Introduction
Human guanylate-binding protein 2 (hGBP2) and murine
(m)GBP2 were first isolated from human fibroblasts in 1982
(1) and macrophages in 1998
(2). GBP2 is a member of the
IFN-inducible GTPase family that serves notable roles in cellular
signaling. hGBP2 is reported to be highly expressed in various
immune cells, including monocytes, lymphocytes and natural killer
cells (3). Under basal conditions,
hGBP2 and mGBP2 are primarily distributed diffusely in the
cytoplasm and nucleus. However, upon stimulation by IFN-γ and in
the GTP-bound state, such as hGBP1 and hGBP5, it can translocate to
the Golgi apparatus (4,5). The genes encoding hGBP1-7 are located
on human chromosome 1, whilst the genes encoding mGBP1-11 are
located on mouse chromosome 3 (3),
where mGBP2 is mapped to the distal end and is putatively
associated with mGBP1 (2).
The inflammasome is a multi-protein complex that
functions to eliminate abnormal cells and amplify inflammation. It
is comprised of various sensor proteins, such as the NLR family
pyrin domain-containing 3 [NLRP3; which primarily recognizes
lipopolysaccharide (LPS)-induced signals], absent in melanoma 2
(AIM2; which detects intracellular double-stranded DNA), adaptor
proteins, such as the apoptosis-associated speck-like protein
containing a caspase recruitment domain (ASC) and caspase effector
proteins. It also senses pathogen-associated molecular patterns
(PAMPs) or damage-associated molecular patterns (DAMPs), thereby
triggering pyroptosis, a type of programmed cell death
characterized by gasdermin-D-mediated plasma membrane perforation
and release of inflammatory cytokines. hGBP2 activates inflammasome
assembly and pyroptosis primarily by binding and delivering LPS
into the cytoplasm (6).
Additionally, hGBP2 facilitates inflammasome assembly through
direct binding to ASC and cooperates with hGBP5 (7).
In the context of cancer, paclitaxel (PTX), which
has become an important chemotherapeutic agent since its discovery
in 1967, is a natural diterpenoid compound that can be isolated
from Taxus brevifolia. hGBP2 cooperates with PTX through the
MCL-1 apoptosis regulator, myeloid cell leukemia-1 (MCL-1)/Bcl-2
antagonist/killer 1 (Bak) pathway (8) to activate the vascular endothelial
growth factor (VEGF) pathway, thereby promoting angiogenesis and
oxygen supply to tumor. Immunologically ‘cold’ tumors are
characterized by poor responses to immunotherapies (9) and host a tumor immunosuppressive
microenvironment (TIME) lacking infiltrating immune cells. Such
tumors typically exhibit rapid progression and poor prognosis
(10), posing a major challenge in
cancer immunotherapy. Notably, hGBP2 may offer a promising
therapeutic strategy for ‘cold’ tumors by activating a number of
immune checkpoints, such as programmed cell death protein 1
(PD-1)/programmed death-ligand 1 (PD-L1) and lymphocyte activating
3 (LAG3) (11), thereby remodeling
the TIME and inhibiting the activation of CD8+ T cells
and CD4+ T cells (12).
Structure and hydrolase function of
hGBP2
Structure of hGBP2
hGBP2 is a 67 kDa GTPase that is comprised of three
distinct domains (13). The
N-terminal region contains a large GTPase domain (LG; residues
1–309), followed by the middle domain (MD; residues 310–480) in the
central region and the α12/13 helical domain (HD; residues 480–580)
at the C-terminus. In its crystalline state, hGBP2 adopts a
bi-molecular asymmetric unit configuration.
The LG domain serves as the nucleotide-binding site
and is characterized by five conserved motifs involved in GTP
binding and hydrolysis (Table I)
(14). The G1/P-loop motif has a
GxxxxGKS/T sequence (15) that
encloses the β-phosphate of GTP. By contrast, the G2/SWITCH I motif
serves a critical role in magnesium ion binding, which is a key
feature for all GTPases, since it stabilizes the GTP molecule
through coordination bonds with the phosphate groups. The G3/SWITCH
II motif, with its conserved DTEG residue sequence, forms hydrogen
bonds with the phosphate, aiding in its release. The G4 motif
interacts specifically with the guanine base with its acidic Asp
residue in the RDF residue sequence, facilitating the binding of
GTP to the domain. The primary non-conserved region, located
between residue 240 and 280, corresponds to a flexible region
involved in membrane interactions and is situated within the G5
region. The G1-G5 motifs collectively constitute the
‘nucleotide-binding pocket’ for GTP (Table I).
 | Table I.Key motifs (G1-G5) of the human
guanylate-binding protein 2 hydrolysis site. |
Table I.
Key motifs (G1-G5) of the human
guanylate-binding protein 2 hydrolysis site.
| Motif
designation | Sequence | Corresponding
residue numbers |
|---|
| G1/P-loop | GLYRTGKS | 45-52 |
| G2/SWITCH I | TVKSHT | 70-75 |
| G3/SWITCH II | DTEG | 97–100 |
| G4 | RDF | 181-183 |
| G5/Guanine cap |
WPAPKKYLAHLEQLKEEE | 236-255 |
The MD domain consists of 5 α-helices. The first
helix group is formed of the N-terminal halves of α7, α8 and α9,
where the second helix group is formed of the C-terminal half of
α9, α10 and α11. By contrast, the α12/α13 domain contains a
conserved CaaX sequence, where isoprenylation occurs, promoting the
localization of hGBP2 on the membrane as a hydrophobic group.
Another feature of α12/α13 domain is its interaction with LG
domain. The α12/α13 domain is mainly negatively charged, while the
LG domain mainly carries positive charge. This enables the two
domains to contact through electrostatic interactions (13,16),
where the α12/α13 domain is in a tightened state.
Hydrolase function of hGBP2
The hydrolase function of hGBP2 is essential for a
number of mechanisms and processes. The membrane localization of
hGBP2 requires homodimerization, which is dependent on the
transition state of GTP hydrolysis (17).
The hGBP2 hydrolysate, which contains >75% GDP
(18,19), yields only a small amount of GMP as
the final product (13,16), which is obtained by the hydrolysis
of GTP instead of GDP. This distinct characteristic, differing from
hGBP1, is primarily attributed to the LG domain of hGBP2 rather
than changes in the MD or HD domains. Specifically, this is due to
differences in the adjustment of the active site in the LG domain
of hGBP2 following the first hydrolysis event. In terms of
tetramerization, hGBP2, unlike hGBP1 but like mGBP2 (18), can extensively tetramerize, although
the tetramerization of hGBP2 does not contribute to the formation
of GMP (20,21).
When hGBP2 binds GTP, GTP enters the nucleotide
binding pocket formed by the G1-G5 motif, where hGBP2 undergoes
homodimerization at the same time with the participation of LG and
MD domains (22,23). The LG domain of hGBP2 is dimerized
only 50% of the time in the presence of GTP, suggesting that the
C-terminal domain of hGBP2 also participates in dimerization by
providing a dimerization interface or otherwise stabilizing hGBP2
dimers in the GTP state (16). In
addition, residue T75 in the G2/SWITCH motif and S52 in the G1
motif are involved in Mg2+ coordination (13), implying that Mg2+ may
enter the nucleotide binding pocket and bind to the phosphate group
and surrounding amino acid residues at this stage, which are
necessary for hGBP2 activity. It has previously been reported that
Mg2+ is necessary for the activation of hGBP2 at Ph 8.0,
but not pH 6.0 (16), which
suggests that Mg2+ and H3O+ may
have a functional substitution relationship. However, this
interaction requires further validation.
In the hGBP2-GDP conformation following phosphate
group dissociation, the N-O bond of the G1/P-loop motif interacts
with the β-phosphate. Concurrently, residue S52 forms N-O bonds
with both the α- and β-phosphates, facilitating nucleotide
positioning and binding (24,25).
The Y47 side chain undergoes movement and participates in dimer
interface formation, whilst the displacement of Y47 and R48 in the
G1 motif induces notable movement of the G2 motif, with some Cα
atoms shifting >10 Å (26). The
G4/RD motif interacts with the guanine moiety through hydrogen
bonding. W238 in the G5 motif undergoes allosteric rearrangement
and interacts with W238 from another hGBP2 molecule (13). Consequently, the guanine base and
ribose are enclosed within the space formed by the G1, G4 and G5
motifs. At the opposite end of the GDP, the diphosphate group is
tightly positioned within the pocket formed by the G1/P-loop,
G2/SWITCH I and G3/SWITCH II motifs. The β-phosphate further
coordinates with the conserved residues K51, S52 and E99. When the
E99-K51 interaction is absent, SWITCH I remains open. However, when
these residues engage through hydrogen bonding, SWITCH I closes,
simultaneously positioning T75 in the G2 motif for optimal
interaction with Mg2+ (16).
Regulation of hGBP2 and mGBP2
Structural foundation of hGBP2 and
mGbp2 genes
The hGBP2 gene, as an IFN-stimulated gene
(ISG), contains a promoter region featuring two transcription
factor binding sequences, namely the IFN-stimulated response
element (ISRE), located at +42 (27) and the γ-activated sequence (GAS)
(27,28), located at +53 and at −532 (27). Notably, GAS exhibits binding
affinity for all STAT factors except for STAT2. Both sequences can
independently respond to IFN signaling, whilst their combined
presence results in enhanced transcriptional activity (29). By contrast, the mGbp2 gene is
also an ISG, which contains one ISRE, located at −2 (27) and two GASs, located at −487 and −735
(27). However, Ramsauer et
al (28) previously reported
that the mGbpP2 gene has two ISREs, located at −30 and −440
and one GAS located at −530 (28).
Molecular regulatory mechanisms
IFN-independent hGBP2 and mGbp2 gene expression
under normal conditions
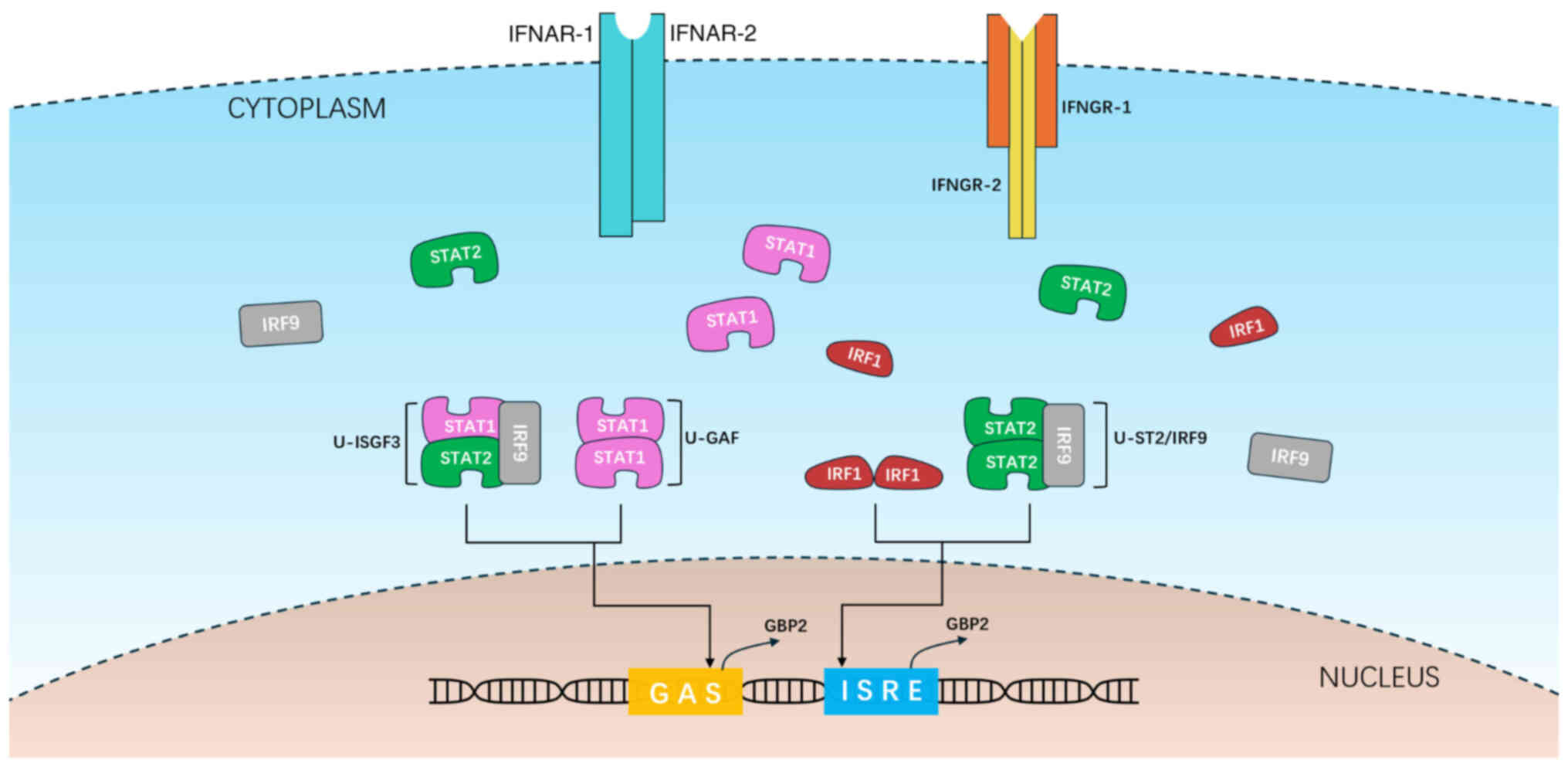
STAT1, STAT2 and IFN-regulatory factor (IRF) 9 can
form four types of oligomers. They are STAT1-STAT1 (binding to GAS,
referred to as U-STAT1), STAT1-STAT2-IRF9 (binding to GAS, referred
to as U-ISGF3), STAT2-STAT2-IRF9 (binding to ISRE, referred to as
U-ST2) and IRF1-IRF1 (binding to ISRE) (29). These oligomers subsequently localize
to hGBP2 or mGbp2 genes and activate their
transcription (Fig. 1). In mice,
the IRF1 dimer has been demonstrated to directly bind to the
transcription complex containing RNA polymerase II to activate
mGbp2 gene transcription (28), whilst STAT1 dimers not only bind to
GAS on the hGBP2 gene promoter but also promote the
acetylation of histone 4 in the hGBP2 gene, thereby
providing active chromatin. Additionally, previous studies have
shown that p53 can form a complex with IRF1 to upregulate hGBP2
expression (30,31).
Highly activated phase under
stimulation by IFN-I, IFN-II and IRF1
Upon pathogen invasion or intracellular
abnormalities, the corresponding PAMPs or DAMPs are recognized by a
series of pattern recognition receptors (PRRs). This type of
recognition triggers the phosphorylation and activation of IRFs,
thereby promoting the expression of IFN-I and IFN-II (the former
mainly includes subtypes IFN-α and IFN-β) through the classical
secretory pathway. These IFNs are secreted extracellularly and bind
to their corresponding respective receptors. Mitochondrial outer
membrane permeabilization (MOMP) under radiation conditions and
apoptosis can lead to the release of mitochondrial double-stranded
(ds)DNA, which activates the cyclic GMP-AMP synthase/stimulator of
IFN genes pathway, subsequently inducing IFN-β and upregulating
mGBP2 expression (32).
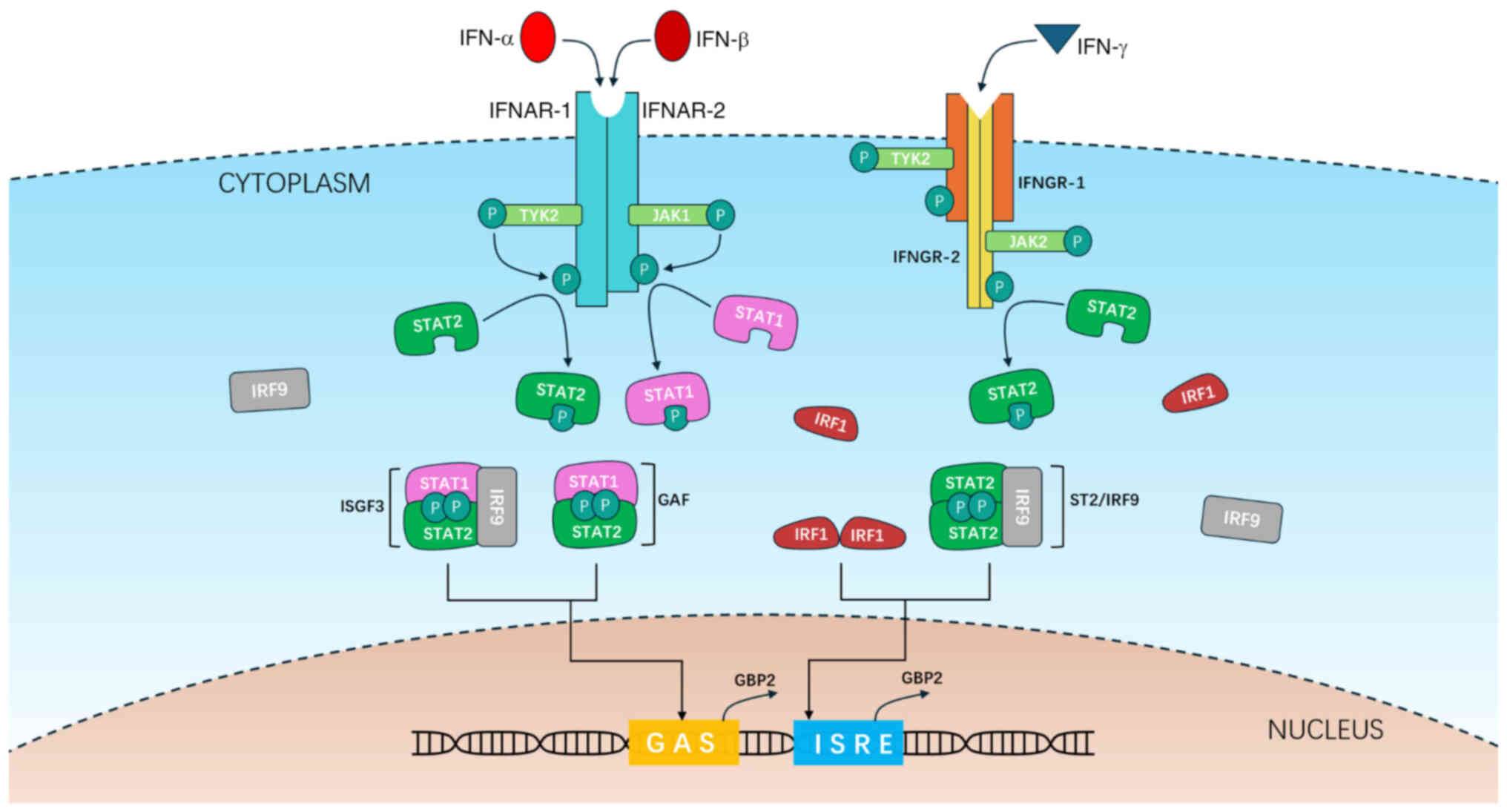
IFN-I can bind to the ubiquitously expressed IFN-α/β
receptor (consisting of the IFNAR1 and IFNAR2 subunits), leading to
the dimerization of the receptor subunits. This dimerization,
through juxtaposition and trans-phosphorylation, enhances the
kinase activity of Janus kinase (JAK) 1 and tyrosine kinase (TYK)
2. Subsequently, JAK1 and TYK2 phosphorylate tyrosine residues on
IFNAR1 and IFNAR2, which serve as binding sites for STAT1 and STAT2
(33). Phosphorylation of STAT1 at
Tyr701 and STAT2 at Tyr690 then occurs. By contrast, IFN-II,
specifically IFN-γ, interacts with a tetrameric receptor complex
composed of two IFNGR1 subunits and two IFNGR2 subunits. This
receptor is associated with JAK1 and JAK2 kinases, which
exclusively phosphorylate the STAT1 protein.
Similar to the pathway under normal conditions,
phosphorylated (p-)STAT1, p-STAT2 and IRF1 can form four types of
oligomers: p-STAT1-p-STAT1 (also known as γ-IFN activation factor,
binds to GAS), p-STAT1-p-STAT2-IRF9 (binds GAS), pSTAT1-pSTAT2-IRF9
(also known as IFN-stimulated gene factor, binds to ISRE) and
IRF1-IRF1 (binds ISRE). These oligomers subsequently localize to
the hGBP2 or mGbp2 gene and activate its
transcription through a similar mechanism (Fig. 2), exhibiting higher activity
compared with their non-phosphorylated counterparts.
Function of hGBP2 and mGBP2 in
inflammation
Activation of the inflammasome pathway
by hGBP2 and mGBP2
The inflammasome pathway can be divided into two
types, namely the canonical pathway and the non-canonical pathway.
Their objective is to induce pyroptosis, which is mediated by the
formation of plasma membrane pores through gasdermin-D. This leads
to membrane rupture and alerting other cells, with the release of
cytokines IL-1β and IL-18 through the gasdermin-D-formed membrane
pores. hGBP2 and mGBP2 primarily serve supporting roles in
inflammasome assembly.
In the canonical pathway, activation is primarily
mediated by PRRs, such as nucleotide-binding oligomerization
domain-like receptors, such as NLRP3, or AIM2-like receptors,
including AIM. PAMPs or DAMPs are recognized by PRRs, such as NLRP3
or AIM2. During this process, mGBP2 can facilitate the release of
dsDNA by lysing pathogen-containing vacuoles or pathogens, enabling
AIM2 recognition (7,34,35).
Upon activation, NLRP3 or AIM2 interacts with the adaptor protein
ASC through their pyrin domains (36). In this step, mGBP2 and mGBP5 can
form a heterocomplex, where mGBP2 binds ASC and mGBP5 binds NLRP3.
This mGBP2-mGBP5 complex brings NLRP3 and ASC together, thereby
promoting the assembly of the NLRP3 inflammasome (7). ASC in turn recruits caspase-1 through
its caspase recruitment domain, forming the inflammasome.
Subsequently, caspase-1 within the inflammasome cleaves
gasdermin-D, releasing its N-terminal fragment to form plasma
membrane pores and induce pyroptosis. Caspase-1 also processes
pro-IL-1β and pro-IL-18 into their active forms, which are then
released through the membrane pores to alert neighboring cells
(7).
In the non-canonical pathway, activation in humans
is primarily mediated by hGBPs through PRRs while in mice, it is
mediated by caspase-11 with the assistance of mGBP2. In humans,
when bacteria enter cells, hGBP1 binds bacterial LPS and recruits
hGBP2, hGBP3 and hGBP4 to the surface of bacteria, especially
gram-negative bacteria, forming a coating (6,37).
hGBP2 and hGBP4 expose the lipid moiety of LPS, recruiting
caspase-4 to the bacterial surface for LPS binding, while hGBP3
regulates caspase-4 activation (6,37).
Activated caspase-4 then cleaves pro-IL-1β, pro-IL-18 and
gasdermin-D, leading to pyroptosis and cytokine release.
In mice, caspase-11 directly recognizes the
mGBP2-LPS complex, oligomerizes and activates, gaining the ability
to cleave gasdermin-D and induce pyroptosis (38). Additionally, the formation of plasma
membrane pores causes potassium ion efflux due to the intracellular
potassium gradient, further promoting NLRP3 inflammasome assembly
and creating a positive-feedback amplification loop (39). In this process, mGBP2 assists
caspase-11 in LPS recognition. A previous study has reported that
mGBP2 can interact with gasdermin-D and mGBP3, potentially serving
as a novel component of this pathway, although the precise
mechanisms remain unclear and require further investigation
(7).
The severe consequences of pyroptosis, such as
further inflammation cascade caused by the release of IL-18 and
IL-1β (40), may explain the need
for the continuity and regulation of the GBP2/caspase pathway. This
type of regulation allows for the existence of multiple regulatory
checkpoints before caspase activation, increasing the difficulty of
activation and preventing unnecessary triggering (6).
Role of hGBP2 and mGBP2 in
inflammatory diseases
The expression of hGBP2 is increased in the kidney
tissues of patients with lupus nephritis, particularly in the
glomeruli and renal tubulointerstitium (41). In diabetic nephropathy, macrophages
at the injury site are predominantly of the M1 subtype, where hGBP2
can promote the polarization of macrophages toward the
pro-inflammatory M1 subtype through the Notch 1 pathway. Through
the hGBP2-mediated pathway, macrophages can be induced into the M1
phenotype by LPS or IFN-γ (42).
In allergic rhinitis (AR), mGBP2 can alleviate
oxidative stress and abnormal lipid metabolism by inhibiting the
hypoxia-inducible factor-1 (HIF-1) pathway. It can also inhibit
mitochondrial fission whilst maintaining mitochondrial fusion to
mitigate oxidative stress-induced damage to cells (43). A previous study has demonstrated
that the overexpression of mGBP2 can notably reduce inflammatory
cell infiltration into the nasal mucosa and markedly decrease the
levels of various factors, such as total cholesterol, low-density
lipoprotein-cholesterol, TNF-α, IL-5, IFN-γ and trimethylamine
N-oxide, in AR mouse models whilst increasing high-density
lipoprotein-cholesterol levels (44). However, the underlying mechanisms
require further investigation.
LPS-induced macrophages-derived exosomes (L-Exo) can
be transported from macrophages to lung epithelial cells, resulting
in damage to the alveolar epithelial tissue. The mGBP2 content in
L-Exo is higher compared with that in control Exo (45), leading to the hypothesis that the
mGBP2 contained in L-Exo can trigger pyroptosis in the lung
epithelial tissue, thereby causing injury. This may represent a
potential therapeutic approach for sepsis-associated acute lung
injury (46). In depression-like
behaviors induced by neuroinflammation, reducing the expression of
mGBP2 may also alleviate symptoms (47).
Role of hGBP2 and mGBP2 in oncogenesis and
cancer
hGBP2 and mGBP2 regulate cancer
progression through multiple signaling pathways
Previous studies have demonstrated that in
glioblastoma (GBM) and low-grade glioma cells, hGBP2 can directly
interact with kinesin family member 22 (KIF22) to
post-transcriptionally regulate and elevate its levels, thereby
enhancing KIF22-mediated EGFR internalization and signaling
(Table II) (48–63).
This in turn fosters cell proliferation (64). Unc-51 like autophagy activating
kinase 1 (ULK1) is another critical regulator of autophagy
(65), phosphorylating and inducing
autophagy under low-nutrient conditions. hGBP2 can activate the
ULK1 complex by suppressing PI3K/Akt/mTORC1 pathway (66), thereby enhancing cellular autophagy
(66). Notably, the outcomes of
this function may vary, where it may exert tumor-suppressive
effects during early-stage cancer but potentially promote cancer
progression at advanced stages (67).
 | Table II.GBP2 expression level across
different cancer types and corresponding mechanisms. |
Table II.
GBP2 expression level across
different cancer types and corresponding mechanisms.
| Tumor type | Cells or databases
involved | Species | GBP2 expression
level | Mechanisms | (Refs.) |
|---|
| GBM | Glioblastoma cell
lines, U87, U251, SNB19, GSC11 and G91; database, TCGA | Human | Upregulated | Expression of
various carcinogenesis-related genes in GBM, particularly in
mesenchymal subtype, including fibronectin 1, MMP-1, MMP-3, MMP-14,
urokinase, secreted protein acidic and rich in cysteine, TGFB1,
macrophage colony-stimulating factor 1, CD44, IL-8, monocyte
chemoattractant protein 1 and IL6, are elevated with hGBP2
involvement. Furthermore, cell proliferation is accelerated, the
block of G0/G1 phase of cell cycle is
prevented and apoptosis is decreased, due to hGBP2-activated
kinesin family member 22/EGFR signaling pathway | (48–51) |
| BC, particularly
triple-negative BC | Primary BC cells
isolated from tumor tissues from patients with BC and normal breast
tissue | Human | Downregulated | The methylation of
hGBP2 gene promoter decreases the levels of hGBP2,
inhibiting the promotion of the immune response of CD8+
T cells, thus facilitating the development of BC | (52–54) |
| SKCM | Human melanoma cell
lines, A2058, A375; mouse melanoma cell lines, B16 and B16F10;
human epidermal cell line, NHEK | Human and
mouse | Downregulated | It has been
revealed that overexpressing hGBP2 can upregulate E-cadherin and
downregulate N-cadherin and vimentin, inhibiting the EMT, invasion
and proliferation of SKCM. While whether hGBP2 downregulation,
which occurs in SKCM cells, can facilitate EMT remains
unstudied.hGBP2 also markedly downregulates the Wnt/β-catenin
signaling pathway and related proteins, including transcription
factor 4, c-Myc and cyclin D1, thus inhibiting cell proliferation,
migration, invasion and promoting cell apoptosis. The mechanisms of
mGBP2 in SKCM is unstudied | (51,55) |
| ccRCC | Primary ccRCC cells
isolated from patient tumor tissue and normal kidney tissue;
monocyte cell line, THP-1; common RCC cell lines, 786-O, 769-P,
CAKI-1, A498 and ACHN; renal tubular epithelium cell line, HK-2;
databases, Clinical Proteomic Tumor Analysis Consortium and
TCGA | Human | Upregulated | hGBP2 activates the
polarization of M0 macrophages toward the M2 macrophages through
JAK/STAT3-dependent upregulation on IL-18 secretion.
Simultaneously, M2 macrophages can induce the expression of GBP2 in
tumor cells by secreting IL-10 and TGF-β, which in turn activates
the expression of hGBP2, forming a loop. hGBP2 can also enhance the
levels of regulatory T cells and exhausted T cells, facilitating
ccRCC tumor immune evasion and proliferation | (51,56,57) |
| PAAD | Databases, TCGA and
GEO; primary PAAD cells isolated from tumor tissue from patients
with PAAD and adjacent normal tissue | Human | Upregulated | hGBP2 levels are
markedly elevated in active CD4 memory T cells, resting dendritic
cells and M1 macrophages, indicating that these three types of
cells can possibly induce the expression of hGBP2. Subsequently,
hGBP2 can further regulate the TIME | (11,51) |
| Thyroid
carcinoma | Database, TCGA | Human | Upregulated | Remains
unstudied | (51) |
| Uveal melanoma | Database, TCGA | Human | Upregulated | Remains
unstudied | (51) |
| Hepatocellular
carcinoma | Databases, TCGA and
GEO | Human | Upregulated | hGBP2 can induce
tumor cell infiltration, especially by upregulating macrophages and
downregulating Th17 and neutrophils in the TIME | (51,58) |
| ESCC | Human ESCC cell
lines, TE-1, TE-7, TE-10 and TE-13 | Human | Upregulated | hGBP2 is involved
in a p53/IRF-1/hGBP2 pathway and is thus overexpressed in ESCC.
Downstream mechanisms remain unstudied | (30) |
| CRC | Databases: GEO,
UCSC Xena Browser, TCGA; human MSS CRC cell lines, HT29 and SW480;
murine MSS CRC cell line, CT26 | Human and
mouse | The level of hGBP2
varies: Upregulated in MSS CRC of IC and dMMR/MSI CRC;
downregulated in pMMR/MSS CRC of non-IC | hGBP2 can activate
STAT1 by competing with SHP1, which inhibits the phosphorylation
and activation of STAT1, for binding to STAT1 | (59,60) |
| Gastric cancer | Databases: GEO,
TIDE, TCGA, UCSC Xena Browser, KEGG | Human | Upregulated | Remains
unstudied | (61) |
| HNSCC | Databases,
ONCOMINE, TCGA, HPA | Human | Stably upregulated
across different stages of HNSCC | Remains
unstudied | (31) |
| OC | Human cervical
cancer cell line, HeLa; murine breast cancer cell line, 4T1; murine
macrophage cell line, RAW264.7; murine preadipocyte cell line,
3T3-L1; murine ovarian cancer cell line, ID8 | Human and
mouse | Downregulated | mGBP2/Pin1/NF-κB
pathway, which is possibly inhibited in OC, induces the
polarization of M0 to M1, thus the anticancer macrophage subtype M1
is at low levels in OC | (62) |
| Prostate
cancer | Databases, KEGG,
GEO, TCGA | Human | Upregulated | Remains
unstudied | (63) |
hGBP2 can induce the assembly of inflammasomes,
leading to a positive feedback loop of inflammatory responses. This
forms the hGBP2/STAT3/fibronectin (FN1) pathway (49,53),
where hGBP2 induces FN1, which is an extracellular glycoprotein
involved in cell migration (68),
on both transcriptional and translational levels to enhance
glioblastoma migration and invasion without influencing
proliferation in vitro (49). In addition, STAT3 contributes to
maintaining the mesenchymal subtype of GBM (50) and is indispensable for hGBP2-induced
FN1 elevation (49). FN1 can also
promote MMP-9 secretion in a focal adhesion kinase (FAK)- and
Ras-dependent manner (69) or
through the FN1/MMP-2/MMP-9 pathway (70). However, hGBP2 can induce FN1 in
vivo but lacks this capability in vitro (49). Furthermore, MMP-9 is an inducer of
epithelial-mesenchymal transition (EMT) (71). Thus, hGBP2 should activate, rather
than inhibit, MMP-9 through the STAT3/FN1/FAK or STAT3/FN1/MMP-2
signaling pathways. However, the regulatory effect of hGBP2 on
migration has not been experimentally demonstrated.
Bak is a member of the Bcl-2 family that is crucial
for the activation of apoptosis. It promotes MOMP, thereby
triggering the apoptotic process (72). hGBP2 acts as a dual stimulator of
Bak. It can not only release Bak but can also promote its
expression (8). The LG domain of
hGBP2 specifically binds to the BH3 domain of Mcl-1, a member of
the Bcl-2 family, thereby preventing the interaction between Bak
and Mcl-1. This then releases Bak, which oligomerizes to induce
MOMP, ultimately promoting apoptosis (8). Simultaneously, hGBP2 upregulates the
expression of Bak through its inhibitory effect on the PI3K/Akt
pathway (8). In summary, hGBP2 can
promote MOMP by activating Bak and enhancing its expression,
thereby facilitating apoptosis. PTX, as an anticancer drug,
occupies the upstream position in the hGBP2/Mcl-1/Bak pathway.
Through this pathway, hGBP2 enhances cellular sensitivity to PTX
(8,66). Notably, hGBP2 upregulation is a
common feature of PTX treatment and other chemotherapeutic agents
for hematologic malignancies (such as doxorubicin, cytarabine,
vincristine, etoposide and IFN-γ). However, among these
antineoplastic agents, only PTX possesses the unique ability to
elevate Bak levels (8).
In breast cancer (BC) cells, hGBP2 has been
demonstrated to directly interact with dynamin-related protein 1
(Drp1) in a K51-dependent manner, preventing its translocation from
the cytoplasm to the mitochondria. This interaction inhibits
Drp1-dependent mitochondrial fission and elongation, thereby
blocking mitochondrial division and cell metastasis in cancer cells
(73). Notably, the activity of
hGBP2 binding to Drp1 is influenced by all three major structural
domains of hGBP2, where it is possible that hGBP2 and Drp1 bind to
each other to form hetero-oligomers, although further
investigations are required (73).
hGBP2 also exerts its anticancer effects by inhibiting the
Wnt/β-catenin/EMT pathway (55,71,74)
and cancer cell metastasis and invasion, which has been
demonstrated in cancers such as skin cutaneous melanoma (SKCM) and
colon cancer, but the specific molecular mechanisms remain unclear.
PTX primarily inhibits tumor metastasis and progression by blocking
angiogenesis in tumor tissues, whilst hGBP2 enhances the
sensitivity of colon cancer cells to PTX by inhibiting the
Wnt/β-catenin pathway (75,76). Additionally, a previous study has
indicated that inhibiting the Wnt/β-catenin pathway in BC and
triple-negative BC (TNBC) can promote ferroptosis in BC cells and
suppress cell cycle and growth regulatory proteins, such as cyclin
D1 and c-Myc. Therefore, it can be speculated that this may be a
potential function of hGBP2, warranting further investigation.
In mouse dendritic cells, mGBP2 has been reported to
interact with the Akt-p110 complex, inhibiting the phosphorylation
and activation of Akt (77,78). This in turn prevents the
phosphorylation of TSC complex subunit 2, disrupting its ability to
form a complex with tuberous sclerosis 1 that inhibits mTORC1
(77–80). VEGF promotes endothelial cell
proliferation and migration by binding to VEGFRs on endothelial
cells, thereby stimulating angiogenesis. Under basal conditions,
the HIF-1α subunit is hydroxylated by prolyl hydroxylases and
recognized by the von Hippel-Lindau protein complex, leading to its
ubiquitination and degradation. Under hypoxic conditions, however,
the HIF-1α subunit is stabilized, where it translocates to the
nucleus and dimerizes with the HIF-1β subunit to form the HIF-1
transcription factor, which then translocates to the nucleus and
binds to hypoxia-response element in the promoter of VEGF gene
(81,82), regulating VEGF transcription
(43). In oxygen-induced
retinopathy mice model, mGBP2 has been shown to inhibit the
HIF-1α/VEGF pathway by suppressing mTORC1 (78), thereby attenuating angiogenesis.
This reduces blood supply to cancer lesions, maintaining hypoxic
conditions and potentially curbing cancer progression.
mGBP2 can activate MMP-9 through distinct pathways,
thereby influencing collagen degradation near the basement membrane
(33), extracellular matrix
remodeling, angiogenesis in cancer lesions and cancer cell invasion
and metastasis (77). Previous
studies in NIH 3T3 fibroblasts have shown that mGBP2 can inhibit
Rac, disrupting its regulation of the cytoskeleton. This inhibition
suppresses the TNF-α-mediated activation of the NF-κB pathway,
thereby reducing NF-κB-induced MMP-9 transcription (83). mGBP2 has been demonstrated in NIH
3T3 fibroblasts to directly interact with the p65 (RelA) protein in
the NF-κB pathway, preventing its binding to the MMP-9 promoter and
subsequently lowering MMP-9 expression (83). In ovarian cancer, mGBP2 promotes the
recruitment of the peptidyl-prolyl cis-trans isomerase
NIMA-interacting 1 (Pin1) protein, which activates the NF-κB
pathway (62). Based on this, it
can be hypothesized that mGBP2 may upregulate MMP-9 expression
through the Pin1/NF-κB and Rac/NF-κB pathways, although further
experimental validations are required.
hGBP2 and mGBP2 modulate cancer
progression by regulating the TIME
hGBP2 exerts a dual influence on the TIME. It can
promote immune activation, transforming the TIME into a ‘hot’
state, thereby addressing the immunologically ‘cold’ tumors, such
as TNBC and proficient mismatch repair/microsatellite stable
colorectal cancer (CRC) of the immune class (59). Simultaneously, it can also reduce
immune cell infiltration through certain mechanisms, enhancing
tumor immune evasion. Additionally, the expression of immunotherapy
biomarkers is positively associated with hGBP2 expression (61). In CRC, hGBP2 expression is
positively associated with CD8+ T cell infiltration,
CD8, PD-L1, C-X-C motif chemokine ligand (CXCL) 9, CXCL10, CXCL11,
CXCL13, HLA-I expression, antigen processing and presentation
machinery and a variety of antitumor immunity steps, including the
release of cancer cell antigens, cancer antigen presentation,
priming and activation and trafficking of immune cells to tumors
(59). By contrast, it is
negatively associated with the cell count of cytokeratin (59). However, specific mechanisms of the
regulation of hGBP2 in CRC require further study.
In gastric cancer, hGBP2 is reported to be
positively associated with an inflamed TIME, immunophenoscore
(IPS), abundance of PD-1+ cells and the expression of
immunotherapy biomarkers, such as PD-L1, PD-L2, IFN-γ, CD8A,
secreted and transmembrane protein 1 and IFN-induced transmembrane
protein 3 (61). In GBM, it has
been demonstrated that urokinase, secreted protein acidic and rich
in cysteine, TGFB1, FN1 and colony-stimulating factor 1 are induced
by elevated hGBP2 expression (49).
In SKCM, previous studies have demonstrated that hGBP2 expression
is positively associated with the infiltration of B cells,
CD8+ T cells, CD4+ T cells, macrophages,
neutrophils and dendritic cells (11,12,57,61),
This may serve a role in therapies that block PD-1/PD-L1
interactions to prevent tumor immune evasion, since CD8+
T cells are the primary effector cells in tumor killing (84). In BC, hGBP2 is positively associated
with T cell infiltration levels and can serve as a marker for T
cell infiltration (53). In clear
cell renal cell carcinoma (ccRCC), hGBP2 promotes the infiltration
of CD8+ T cells, regulator T cells and both M1 and M2
macrophages (57).
From the perspective of reducing immune infiltration
and promoting tumor immune evasion, hGBP2 can exert oncogenic
effects by increasing the phosphorylation of STAT2 and STAT3,
modulating the JAK/STAT signaling pathway and reducing tumor immune
infiltration (51). hGBP2 can also
induce immune checkpoints, such as PD-1/PD-L1, cytotoxic
T-lymphocyte-associated protein 4 (CTLA-4), T cell immunoreceptor
with Ig and ITIM domains, LAG3, indoleamine 2,3-dioxygenase 2 and
V-domain Ig suppressor of T cell activation (VISTA) (11), which are inhibitory immune
checkpoints. Additionally, hGBP2 can promote PD-L1 expression on
the transcriptional level by binding to and phosphorylating STAT1,
thereby reducing tumor immune infiltration and facilitating immune
evasion (85). hGBP2 can also
induce PD-L2; however, the relevant mechanisms remain unstudied
(61). Based on these functions,
hGBP2 will likely be a crucial target for restoring the TIME for
the treatment of immunologically ‘cold’ tumors, such as TNBC.
mGBP2 has been found to upregulate the secretion of
IL-6, IL-12 and TNF-α (77).
Furthermore, mGBP2 promotes the maturation of dendritic cells and
enhances their antigen-presenting capacity, thereby boosting T cell
activation (77). mGBP2 also
promotes the polarization of macrophages into M1 and M2 subtypes by
promoting STAT3 pathway and by activating the NF-κB signaling
pathway (62), while hGBP2 promotes
polarization by stimulating the secretion of IL-18 (56). M2 macrophages, in turn, can enhance
the migration and invasion of tumor cells by secreting IL-10 and
TGF-β, which upregulate the hGBP2/STAT3 and ERK axes (part of the
MAPK signaling pathway) as demonstrated in ccRCC (56).
Notably, a number of downstream factors promoted by
hGBP2 can exhibit both pro-tumor and antitumor effects. CD80 in
pancreatic adenocarcinoma can bind to either CD28 (activates T
cells) or CTLA-4 (inhibits T cells), thereby bidirectionally
modulating immune responses (11).
Notably, hGBP2 is reported to promote the polarization of M0 to M2
in ccRCC by activating the secretion of IL-18 (56) and to M1 in ovarian cancer (62) and in diabetic nephropathy by
activating the Notch 1 signaling pathway (42). By contrast, mGBP2 has only been
reported to promote the polarization of M0 to M1 under the
activation of the PLGA-CpG@ID8-M nano vaccine (62). The underlying mechanisms of these
outcomes and whether mGBP2 can also promote the polarization of M0
to M2 require further investigation.
Regulation of hGBP2 in cancer
In primary CRC, hGBP2 has been found to exhibit a
high mutation rate alongside genes such as G protein subunit β1 and
GATA zinc finger domain containing 2A. However, only hGBP2 showed
an even higher mutation rate in CRC with liver metastasis (60). Additionally, in CRC with liver
metastasis, the hGBP2 gene undergoes methylation at four
specific sites (m1A, m5C, m6A and m7G) (60). Peroxisome proliferator-activated
receptor α, an anticancer factor, can inhibit hGBP2 expression
(58). In BC (52) and SKCM (12), the methylation level of hGBP2
increases, leading to a decrease in hGBP2 expression.
Future directions
Prior reviews on GBP2 have predominantly emphasized
their roles in host defense against bacterial and viral pathogens
(6,86,87),
with limited attention to the regulatory mechanisms of GBP2
expression or its functions in cancer and oncogenesis. Furthermore,
species-specific distinctions-particularly the differences between
hGBP2 and mGBP2-have been overlooked (86,87).
This review bridges these gaps by providing a comprehensive
analysis of the regulation of GBP2 expression and by systematically
comparing hGBP2 and mGBP2 functions in oncogenesis. Whether a
potential functional substitution relationship between
Mg2+ and H3O+ during hGBP2
activation exists and its possible mechanisms could offer further
insight into the hydrolysis of hGBP2, and requires further
investigation. Although hGBP2 can form tetramers extensively, this
tetramerization cannot facilitate the hydrolysis from GTP to GMP,
rendering the physiological relevance of its tetramerization
unknown. Moreover, while the context-dependent dual role of hGBP2
in cancer and oncogenesis has been extensively documented, the
underlying mechanisms have not yet been systematically elucidated.
For instance, although hGBP2 drives M0-to-M1 polarization via the
Notch1 pathway in diabetic nephropathy, it mediates an M0-to-M2
polarization in ccRCC; the determinants of these opposing outcomes
warrant further investigation.
Clinically, agonistic monoclonal antibodies which
promote hGBP2 oligomerization and activation could be exploited to
convert immunologically ‘cold’ tumors into ‘hot’ ones, thereby
augmenting existing immunotherapies. Across multiple cancer types,
hGBP2 abundance is positively associated with levels of
immune-related biomarkers, such as PD-1/PD-L1 and VISTA,
immune-cell infiltration and IPS, revealing its potential as a
robust tumor biomarker for assessing the activation level of TIME
and prognosis. Additionally, the methylation status of the
hGBP2 promoter also has also emerged as a promising
diagnostic indicator in SKCM and BC.
Acknowledgements
Not applicable.
Funding
The present study was Supported by the National Natural Science
Foundation of China (grant no. 82104603), Beijing Natural Science
Foundation (grant no. 7204322), Peking University People's Hospital
Research and Development Fund (grant nos. RDZH2022-04, RS2021-12
and RDX2021-08).
Availability of data and materials
Not applicable.
Authors' contributions
ZL performed project administration, methodology,
data curation and wrote the draft. SP performed visualization and
reviewed the manuscript. JO conceived and designed the review, and
was involved in funding acquisition, supervision, proofreading and
revision of the manuscript. All authors read and approved the final
version of the manuscript. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GBP2
|
guanylate-binding protein 2
|
|
Pin1
|
peptidyl-prolyl cis-trans isomerase
NIMA-interacting 1
|
|
CRC
|
colorectal cancer
|
|
IRF
|
IFN-regulatory factor
|
|
TIME
|
tumor immunosuppressive
microenvironment
|
|
ccRCC
|
clear cell renal cell carcinoma
|
|
SKCM
|
skin cutaneous melanoma
|
|
BC
|
breast cancer
|
|
GBM
|
glioblastoma
|
References
|
1
|
Cheng YS, Colonno RJ and Yin FH:
Interferon induction of fibroblast proteins with guanylate binding
activity. J Biol Chem. 258:7746–7750. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vestal DJ, Buss JE, McKercher SR, Jenkins
NA, Copeland NG, Kelner GS, Asundi VK and Maki RA: Murine GBP-2: A
new IFN-gamma-induced member of the GBP family of GTPases isolated
from macrophages. J Interferon Cytokine Res. 18:977–985. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Quan ST, Jiao WW, Xu F, Sun L, Qi H and
Shen A: Advances in the regulation of inflammasome activation by
GBP family in infectious diseases. Yi Chuan. 45:1007–1017.
2023.PubMed/NCBI
|
|
4
|
Britzen-Laurent N, Bauer M, Berton V,
Fischer N, Syguda A, Reipschläger S, Naschberger E, Herrmann C and
Stürzl M: Intracellular trafficking of guanylate-binding proteins
is regulated by heterodimerization in a hierarchical manner. PLoS
One. 5:e142462010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Modiano N, Lu YE and Cresswell P: Golgi
targeting of human guanylate-binding protein-1 requires nucleotide
binding, isoprenylation, and an IFN-gamma-inducible cofactor. Proc
Natl Acad Sci USA. 102:8680–8685. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kirkby M, Enosi Tuipulotu D, Feng S, Lo
Pilato J and Man SM: Guanylate-binding proteins: Mechanisms of
pattern recognition and antimicrobial functions. Trends Biochem
Sci. 48:883–893. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim BH, Chee JD, Bradfield CJ, Park ES,
Kumar P and MacMicking JD: Interferon-induced guanylate-binding
proteins in inflammasome activation and host defense. Nat Immunol.
17:481–489. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Luo Y, Jin H, Kim JH and Bae J:
Guanylate-binding proteins induce apoptosis of leukemia cells by
regulating MCL-1 and BAK. Oncogenesis. 10:542021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu YT and Sun ZJ: Turning cold tumors
into hot tumors by improving T-cell infiltration. Theranostics.
11:5365–5386. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Galon J and Bruni D: Approaches to treat
immune hot, altered and cold tumours with combination
immunotherapies. Nat Rev Drug Discov. 18:197–218. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu B, Huang R, Fu T, He P, Du C, Zhou W,
Xu K and Ren T: GBP2 as a potential prognostic biomarker in
pancreatic adenocarcinoma. PeerJ. 9:e114232021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang S, Chen K, Zhao Z, Zhang X, Xu L,
Liu T and Yu S: Lower expression of GBP2 associated with less
immune cell infiltration and poor prognosis in skin cutaneous
melanoma (SKCM). J Immunother. 45:274–283. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang JH: Structural study of human
guanylate binding protein GBP2. 2019.
|
|
14
|
Ban T, Heymann JA, Song Z, Hinshaw JE and
Chan DC: OPA1 disease alleles causing dominant optic atrophy have
defects in cardiolipin-stimulated GTP hydrolysis and membrane
tubulation. Hum Mol Genet. 19:2113–2122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Daumke O and Praefcke GJK: Mechanisms of
GTP hydrolysis and conformational transitions in the dynamin
superfamily. Biopolymers. 109:e230792018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Roy S, Wang B, Roy K, Tian Y, Bhattacharya
M, Williams S and Yin Q: Crystal structures reveal
nucleotide-induced conformational changes in G motifs and distal
regions in human guanylate-binding protein 2. Commun Biol.
8:2822025. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kravets E, Degrandi D, Ma Q, Peulen TO,
Klümpers V, Felekyan S, Kühnemuth R, Weidtkamp-Peters S, Seidel CA
and Pfeffer K: Guanylate binding proteins directly attack
Toxoplasma gondii via supramolecular complexes. Elife.
5:e114792016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kravets E, Degrandi D, Weidtkamp-Peters S,
Ries B, Konermann C, Felekyan S, Dargazanli JM, Praefcke GJ, Seidel
CA, Schmitt L, et al: The GTPase activity of murine
Guanylate-binding protein 2 (mGBP2) controls the intracellular
localization and recruitment to the parasitophorous vacuole of
toxoplasma gondii. J Biol Chem. 287:27452–27466. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Neun R, Richter MF, Staeheli P and
Schwemmle M: GTPase properties of the interferon-induced human
Guanylate-binding protein 2. FEBS Lett. 390:69–72. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rajan S, Pandita E, Mittal M and Sau AK:
Understanding the lower GMP formation in large GTPase hGBP-2 and
role of its individual domains in regulation of GTP hydrolysis.
FEBS J. 286:4103–4121. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Honkala AT, Tailor D and Malhotra SV:
Guanylate-binding protein 1: An emerging target in inflammation and
cancer. Front Immunol. 10:31392019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cui W, Braun E, Wang W, Tang J, Zheng Y,
Slater B, Li N, Chen C, Liu Q, Wang B, et al: Structural basis for
GTP-induced dimerization and antiviral function of
guanylate-binding proteins. Proc Natl Acad Sci USA.
118:e20222691182021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Abdullah N, Balakumari M and Sau AK:
Dimerization and its role in GMP formation by human guanylate
binding proteins. Biophys J. 99:2235–2244. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Walker JE, Saraste M, Runswick MJ and Gay
NJ: Distantly related sequences in the alpha- and beta-subunits of
ATP synthase, myosin, kinases and other ATP-requiring enzymes and a
common nucleotide binding fold. EMBO J. 1:945–951. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wittinghofer A and Vetter IR:
Structure-function relationships of the G domain, a canonical
switch motif. Annu Rev Biochem. 80:943–971. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ghosh A, Praefcke GJ, Renault L,
Wittinghofer A and Herrmann C: How guanylate-binding proteins
achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature. 440:101–104. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Olszewski MA, Gray J and Vestal DJ: In
silico genomic analysis of the human and murine Guanylate-binding
protein (GBP) gene clusters. J Interferon Cytokine Res. 26:328–352.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ramsauer K, Farlik M, Zupkovitz G, Seiser
C, Kröger A, Hauser H and Decker T: Distinct modes of action
applied by transcription factors STAT1 and IRF1 to initiate
transcription of the IFN-gamma-inducible gbp2 gene. Proc Natl Acad
Sci USA. 104:2849–2854. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Michalska A, Blaszczyk K, Wesoly J and
Bluyssen HAR: A positive feedback amplifier circuit that regulates
interferon (IFN)-Stimulated gene expression and controls type I and
type II IFN responses. Front Immunol. 9:11352018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guimarães DP, Oliveira IM, de Moraes E,
Paiva GR, Souza DM, Barnas C, Olmedo DB, Pinto CE, Faria PA, De
Moura Gallo CV, et al: Interferon-inducible guanylate binding
protein (GBP)-2: A novel p53-regulated tumor marker in esophageal
squamous cell carcinomas. Int J Cancer. 124:272–279. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu ZH, Cai F and Zhong Y: Comprehensive
analysis of the expression and prognosis for GBPs in head and neck
squamous cell carcinoma. Sci Rep. 10:60852020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Du CH, Wu YD, Yang K, Liao WN, Ran L, Liu
CN, Zhang SZ, Yu K, Chen J, Quan Y, et al: Apoptosis-resistant
megakaryocytes produce large and hyperreactive platelets in
response to radiation injury. Mil Med Res. 10:662023.PubMed/NCBI
|
|
33
|
Mondal S, Adhikari N, Banerjee S, Amin SA
and Jha T: Matrix metalloproteinase-9 (MMP-9) and its inhibitors in
cancer: A minireview. Eur J Med Chem. 194:1122602020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ngo CC and Man SM: Mechanisms and
functions of guanylate-binding proteins and related
interferon-inducible GTPases: Roles in intracellular lysis of
pathogens. Cell Microbiol. 192017.doi: 10.1111/cmi.12791.
|
|
35
|
Meunier E and Broz P: Interferon-inducible
GTPases in cell autonomous and innate immunity. Cell Microbiol.
18:168–180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fu J and Wu H: Structural mechanisms of
NLRP3 inflammasome assembly and activation. Annu Rev Immunol.
41:301–316. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wandel MP, Kim BH, Park ES, Boyle KB,
Nayak K, Lagrange B, Herod A, Henry T, Zilbauer M, Rohde J, et al:
Guanylate-binding proteins convert cytosolic bacteria into
caspase-4 signaling platforms. Nat Immunol. 21:880–891. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kayagaki N, Stowe IB, Lee BL, O'Rourke K,
Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT,
et al: Caspase-11 cleaves gasdermin D for non-canonical
inflammasome signalling. Nature. 526:666–671. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huang S, Dong W, Lin X, Xu K, Li K, Xiong
S, Wang Z, Nie X and Bian JS: Disruption of the
Na+/K+-ATPase-purinergic P2X7 receptor complex in microglia
promotes Stress-induced anxiety. Immunity. 57:495–512.e11. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Garlanda C, Dinarello CA and Mantovani A:
The interleukin-1 family: Back to the future. Immunity.
39:1003–1018. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Y, Liao Y, Hang Q, Sun D and Liu Y:
GBP2 acts as a member of the interferon signalling pathway in lupus
nephritis. BMC Immunology. 23:442022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li X, Liu J, Zeng M, Yang K, Zhang S, Liu
Y, Yin X, Zhao C, Wang W and Xiao L: GBP2 promotes M1 macrophage
polarization by activating the notch1 signaling pathway in diabetic
nephropathy. Front Immunol. 14:11276122023. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Schori C, Trachsel C, Grossmann J, Barben
M, Klee K, Storti F, Samardzija M and Grimm C: A chronic hypoxic
response in photoreceptors alters the vitreous proteome in mice.
Exp Eye Res. 185:1076902019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
An Y, Xu J, Hu X, Xu M, Yang X and Liu T:
GBP2 regulates lipid metabolism by inhibiting the HIF-1 pathway to
alleviate the progression of allergic rhinitis. Cell Biochem
Biophys. 83:1689–1701. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang G, Jin S, Ling X, Li Y, Hu Y, Zhang
Y, Huang Y, Chen T, Lin J, Ning Z, et al: Proteomic profiling of
LPS-induced Macrophage-derived exosomes indicates their involvement
in acute liver injury. Proteomics. 19:e18002742019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Huang W, Zhang Y, Zheng B, Ling X, Wang G,
Li L and Meng Y: GBP2 upregulated in LPS-stimulated
macrophages-derived exosomes accelerates septic lung injury by
activating epithelial cell NLRP3 signaling. Int Immunopharmacol.
124:1110172023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gao R, Ali T, Liu Z, Li A, Hao L, He L, Yu
X and Li S: Ceftriaxone averts neuroinflammation and relieves
depressive-like behaviors via GLT-1/TrkB signaling. Biochem Biophys
Res Commun. 701:1495502024. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ren Y, Yang B, Guo G, Zhang J, Sun Y, Liu
D, Guo S, Wu Y, Wang X, Wang S, et al: GBP2 facilitates the
progression of glioma via regulation of KIF22/EGFR signaling. Cell
Death Discov. 8:2082022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yu S, Yu X, Sun L, Zheng Y, Chen L, Xu H,
Jin J, Lan Q, Chen CC and Li M: GBP2 enhances glioblastoma invasion
through Stat3/fibronectin pathway. Oncogene. 39:5042–5055. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Verdugo E, Puerto I and Medina MÁ: An
update on the molecular biology of glioblastoma, with clinical
implications and progress in its treatment. Cancer Commun (Lond).
42:1083–111. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Meng K, Li YY, Liu DY, Hu LL, Pan YL,
Zhang CZ and He QY: A five-protein prognostic signature with GBP2
functioning in immune cell infiltration of clear cell renal cell
carcinoma. Comput Struct Biotechnol J. 21:2621–2630. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Rahvar F, Salimi M and Mozdarani H: Plasma
GBP2 promoter methylation is associated with advanced stages in
breast cancer. Genet Mol Biol. 43:e201902302020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Godoy P, Cadenas C, Hellwig B, Marchan R,
Stewart J, Reif R, Lohr M, Gehrmann M, Rahnenführer J, Schmidt M
and Hengstler JG: Interferon-inducible guanylate binding protein
(GBP2) is associated with better prognosis in breast cancer and
indicates an efficient T cell response. Breast Cancer. 21:491–499.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li NN, Qiu XT, Xue JS, Yi LM, Chen ML and
Huang ZJ: Predicting the prognosis and immunotherapeutic response
of Triple-negative breast cancer by constructing a prognostic model
based on CD8+ T Cell-related immune genes. Biomed Environ Sci.
37:581–593. 2024.PubMed/NCBI
|
|
55
|
Ji G, Luo B, Chen L, Shen G and Tian T:
GBP2 is a favorable prognostic marker of skin cutaneous melanoma
and affects its progression via the Wnt/β-catenin pathway. Ann Clin
Lab Sci. 51:772–782. 2021.PubMed/NCBI
|
|
56
|
Zheng W, Ye S, Liu B, Liu D, Yan R, Guo H,
Yu H, Hu X, Zhao H, Zhou K and Li G: Crosstalk between GBP2 and M2
macrophage promotes the ccRCC progression. Cancer Science.
115:3570–3586. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tian Y, Wang H, Guan W, Tu X, Zhang X, Sun
Y, Qian C, Song X, Peng B and Cui X: GBP2 serves as a novel
prognostic biomarker and potential immune microenvironment
indicator in renal cell carcinoma. Mol Carcinog. 61:1082–1098.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
AmeliMojarad M, AmeliMojarad M and Cui X:
Weighted gene co-expression network analysis identified GBP2
connected to PPARα activity and liver cancer. Sci Rep.
14:207452024. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang H, Zhou Y, Zhang Y, Fang S, Zhang M,
Li H, Xu F, Liu L, Liu J, Zhao Q and Wang F: Subtyping of
microsatellite stability colorectal cancer reveals guanylate
binding protein 2 (GBP2) as a potential immunotherapeutic target. J
Immunother Cancer. 10:e0043022022. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Qi F, Gao N, Li J, Zhou C, Jiang J, Zhou
B, Guo L, Feng X, Ji J, Cai Q, et al: A multidimensional
recommendation framework for identifying biological targets to aid
the diagnosis and treatment of liver metastasis in patients with
colorectal cancer. Mol Cancer. 23:2392024. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang Y, Pan J, An F, Chen K, Chen J, Nie
H, Zhu Y, Qian Z and Zhan Q: GBP2 is a prognostic biomarker and
associated with immunotherapeutic responses in gastric cancer. BMC
Cancer. 23:9252023. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Xiong J, Huang J, Xu H, Wu Q, Zhao J, Chen
Y, Fan G, Guan H, Xiao R, He Z, et al: CpG-based nanovaccines
enhance ovarian cancer immune response by Gbp2-mediated remodeling
of tumor-associated macrophages. Adv Sci (Weinh). 12:e24128812025.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Feng D, Zhu W, Shi X, Wang Z, Wei W, Wei
Q, Yang L and Han P: Immune-related gene index predicts metastasis
for prostate cancer patients undergoing radical radiotherapy. Exp
Hematol Oncol. 12:82023. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Uribe ML, Marrocco I and Yarden Y: EGFR in
cancer: Signaling mechanisms, drugs, and acquired resistance.
Cancers (Basel). 13:27482021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Kim J, Kundu M, Viollet B and Guan KL:
AMPK and mTOR regulate autophagy through direct phosphorylation of
Ulk1. Nat Cell Biol. 13:132–141. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhang W, Tang X, Peng Y, Xu Y, Liu L and
Liu S: GBP2 enhances paclitaxel sensitivity in triple-negative
breast cancer by promoting autophagy in combination with ATG2 and
inhibiting the PI3K/AKT/mTOR pathway. Int J Oncol. 64:342024.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Li X, He S and Ma B: Autophagy and
autophagy-related proteins in cancer. Mol Cancer. 19:122020.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhang H, Sun Z, Li Y, Fan D and Jiang H:
MicroRNA-200c binding to FN1 suppresses the proliferation,
migration and invasion of gastric cancer cells. Biomed
Pharmacother. 88:285–292. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Shibata K, Kikkawa F, Nawa A, Thant AA,
Naruse K, Mizutani S and Hamaguchi M: Both focal adhesion kinase
and c-Ras are required for the enhanced matrix metalloproteinase 9
secretion by fibronectin in ovarian cancer cells. Cancer Res.
58:900–1093. 1998.PubMed/NCBI
|
|
70
|
Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin
L, Chen WM, Han L, Zhang EB, Kong R, et al: Decreased expression of
the long non-coding RNA FENDRR is associated with poor prognosis in
gastric cancer and FENDRR regulates gastric cancer cell metastasis
by affecting fibronectin1 expression. J Hematol Oncol. 7:632014.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Xue W, Yang L, Chen C, Ashrafizadeh M,
Tian Y and Sun R: Wnt/β-catenin-driven EMT regulation in human
cancers. Cell Mol Life Sci. 81:792024. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Peña-Blanco A and García-Sáez AJ: Bax, Bak
and beyond-mitochondrial performance in apoptosis. FEBS J.
285:416–431. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhang J, Zhang Y, Wu W, Wang F, Liu X,
Shui G and Nie C: Guanylate-binding protein 2 regulates
Drp1-mediated mitochondrial fission to suppress breast cancer cell
invasion. Cell Death Dis. 8:e31512017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liu W, Chen Y, Xie H, Guo Y, Ren D, Li Y,
Jing X, Li D, Wang X, Zhao M, et al: TIPE1 suppresses invasion and
migration through down-regulating Wnt/β-catenin pathway in gastric
cancer. J Cell Mol Med. 22:1103–1117. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wang J, Min H, Hu B, Xue X and Liu Y:
Guanylate-binding protein-2 inhibits colorectal cancer cell growth
and increases the sensitivity to paclitaxel of paclitaxel-resistant
colorectal cancer cells by interfering Wnt signaling. J Cell
Biochem. 121:1250–129. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Song JX, Wang Y, Hua ZP, Huang Y, Hu LF,
Tian MR, Qiu L, Liu H and Zhang J: FATS inhibits the Wnt pathway
and induces apoptosis through degradation of MYH9 and enhances
sensitivity to paclitaxel in breast cancer. Cell Death Dis.
15:8352024. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zhang SW, Feng TB, Ning YL, Zhang XH and
Qi CJ: Guanylate-binding protein 2 regulates the maturation of
mouse dendritic cells induced by β-glucan. Xi Bao Yu Fen Zi Mian Yi
Xue Za Zhi. 33:1153–1159. 2017.(In Chinese). PubMed/NCBI
|
|
78
|
Xu X, Ding X, Wang Z, Ye S, Xu J, Liang Z,
Luo R, Xu J, Li X and Ren Z: GBP2 inhibits pathological
angiogenesis in the retina via the AKT/mTOR/VEGFA axis. Microvasc
Res. 154:1046892024. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Du F, Liu M, Wang J, Hu L, Zeng D, Zhou S,
Zhang L, Wang M, Xu X, Li C, et al: Metformin coordinates with
mesenchymal cells to promote VEGF-mediated angiogenesis in diabetic
wound healing through Akt/mTOR activation. Metabolism.
140:1553982023. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Inoki K, Li Y, Zhu T, Wu J and Guan KL:
TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR
signalling. Nat Cell Biol. 4:648–6457. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ruchko MV, Gorodnya OM, Pastukh VM, Swiger
BM, Middleton NS, Wilson GL and Gillespie MN: Hypoxia-induced
oxidative base modifications in the VEGF hypoxia-response element
are associated with transcriptionally active nucleosomes. Free
Radic Biol Med. 46:352–359. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wenger RH, Stiehl DP and Camenisch G:
Integration of oxygen signaling at the consensus HRE. Sci STKE.
2005:re122005. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Balasubramanian S, Fan M, Messmer-Blust
AF, Yang CH, Trendel JA, Jeyaratnam JA, Pfeffer LM and Vestal DJ:
The interferon-gamma-induced GTPase, mGBP-2, inhibitsc (TNF-alpha)
induction of matrix metalloproteinase-9 (MMP-9) by inhibiting
NF-kappaB and Rac protein. J Biol Chem. 286:20054–20064. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Raskov H, Orhan A, Christensen JP and
Gögenur I: Cytotoxic CD8+ T cells in cancer and cancer
immunotherapy. Br J Cancer. 124:359–367. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Ye S, Li S, Qin L, Zheng W, Liu B, Li X,
Ren Z, Zhao H, Hu X, Ye N and Li G: GBP2 promotes clear cell renal
cell carcinoma progression through immune infiltration and
regulation of PD-L1 expression via STAT1 signaling. Oncol Rep.
49:492023. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Kutsch M and Coers J: Human guanylate
binding proteins: Nanomachines orchestrating host defense. FEBS J.
288:5826–5849. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Chakraborty S, Kasirajan A, Mariappan V,
Green SR and Pillai AKB: Guanylate binding proteins (GBPs) as novel
therapeutic targets against single-stranded RNA viruses. Mol Biol
Rep. 52:7802025. View Article : Google Scholar : PubMed/NCBI
|