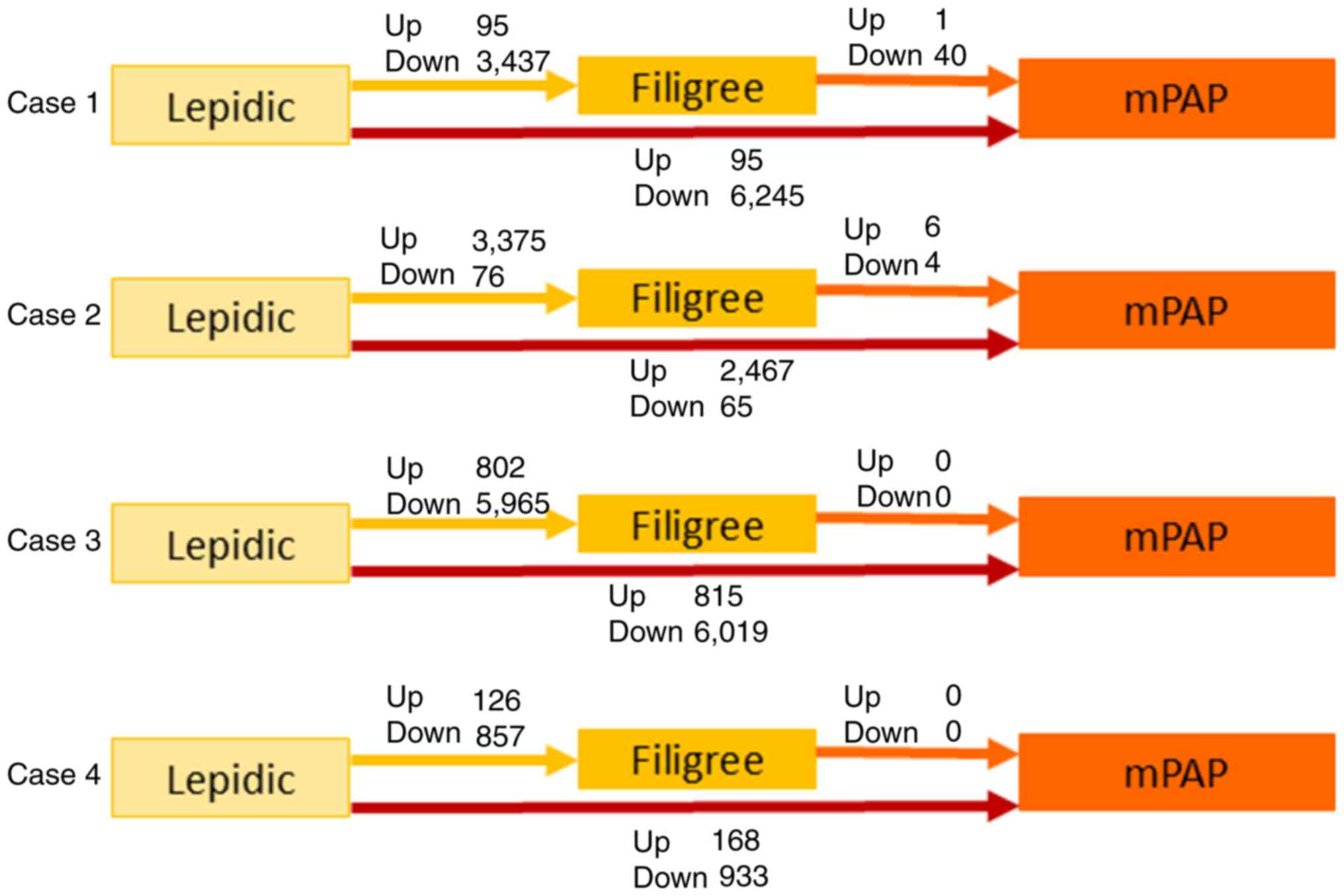
The number of differentially expressed genes was
considerably higher during the transition from lepidic to filigree
than from filigree to mPAP among all four cases (Fig. 2). This result suggests that the gene
expression profile primarily changes during the early stages of the
progression.
Differentially expressed genes during
the lepidic-filigree-mPAP progression
Among the four cases, three genes (CRABP2,
CEACAM5, and MUC21) were commonly upregulated during the
transition from lepidic to mPAP elements (Table SI, Table SII, Table SIII). Notably, CRABP2 was
upregulated in the early stage (from lepidic to filigree elements).
Conversely, 14 genes were commonly downregulated during the
transition from lepidic to mPAP elements, whereas 6 genes were
downregulated during the transition from lepidic to filigree
elements (Table SIV, Table V, Table SVI).
Immunohistochemical expression of
CRABP2, CEACAM5, and MUC21 in different histological elements
Among the identified genes, we focused on
three-CRABP2, CEACAM5, and MUC21- and examined their
protein expression in a larger cohort of LUAD cases using
immunohistochemical analysis (Fig.
3). CRABP2 was preferentially expressed both in filigree and
mPAP elements (Fig. 3E and F).
Conversely, CEACAM5 and MUC21 expression was specific to mPAP
elements, with positive rates were 50.6% (39/77) and 66.2% (51/77),
respectively (Fig. 3I and L).
Positive immunohistochemical signals for CEACAM5 and MUC21 revealed
predominantly negative staining in the filigree element, with
positive rates of 38.7% (60/155) and 21.3% (33/155), respectively
(Fig. 3H and K). To confirm this
observation, we semi-quantified the immunohistochemical expression
levels in the different histological elements, including lepidic,
filigree, acinar, papillary, mPAP, and solid elements, using a
scoring system (as described in the Materials and Methods).
Representative images of tumors with varying immunohistochemical
scores are presented in Fig. 4.
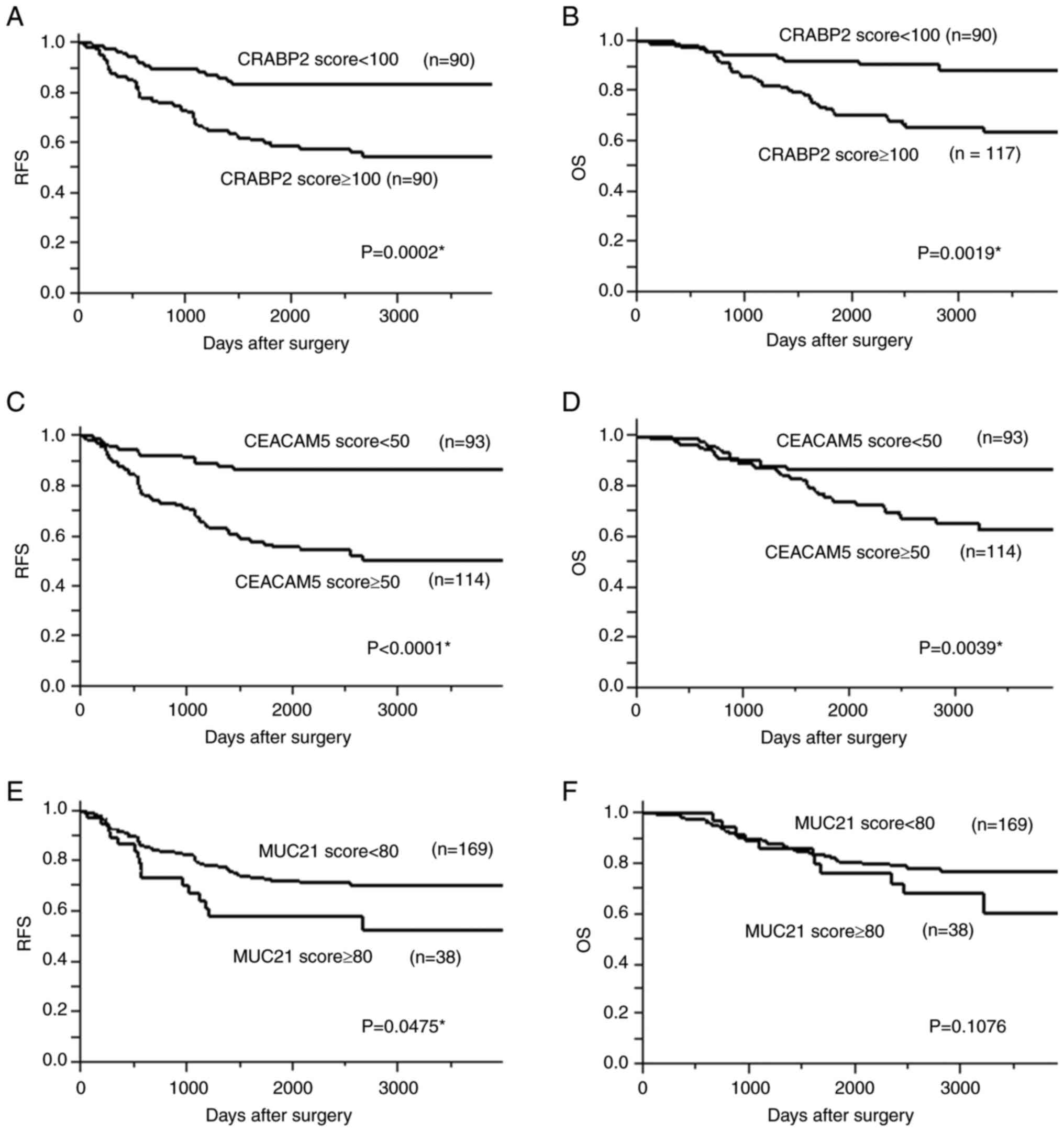
Consistent with our initial observation, the scores of CRABP2 were
significantly higher in filigree than lepidic elements; however, no
significant difference was observed between mPAP and filigree
elements (Fig. 5A). The scores of
CEACAM5 and MUC21 were significantly higher in mPAP than in
filigree elements, and the scores of these were significantly
higher in filigree than in lepidic elements (Fig. 5B and C). The increase in the CRABP2
score between lepidic and filigree elements was considerably larger
than that in CEACAM5 or MUC21 expression scores (Fig. 5). This result suggests that CRABP2
may play a role in the earlier stages of the lepidic-filigree-mPAP
progression. High scores of CRABP2 and CEACAM5 were also observed
in solid elements, another high-grade element, as well as mPAP
elements (Fig. 5A and B). In some
cases, CRABP2 and CEACAM5 expression was observed in the acinar and
papillary elements. However, the expression scores in these
elements were lower than those in filigree and/or mPAP elements
(Fig. 5A and B). The expression
score of MUC21 was consistently lower in the papillary, acinar, and
solid elements than in the filigree and mPAP components (Fig. 5). The expression of CRABP2,
CEACAM15, and MUC21 was not detected in non-neoplastic bronchiolar
and alveolar epithelial cells.
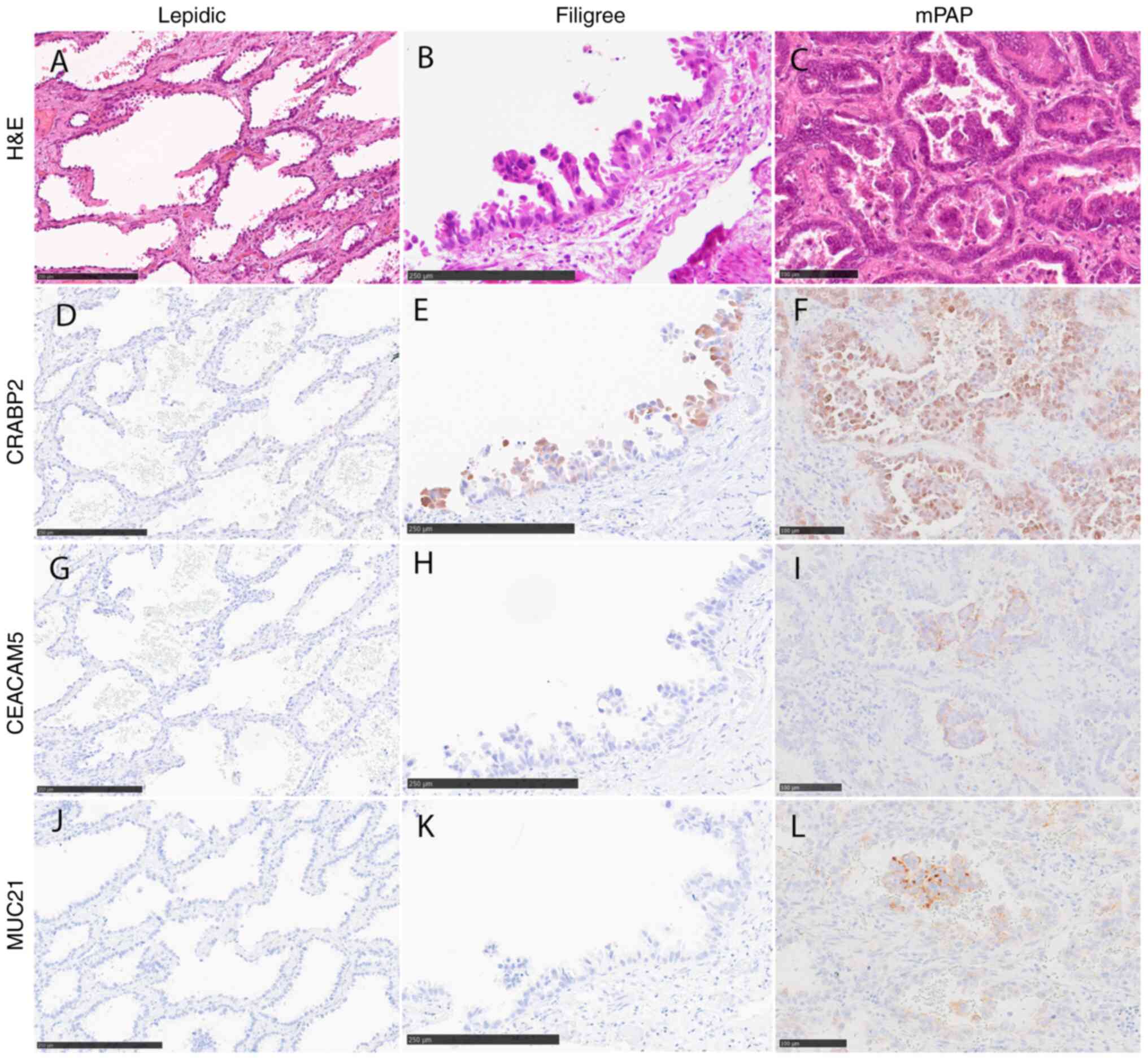 | Figure 3.Representative images of CRABP2,
CEACAM5 and MUC21 immunohistochemical staining. (A) Lepidic, (B)
filigree and (C) mPAP elements stained with H&E. (D) CRABP2,
(G) CEACAM5 and (J) MUC21 were rarely expressed in lepidic
elements. CRABP2 was expressed in (E) filigree and (F) mPAP
elements. (I) CEACAM5 and (L) MUC21 were expressed only in mPAP
elements. (H) CEACAM5 and (K) MUC21 were rarely expressed in
filigree elements. Scale bar, 250 µm (A, B, D, E, G, H, J and K) or
100 µm (C, F, I and L). CEACAM5, carcinoembryonic antigen-related
cell adhesion molecule 5; CRABP2, cellular retinoic acid binding
protein 2; mPAP, conventional/overt micropapillary; MUC21, mucin
21. |
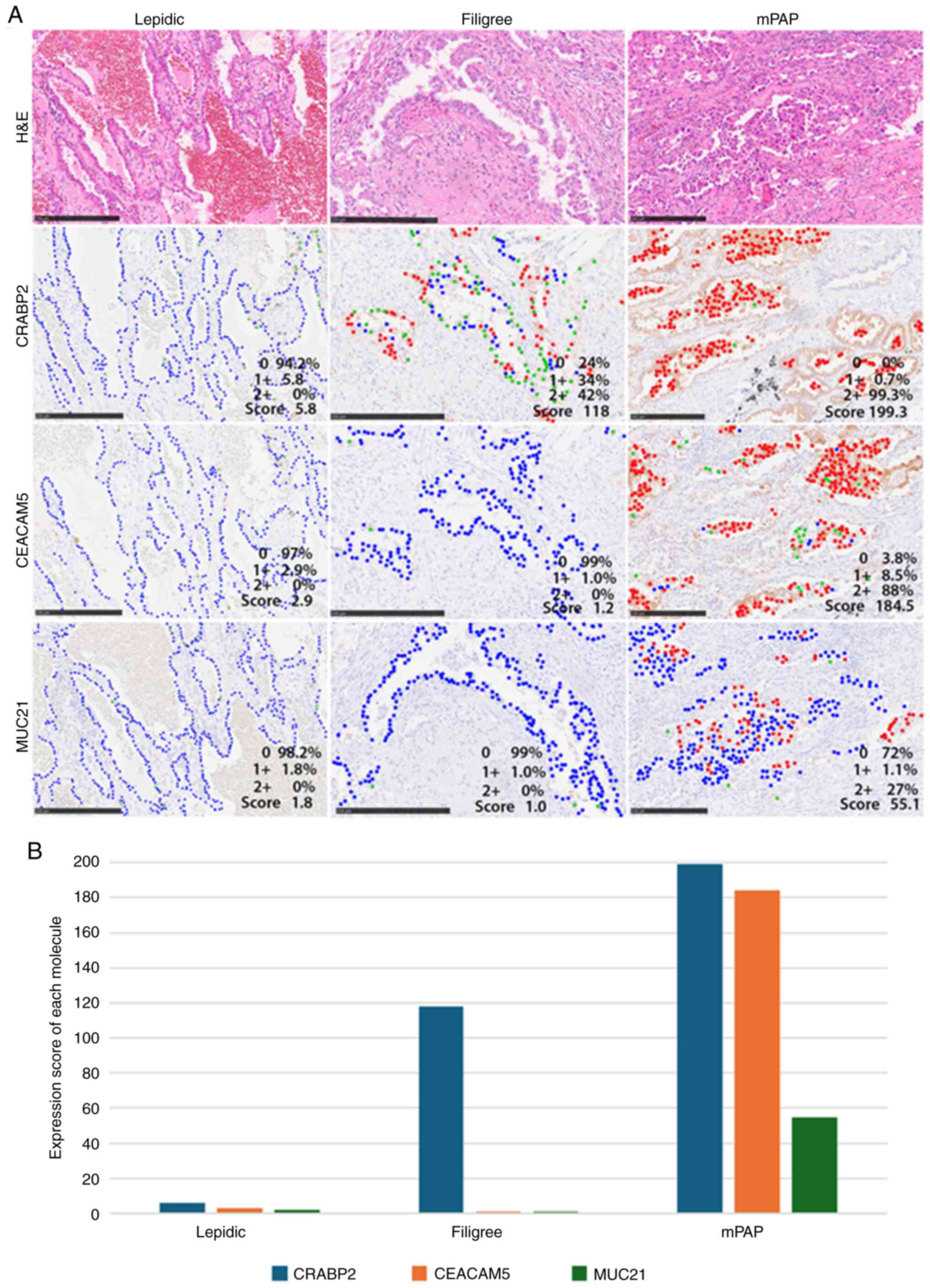 | Figure 4.Representative images of tumors with
various immunohistochemical scores. Red, green and blue dots
indicate tumor cells with strong, weak and no signals (intensity,
2, 1 and 0, respectively) (A) Lepidic elements (left) rarely
exhibited positive signals for each antibody (CRABP2, CEACAM5 and
MUC21 scores: 5.8, 2.9 and 1.8). Filigree elements (center) showed
some strong and weak CRABP2 signals in most neoplastic cells
(score, 118=1×34+2×42). Signals for CEACAM5 and MUC21 were rarely
positive (scores, 1.2 and 1.0, respectively). mPAP elements (right)
exhibited stronger signals in most neoplastic cells for CRABP2,
CEACAM5 and MUC21 than the other elements (lepidic and filigree)
(scores, 199.3=1×0.7+2×99.3, 184.5=1×8.5+2×88 and 55.1=1×1.1+2×27,
respectively). Scale bar, 250 µm. (B) All scores were low in
lepidic elements. Only the expression score for CRABP2 was high in
the filigree element, whereas the scores for all three proteins
were high in mPAP elements. CEACAM5, carcinoembryonic
antigen-related cell adhesion molecule 5; CRABP2, cellular retinoic
acid binding protein 2; mPAP, conventional/overt micropapillary;
MUC21, mucin 21. |
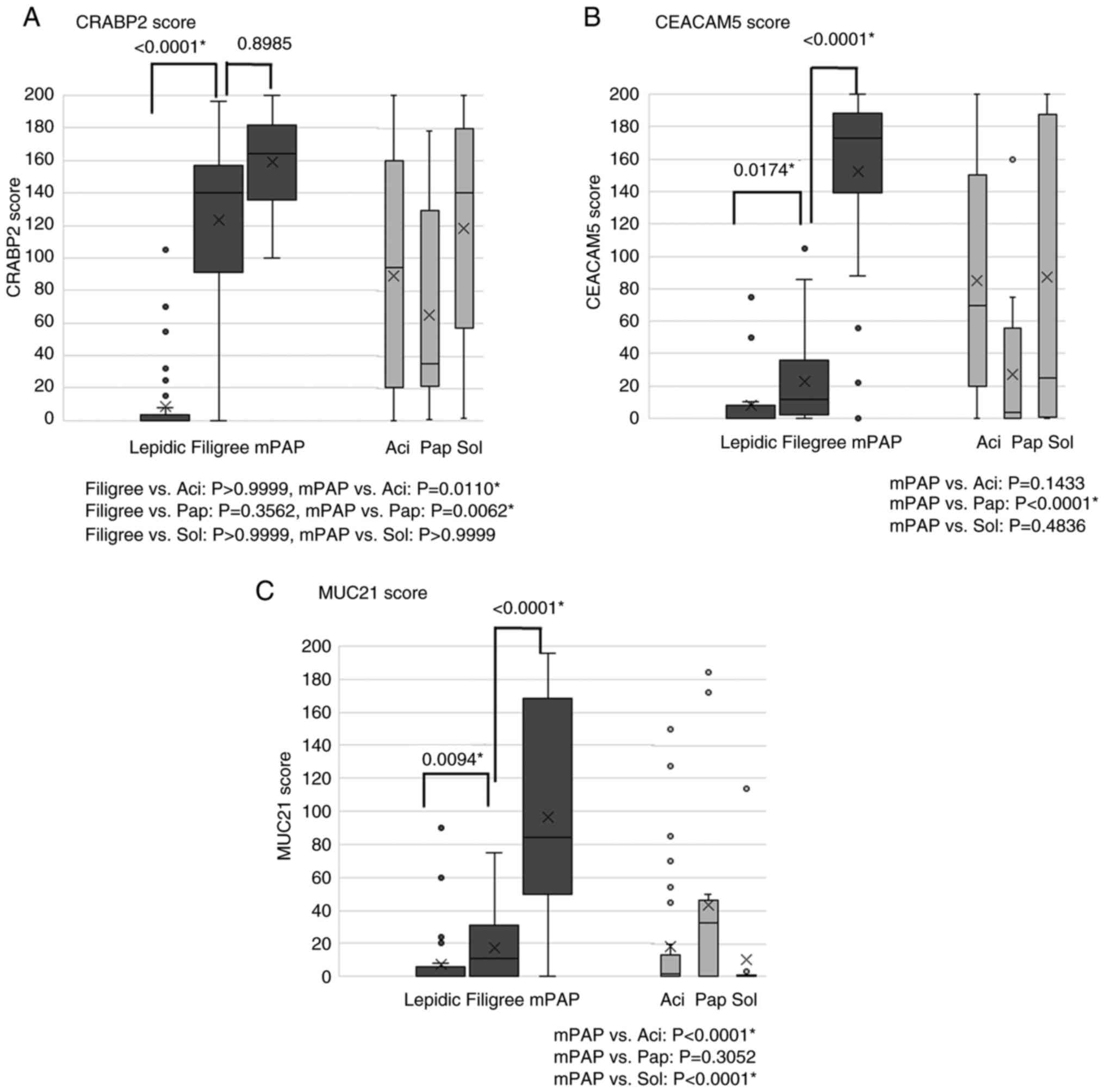 | Figure 5.Association between histological
elements and CRABP2, CEACAM5 and MUC21 immunohistochemical scores.
(A) The CRABP2 score was significantly higher in filigree elements
than in lepidic elements (P<0.0001) but no significant
difference was observed between mPAP elements and filigree elements
(P=0.8985). (B) The CEACAM5 score was significantly higher in
filigree elements than in lepidic elements (P=0.0174) and
significantly higher in mPAP elements than in filigree elements
(P<0.0001). This difference was more significant between
filigree and mPAP elements than between filigree and lepidic
elements. (C) The MUC21 score was significantly higher in filigree
elements than in lepidic elements (P=0.0094) and significantly
higher in mPAP elements than in filigree elements (P<0.0001).
This difference was more significant between filigree and mPAP
elements than between filigree and lepidic elements.
Box-and-whisker plots show immunohistochemical scores for 50
surgically resected lung adenocarcinoma cases (median, thick line;
25th-75th percentiles, box; 10th-90th percentiles, whiskers;
outliers, dots). P<0.05 (Kruskal-Wallis test with Dunn's
multiple comparison test). *Statistically significant. Aci, acinar;
CEACAM5, carcinoembryonic antigen-related cell adhesion molecule 5;
CRABP2, cellular retinoic acid binding protein 2; mPAP,
conventional/overt micropapillary; MUC21, mucin 21; Pap, papillary;
Sol, solid. |
Cut-off values for ‘high’ and ‘low’
immunohistochemical scores
The optimal cut-off immunohistochemical scores for
CRABP2, CEACAM5, and MUC21, determined using ROC curves, were 100,
50, and 80, respectively. We then categorized the samples as ‘high’
or ‘low expressors’ in subsequent correlation analyses.
Relationship among CRABP2, CEACAM5,
and MUC21 immunohistochemical levels and clinicopathological
characteristics
The clinicopathological characteristics of the 207
LUAD cases are presented in Table
I. High CRABP2, CEACAM5, and MUC21 expression levels were
significantly related to EGFR mutations (Table I). Additionally, high MUC21
expression was significantly associated with a non-smoking status
(Table I).
 | Table I.Relationships between CRABP2, CEACAM5
and MUC21 expression and clinicopathological factors in lung
adenocarcinoma. |
Table I.
Relationships between CRABP2, CEACAM5
and MUC21 expression and clinicopathological factors in lung
adenocarcinoma.
|
| CRABP2 | CEACAM5 | MUC21 |
|---|
|
|
|
|
|
|---|
| Clinicopathological
factors | Low expression
(N=90) | High expression
(N=117) | P-value | Low expression
(N=93) | High expression
(N=114) | P-value | Low expression
(N=169) | High expression
(N=38) | P-value |
|---|
| Age |
|
| 0.7698 |
|
| 0.6693 |
|
| 0.4535 |
| Young
(≤65 years), % (n) | 36.7 (33) | 34.2 (40) |
| 37.6 (35) | 33.3 (38) |
| 36.7 (62) | 28.9 (11) |
|
| Older
(>65 years), % (n) | 63.3 (57) | 65.8 (77) |
| 62.4 (58) | 66.7 (76) |
| 63.3 (107) | 71.1 (27) |
|
| Median,
years (range) | 69 (47–84) | 69 (36–86) |
| 67 (38–85) | 69 (36–86) |
| 68 (36–86) | 72 (47–83) |
|
| Sex, % (n) |
|
| 0.3247 |
|
| 0.5758 |
|
| 0.1502 |
|
Female | 58.9 (53) | 51.3 (60) |
| 57.0 (53) | 52.6 (60) |
| 52.1 (88) | 65.8 (25) |
|
|
Male | 41.1 (37) | 48.7 (57) |
| 43.0 (40) | 47.4 (54) |
| 47.9 (81) | 34.2 (13) |
|
| Smoking status, %
(n) |
|
| 0.7799 |
|
| 0.2645 |
|
| 0.0122a |
| Never
smoked | 51.1 (46) | 48.7 (57) |
| 45.2 (42) | 53.5 (61) |
| 45.6 (77) | 68.4 (26) |
|
|
Smoking | 48.9 (44) | 51.3 (60) |
| 54.8 (51) | 46.5 (53) |
| 54.4 (92) | 31.6 (12) |
|
| Tumor size, %
(n) |
|
| 0.3961 |
|
| 0.1194 |
|
| >0.9999 |
| ≤20
mm | 45.6 (41) | 39.3 (46) |
| 48.4 (45) | 36.8 (42) |
| 42.0 (71) | 42.1 (16) |
|
| >20
mm | 54.4 (49) | 60.7 (71) |
| 51.6 (48) | 63.2 (72) |
| 58.0 (98) | 57.9 (22) |
|
| Lymphatic invasion,
% (n) |
|
|
<0.0001a |
|
| 0.0171a |
|
| 0.0112a |
|
Present | 38.9 (35) | 66.7 (78) |
| 45.2 (42) | 62.3 (71) |
| 50.3 (85) | 73.7 (28) |
|
|
Absent | 61.1 (55) | 33.3 (39) |
| 54.8 (51) | 37.7 (43) |
| 49.7 (84) | 26.3 (10) |
|
| Vascular invasion,
% (n) |
|
| 0.0007a |
|
| 0.0334a |
|
| 0.5646 |
|
Present | 17.8 (16) | 40.2 (47) |
| 22.6 (21) | 36.8 (42) |
| 29.6 (50) | 34.2 (13) |
|
|
Absent | 82.2 (74) | 59.8 (70) |
| 77.4 (72) | 63.2 (72) |
| 70.4 (119) | 65.8 (25) |
|
| pStage, % (n) |
|
| 0.0020a |
|
| 0.0117a |
|
| 0.0343a |
| IA,
IB | 91.1 (82) | 74.4 (87) |
| 89.2 (83) | 75.4 (86) |
| 84.6 (143) | 68.4 (26) |
|
| IIA,
IIB, IIIA, IIIB | 8.9 (8) | 25.6 (30) |
| 10.8 (10) | 24.6 (28) |
| 15.4 (26) | 31.6 (12) |
|
| Histological
subtype, % (n) |
|
| 0.0050a |
|
| 0.0011a |
|
| 0.1858 |
|
Lepidicb | 63.3 (57) | 45.3 (53) |
| 65.6 (61) | 43.0 (49) |
| 55.0 (93) | 44.7 (17) |
|
|
Acinar | 21.1 (19) | 31.6 (37) |
| 16.1 (15) | 36.0 (41) |
| 26.6 (45) | 28.9 (11) |
|
|
Papillary | 3.3 (3) | 11.1 (13) |
| 5.4 (5) | 9.6 (11) |
| 5.9 (10) | 15.8 (6) |
|
|
Micropapillaryc | 0.0 (0) | 1.7 (2) |
| 0.0 (0) | 1.8 (2) |
| 0.6 (1) | 2.6 (1) |
|
|
Solid | 5.6 (5) | 9.4 (11) |
| 6.5 (6) | 8.8 (10) |
| 7.7 (13) | 7.9 (3) |
|
|
Mucinous | 6.7 (6) | 0.9 (1) |
| 6.5 (6) | 0.9 (1) |
| 4.1 (7) | 0.0 (0) |
|
| Cytological
subtype, % (n) |
|
| 0.0117a |
|
| 0.1114 |
|
| 0.1987 |
|
TRU | 87.8 (79) | 91.5 (107) |
| 88.2 (82) | 91.2 (104) |
| 88.2 (149) | 97.4 (37) |
|
|
Non-TRU/BSE | 10.0 (9) | 1.7 (2) |
| 8.6 (8) | 2.6 (3) |
| 6.5 (11) | 0.0 (0) |
|
|
Unclassifiable | 2.2 (2) | 6.8 (8) |
| 3.2 (3) | 6.1 (7) |
| 5.3 (9) | 2.6 (1) |
|
| EGFR, %
(n) |
|
| 0.0377a |
|
| 0.0498a |
|
| 0.0188a |
|
Mutated | 45.6 (41) | 59.0 (69) |
| 45.2 (42) | 59.6 (68) |
| 49.1 (83) | 71.1 (27) |
|
|
Wild-type | 54.4 (49) | 41.0 (48) | | 54.8 (51) | 40.4 (46) | | 50.9 (86) | 28.9 (11) |
|
Relationship between CRABP2, CEACAM5,
and MUC21 levels and highly malignant pathological factors
High CRABP2, CEACAM5, and MUC21 expression levels
were significantly associated with lymphatic canal invasion
[CRABP2, 78 (66.7%) of 117 vs. 35 (38.9%) of 90; Pearson ×2 test,
P<0.0001; CEACAM5, 71 (62.3%) of 114 vs. 42 (45.2%) of 93,
P=0.0171; MUC21, 28 (73.7%) of 38 vs. 85 (50.3%) of 169, P=0.0112;
Table I]. Both CRABP2 and CEACAM5
were significantly associated with vascular invasion [47 (40.2%) of
117 vs. 16 (17.8%) of 90; P=0.0007 Pearson ×2 tests and 42 (36.8%)
of 114 vs. 21 (22.6%) of 93; P=0.0334, respectively (Table I)]. These results support the notion
that frequent lymphatic canal invasion is a biological basis for
the aggressiveness of mPAP elements (1,7–10).
Relationships between CRABP2, CEACAM5,
and MUC21 levels and postoperative outcomes
Individuals with high CRABP2, CEACAM5, and MUC21
expression had significantly worse 5-year RFS rates [CRABP2: 50.7%
vs. 73.8%, P=0.0002; CEACAM5: 55.6% vs. 86.5%, P<0.0001; MUC21:
5 58.3% vs. 72.2%, P=0.0475; (Fig.
6)]. Individuals with high CRABP2 and CEACAM5 expression had
significantly worse 5-year OS rates [72.3% vs. 92.0%, P=0.0019, and
75.1% vs. 86.9%, P=0.0039, respectively]. All P-values were
determined using log-rank tests.
In addition, multivariate analysis revealed CRABP2
(P=0.0325) and CEACAM5 expression (P=0.0002) as independent
predictors of disease recurrence and stage (Table II).
 | Table II.Multivariate analysis performed using
the Cox proportional hazards model. |
Table II.
Multivariate analysis performed using
the Cox proportional hazards model.
|
| Recurrence-free
survival | Overall
survival |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| CRABP2 |
|
|
|
|
|
|
|
Expression score (≤100 vs.
>100) | 1.894 | 1.05–3.59 | 0.0325a | 1.804 | 0.93–3.75 | 0.0842 |
| Sex
(female vs. male) | 1.100 | 0.61–2.01 | 0.7547 | 1.373 | 0.70–2.73 | 0.3588 |
| Smoking
status (never smoked vs. smoking) | 1.077 | 0.59–1.98 | 0.8107 | 1.116 | 0.56–2.25 | 0.7555 |
| Tumor
size (≤20 vs. >20 mm) | 1.193 | 0.66–2.13 | 0.5520 | 1.095 | 0.58–2.05 | 0.7769 |
|
Vascular invasion (absent vs.
present) | 1.385 | 0.74–2.64 | 0.3149 | 1.222 | 0.58–2.62 | 0.5989 |
|
Lymphatic invasion (absent vs.
present) | 1.128 | 0.57–2.23 | 0.7269 | 0.691 | 0.32–1.45 | 0.3315 |
| pStage
(I vs. II, III) | 4.859 | 2.65–8.88 | 0.0090a | 5.778 | 3.02–11.1 |
<0.0001a |
| CEACAM5 |
|
|
|
|
|
|
|
Expression score (≤50 vs.
>50) | 3.045 | 1.67–5.95 | 0.0002a | 1.547 | 0.82–3.08 | 0.1849 |
| Sex
(female vs. male) | 1.023 | 0.56–1.87 | 0.9398 | 1.346 | 0.68–2.69 | 0.3928 |
| Smoking
status (never smoked vs. smoking) | 1.256 | 0.69–2.30 | 0.4578 | 1.191 | 0.60–2.42 | 0.6228 |
| Tumor
size (≤20 vs. >20 mm) | 1.308 | 0.72–2.35 | 0.3736 | 1.103 | 0.58–2.06 | 0.7605 |
|
Vascular invasion (absent vs.
present) | 1.344 | 0.71–2.60 | 0.3697 | 1.192 | 0.56–2.58 | 0.8480 |
|
Lymphatic invasion (absent vs.
present) | 1.191 | 0.60–2.35 | 0.6118 | 0.759 | 0.36–1.58 | 0.4643 |
| pStage
(I vs. II, III) | 4.720 | 2.55–8.70 |
<0.0001a | 5.795 | 2.99–11.3 |
<0.0001a |
| MUC21 |
|
|
|
|
|
|
|
Expression score (≤80 vs.
>80) | 1.135 | 0.59–2.09 | 0.6971 | 1.100 | 0.52–2.22 | 0.7995 |
| Sex
(Female vs. Male) | 1.127 | 0.62–2.07 | 0.6974 | 1.383 | 0.70–2.77 | 0.3526 |
| Smoking
status (never smoked vs. smoking) | 1.163 | 0.62–2.19 | 0.6355 | 1.165 | 0.57–2.45 | 0.6809 |
| Tumor
size (≤20 vs. >20 mm) | 1.169 | 0.64–2.10 | 0.6047 | 1.099 | 0.57–2.07 | 0.7735 |
|
Vascular invasion (absent vs.
present) | 1.401 | 0.74–2.70 | 0.3055 | 1.197 | 0.56–2.60 | 0.6426 |
|
Lymphatic invasion (absent vs.
present) | 1.245 | 0.63–2.47 | 0.5268 | 0.768 | 0.35–1.63 | 0.4925 |
| pStage
(I vs. II, III) | 5.222 | 2.75–9.85 |
<0.0001a | 6.475 | 3.28–12.8 |
<0.0001a |
A sub-analysis of stage I LUAD revealed that
individuals with high CRABP2 and CEACAM5 expression had
significantly worse 5-year RFS rates of 71.9% vs. 88.3% (P=0.009)
and 68.6% vs. 91.1% (P=0.002), respectively (Fig. S1). Moreover, individuals with high
CRABP2 and CEACAM5 expression had significantly worse 5-year OS
rates of 72.7% vs. 92.0% (P=0.033) and 87.2% vs. 92.6% (P=0.0345),
respectively. All P-values were determined using log-rank tests.
These results indicate that CRABP2 and CEACAM5 could serve as
excellent prognostic markers for recurrence and mortality in the
early stages of LUAD.
Discussion
In the present study, we identified three
molecules-CRABP2, CEACAM5, and MUC21-that play crucial roles in
promoting the lepidic-filigree-mPAP progression. These molecules
may be important in conferring aggressiveness to micropapillary, as
their high expression levels were associated with lymphatic canal
invasion, high recurrence rates, and poor 5-year survival rates.
However, the stage at which these molecules exert their effects
appears to vary. Specifically, CRABP2 is likely involved in the
early transition from lepidic to filigree, whereas CEACAM5 and
MUC21 are implicated in the later transition from filigree to mPAP
elements. No significant differences in CEACAM5 and
CRABP2 mRNA expression levels were observed between the
filigree and mPAP elements. However, an increase in the protein
expression of CEACAM5 and CRABP2 was observed between these
elements in the immunohistochemical analysis. This discrepancy may
be attributed to post-translational modifications.
To regulate the transcription of downstream genes,
CRABP2 transports retinoic acid to the nucleus, where it binds to
transcription factors. CRABP2 expression has been detected in lung
cancer (14), breast cancer
(15), and glioblastoma (16). Furthermore, CRABP2 enhances the
migration, invasion, and anoikis resistance of lung cancer cells
via the HuR and integrin β1/FAK/ERK signaling pathways and
promotes metastasis in vivo (14). Integrins play an important role in
cell adhesion, and CRABP2 overexpression activates the
integrin β1/FAK/ERK signaling pathway. These data suggest that
CRABP2 is involved in decreasing tumor cell adhesion and promoting
the morphological progression to filigree and mPAP patterns during
the early stages of LUAD development. CRABP2 is reportedly
significantly upregulated in LUAD with micropapillary elements
(17) and in small invasive
adenocarcinoma (18). Moreover,
high CRABP2 levels are correlated with lymph node metastases, poor
OS, and increased recurrence (18).
Collectively, these findings further support our findings.
The surface glycoprotein CEACAM5 is involved in cell
adhesion, intracellular signaling, and tumor progression. This
protein promotes cell proliferation and invasion via
p38/Smad2/3 signaling in non-small-cell lung cancer (NSCLC)
(19). The induced overexpression
of CEA is associated with anoikis, a form of apoptosis caused by
detachment from the cell matrix, which enhances metastasis of
colorectal cancer (20). These data
implicate CEACAM5 in the morphological progression of the
micropapillary pattern (21,22).
MUC21 is a transmembrane mucin expressed in various
human neoplasms, including lung carcinomas (13,23).
It is associated with the aggressive behavior of neoplastic cells
(13). We have previously revealed
that MUC21, characterized by short glycosylated sugar chains, is
associated with highly malignant histological components, such as
micropapillary elements (13).
Moreover, mouse Muc21/epiglycanin disrupts cell-extracellular
matrix interactions and interferes with intercellular adhesions,
suggesting that Muc21/epiglycanin inhibits surface integrins and
intercellular adhesion molecules (24,25).
Miyoshi et al (26) reported
that the micropapillary element exhibits a loss of
integrin-mediated adhesion to the basal membrane, which may help
explain the mechanism by which MUC21 contributes to the
micropapillary morphology.
In the present study, we identified CRABP2 and
CEACAM5 as independent prognostic markers for LUAD recurrence.
Clinically, these molecules have various applications. First,
immunohistochemical examination of CRABP2 and CEACAM5 in
preoperative biopsied tissue or surgical resected tissue serves as
predictive markers to determine the suitability of neo-adjuvant or
adjuvant chemotherapy. Second, the plasma concentrations of these
proteins may be used as a screening assay for the early detection
of lung cancer (27,28). Increased plasma CRABP2 and CEACAM5
levels are correlated with decreased survival rates in patients
with LUAD (28,29). Third, targeting these molecules for
molecular therapy is becoming increasingly feasible. Specifically,
CEACAM5 has recently emerged as a promising target for
antibody-drug conjugate therapy of NSCLC. Tusamitamab ravtansine
(SAR408701), a humanized antibody-drug conjugate targeting CEACAM5,
is in clinical development for nonsquamous NSCLC with high CEACAM5
expression (30).
This study has some limitations. First, the quality
of RNA extracted from FFPE specimens may deteriorate due to
long-term storage or poor storage conditions (such as high
temperature and humidity). Japan Pathology Quality Assurance System
recommends using FFPE specimens within 2.5 to 3 years of
preparation to ensure reliable genetic testing. We use FFPE
specimens prepared within 3 years for the Visium Spatial Gene
Expression Profiling System. Additionally, these FFPE specimens
were stored at temperatures below 25°C. Second, the spatial
resolution of the Visium platform was insufficient to analyze the
microenvironment surrounding tissue elements (such as lepidic,
filigree, and mPAP elements) each in LUAD. However, in recent
years, the Xenium spatial transcriptomics platform, which offers
higher resolution than the Visium spatial transcriptomics platform,
has emerged. This technology will enable the analysis of the
microenvironment for different tissue elements of LUAD in the
future.
In conclusion, we investigated the potential
molecular basis of the lepidic-filigree-mPAP progression using the
Visium Spatial Gene Expression Solution and identified three
molecules that promote this progression. These molecules may have
clinical utility as prognostic markers and as potential targets of
molecular therapy.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Japanese Ministry of
Education, Culture, Sports and Science, Tokyo, Japan (grant nos.
21K15387 and 22K15410).
Availability of data and materials
The raw sequencing data generated in the present
study may be found in the National Center for Biotechnology
Information Gene Expression Omnibus database under accession number
GSE 300676 or at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE300676.
The other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
MM and KO wrote the majority of the manuscript. TW
and HA collected patient information, compiled a clinical database
and conducted formal analysis. TW and HA made substantial
contributions to analysis and interpretation of data. TK was
responsible for the statistical analysis. MM and KO designed the
study and suggested the contents of the manuscript. MM, CK and KO
examined the tissue sections and made pathological diagnoses. MM
and CK contributed to funding acquisition. TS and HM cut tissue
sections and stained them with hematoxylin and eosin. DM and KF
performed Visium experiments, and analyzed and interpreted the
data. MM and KO confirm the authenticity of all the raw data. All
authors have read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committees of Kanagawa Prefectural Cardiovascular and Respiratory
Center (Yokohama, Japan; approval no. KCRC-17-0016; October 1,
2018). All patients provided written informed consent for the use
of their samples for research purposes. For prospective
observational studies, written informed consent has been obtained
from study participants. For retrospective observational studies,
existing specimens obtained for clinical purposes (pathological
specimens not additionally collected for research purposes) were
used. In principle, informed consent was sought for these
specimens; however, when obtaining informed consent was difficult,
opt-out consent was taken.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CEACAM5
|
carcinoembryonic antigen-related cell
adhesion molecule 5
|
|
CRABP2
|
cellular retinoic acid binding protein
2
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
LUAD
|
lung adenocarcinoma
|
|
mPAP
|
conventional/overt micropapillary
|
|
MUC21
|
mucin 21
|
|
OS
|
overall survival
|
|
RFS
|
recurrence-free survival
|
References
|
1
|
Makimoto Y, Nabeshima K, Iwasaki H,
Miyoshi T, Enatsu S, Shiraishi T, Iwasaki A, Shirakusa T and
Kikuchi M: Micropapillary pattern: A distinct pathological marker
to subclassify tumours with a significantly poor prognosis within
small peripheral lung adenocarcinoma (</=20 mm) with mixed
bronchioloalveolar and invasive subtypes (Noguchi's type C
tumours). Histopathology. 46:677–684. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miyoshi T, Satoh Y, Okumura S, Nakagawa K,
Shirakusa T, Tsuchiya E and Ishikawa Y: Early-stage lung
adenocarcinomas with a micropapillary pattern, a distinct
pathologic marker for a significantly poor prognosis. Am J Surg
Pathol. 27:101–109. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chao L, Yi-Sheng H, Yu C, Li-Xu Y, Xin-Lan
L, Dong-Lan L, Jie C, Yi-Lon W and Hui LY: Relevance of EGFR
mutation with micropapillary pattern according to the novel
IASLC/ATS/ERS lung adenocarcinoma classification and correlation
with prognosis in Chinese patients. Lung Cancer. 86:164–169. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Matsumura M, Okudela K, Kojima Y, Umeda S,
Tateishi Y, Sekine A, Arai H, Woo T, Tajiri M and Ohashi K: A
histopathological feature of EGFR-mutated lung adenocarcinomas with
highly malignant potential-An implication of micropapillary
element. PLOS One. 11:e01667952016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Husain A, Farver C and Nicholson A:
Adenocarcinoma of the lung. WHO classification of tumours. Borczuk
A, Chan J, Cooper W, Dacic S, Kerr K, Lantuejoul S and Marx A: 5th
edition. IARC; Lyon: 2021
|
|
6
|
Fukutomi T, Hayashi Y, Emoto K, Kamiya K,
Kohno M and Sakamoto M: Low papillary structure in lepidic growth
component of lung adenocarcinoma: A unique histologic hallmark of
aggressive behavior. Hum Pathol. 44:1849–1858. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gracia Villacampa E, Larsson L, Mirzazadeh
R, Kvastad L, Andersson A, Mollbrink A, Kokaraki G, Monteil V,
Schultz N, Appelberg KS, et al: Genome-wide spatial expression
profiling in formalin-fixed tissues. Cell Genom. 1:1000652021.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xie L, Kong H, Yu J, Sun M, Lu S, Zhang Y,
Hu J, Du F, Lian Q, Xin H, et al: Spatial transcriptomics reveals
heterogeneity of histological subtypes between lepidic and acinar
lung adenocarcinoma. Clin Transl Med. 14:e15732024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takano Y, Suzuki J, Nomura K, Fujii G,
Zenkoh J, Kawai H, Kuze Y, Kashima Y, Nagasawa S, Nakamura Y, et
al: Spatially resolved gene expression profiling of tumor
microenvironment reveals key steps of lung adenocarcinoma
development. Nat Commun. 15:106372024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sobue T, Yamamoto S, Hara M, Sasazuki S,
Sasaki S and Tsugane S: Cigarette smoking and subsequent risk of
lung cancer by histologic type in middle-aged Japanese men and
women: the JPHC study. Int J Cancer. 99:245–251. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Suzuki M, Shinozaki-Ushiku A, Yuhara S,
Nagano M, Sato M and Ushiku T: The prognostic impact of
extra-alveolar invasion in lung adenocarcinoma. Lung Cancer.
205:1086122025. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kawai H, Miura T, Kawamatsu N, Nakagawa T,
Shiba-Ishii A, Yoshimoto T, Amano Y, Kihara A, Sakuma Y, Fujita K,
et al: Expression patterns of HNF4a, tTF-1, and SMARCA4 in lung
adenocarcinomas: Impacts on clinicopathological and genetic
features. Virchows Archiv. 486:343–354. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Matsumura M, Okudela K, Nakashima Y,
Mitsui H, Denda-Nagai K, Suzuki T, Arai H, Umeda S, Tateishi Y,
Koike C, et al: Specific expression of MUC21 in micropapillary
elements of lung adenocarcinomas-Implications for the progression
of EGFR-mutated lung adenocarcinomas. PLoS One. 14:e02152372019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu JI, Lin YP, Tseng CW, Chen HJ and Wang
LH: Crabp2 promotes metastasis of lung cancer cells via HuR and
integrin β1/FAK/ERK signaling. Sci Rep. 9:8452019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Feng X, Zhang M, Wang B, Zhou C, Mu Y, Li
J, Liu X, Wang Y, Song Z and Liu P: CRABP2 regulates invasion and
metastasis of breast cancer through hippo pathway dependent on ER
status. J Exp Clin Cancer Res. 38:3612019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu RZ, Li S, Garcia E, Glubrecht DD, Poon
HY, Easaw JC and Godbout R: Association between cytoplasmic CRABP2,
altered retinoic acid signaling, and poor prognosis in
glioblastoma. Glia. 64:963–976. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu L, Su H, Zhao S, Si H, Xie H, Ren Y,
Gao J, Wang F, Xie X, Dai C, et al: Development of the semi-dry
dot-blot method for intraoperative detecting micropapillary
component in lung adenocarcinoma based on proteomics analysis. Br J
Cancer. 128:2116–2125. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dai T, Adachi J, Dai Y, Nakano N, Yamato
M, Kikuchi S, Usui S, Minami Y, Tomonaga T, Noguchi M, et al:
In-depth proteomics reveals the characteristic developmental
profiles of early lung adenocarcinoma with epidermal growth factor
receptor mutation. Cancer Med. 12:10755–10767. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang X, Han X, Zuo P, Zhang X and Xu H:
CEACAM5 stimulates the progression of non-small-cell lung cancer by
promoting cell proliferation and migration. J Int Med Res.
48:3000605209594782020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Q, Li Y, Li J, Ma Y, Dai W, Mo S, Xu Y,
Li X and Cai S: FBW7 suppresses metastasis of colorectal cancer by
inhibiting HIF1α/CEACAM5 functional axis. Int J Biol Sci.
14:726–735. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Süer H, Erus S, Cesur EE, Yavuz Ö,
Ağcaoğlu O, Bulutay P, Önder TT, Tanju S and Dilege Ş: Combination
of CEACAM5, EpCAM and CK19 gene expressions in mediastinal lymph
node micrometastasis is a prognostic factor for non-small cell lung
cancer. J Cardiothorac Surg. 18:1892023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Odintsov I and Sholl LM: Prognostic and
predictive biomarkers in non-small cell lung carcinoma. Pathology.
56:192–204. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yoshimoto T, Matsubara D, Soda M, Ueno T,
Amano Y, Kihara A, Sakatani T, Nakano T, Shibano T, Endo S, et al:
Mucin 21 is a key molecule involved in the incohesive growth
pattern in lung adenocarcinoma. Cancer Sci. 110:3006–3011. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yi Y, Kamata-Sakurai M, Denda-Nagai K,
Itoh T, Okada K, Ishii-Schrade K, Iguchi A, Sugiura D and Irimura
T: Mucin 21/epiglycanin modulates cell adhesion. J Biol Chem.
285:21233–21240. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Denda-Nagai K, Ishii-Schrade KB, Tian Y,
Okada K and Irimura T: Immunohistochemistry of mucin. Glycoscience
protocols (GlycoPODv2). Nishihara S, Angata K, Aoki-Kinoshita KF
and Hirabayashi J: Japan Consortium for Glycobiology and
Glycotechnology; Saitama: pp. p212021, PubMed/NCBI
|
|
26
|
Miyoshi T, Shirakusa T, Ishikawa Y,
Iwasaki A, Shiraishi T, Makimoto Y, Iwasaki H and Nabeshima K:
Possible mechanism of metastasis in lung adenocarcinomas with a
micropapillary pattern. Pathol Int. 55:419–424. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Djureinovic D, Pontén V, Landelius P, Al
Sayegh S, Kappert K, Kamali-Moghaddam M, Micke P and Ståhle E:
Multiplex plasma protein profiling identifies novel markers to
discriminate patients with adenocarcinoma of the lung. BMC Cancer.
19:7412019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lung Cancer Cohort Consortium (LC3), . The
blood proteome of imminent lung cancer diagnosis. Nat Commun.
14:30422023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim DJ, Kim WJ, Lim M, Hong Y, Lee SJ,
Hong SH, Heo J, Lee HY and Han SS: Plasma CRABP2 as a novel
biomarker in patients with non-small cell lung cancer. J Korean Med
Sci. 33:e1782018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lefebvre AM, Adam J, Nicolazzi C, Larois
C, Attenot F, Falda-Buscaiot F, Dib C, Masson N, Ternès N, Bauchet
AL, et al: The search for therapeutic targets in lung cancer:
Preclinical and human studies of carcinoembryonic antigen-related
cell adhesion molecule 5 expression and its associated molecular
landscape. Lung Cancer. 184:1073562023. View Article : Google Scholar : PubMed/NCBI
|