Introduction
Ovarian cancer (OC) has the greatest fatality rate
among all global gynecological malignancies, with epithelial OC
being the most frequently occurring type (1,2). Due
to a lack of accurate early screening techniques for OC, diagnosis
is delayed and ~50% of patients die within 5 years of diagnosis
(3,4). Despite recent therapeutic
breakthroughs, OC still has poor results due to the unknown gene
regulation network that underpins its etiology (5). Notably, there is an abundance of
clinical prognostic indicators for OC, such as CA125; however, the
identification of meaningful biomarkers for early detection and
improving treatment outcomes in OC is required.
Histone acetyltransferases (HATs) and deacetylases
are capable of controlling DNA transcription by regulating histone
acetylation and deacetylation. Overexpression or inappropriate
recruitment of HATs have been associated with the development and
progression of malignant tumors (6). HAT1 can only acetylate newly generated
histone H4, and was the first HAT discovered and remains one of the
insufficiently studied members of the HAT family (7). HAT1 is upregulated in numerous types
of solid tumors, including lung, breast, esophageal and liver
cancer, and functions as an oncogene to promote carcinogenesis
(8–11). It has also been revealed that HAT1
operates as a transcription factor (TF), modulating cancer cell
metabolism and treatment resistance (12,13).
To date, to the best of our knowledge, the role and
function of HAT1 in OC have not been investigated. The present
study aimed to elucidate the HAT1 expression levels in specimens
from patients with OC and healthy human samples. Then the molecular
mechanism of upstream regulator of HAT1 will be studied, as well as
the function of HAT1 in regulating OC cell proliferation and colony
formation activities. Furthermore, the role of HAT1 in regulation
of cell cycle pathway in OC cells will be analyzed, and whether
knockdown of HAT1 could decrease cell cycle-related protein
expression levels in OC cells. Thus, the present results may
contribute to a better understanding of the potential role of HAT1
in OC, which could provide new biomarkers for novel therapeutic
strategies in OC.
Materials and methods
Cell culture and reagents
Human OC cell lines OVCAR3, HEY, A2780 and SKOV3,
and the immortalized ovarian epithelial cell line IOSE386 were
grown in RPMI-1640 medium with 10% FBS (both Gibco; Thermo Fisher
Scientific, Inc.), 100 IU/ml penicillin and 100 µg/ml streptomycin
(Thermo Fisher Scientific, Inc.). The cells were cultured in a 37°C
incubator with 5% CO2, and cells were detached using
0.25% trypsin and passaged every 2 days. Small interfering RNA
(siRNA) transfection was used to silence HAT1 expression in
3×106 HEY and SKOV3 cells at 150 nM concentration, and a
siRNA transfection reagent (cat. no. sc-29528, Santa Cruz
Biotechnology, Inc.) was used to transfect cells at room
temperature for 48 h according to the manufacturer's guidelines.
siRNAs including scrambled siRNA (siSCR) and siHAT1 were purchased
from the Dharmacon (Revvity, Inc.). The ON-TARGETplus Human HAT1
siRNA-SMART pool contained four target sequences of HAT1 together
in the tube: 5′-GCUACAGACUGGAUAUUAAUU-3′,
5′-CAGAUGAACCAAAUAGAAAUU-3′, 5′-CAACACAGCAAUUGAACUAUU-3′ and
5′-AGGAACUAGUGGAAGAUUAUU-3′; the siRNA also contained four target
sequences of siSCR: 5′-UGGUUUACAUGUCGACUAA-3′,
5′-UGGUUUACAUGUUGUGUGA-3′, 5′-UGGUUUACAUGUUUUCUGA-3′ and
5′-UGGUUUACAUGUUUUCCUA-3′. The FOXA1 overexpression plasmid was
synthesized by YouBio Biotechnology. A total of 1 µg pcDNA3-FOXA1
plasmid and 1 µg pcDNA3-Flag control plasmid was transfected into
3×106 HEY cells for 48 h using the FuGENE® HD
transfection reagent at room temperature according to the
manufacturer's instructions (Roche Diagnostics).
Western blot analysis
Lysates from whole cells (IOSE386, OVCAR3, HEY,
A2780 and SKOV3) were extracted using RIPA buffer (Thermo Fisher
Scientific, Inc.) with a protease inhibitor cocktail. Cell
suspensions were homogenized and centrifuged at 16,000 × g for 20
min at 4°C to extract the supernatant. Equal amounts of protein (15
µg) were separated by SDS-PAGE (Upper layer 4% stacking gel and
lower layer 8% separating gel) and transferred to a PVDF membrane,
which was blocked with 5% non-fat milk for 2 h at room temperature.
Subsequently, the membrane was treated with the indicated primary
antibodies at 4°C overnight and then horseradish
peroxidase-conjugated secondary antibodies (1:2,000; Cat.31430,
Cat.31460; Thermo Fisher Scientific, USA) for another 2 h at room
temperature. Signals were detected using an enhanced
chemiluminescence reagent from Pierce (Thermo Fisher Scientific,
USA) and analyzed with a ImageQuant LAS 4000 and ImageQuant TL 8.1
software (GE Healthcare). The following primary antibodies were
used in the present study: Anti-HAT1 (1:1,000; cat no. 11432-1-AP;
Proteintech Group, Inc.), anti-CDK2 (1:1,000; cat no. 10122-1-AP;
Proteintech Group, Inc.), anti-FOXA1 (1:1,000; cat no. 20411-1-AP;
Proteintech Group, Inc.), anti-CDK4 (1:1,000; cat no. 12790; Cell
Signaling Technology, Inc.), anti-cyclin E (1:1,000; cat no. 20808;
Cell Signaling Technology, Inc.) and anti-GAPDH (1:5,000; cat no.
60004-1-Ig; Proteintech Group, Inc.).
RNA isolation and reverse
transcription-quantitative PCR (qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was adopted for total mRNA isolation from
cell lines (IOSE386, OVCAR3, HEY, A2780 and SKOV3). mRNA was
reverse transcribed with PrimeScript™ RT Master Mix
(Takara Bio, Inc.) as follows: Primer annealing at 25°C, 5 min;
Reverse transcription: 42°C, 30–60 min; Enzyme inactivation: 85°C 5
min; and Hold: 4°C. qPCR was performed on QuantStudio 5 qPCR
equipment (Applied Biosystems; Thermo Fisher Scientific, Inc.) with
Universal SYBR Green Master reagent, and the cycling conditions
were: Initial denaturation: 95°C, 3 min; PCR cycles (40 cycles):
Denaturation: 95°C, 15 sec and Annealing/extension: 60°C, 30 sec
(with fluorescence acquisition); and Melting curve analysis:
60–95°C. The relative gene expression levels were normalized to
those of the internal control, GAPDH, using the 2−ΔΔCq
method (14). The primer sequences
were as follows: HAT1, forward, 5′-AAGCCATTCGGAACCTTACTTC-3′,
reverse 5′-AGTGCCATCTTTCATCATCCAC-3′; and GAPDH, forward
5′-GGAGCGAGATCCCTCCAAAAT-3′, reverse
5′-GGCTGTTGTCATACTTCTCATGG-3′.
Colony formation assay
500 Cells (HEY and SKOV3 cells) were trypsinized,
assessed and cultivated for 2 weeks in medium containing 10% FBS.
Subsequently, the cells were washed in PBS, fixed with 4%
paraformaldehyde for 15 min and stained with 1% crystal violet for
30 min at 37°C. The colonies visible at the plates were counted
with ImageJ software v1.41 (National Institutes of Health) and
images were captured.
Cell viability assay
2000 HEY and SKOV3 cells were trypsinized, measured
and seeded into 96-well plates with medium containing 10% FBS. Each
well received 10 µl Cell Counting Kit-8 (CCK-8) solution (Dojindo
Laboratories, Inc.) and the cells were cultured in an incubator at
37°C for 1 h. At various time points (24, 48, 72, 96, 120 h), the
absorbance was measured at 450 nm with a microplate reader and
analyzed (Bio-Rad Laboratories, Inc.).
Cell cycle assay
A total of 5×106 HEY and SKOV3 cells were
collected, washed with PBS and were subsequently fixed with 70%
cold ethanol at 4°C overnight. The cells were then incubated with
500 µl PI/RNase A staining solution (Nanjing KeyGen Biotech Co.,
Ltd.) for 1 h at room temperature in the dark, according to the
manufacturer's instructions, and were examined using a BD FACSCanto
flow cytometer (BD Biosciences) and analyzed using FlowJo software
version 4.5 (Tree Star, Inc.).
EdU staining
The EdU reagent was used to measure cell
proliferation (Guangzhou RiboBio Co., Ltd.). A total of
2×104 HEY cells were cultured in 96-well plates. Cells
were then treated with 50 µM EdU solution for an additional 2 h in
37°C. After washing with PBS, the cells were fixed at room
temperature with 4% paraformaldehyde for 30 min and then
permeabilized with 0.5% Triton X-100 for 10 min. Cells were exposed
to 200 µl Apollo staining solution in the EdU kit for 30 min at
room temperature in the dark. Finally, the DNA was stained with 200
µl 1 X Hoechst 33342 for 30 min at room temperature in the dark.
Cells were then examined under a fluorescence microscope with
excitation wavelength at 567 nm.
Cell survival assay
HEY and SKOV3 cells were seeded into 96-well plates
(5×103/well) and treated with 0.0, 0.5, 1.0, 5, 10, 20,
30 and 50 µM) of HAT1 inhibitor JG-2016 or DMSO as indicated
(Cat.HY-154944; MedChemExpress) for 72 h at room temperature. OD
values were determined using a luminometer at an absorbance of 450
nm with the CCK-8 Kit as aforementioned (Dojindo Laboratories,
Inc.), and cell viability levels were calculated based on OD
values.
Luciferase reporter assay
The potential transcriptional factor FOXA1 binding
sites of the HAT1 promoter region were analyzed through the UCSC
Genome Browser (genome.ucsc.edu/) and TF database JASPAR
(https://jaspar.elixir.no/). The pmiRGLO
vectors (Promega Corporation) containing HAT1 wild-type (WT) and
HAT1 mutant (MUT) sequences were constructed and Sanger sequenced.
Subsequently, the HEY cells were co-transfected with pmiRGLO WT/MUT
vectors and an empty or FOXA1 vector using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Following 48 h of transfection, the Luciferase
Assay Kit (Promega Corporation) was used to assess the activities
of firefly and Renilla luciferase, and the firefly
luciferase activity levels were adjusted to Renilla
luciferase.
Bioinformatics analysis
The HAT1 protein expression data from healthy human
tissue and OC tissues was derived from antibody-based protein
profiling using immunohistochemistry from the Human Protein Atlas
database (https://www.proteinatlas.org/). The mRNA and protein
expression levels of HAT1 in healthy human tissue (n=133) and OC
tissues (n=374) were analyzed using The Cancer Genome Atlas (TCGA
data (https://www.cancer.gov/ccg/research/genome-sequencing/tcga)
through the TNMplot analysis platform (tnmplot.com/analysis/), the University of Alabama at
Birmingham Cancer data analysis Portal (UALCAN) and Clinical
Proteomic Tumor Analysis Consortium (ualcan.path.uab.edu/index.html; gdc.cancer.gov/about-gdc/contributed-genomic-data-cancer-research/clinical-proteomic-tumor-analysis-consortium-cptac).
Survival analysis was performed using the Kaplan-Meier method and
log-rank testing to analyze the prognostic value of the HAT1 gene
(kmplot.com/analysis/; http://www.gsea-msigdb.org/gsea/index.jsp); the
patient samples were split into two groups according to various
quantile expressions of HAT1 (Cutoff value used in analysis: 1383),
and the hazard ratios with 95% confidence intervals and log-rank
P-values were calculated; the Kaplan-Meier survival plots with the
number at risk, hazard ratios and log-rank P-values were obtained
using the Kaplan-Meier plotter website. The LinkedOmics database
was used to perform HAT1 gene and pathway analysis using both
over-representation analysis and gene set enrichment analysis
(linkedomics.org; cutoff value, 2000). The binding
regions of FOXA1 in HAT1 promoter region were analyzed by Cistrome
DB (cistrome.org/db/#/) with the public
chromatin immunoprecipitation-sequencing data downloaded from the
Gene Expression Omnibus (GEO) datasets GSM 1538430/803461/1858655.
The binding peaks of FOXA1 in HAT1 promoter indicated that FOXA1
transcriptionally regulated HAT1 expression. The cBioPortal
database (cbioportal.org/) was used to analyze the
correlation between FOXA1 and HAT1 through Pearson's correlation
coefficient. Transcriptional factor FOXA1 was enriched at the HAT1
promoter region as suggested by Cistrome DB. The association
between HAT1 and regulatory proteins of DNA replication and
pyrimidine metabolism pathways in OC tissues were analyzed through
cBioPortal database (https://www.cbioportal.org/).
Statistical analysis
Experiments were repeated at least three times and
data are presented as the mean ± SD, with results analyzed using
GraphPad 8.0 software (Dotmatics). The differences between two
groups were analyzed via unpaired Student's t-test. HAT1 expression
levels between the OC cell lines and the normal IOSE386 cell line
were analyzed with one-way ANOVA and Dunnett's post hoc test. The
significance of differences in HAT1 expression in the
bioinformatics analysis was determined using Welch's t-test, or
one-way ANOVA with Dunnett's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
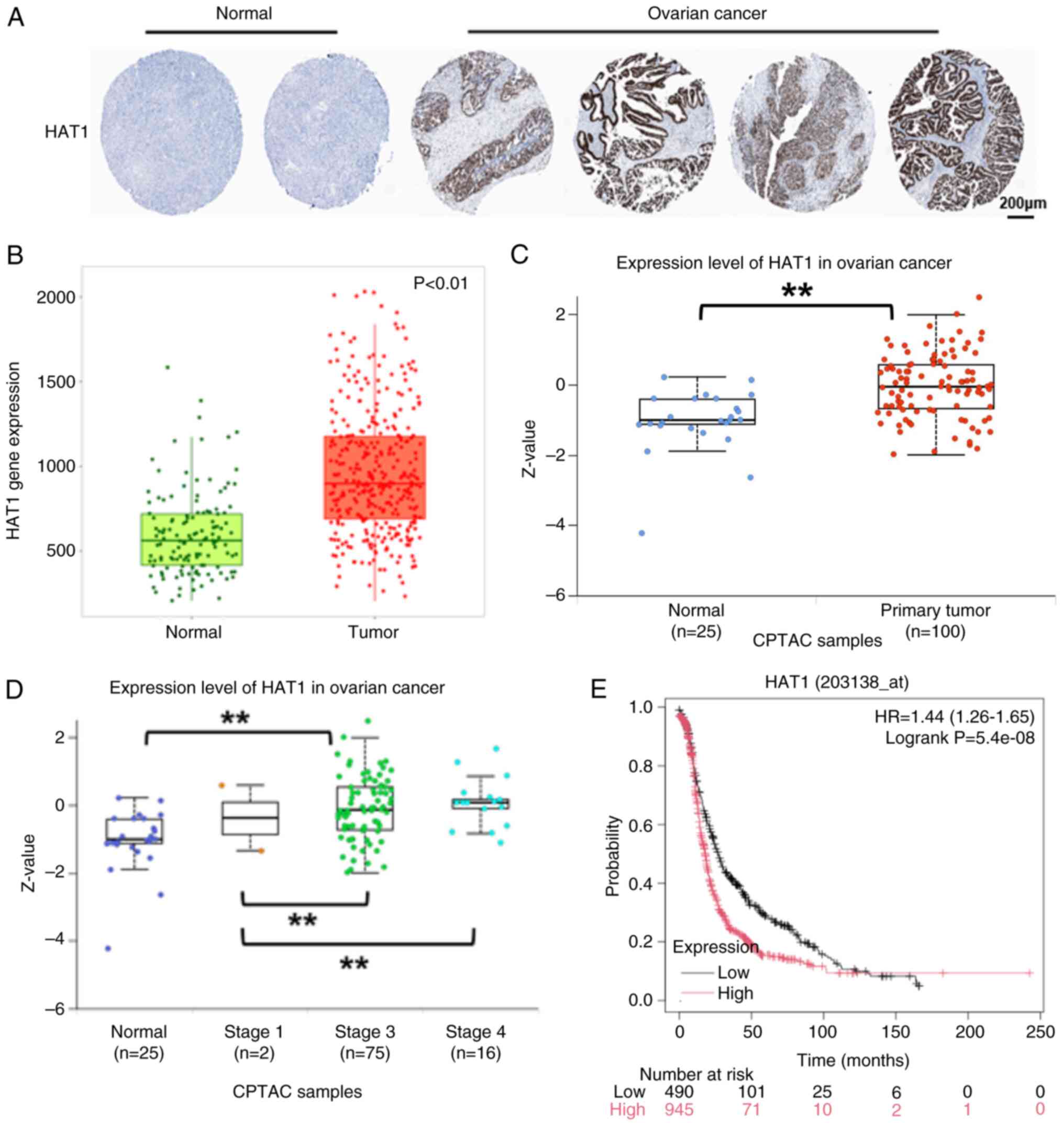
Expression levels of HAT1 are
upregulated in OC tissues and higher HAT1 levels indicate poor
prognosis
HAT1 was the first HAT) discovered; however, its
biological functions together with the cell mechanisms involved in
cancer progression are poorly characterized. To identify the
potential role of HAT1 in OC, the immunohistochemistry staining
results of HAT1 in OC tissue were analyzed in the Human Protein
Atlas database, and HAT1 staining was found to be higher in OC
compared with in healthy human specimens (Fig. 1A). Through TCGA) database analysis,
the OC tissues were found to have higher expression levels of HAT1
compared with healthy human tissues (Fig. 1B). In addition, HAT1 protein levels
were increased in OC samples (Fig.
1C) and were associated with tumor grades (Fig. 1D). The association of HAT1 with
overall survival was analyzed using the Kaplan-Meier plotter
website and the results showed that higher HAT1 levels indicated
shorter survival rates (Fig. 1E).
These findings suggested that HAT1 may act as a tumor promoter in
OC.
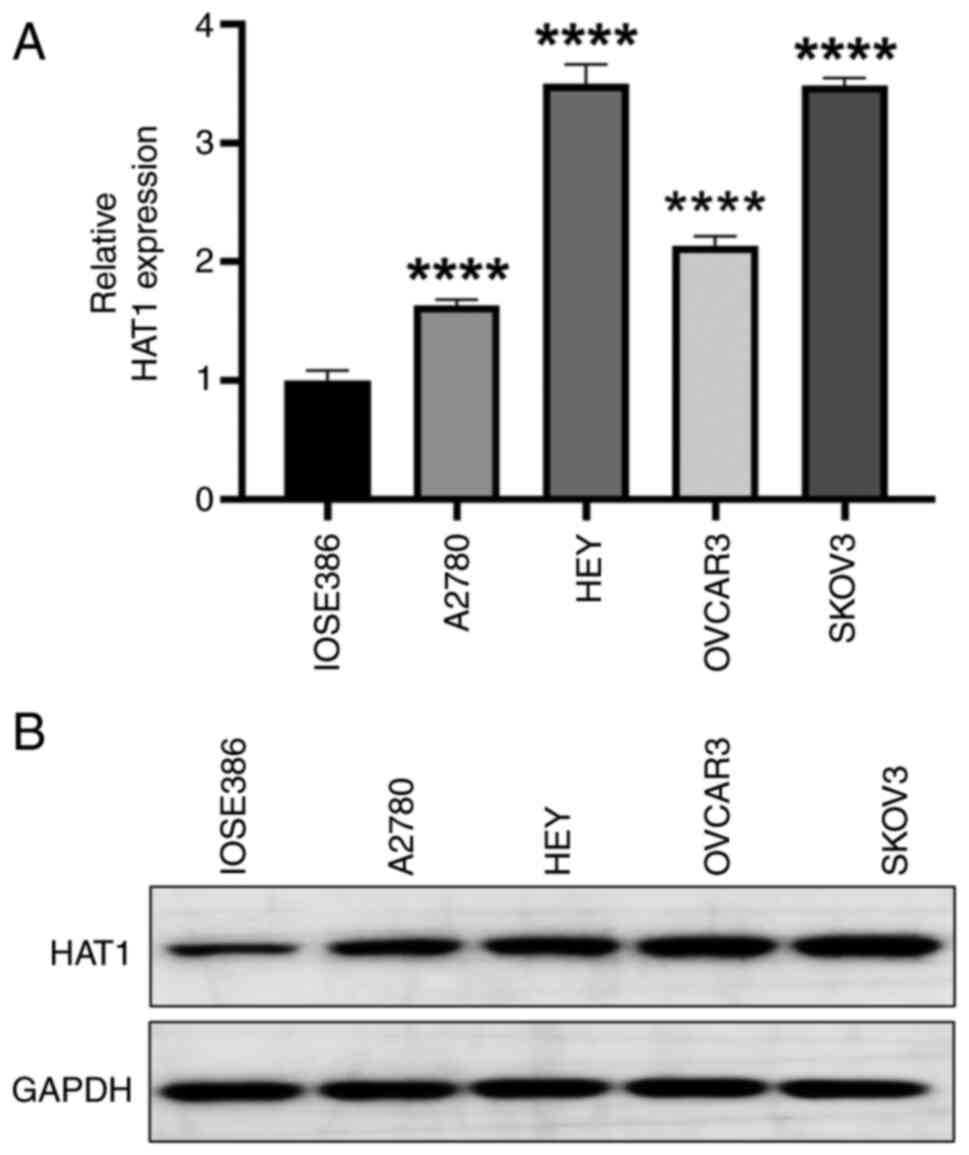
HAT1 levels are increased in OC cell
lines
To determine the role of HAT1 in OC development, the
mRNA expression levels of HAT1 were assessed in different OC cell
lines: A2780, HEY, OVCAR3 and SKOV3. The results showed that HAT1
expression levels were generally upregulated in OC cell lines
compared with normal IOSE386 cell line, especially in HEY and SKOV3
cells, which were used in subsequent study (Fig. 2A). Similarly, the protein levels of
HAT1 were increased in OC cells (Fig.
2B). Thus, HAT1 may induce ovarian tumorigenesis.
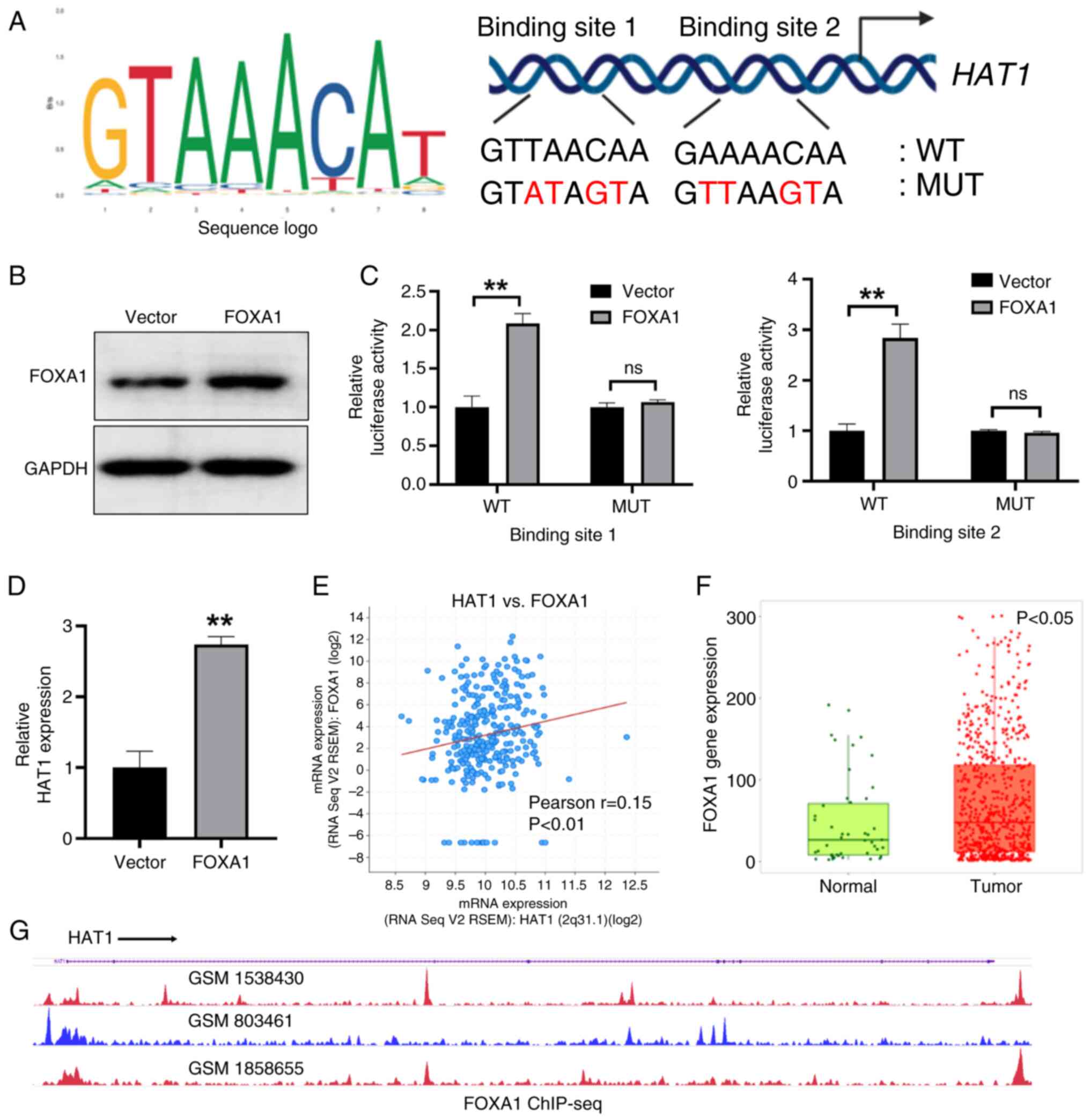
FOXA1 transcriptionally regulates HAT1
expression in OC
TFs are important for the modulation of various
signaling pathways associated with cell homeostasis and disease
conditions (15). To identify the
upstream regulator of HAT1 in OC, the TF database was used to
predict the potential TFs of HAT1 and it was revealed that the TF
FOXA1 may regulate HAT1. Among cancer-related TFs, FOXA1 is an
important molecule that regulates multiple aspects of cancer cells,
including cell proliferation and cancer metastasis. The potential
FOXA1 binding sites of the HAT1 promoter region were then analyzed
through the UCSC Genome Browser and JASPAR databases (Fig. 3A). Subsequently, WT and MUT HAT1
luciferase reporter plasmids were constructed (Fig. 3A). Overexpression of FOXA1 could
promote the activities of HAT1 WT plasmids, as determined through
the luciferase reporter assay; however, this was not observed with
HAT1 MUT plasmids (Fig. 3B and C).
Compared with the vector control, overexpression of FOXA1
significantly increased HAT1 expression levels (Fig. 3D). In addition, the cBioPortal
database was adopted to analyze the correlation between FOXA1 and
HAT1, and a positive association was observed between FOXA1 and
HAT1 levels in OC tissues from TCGA data in the cBioPortal database
(Fig. 3E). FOXA1 expression levels
were also upregulated in OC tissues as suggested in TCGA database
(Fig. 3F). Subsequently, the
chromatin immunoprecipitation-sequencing data conveyed that FOXA1
was enriched in the promoter region of HAT1 (Fig. 3G). In conclusion, these data showed
that FOXA1 functioned as a TF of HAT1.
HAT1 is involved in DNA replication
and pyrimidine metabolism pathways
To explore the regulatory pathways associated with
HAT1 expression levels, OC data from TCGA database were downloaded
and analyzed, before being separated into HAT1 high- and
low-expression groups section. The LinkedOmics database suggested
that differentially expressed genes of HAT1 were enriched in
several biological processes, such as ‘DNA replication’,
‘pyrimidine metabolism’ and ‘RNA transport’ (Fig. 4A). The gene set enrichment analysis
results showed that the HAT1 high-expression group was positively
associated with the ‘DNA replication’ and ‘pyrimidine metabolism’
pathways (Fig. 4B). Subsequently,
the association between HAT1 and regulatory proteins of DNA
replication and pyrimidine metabolism pathways in OC tissues were
analyzed, and the expression levels of HAT1 were revealed to be
positively correlated with proliferating cell nuclear antigen
(PCNA), replication protein A1 and DNA polymerase α catalytic
subunit from the cell cycle pathway (Fig. 4C). Furthermore, HAT1 levels were
positively correlated with thymidine kinase 1,
ribonucleoside-diphosphate reductase subunit M (RRM)1 and RRM2,
which are involved in the pyrimidine metabolism pathway (Fig. 4D).
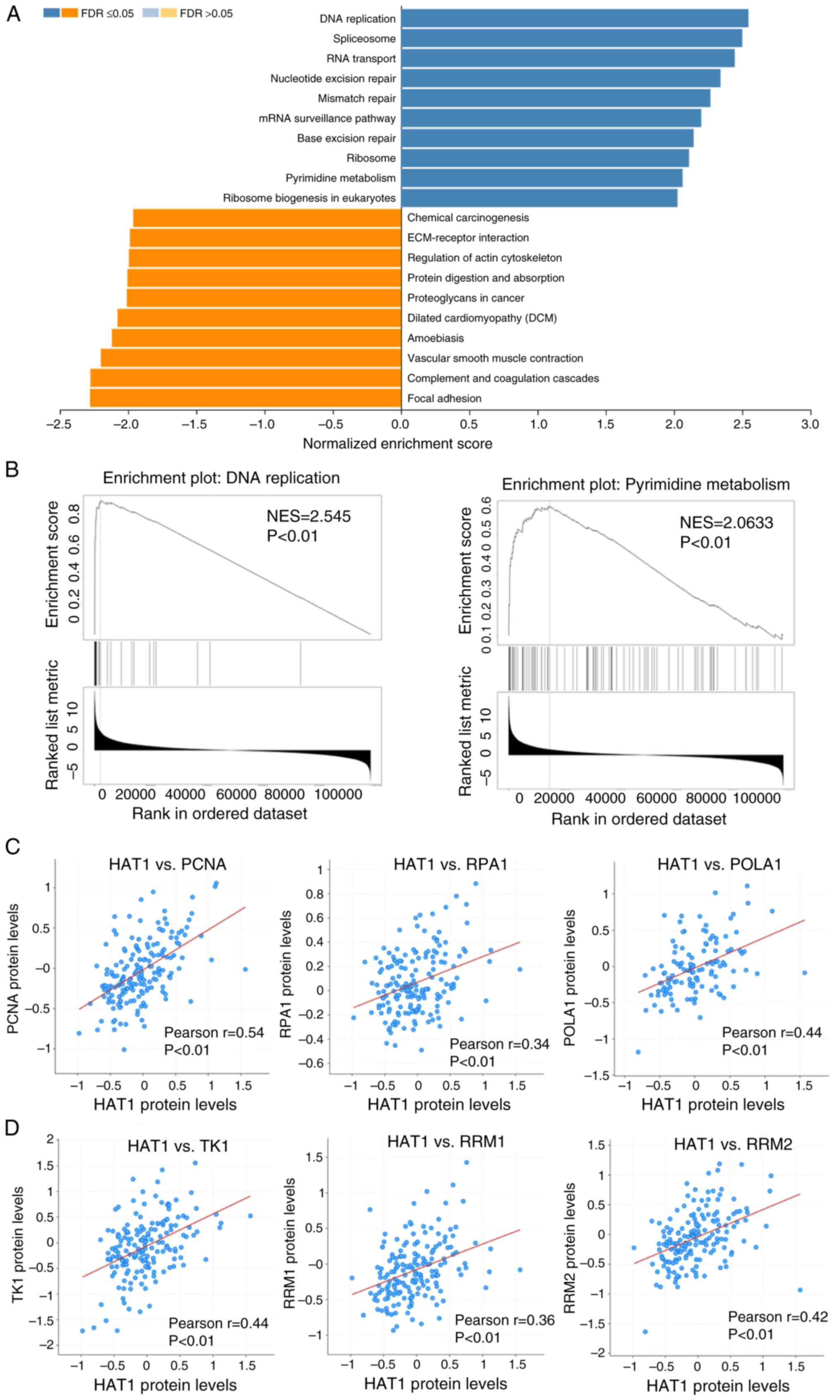 | Figure 4.Relationship between HAT1 and DNA
replication and pyrimidine metabolism pathways. (A) LinkedOmics
database suggested that differentially expressed genes of HAT1 were
enriched in several biological processes, such as ‘DNA
replication’, ‘pyrimidine metabolism’ and ‘RNA transport’. (B) Gene
set enrichment analysis revealed that genes altered by HAT1 were
positively associated with ‘DNA replication’ as well as ‘pyrimidine
metabolism’ pathways. (C) cBioPortal database was used to analyze
the correlations between HAT1 and regulatory proteins of the DNA
replication pathway. Pearson's rank correlation between HAT1 and
DNA replication-related proteins (PCNA, RPA1 and POLA1) was
analyzed in ovarian cancer tissues. (D) Pearson's rank correlation
between HAT1 and pyrimidine metabolism-related proteins (TK1, RRM1
and RRM2) was analyzed in ovarian cancer tissues as suggested in
the cBioPortal database. HAT1, histone acetyltransferase 1; PCNA,
proliferating cell nuclear antigen; RPA1, replication protein A1;
POLA1, DNA polymerase α catalytic subunit; TK1, thymidine kinase 1;
RRM, ribonucleoside-diphosphate reductase subunit M; NES,
normalized enrichment score; FDR, false discovery rate. |
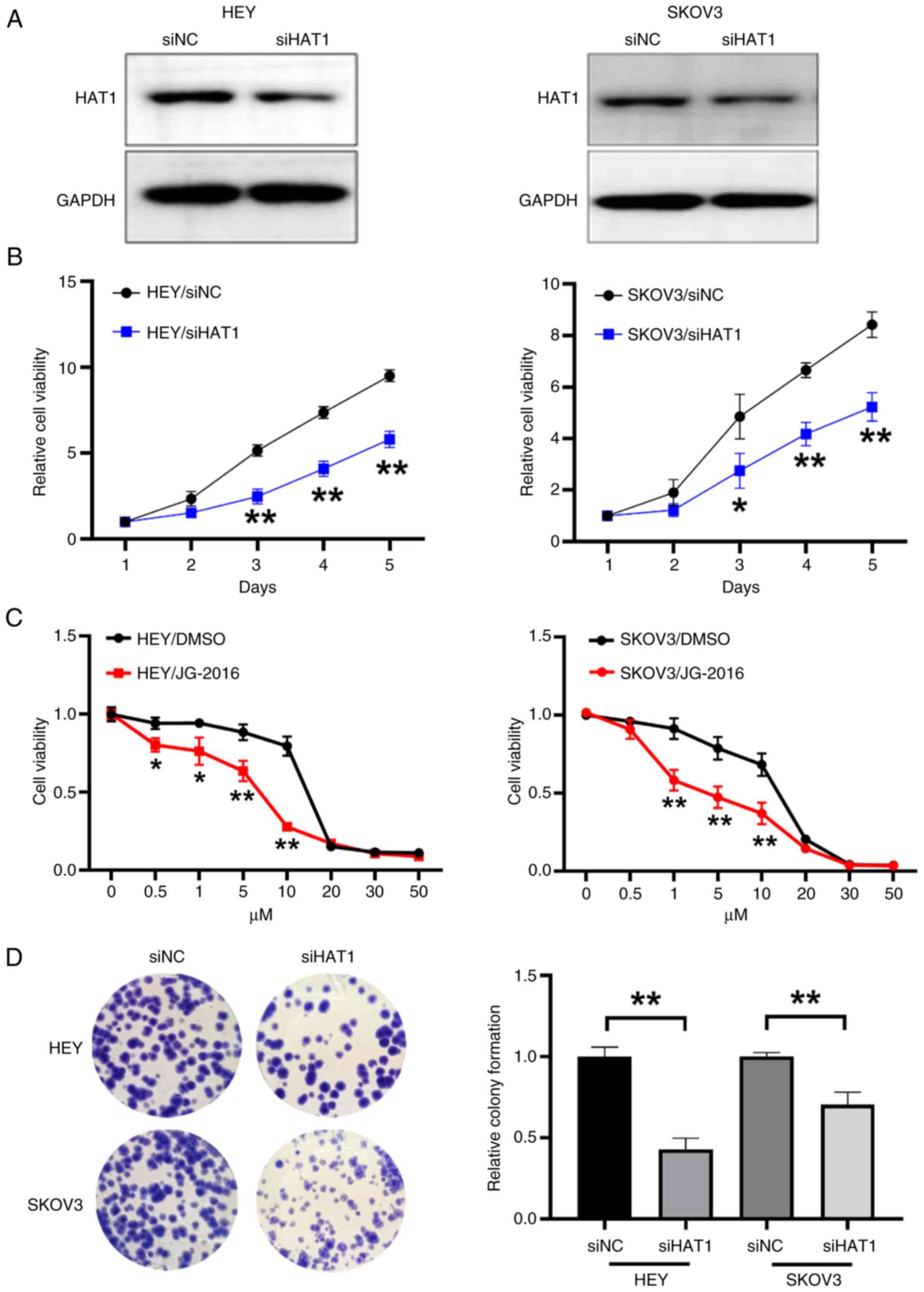
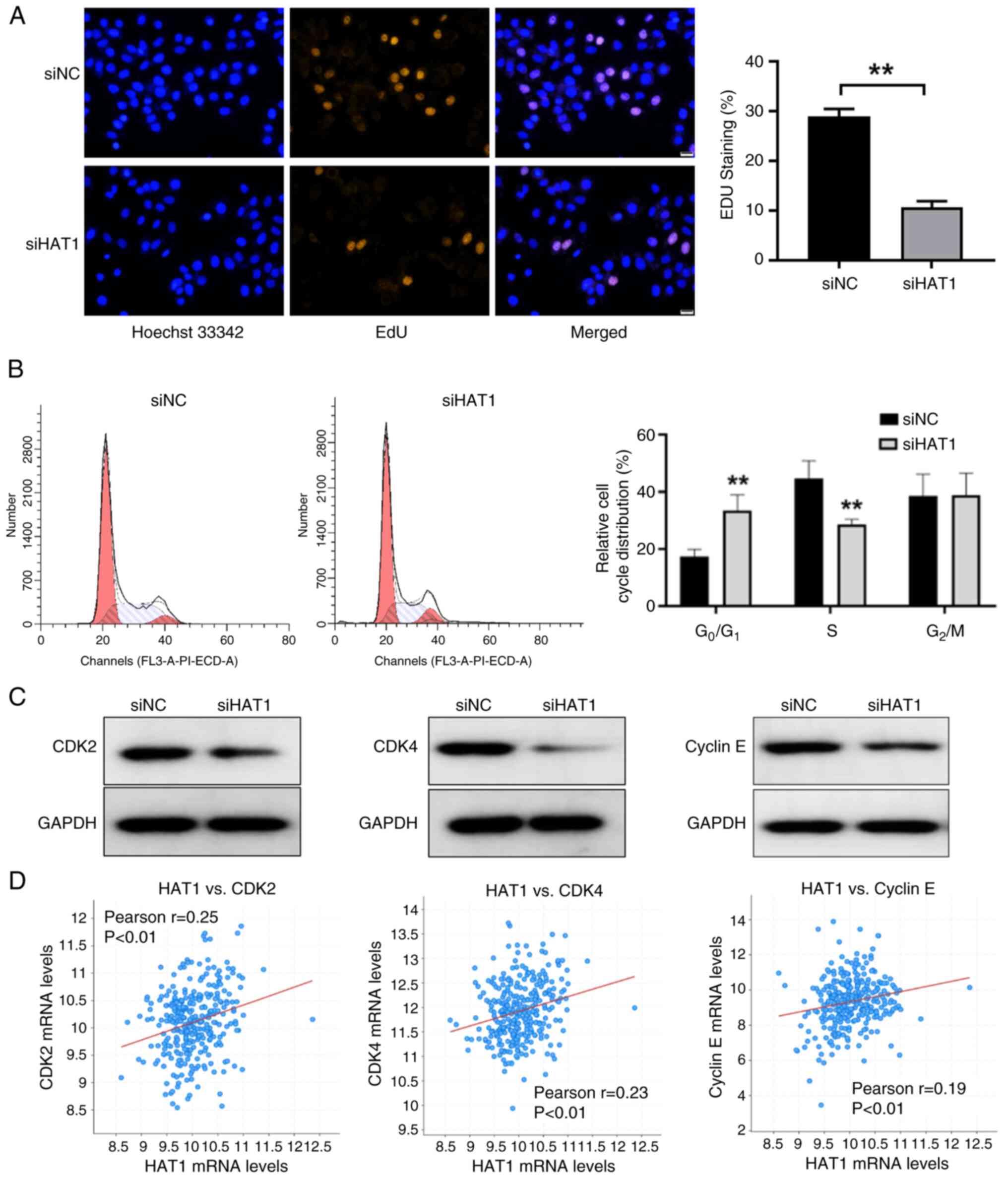
HAT1 induces OC cell viability and
colony formation
To confirm the tumor-promoting role of HAT1, HEY and
SKOV3 OC cell lines were used to knockdown HAT1 expression
(Fig. 5A). As determined by CCK-8
assay, HAT1 knockdown markedly suppressed cell proliferation
(Fig. 5B). In the present study,
cells were also treated with the HAT1 inhibitor JG-2016, and the
results showed that JG-2016 significantly inhibited cell
proliferation (Fig. 5C). In
addition, inhibition of HAT1 reduced colony formation activities in
HEY and SKOV3 cell lines (Fig. 5D).
Thus, the present results indicated that HAT1 contributed to
tumorigenic growth.
HAT1 expression is associated with
CDK2, CDK4 and cyclin E levels
Human HEY OC cells were transfected with negative
control and HAT1 siRNAs. These cells were then stained with an EdU
probe, which functions as a cell proliferation marker, and the
results showed that HAT1 knockdown significantly suppressed EdU
activities (Fig. 6A). The cell
cycle regulatory pathway integrates into other hallmarks of cancer,
including metabolism remodeling and immune escape, and promotes
tumorigenesis (16). In the present
study, inhibition of HAT1 suppressed cell cycle progression and
reduced the percentage of cells at S phase (Fig. 6B). Cell cycle-related proteins
levels were then analyzed and it was revealed that suppression of
HAT1 decreased CDK2, CDK4 and cyclin E levels (Fig. 6C). Furthermore, positive
associations were shown between HAT1 and CDK2, CDK4 and cyclin E
levels in OC tissues from TCGA data obtained from the cBioPortal
database (Fig. 6D). Thus, HAT1 was
suggested to regulate the cell cycle pathway in OC.
Discussion
OC has become one of the most frequently diagnosed
cancers in female patients worldwide, with an expected 313,000
cases and 207,000 mortalities in 2020, ranking as the third highest
cause of death among gynecological cancers (17). The high mortality rate is partly due
to the heterogeneity of the tumor combined with the absence of
symptoms at the early stages of OC, which limits early diagnosis,
resulting in most patients presenting with advanced stages of OC
when identified (18). Histone
modifications, as major chromatin regulators, play an important
role in the etiology of numerous diseases, especially cancer
(19). Acetylation, specifically
lysine acetylation, is a dynamic epigenetic process with a notable
role in cell cycle, cell survival or to cellular processes, which
capable of targeting both histone and non-histone proteins
(16,20). Epigenetic modification via
acetylation regulates gene transcriptional processes and protein
stability, activity and localization (21). HAT1 was the first identified HAT;
however, its biological significance remains to be elucidated.
Several studies have highlighted the importance of HAT1 in
regulating cellular processes that are altered in a variety of
diseases, including cancer (13,22,23).
HAT1 protein knockdown or knockout in various pre-clinical models
has conveyed that HAT1 is likely a new potential therapeutic target
in OC treatment (13,22,23).
In the present study, the mRNA and protein expression levels of
HAT1 were found to be elevated in OC tissues, and elevated HAT1
levels were associated with shorter survival rate.
FOXA1 belongs to the FOXA TF family and has a
forkhead (or winged helix) DNA-binding domain of ~100 amino acids.
FOXA1 functions as a TF and attaches to the chromosome to induce
nucleosome remodeling, which allows other TFs to bind to the
chromosome and perform tissue-specific transcriptional programs
(24–28). FOXA1 can be pro- or antitumorigenic
in various human malignancies (29). Up to 80% of estrogen
receptor-positive breast cancer cases are FOXA1-positive, and FOXA1
expression is linked with improved prognosis (29). Furthermore, FOXA1 upregulation has
been shown to be inversely linked with interferon signature and
activated tumor-specific CD8+T cells in patients with
prostate cancer, and to increase cancer immunoresistance and
resistance to chemotherapy in nude mice and patients with prostate
cancer (30). FOXA1 acts as a tumor
suppressor during its development of endometrial cancer (31). In the present study, the TF FOXA1
was shown to regulate HAT1 levels, and there was a positive
association between FOXA1 and HAT1 in OC.
The functional importance of HAT1 has been
investigated in a number of cell models to improve understanding of
its biological function. Previous investigation has suggested that
HAT1 is briefly localized on chromatin, adjacent to DNA replication
sites, where it plays a regulatory role in replication fork
stalling (32). According to
additional research, the loss of HAT1 enhances genomic instability,
making cells vulnerable to DNA double-strand breaks (33). The significance of HAT1 in
replication and chromatin assembly has previously been shown
(34).
In the present study, the expression levels of HAT1
were positively correlated with PCNA in the DNA replication
pathway. PCNA is the eukaryotic DNA sliding clamp that interacts
with cell cycle proteins, including cyclins and CDKs, involved in
DNA replication (35). A previous
study has shown that PCNA is a key factor in DNA replication during
the cell cycle and is directly associated with prostate tissue
proliferation (36). PCNA
inhibition induces S-phase arrest of human prostatic epithelial
cancer cell lines (37). PCNA also
interacts with the cyclin-CDK complex in several eukaryotic cells
(38). It has been noted that CDK2
could directly interact with PCNA in numerous cells, supporting the
linkage between cell cycle checkpoints and PCNA (39). Thus, PCNA is considered an accessory
protein in cellular processes, contributing to cell division, DNA
replication and various cell cycle controls. In the present study,
HAT1 was suggested to regulate the cell cycle pathway in OC. The
cell cycle is a series of interconnected events that allow the cell
to continue growing and proliferating. Cell cycle arrest serves as
a survival mechanism that enables cancer cells to compensate for
their own DNA that has been damaged. CDKs are key components of the
cell cycle machinery that, when activated, enable the cell to move
from one phase to the next (40).
Cyclins positively regulate CDKs, whereas naturally generated CDK
inhibitors (CDKIs) adversely regulate them. Cancer is characterized
by cell cycle dysregulation, with cells upregulating cyclins or
lacking CDKIs continuing to grow abnormally (41). The cell cycle also protects the cell
against DNA damage. The present research conveyed that suppression
of HAT1 decreased CDK2, CDK4 and cyclin E levels; thus, HAT1 may be
a potential therapeutic target in OC.
Notably, findings have revealed the importance of
HAT1 in a variety of physiological processes, encouraging
additional research to determine its biological function (12,42).
HAT1 has been linked to a variety of cellular functions, including
proliferation, DNA replication and cellular metabolism (12,42).
The JG-2016 compound has shown relative specificity toward HAT1
compared with other acetyltransferases, suppressing the
proliferation of human cancer cell lines and interfering with tumor
growth (43). The present study is,
to the best of our knowledge, the first report of a small-molecule
inhibitor of the HAT1 enzyme complex and represents a step toward
targeting the HAT1 pathway for cancer therapy. In the present
study, the inhibition of HAT1 was discovered to promote cell cycle
arrest, and reduce CDK2, CDK4 and cyclin E levels in OC cells.
Thus, the present work highlighted the importance of FOXA1 as an
upstream regulator mediating HAT1 expression levels and HAT1 as a
potential oncogene in OC. Therefore, specifically inhibiting HAT1
expression may provide a unique strategy for OC treatment.
Acknowledgements
Not applicable.
Funding
The present work was supported by the Science and Technology
Research Project of Henan province (grant no. LHGJ20220532).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XH conceptualized and designed the study, performed
experiments and wrote the manuscript. JLi, YZ, LZ, LY and XO
developed methodology, analyzed data and performed experiments. LL
and JLiu performed experiments, analyzed data and confirm the
authenticity of all the raw data. All authors have read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Obermair A, Beale P, Scott CL, Beshay V,
Kichenadasse G, Simcock B, Nicklin J, Lee YC, Cohen P and Meniawy
T: Insights into ovarian cancer care: report from the ANZGOG
ovarian cancer webinar series 2020. J Gynecol Oncol. 32:e952021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xia CF, Dong XS, Li H, Cao M, Sun D, He S,
Yang F, Yan X, Zhang S, Li N and Chen W: Cancer statistics in China
and United States, 2022: Profiles, trends, and determinants. Chin
Med J (Engl). 135:584–590. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Garrido MP, Fredes AN, Lobos-Gonzalez L,
Valenzuela-Valderrama M, Vera DB and Romero C: Current treatments
and new possible complementary therapies for epithelial ovarian
cancer. Biomedicines. 10:772021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jayde V, White K and Blomfield P: Symptoms
and diagnostic delay in ovarian cancer: A summary of the
literature. Contemp Nurse. 34:55–65. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Konstantinopoulos PA and Matulonis UA:
Clinical and translational advances in ovarian cancer therapy. Nat
Cancer. 4:1239–1257. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Orr JA and Hamilton PW: Histone
acetylation and chromatin pattern in cancer. A review. Anal Quant
Cytol Histol. 29:17–31. 2007.PubMed/NCBI
|
|
7
|
Makowski AM, Dutnall RN and Annunziato AT:
Effects of acetylation of histone H4 at lysines 8 and 16 on
activity of the Hat1 histone acetyltransferase. J Biol Chem.
276:43499–43502. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jin X, Tian SH and Li PP: Histone
acetyltransferase 1 promotes cell proliferation and induces
cisplatin resistance in hepatocellular carcinoma. Oncol Res.
25:939–946. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Klett-Mingo JI, Pinto-Díez C,
Cambronero-Plaza J, Carrión-Marchante R, Barragán-Usero M,
Pérez-Morgado MI, Rodríguez-Martín E, Toledo-Lobo MV, González VM
and Martín ME: Potential therapeutic use of aptamers against HAT1
in lung cancer. Cancers (Basel). 15:2272022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xue L, Hou J, Wang Q, Yao L, Xu S and Ge
D: RNAi screening identifies HAT1 as a potential drug target in
esophageal squamous cell carcinoma. Int J Clin Exp Patho.
7:3898–3907. 2014.PubMed/NCBI
|
|
11
|
Sarkar T, Dhar S, Chakraborty D, Pati S,
Bose S, Panda AK, Basak U, Chakraborty S, Mukherjee S, Guin A, et
al: FOXP3/HAT1 axis controls treg infiltration in the tumor
microenvironment by inducing CCR4 expression in breast cancer.
Front Immunol. 13:7405882022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun Y, Ren D, Zhou Y, Shen J, Wu H and Jin
X: Histone acetyltransferase 1 promotes gemcitabine resistance by
regulating the PVT1/EZH2 complex in pancreatic cancer. Cell Death
Dis. 12:8782021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang G, Yuan Y, Yuan H, Wang J, Yun H,
Geng Y, Zhao M, Li L, Weng Y, Liu Z, et al: Histone
acetyltransferase 1 is a succinyltransferase for histones and
non-histones and promotes tumorigenesis. Embo Rep. 22:e509672021.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Weidemüller P, Kholmatov M, Petsalaki E
and Zaugg JB: Transcription factors: Bridge between cell signaling
and gene regulation. Proteomics. 21:e20000342021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hanahan D: Hallmarks of cancer: New
dimensions. Cancer Discov. 12:31–46. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
18
|
Webb PM and Jordan SJ: Epidemiology of
epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol.
41:3–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shirvaliloo M: The landscape of histone
modifications in epigenomics since 2020. Epigenomics. 14:1465–1477.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shvedunova M and Akhtar A: Modulation of
cellular processes by histone and non-histone protein acetylation.
Nat Rev Mol Cell Bio. 23:329–349. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun LC, Zhang HF and Gao P: Metabolic
reprogramming and epigenetic modifications on the path to cancer.
Protein Cell. 13:877–919. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fan P, Zhao J, Meng Z, Wu H, Wang B, Wu H
and Jin X: Overexpressed histone acetyltransferase 1 regulates
cancer immunity by increasing programmed death-ligand 1 expression
in pancreatic cancer. J Exp Clin Cancer Res. 38:472019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gruber JJ, Geller B, Lipchik AM, Chen J,
Salahudeen AA, Ram AN, Ford JM, Kuo CJ and Snyder MP: HAT1
coordinates histone production and acetylation via H4 promoter
binding. Mol Cell. 75:711–724.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Carroll JS, Liu XS, Brodsky AS, Li W,
Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger
TR, et al: Chromosome-wide mapping of estrogen receptor binding
reveals long-range regulation requiring the forkhead protein FoxA1.
Cell. 122:33–43. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cirillo LA, Lin FR, Cuesta I, Friedman D,
Jarnik M and Zaret KS: Opening of compacted chromatin by early
developmental transcription factors HNF3 (FoxA) and GATA-4. Mol
Cell. 9:279–289. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cirillo LA and Zaret KS: An early
developmental transcription factor complex that is more stable on
nucleosome core particles than on free DNA. Mol Cell. 4:961–969.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Laganière J, Deblois G, Lefebvre C,
Bataille AR, Robert F and Giguère V: From the Cover: Location
analysis of estrogen receptor alpha target promoters reveals that
FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci
USA. 102:11651–11656. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zaret K: Developmental competence of the
gut endoderm: Genetic potentiation by GATA and HNF3/fork head
proteins. Dev Biol. 209:1–10. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Albergaria A, Paredes J, Sousa B, Milanezi
F, Carneiro V, Bastos J, Costa S, Vieira D, Lopes N, Lam EW, et al:
Expression of FOXA1 and GATA-3 in breast cancer: The prognostic
significance in hormone receptor-negative tumours. Breast Cancer
Res. 11:R402009. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
He Y, Wang L, Wei T, Xiao YT, Sheng H, Su
H, Hollern DP, Zhang X, Ma J, Wen S, et al: FOXA1 overexpression
suppresses interferon signaling and immune response in cancer. J
Clin Invest. 131:e1470252021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Abe Y, Ijichi N, Ikeda K, Kayano H,
Horie-Inoue K, Takeda S and Inoue S: Forkhead box transcription
factor, forkhead box A1, shows negative association with lymph node
status in endometrial cancer, and represses cell proliferation and
migration of endometrial cancer cells. Cancer Sci. 103:806–812.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Agudelo Garcia PA, Lovejoy CM, Nagarajan
P, Park D, Popova LV, Freitas MA and Parthun MR: Histone
acetyltransferase 1 is required for DNA replication fork function
and stability. J Biol Chem. 295:8363–8373. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang X, Li L, Liang J, Shi L, Yang J, Yi
X, Zhang D, Han X, Yu N and Shang Y: Histone acetyltransferase 1
promotes homologous recombination in DNA repair by facilitating
histone turnover. J Biol Chem. 288:18271–18282. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Agudelo Garcia PA, Hoover ME, Zhang P,
Nagarajan P, Freitas MA and Parthun MR: Identification of multiple
roles for histone acetyltransferase 1 in replication-coupled
chromatin assembly. Nucleic Acids Res. 45:9319–9335. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kelman Z and O'Donnell M: Structural and
functional similarities of prokaryotic and eukaryotic DNA
polymerase sliding clamps. Nucleic Acids Res. 23:3613–3620. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Taftachi R, Ayhan A, Ekici S, Ergen A and
Ozen H: Proliferating-cell nuclear antigen (PCNA) as an independent
prognostic marker in patients after prostatectomy: A comparison of
PCNA and Ki-67. BJU Int. 95:650–654. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tan Z, Wortman M, Dillehay KL, Seibel WL,
Evelyn CR, Smith SJ, Malkas LH, Zheng Y, Lu S and Dong Z:
Small-molecule targeting of proliferating cell nuclear antigen
chromatin association inhibits tumor cell growth. Mol Pharmacol.
81:811–819. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xiong Y, Zhang H and Beach D: D type
cyclins associate with multiple protein kinases and the DNA
replication and repair factor PCNA. Cell. 71:505–514. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Koundrioukoff S, Jónsson ZO, Hasan S, de
Jong RN, van der Vliet PC, Hottiger MO and Hübscher U: A direct
interaction between proliferating cell nuclear antigen (PCNA) and
Cdk2 targets PCNA-interacting proteins for phosphorylation. J Biol
Chem. 275:22882–22887. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Suski JM, Braun M, Strmiska V and Sicinski
P: Targeting cell-cycle machinery in cancer. Cancer Cell.
39:759–778. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cavalu S, Abdelhamid AM, Saber S, Elmorsy
EA, Hamad RS, Abdel-Reheim MA, Yahya G and Salama MM: Cell cycle
machinery in oncology: A comprehensive review of therapeutic
targets. FASEB J. 38:e237342024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhou P, Peng X, Zhang K, Cheng J, Tang M,
Shen L, Zhou Q, Li D and Yang L: HAT1/HDAC2 mediated ACSL4
acetylation confers radiosensitivity by inducing ferroptosis in
nasopharyngeal carcinoma. Cell Death Dis. 16:1602025. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gaddameedi JD, Chou T, Geller BS,
Rangarajan A, Swaminathan TA, Dixon D, Long K, Golder CJ, Vuong VA,
Banuelos S, et al: Acetyl-click screening platform identifies
small-molecule inhibitors of histone acetyltransferase 1 (HAT1). J
Med Chem. 66:5774–5801. 2023. View Article : Google Scholar : PubMed/NCBI
|