Introduction
Prostate cancer (PCa) is one of the most common
malignancies in men worldwide. According to the 2024 data from the
American Cancer Society, PCa is the second leading cause of
mortality in men, after lung cancer (1). Annually, there are ~1.5 million cases
of PCa and it results in 396,700 mortalities globally (2). With increases in the global and aging
populations, PCa has become a notable public health challenge in
men worldwide (3). The incidence of
PCa in China has markedly increased (4–6) and it
has become one of the most common types of cancer in men in China.
In 2020, in China, ~120,000 novel cases of PCa were reported, which
accounted for 8.16% of cancer diagnoses in men. The number of PCa
mortalities was ~50,000 making up 13.61% of cancer-related
mortalities among men (7,8).
The 5- and 10-year survival rates for PCa depend on
the stage and grade of the cancer at diagnosis. For localized or
regional PCa, when the cancer is confined to the prostate or nearby
areas, the 5-year relative survival rate is ~100%. For metastatic
PCa, in which the cancer has spread to distant parts of the body,
the 5-year survival rate is markedly reduced, ~37% (9). Recurrence is a concern with PCa,
especially in advanced cases with increasing prostate-specific
antigen (PSA) levels often indicating recurrence within 5–10 years
in 20–30% of men treated for PCa. Recurrence is more likely in
cases with aggressive tumor characteristics, increased initial PSA
levels and advanced stage at diagnosis (10).
The high risk of PCa is primarily attributed to its
aggressive metastasis. In cases where cancer types are hidden, it
is difficult for clinicians to diagnose and treat the disease
early. In several cases, by the time PCa is diagnosed, tumor
tissues have metastasized to areas outside the prostate, such as in
the bone (11,12). PCa is associated with multiple
genomic alterations, which result in a high degree of tumor
heterogeneity. However, the specific mechanism underlying its
malignancy remains unclear (13).
Current progress in medical diagnostic imaging, surgery,
chemotherapy and radiotherapy has improved the effective diagnosis,
treatment and management of PCa. Androgen-blocking therapy (ADT),
which targets androgen receptors (ARs), is the first-line treatment
option in clinical practice, further to surgical excision. However,
~2 years of administering ADT to patients with advanced PCa results
in the development of castration-resistant PCa; which is typically
accompanied by elevated serum testosterone and PSA levels (14). Therefore, exploring the internal
mechanisms and therapeutic targets of PCa and developing
corresponding drugs is key to improve the survival and prognosis of
patients and reduce the medical burden of searching for early
diagnostic markers to supplement the diagnostic shortcomings of
PSA.
Cytokines serve an important role in the development
of cancer (15). Further to
abnormal DNA methylation, histone post-translational modifications
(PTMs) and changes in chromatin modification patterns are
associated with carcinogenesis; the latter two have revealed to be
key factors in cancer-related pathways (16). Histone methylation serves a
regulatory role in various cancer cells; this process is catalyzed
by histone methyltransferases (HMTs), which comprise different
families of enzymes that methylate specific residues to alter gene
transcription. For example, lysine HMT (protein lysine
methyltransferase) methylates lysine residues, while histone
arginine methyltransferase (PRMT) methylates arginine. The only
type III PRMT among the nine members of the PRMT family is PRMT7,
which contains only monomethyl arginine (17–22).
PTMs regulated by PRMT7 and its proteins are
associated with tumor growth and metastasis (23). Specifically, PRMT7 expression is
increased in clear cell renal cell carcinoma tissues and leads to
renal cell carcinoma growth through the β-catenin/c-Myc axis
(24). The upregulation of PRMT7
expression may promote breast cancer cell invasion by regulating
MMP9 expression (25). Research on
PRMT7 in breast cancer has indicated its potential as a biomarker
and therapeutic target (26,27).
Although PRMT7 has not been thoroughly studied in other cancer
types, as a member of the PRMT family, it is known to regulate
histone methylation, a PTM associated with cancer (28,29).
In particular, as a member of the PRMT family, PRMT5 can promote
the progression of PCa by influencing key regulatory factors, such
as the AR. This influence on AR, a key regulator in PCa, suggests
that PRMT5 serves a notable role in cancer development, which
highlights its potential as a target for therapeutic intervention
(30,31); therefore, PRMT7 is likely to serve a
key role as an epigenetic regulator in PCa and ultimately affect
tumor progression and prognosis.
In 2014, Vieira et al (32) studied the expression levels of
partial HMT or demethylase in PCa and its relationship with the
occurrence and progression of cancer. Based on this and other
previous studies (28–31) on the PRMT family and malignant
tumors, the molecular mechanism of action of the PRMT7 in PCa were
investigated. By validating the expression level of PRMT7 in
prostate cancer cells and tissues, its correlation with
clinicopathological features and patient survival was analyzed to
further elucidate the potential mechanisms by which PRMT7 promotes
prostate cancer progression.
Materials and methods
Patient samples
In the present study, 159 PCa samples were
prospectively collected from patients diagnosed at the Affiliated
Hospital of Jiangnan University (Wuxi, China) between December 2023
and June 2024. The tissue samples were obtained through standard
hospital procedures. Patients were evaluated through clinical and
histological analyses. The present study strictly adhered to the
following predefined inclusion criteria: i) Histopathologically
confirmed primary prostate adenocarcinoma [International Society of
Urological Pathology (ISUP) grade ≥2]; ii) no prior history of
radiotherapy or systemic therapy; and iii) availability of matched
tumor-normal paired tissue samples for genomic analysis. The
exclusion criteria comprised cases with metastasis at diagnosis (to
avoid confounding by advanced disease biology) and specimens that
did not meet tissue quality standards (RNA integrity number >7).
The age distribution of the cohort (median, 68 years; range, 52–81
years), PSA levels and ISUP grades mirrored regional epidemiology.
The tissue samples were collected for tissue microarray analysis
and the correlation between PRMT7 and the age of the patients,
Gleason score (33), PSA levels and
TNM stage (34) in PCa were
analyzed. Then, seven pairs of matched PCa clinical samples
detected using reverse transcription-quantitative PCR (RT-qPCR)
were also derived from the aforementioned tissues, with normal
tissue samples collected 5 cm away from the tumor margin serving as
negative controls. The present study was approved by the Ethics
Committee of Affiliated Hospital of Jiangnan University (approval
no. LS2023099; Wuxi, China). The present study was conducted in
accordance with the Declaration of Helsinki. Each patient included
in the present study signed a written informed consent form prior
to sample collection.
Bioinformatics analysis
PRMT7 expression data were downloaded from The
Cancer Genome Atlas (TCGA) online database (http://tcga-data.nci.nih.gov/tcga/). R software
(version 4.3.0; http://www.r-project.org) and SPSS (version 17.0; SPSS
Inc.) were used to analyze and process all data.
Immunohistochemistry staining and
scoring
Paraformaldehyde (4%) was used to fix collected
tissue samples at room temperature (25°C) for 24 h.
Paraffin-embedded tissues were cut into 4-µm-thick sections. Tissue
sections were dewaxed, hydrated, then permeabilized with 0.5%
Triton X-100 at room temperature (25°C) for 10 min, blocked with 3%
H2O2 (25°C) for 30 min and 10% goat serum
(25°C) for 30 min. Next, the samples were incubated with antibodies
against PRMT7 from Merck KGaA (1:50; cat. no. HPA044241) at 4°C
overnight. Thereafter, images of the sections were taken using an
Olympus IX73 inverted fluorescence microscope (Olympus
Corporation). The subsequent steps were performed using the
GTVision III Detection System/Mo&Rb (Gene Tech Co., Ltd.)
(35,36). Immunohistochemistry results for
healthy individuals were selected from the Human Protein Atlas
database (https://www.proteinatlas.org/ENSG00000132600-PRMT7/tissue/prostate#).
Cell culture
Human PCa cell lines PC3, DU145, LNCAP, 22RV1 and
WPMY-1 were purchased from the Cell Bank of the Chinese Academy of
Science. DU145 and WPMY-1 cells were cultured in DMEM (Cytiva)
supplemented with 10% FBS (Shenzhen Aipno Biomedical Technology
Co., Ltd.). PC3, LNCAP and 22RV1 cells were cultured in RPMI-1640
(Cytiva) medium supplemented with 10% FBS (Shenzhen Aipno
Biomedical Technology Co., Ltd.). All cells were incubated at 37°C
with 5% CO2.
RT-qPCR
RNA extraction from PC3, DU145, LNCAP, 22RV1 and
WPMY-1 cells (each sample used 3×106 cells) was carried
out using a FastPure® Cell/Tissue Total RNA Isolation
Kit V2 (cat. no. RC112-01; Vazyme Biotech Co., Ltd.) according to
the manufacturer's protocol. RNA was reverse transcribed into cDNA
using a HiScript® III RT SuperMix for qPCR (+gDNA wiper)
(cat. no. R323-01; Vazyme Biotech Co. Ltd.). The primer sequences
used for GAPDH and PRMT7 are listed in Table SI. qPCR was carried out in an
Applied Biosystems 7500 PCR system (Thermo Fisher Scientific, Inc.)
using a ChamQ Universal SYBR® qPCR Master Mix (cat. no.
Q711-02/03, Vazyme Biotech Co. Ltd.). The RT-qPCR protocol
consisted of: Initiation at 95°C for 30 sec, followed by 40 cycles
at 95°C for 10 sec and 60°C for 30 sec. Relative quantification of
gene expression was carried out using the 2−ΔΔCq method
(35), with GAPDH as the endogenous
control. Primer efficiencies were validated to be between 95–105%
prior to analysis. The results were analyzed using a previously
reported method (35).
Plasmid transfection
Small interfering (si)PRMT7 was designed based on
the PRMT7 (NM_019023.5; Infection, http://www.ncbi.nlm.nih.gov/nuccore/NM_019023.5)
target sequence. The sequences of the specific genes used in the
present study were as follows: PRMT7-siRNA (43684–1) sense (S),
5′-CACAUCAUGGACGACAUGAUU-3′ and antisense (AS),
5′-AAUCAUGUCGUCCAUGAUGUG-3′; PRMT7-siRNA (43685–1) S,
5′-GCUAACCACUUGGAAGAUAAA-3′ and AS, 5′-UUUAUCUUCCAAGUGGUUAGC-3′;
PRMT7-siRNA (43686–1) S, 5′-CGAUGACUACUGCGUAUGGUA-3′ and AS,
5′-UACCAUACGCAGUAGUCAUCG-3′; non-silencing siRNA S,
5′-UUCUCCGAACGUGUCACGU-3′ and AS, 5′-ACGUGACACGUUCGGAGAA-3′. They
were synthesized and cloned into GV248 (11.5 kb) vector with
BsmBI sites (purchased from Shanghai Genechem Co., Ltd.),
recombinant vector was detected by DNA sequencing. The final
products were then transfected into Escherichia coli
(>1×108 cfu/µg; purchased from Shanghai Genechem Co.,
Ltd.). The PRMT7-siRNA plasmid was then isolated/purified with the
EndoFree Plasmid Mega Kit (Qiagen GmbH, Germany; Cat. #12381).
Briefly, 4×105 cells were seeded in each well of a
6-well plate. Upon reaching 60–70% confluency, transfections were
carried out using Lipofectamine® 3000 reagent (cat. no.
L3000150, Thermo Fisher Scientific, Inc.) Cells were seeded in
6-well plates at a density of 1×106 cells/well and
cultured overnight to reach 70–80% confluency. For each well, 2.5
µg of plasmid DNA and 5 µl P3000™ were diluted in 125 µl of
Opti-MEM® Reduced-Serum Medium (Gibco; Thermo Fisher
Scientific, Inc.) and mixed gently. Separately, 7.5 µl of
Lipofectamine™ 3000 was diluted in 125 µl of Opti-MEM®.
The diluted DNA was then combined with the diluted
Lipofectamine® 3000 (1:1 ratio) and incubated for 15 min
at room temperature to form DNA-lipid complexes. The mixture was
added dropwise to the cells, followed by gentle swirling. The cells
were cultured at 37°C for an additional 48 h. The transfection
efficiency was >80% based on green protein fluorescence.
Other assays
Western blot analysis, cell proliferation and colony
formation assays, apoptosis analysis and cell cycle detection,
wound healing, cell migration and invasion assays were carried out
as previously described (35,36).
The antibodies that were used are: anti-PRMT7 (1:1,000; cat. no.
ab181214; Abcam), anti-GAPDH (1:2,500; cat. no. ab9485; Abcam),
anti-β-actin (1:2,000; cat. no. GB12001; Wuhan Servicebio
Technology); anti-cyclin D2 (CCND2; 1:2,000; cat. no. 67048-1-Ig)
and anti-retinoblastoma (RB1; 1:2,000; cat. no. 67521-1-Ig) were
purchased from Wuhan Sanying Biotechnology; anti-CDK6 (1:1,000;
cat. no. A0705; Abclonal Biotech Co., Ltd.); and anti-Yin-yang 1
(YY1; 1:1,000; cat. no. GB111880), antitumor protein p53 (TP53;
1:1,000; cat. no. GB111740), anti-rabbit IgG (1:5,000; cat. no.
GB23303) and anti-mouse IgG (1:5,000; cat. no. GB23301) were
purchased from Wuhan Servicebio Technology Co., Ltd.
Transcriptome sequencing and
analysis
Total RNA was isolated from all samples (4 PRMT7-KD
replicates and 4 Control replicates) using TRIzol™ reagent (cat.
no. 15596026CN, Thermo Fisher Scientific, Inc.). RNA integrity was
assessed using the Agilent Bioanalyzer 2100 system (RNA Nano 6000
Assay Kit; Agilent Technologies), and purity was confirmed using a
NanoPhotometer® spectrophotometer (IMPLEN). Only
high-quality RNA samples (RNA integrity Number >8.0; OD260/280
ratio ~2.0) were used for library construction. Sequencing
libraries were prepared from 1 µg of total RNA per sample using the
NEBNext® Ultra™ RNA Library Prep Kit for
Illumina® (New England BioLabs, Inc.) following the
manufacturer's protocol. Briefly, poly(A)+ mRNA was enriched using
poly-T oligo-attached magnetic beads and fragmented. First-strand
cDNA synthesis was performed using random hexamer priming and
M-MuLV Reverse Transcriptase (New England BioLabs, Inc.), followed
by second-strand synthesis with DNA Polymerase I and RNase H. cDNA
fragments were end-repaired, adenylated, and ligated to NEBNext
adaptors. Libraries were size-selected (~250-300 bp) using AMPure
XP beads (Beckman Coulter Inc.) and amplified by PCR with index
primers. Library quality and concentration were validated using the
Agilent Bioanalyzer 2100 system. Indexed libraries were pooled and
clustered on a cBot Cluster Generation System (TruSeq PE Cluster
Kit v3-cBot-HS; Illumina, Inc.). Paired-end sequencing (2×150 bp)
was performed on an Illumina NovaSeq platform. Raw sequencing reads
(FASTQ format) were quality-filtered using custom Perl scripts to
remove adapter sequences, poly-N reads, and low-quality reads
(Q<20), yielding clean reads. Quality metrics (Q20, Q30, GC
content) were calculated for clean data. Clean reads were aligned
to the human reference genome (GRCh38/hg38; http://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000001405.26/)
using HISAT2 (v2.0.5; http://daehwankimlab.github.io/hisat2/) with
splice-site information derived from gene model annotations.
Transcript assembly and read quantification per gene were performed
using StringTie (v1.3.3b; http://ccb.jhu.edu/software/stringtie/). Gene
expression levels were quantified as Fragments Per Kilobase of
transcript per Million mapped reads (FPKM). Differentially
expressed genes (DEGs) between PRMT7-KD and Control groups (4 vs. 4
replicates) were identified using DESeq2 (v1.16.1) in R. Genes with
an adjusted P-value (Benjamini-Hochberg FDR) <0.05 were
considered statistically significant DEGs. Gene Ontology (GO)
enrichment analysis (Biological Process, Molecular Function,
Cellular Component), Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway enrichment analysis, and Disease Ontology (DO) enrichment
analysis of the identified DEGs were performed using the
clusterProfiler R package. Gene length bias was corrected.
Terms/pathways with a corrected P-value (FDR) <0.05 were
considered significantly enriched. The Search Tool for the
Retrieval of Interacting Genes/Proteins database (https://cn.string-db.org/) was used to predict the
possible cell cycle-related regulatory proteins downstream of
PRMT7.
Statistical analysis
Data were analyzed using SPSS software (version
17.0; SPSS, Inc.). All data were presented as the mean ± SEM. The
normality of the data was analyzed using the Shapiro-Wilk test.
P<0.05 was considered to indicate normally distributed data.
Homogeneity of variance was assessed using Levene's test, with
P>0.05 considered to indicate homogenous data. For data that met
normal distribution assumptions and equal variances, an unpaired
Student's t-test was used to analyze the differences between two
groups and one-way ANOVA and Bonferroni's post hoc test for
multiple comparisons. All P-values derived from high-throughput
analyses (for example, differential gene expression and pathway
enrichment) were adjusted for multiple comparisons using the
Benjamini-Hochberg false discovery rate (FDR) method with a
significance threshold of FDR <0.05. P<0.05 was considered to
indicate a statistically significant difference. Images were
generated using GraphPad Prism 8 (Dotmatics) and Illustrator CC
2018 software (v22.0; Adobe Systems, Inc.). Principal Component
Analysis (PCA) was performed to reduce dimensionality and identify
patterns. The raw dataset was standardized by mean-centering and
scaling to unit variance. The covariance matrix of the standardized
data was computed and decomposed into its eigenvalues and
eigenvectors. Principal Components (PCs) were selected based on
eigenvalues sorted in descending order, retaining components
explaining >80% cumulative variance. The standardized data was
projected onto the orthogonal axes defined by the selected PCs.
Analysis was implemented using: Python, scikit-learn (v1.2.2) with
PCA(svd_solver=‘auto’); R, stats::prcomp() (R v4.3.0); MATLAB,
pca() (MATLAB R2023a).
Results
PRMT7 is highly expressed in PCa
To explore whether there is a potential association
between PRMT7 and PCa, relevant data were searched and analyzed in
TCGA database. Significant differences in PRMT7 expression were
identified in cancerous and adjacent tissues and in terms of the
age of patients with PCa, Gleason score, PSA levels and TNM stage
(Fig. 1A-H), which indicated that
PRMT7 may be a key gene in PCa development. To further validate the
aforementioned findings, tissues were collected from 159 patients
with PCa and the correlation between PRMT7 expression and
clinicopathological parameters were analyzed (including age,
Gleason score, PSA level and TNM stage) using tissue microarray
technology. PRMT7 was highly expressed in patients with PCa
(Figs. 1I, SIA). Significant PRMT7 expression
differences were also identified among different PSA levels in
patients with PCa, although there were no significant differences
in PRMT7 expression associated with age, Gleason score or TNM stage
(Table I). This suggested that high
PRMT7 expression is associated with worse prognosis in PCa.
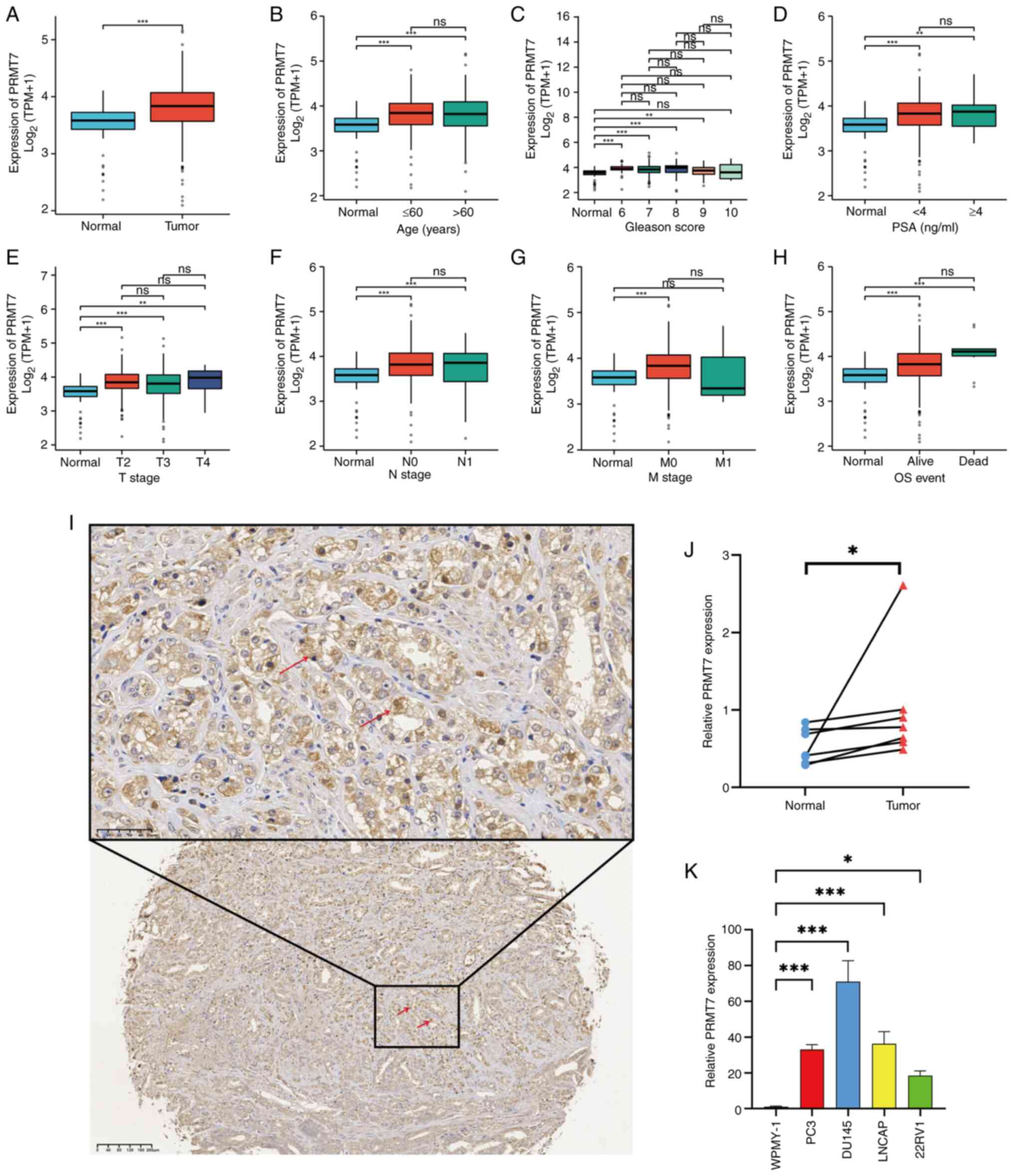 | Figure 1.Differential gene expression patterns
of PRMT7 in PCa. (A) The correlation between PRMT7 expression in
cancerous and adjacent tissues of patients with PCa in TCGA
database. (B) The correlation between PRMT7 expression and age of
patients with PCa in TCGA database. (C) The correlation between
PRMT7 expression and Gleason score of patients with PCa in TCGA
database. (D) The correlation between PRMT7 expression and PSA
levels of patients with PCa in TCGA database. The correlation
between PRMT7 expression and (E) T, (F) N and (G) M stage of
patients with PCa in TCGA database. (H) The correlation between
PRMT7 expression and OS event of patients with PCa in TCGA
database. (I) Representative image of PRMT7 immunohistochemical
staining of PCa tissues and adjacent normal tissues (magnification,
50 and 100×; scale bar, 40 and 10 µm). The red arrows indicate the
PRMT7 positive areas. (J) The expression levels of PRMT7 in PCa
tissues was detected using RT-qPCR. The blue circle represents the
normal tissue around the cancer, while the red arrow represents the
tumor tissue. (K) The expression level of PRMT7 in PCa cell lines
and normal prostate cells was detected using RT-qPCR. *P<0.05,
**P<0.01 and ***P<0.001. PRMT7, protein arginine
methyltransferase 7; PCa, prostate cancer; PSA, protein specific
antigen; TPM, transcript per million; TCGA, The Cancer Genome
Atlas; RT-qPCR, reverse transcription-quantitative PCR; ns, not
significant. |
 | Table I.Correlation between PRMT7 expression
and clinicopathological factors. |
Table I.
Correlation between PRMT7 expression
and clinicopathological factors.
|
|
| PRMT7
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Covariates | Total, n (%) | Low, n (%) | High, n (%) | P-value | Statistical
test |
|---|
| Age, years |
|
|
|
|
|
|
<70 | 91 (57.23) | 40 (56.34) | 51 (57.95) | 0.838 | χ2 |
|
≥70 | 68 (42.77) | 31 (43.67) | 37 (42.05) |
|
|
| PSA value |
|
|
|
|
|
|
<10 | 64 (40.25) | 22 (34.40) | 42 (65.60) | 0.032a | χ2 |
|
≥10 | 95 (59.75) | 49 (51.60) | 46 (48.40) |
|
|
| Gleason score |
|
|
|
|
|
| ≤7 | 114 (71.70) | 52 (73.24) | 62 (70.45) | 0.698 | χ2 |
|
>7 | 45 (28.30) | 19 (26.76) | 26 (29.55) |
|
|
| TNM stage |
|
|
|
|
|
|
T1-T2 | 157 (98.74) | 69 (97.18) | 88 (100) | 0.198 | Fisher's exact
test |
|
T3-T4 | 2 (1.26) | 2 (2.82) | 0 (0.00) |
|
|
To verify the differential upregulation of PRMT7
expression in PCa tissues, PRMT7 mRNA expression levels were
measured in seven pairs of PCa clinical specimens. PRMT7
mRNA expression levels were elevated in PCa tissues (Fig. 1J). Furthermore, compared with those
in WPMY-1 cells (a normal prostate cell line), PRMT7 mRNA
levels were significantly increased in the PCa cell lines (PC3,
DU145, LNCAP and 22RV1; Fig. 1K).
Taken together, the present study results suggested that
PRMT7 is highly expressed in PCa.
PRMT7 promotes PCa cell proliferation,
migration and invasion, and affects the cell cycle and
apoptosis
To explore the biological mechanism underlying the
effect of PRMT7 expression on PCa cells, siRNA-mediated PRMT7
silencing was performed to observe the biological changes in cancer
cells. A total of three siRNA-coding clones were designed based on
the PRMT7 sequence; they could be used for both stable and
transient expression in vertebrate cells to decrease the expression
levels of PRMT7. Subsequently, RT-qPCR was performed to verify the
silencing efficiency of the three siRNAs in PC3 and DU145 cells.
Analysis suggested that after 3 days of siRNA (43684-1, 43685-1 and
43686-1) transfection, the mRNA and protein expression levels of
PRMT7 in the experimental group (si-43686-1) was reduced (Fig. 2A). SiRNA 43686-1 demonstrated high
silencing efficiency in PC3 and DU145 cells; therefore, siRNA
43686-1 was selected for subsequent experiments. Knockdown
efficiency was also evaluated using western blotting (Fig. 2B).
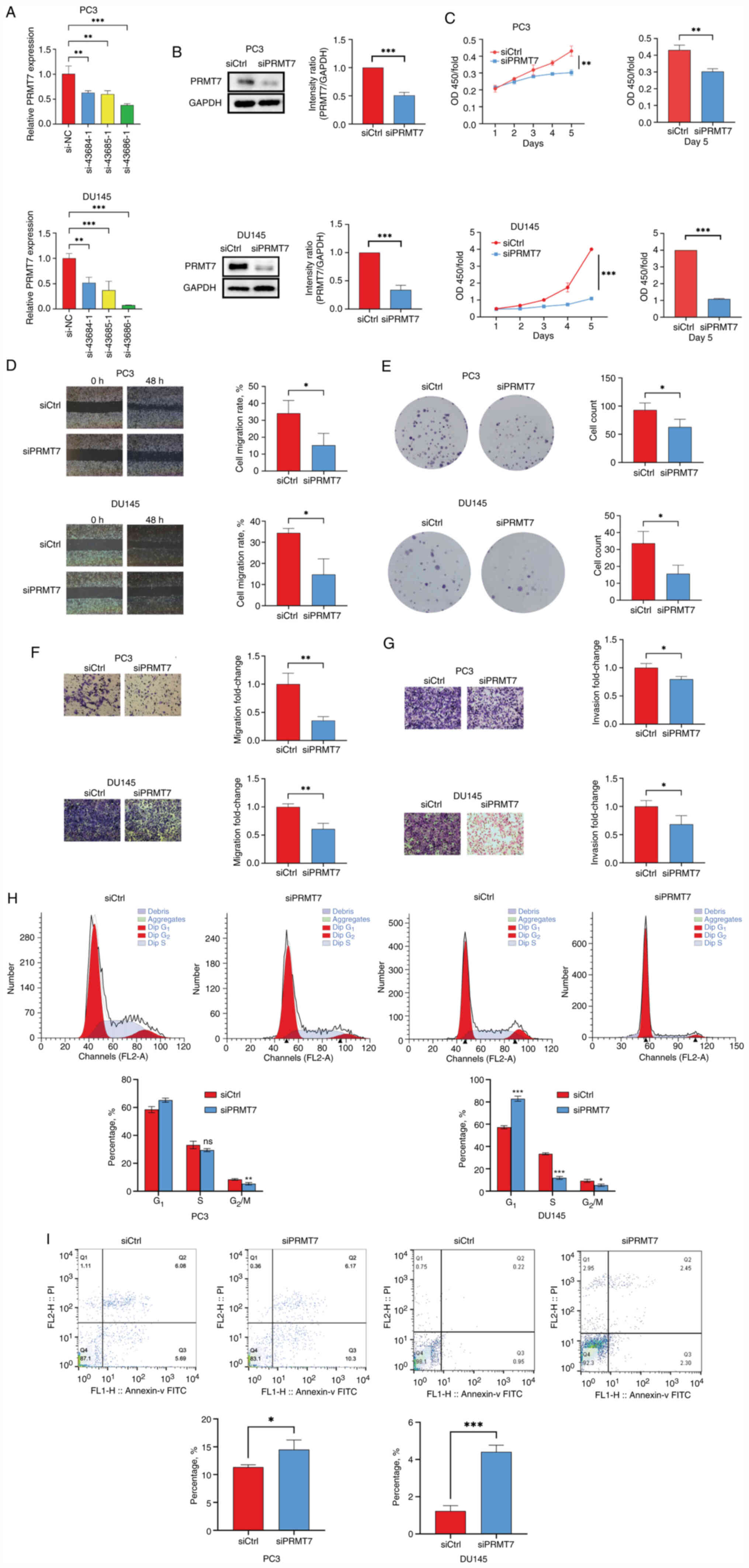 | Figure 2.Function of PRMT7 in the PC3 and
DU145 cell lines. (A) The inhibition efficiency of siPRMT7 in cell
lines was determined using RT-qPCR. (B) Western blotting was used
to detect the protein expression levels of PRMT7 in the siPRMT7 and
siCtrl groups. (C) Cell proliferation in the siCtrl and siPRMT7
groups was assessed using a Cell Counting Kit-8 assay. (D) A wound
healing assay was used to investigate the migratory ability of PCa
cells after PRMT7 knockdown (scale bar, 500 µm). (E) Plate colony
formation assays were performed to determine the proliferation
abilities of PC3 and DU145 cells treated with siCtrl and siPRMT7.
The (F) migration and (G) invasion of PC3 and DU145 cells were
investigated using a Transwell assay after PRMT7 knockdown (scale
bar, 100 µm). (H) PI-FACS detection of the effect of PRMT7
knockdown on the PCa cell cycle. (I) The effect of PRMT7 knockdown
on the apoptosis of PCa cells was detected using Annexin V-APC
single staining. *P<0.05, **P<0.01 and ***P<0.001. PRMT7,
protein arginine methyltransferase 7; PCa, prostate cancer;
RT-qPCR, reverse transcription-quantitative PCR; siRNA, small
interfering RNA; si, siRNA; Ctrl; control; NC, negative control;
CCK-8, Cell Counting Kit-8. |
CCK-8 assays indicated that the proliferation rates
of PC3 and DU145 cells were significantly reduced after PRMT7
knockdown, which suggested that PRMT7 significantly affected cell
proliferation (Fig. 2C). The
results of wound healing and migration assays suggested that PRMT7
expression is significantly associated with the migratory ability
of PC3 and DU145 cells (Fig. 2D).
PRMT7 knockdown also significantly reduced the number of PC3 and
DU145 cell clones (Fig. 2E).
However, no effects associated with proliferation and colony
formation were observed in the PRMT7-knockdown WPMY-1 cell line
(Fig. S1B and C). The results of
the invasion assay suggested that the invasion and metastatic
capacities of PC3 and DU145 cells in the experimental group were
significantly inhibited 3 days after siRNA infection, which
suggested a significant association between these cell properties
and PRMT7 expression (Fig. 2F and
G). After PRMT7 knockdown, the number of PC2 cells in the
G1 phase increased (P<0.05), while that in the
G2/M phase decreased (P<0.01). Similarly, the
proportion of DU145 cells in the G1 phase increased
(P<0.001), whereas that in the S-phase and G2/M
phases decreased (P<0.001 and P<0.05, respectively). These
results suggested that PRMT7 expression is significantly associated
with the cell cycle (Fig. 2H).
Furthermore, the rate of apoptosis in PC3 and DU145 cells was
significantly increased after transfection with siPRMT7 compared
with that in the siCtrl group, which suggested a significant
correlation between PCa with PRMT7 (Fig. 2I). These results confirmed that
PRMT7 knockdown could inhibit PC3 and DU145 cell proliferation,
migration and invasion, as well as the cell cycle and
apoptosis.
PRMT7 influences the cell cycle in PCa
cells
Next, the PRMT7 knockdown plasmid siPRMT7 was
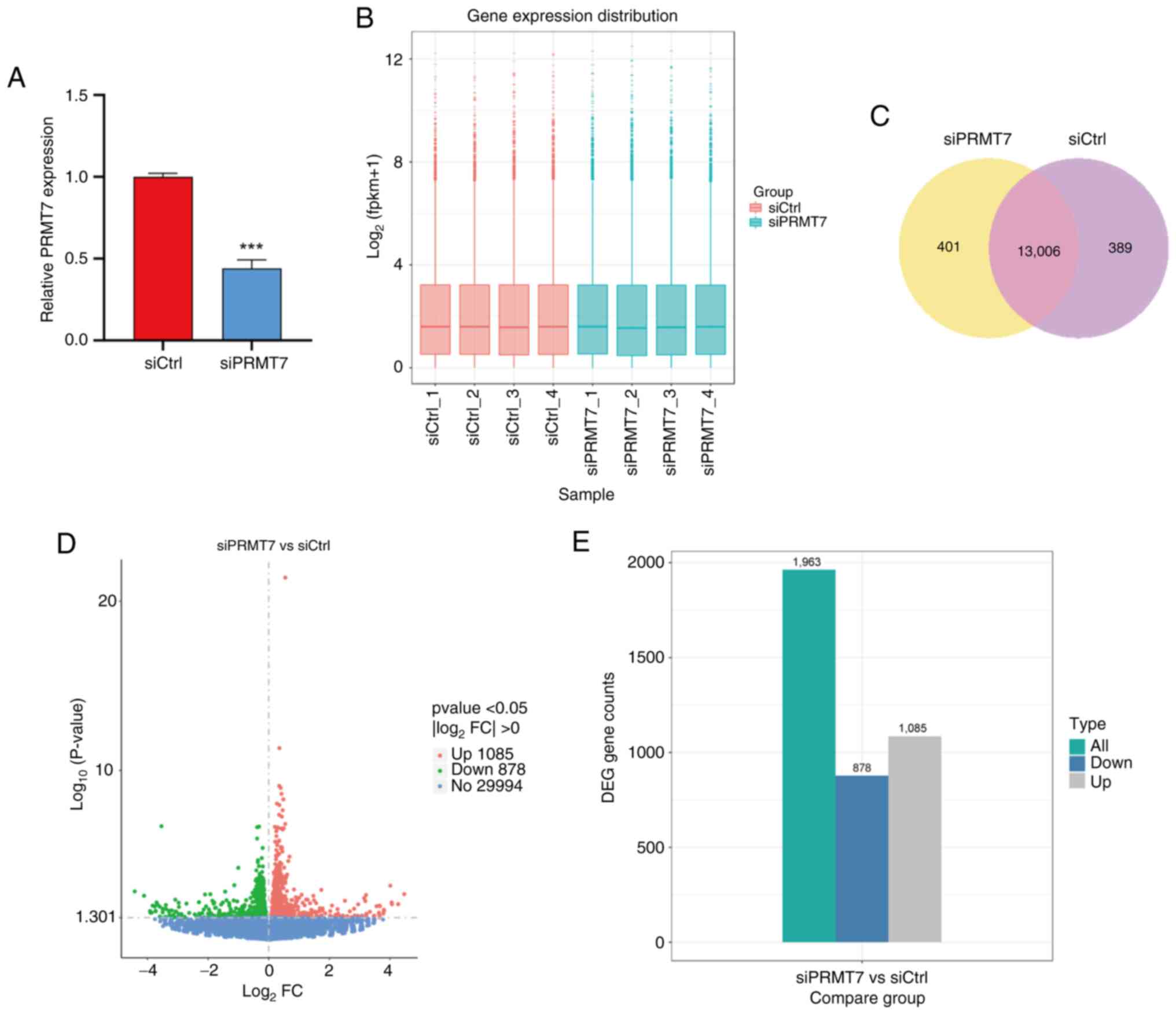
transfected into the PCa cell line PC3 (Fig. 3A). A total of four groups of siCtrl
and siPRMT7 cells were selected for transcriptome sequencing
(Fig. 3B), in which 13,006 genes
were revealed to be co-expressed between the two groups of samples
(Fig. 3C). Differential gene
screening among samples revealed that the expression levels of
1,085 genes were upregulated and expression levels of 878 genes
were downregulated in siPRMT7 cells compared with that in siCtrl
cells (Fig. 3D and E). A
differential gene set was formed by combining the differential
genes after classification. Mainstream hierarchical clustering was
used to perform cluster analysis on the FPKM values of the genes
and conducted homogenization of rows (Z-score; Figs. S2 and S3).
To explore the functional characteristics of the
differentially expressed genes and predict the downstream pathways
of PRMT7, the differentially expressed genes were classified
according to their functions and gene function enrichment analyses
were carried out, including GO, KEGG, DO and other pathway
analyses. DO analysis revealed that the differential genes were
enriched in ‘prostate cancer’ pathways, which confirmed the data
presented in Fig. 4A. Furthermore,
GO functional enrichment analysis demonstrated that the
differentially expressed genes were enriched in the ‘cell cycle’
and ‘cycle checkpoint’ functions (Fig.
4B), which was confirmed by the KEGG pathway enrichment
analysis results. KEGG is a comprehensive database that integrates
genomic, chemical and system function information. A total of 33
DEGs in PC3 cells after PRMT7 knockdown regulate the ‘cell cycle’
function (Fig. 4C and D). Thus,
PRMT7 is involved in cell cycle-related pathways in PCa cells,
which could contribute to the malignant phenotype of PCa.
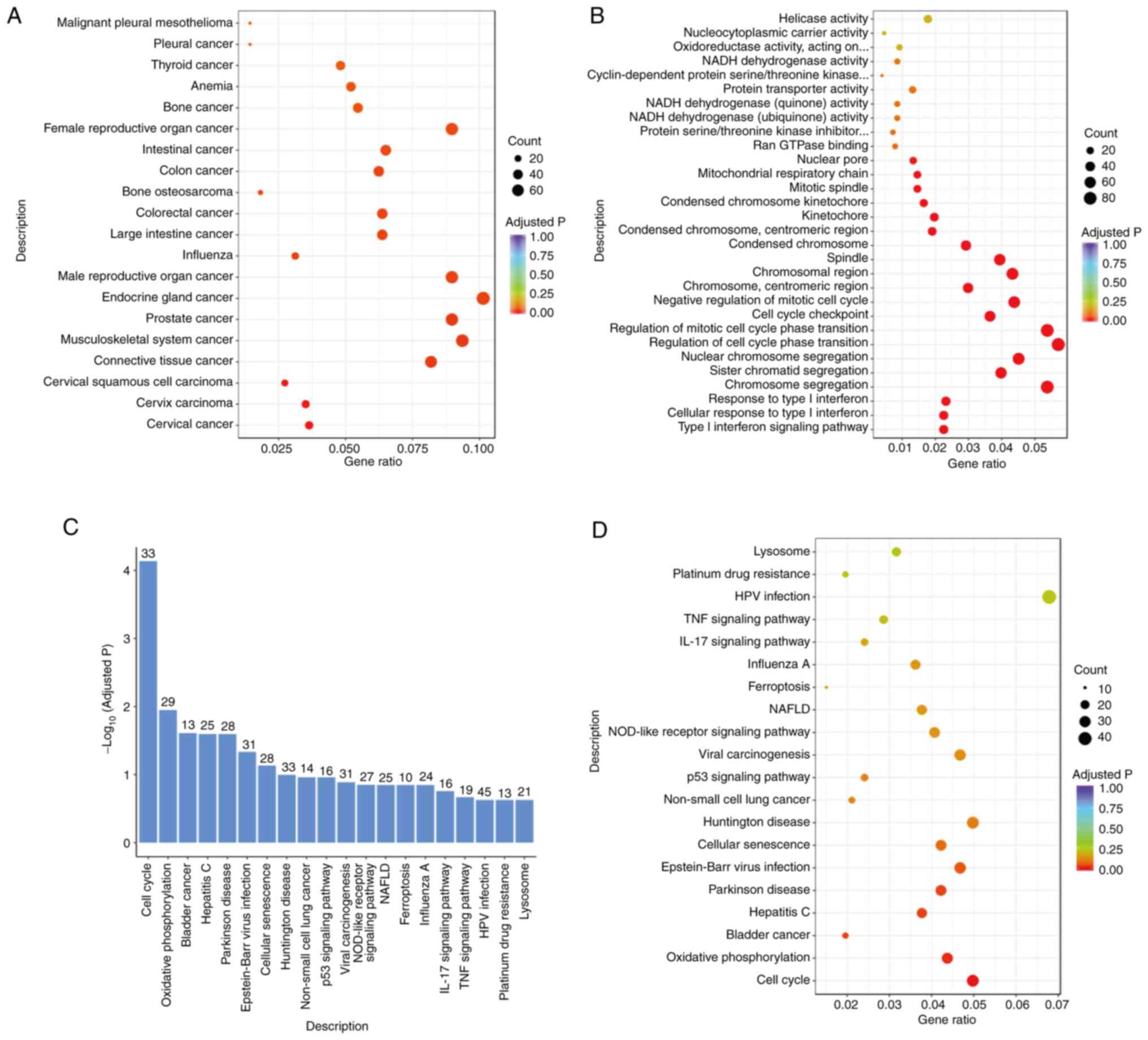 | Figure 4.PRMT7 is involved in the regulation
of cell cycle-related pathways and affects the cell cycle in PCa.
(A) The DO enrichment distribution map of the DEGs associated with
PRMT7. DO is a biomedical database that systematically annotates
associations between human gene functions and diseases, providing
standardized disease descriptors for functional genomics research.
(B) Map of the GO enrichment sites of the DEGs associated with
PRMT7. GO is a comprehensive bioinformatics resource that
systematically characterizes gene functions through three
orthogonal categories: Biological processes, cellular components
and molecular functions. (C) KEGG enrichment analysis of the DEGs
associated with PRMT7. KEGG is an integrated database that
systematically consolidates genomic, chemical and systems
functional information to facilitate biological pathway analysis
and network modeling. (D) KEGG enrichment and distribution map of
the PRMT7 differential genes. PRMT7, protein arginine
methyltransferase 7; DEGs, Differentially expressed genes;
log2FC, log2 fold change; DO, Disease
Ontology; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and
Genomes; NAFLD, Non-alcoholic fatty liver disease; NOD,
Nucleotide-binding Oligomerization Domain. |
PRMT7 promotes the malignant
progression of PCa through the YY1/TP53/CCND2/CDK6/RB1 signaling
axis
To further explore the downstream mechanisms of
PRMT7 in PCa, transcriptome sequencing data were used to map the
differentially expressed genes associated with the cell cycle
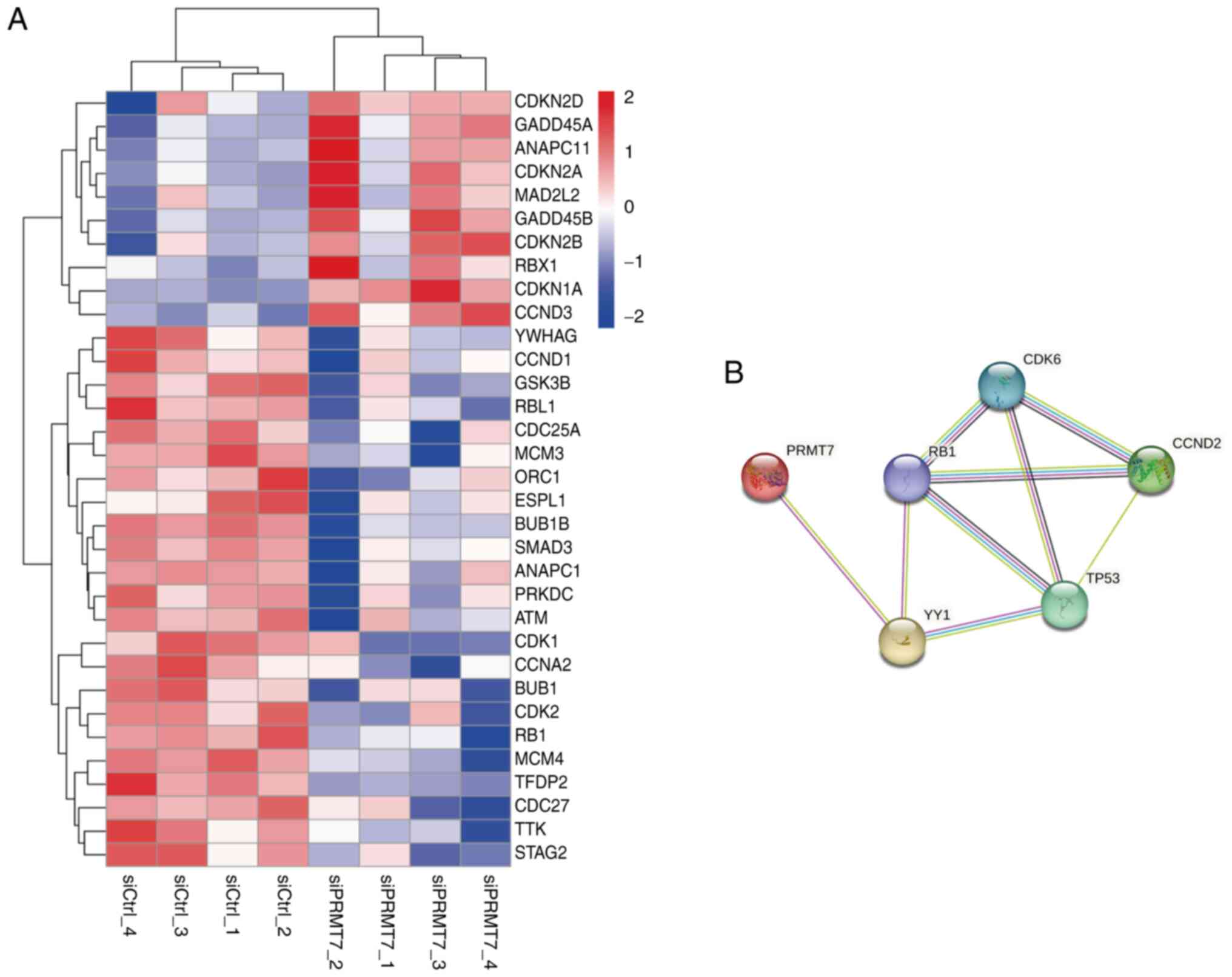
(Fig. 5A). We also predicted the
potential cell cycle-related regulatory proteins downstream of
PRMT7 (Fig. 5B). TP53 is an
important cell cycle-related protein; mutations of TP53 have been
reported to be important carcinogenic factors in PCa (12). To verify the validity of the
predicted PRMT7 cell cycle-related pathways, proteins were
collected and extracted from PC3 cells in the siPRMT7 and siCtrl
groups and the expression levels of proteins involved in the
aforementioned pathways between the two groups were compared using
western blotting. The western blot data indicated that PRMT7
knockdown may modulate the activity of the aforementioned signaling
pathways, as evidenced by altered expression levels of key pathway
markers (Fig. S4). Collectively,
the findings of the present study demonstrated that PRMT7 modulates
the activity of key cell cycle regulators, including ‘YY1’, ‘TP53’,
‘CCND2’, ‘CDK6’ and ‘RB1’ in prostate carcinogenesis, which
suggests its key role in driving cell cycle dysregulation.
Discussion
The occurrence and progression of cancer involves
several cancer-promoting and cancer-suppressing genes. With the
development of epigenetics, numerous tumor-related epigenetic
changes have become the focus of research on cancer-related
mechanisms. PTMs involved in PCa are also a current research focus
(37,38). Yao et al (30) suggested that PRMT5 inhibits the
transcription of CAMK2N1 and promotes PCa progression, which
is regulated by the circSPON2/miR-331-3p axis. PRMT5 is recruited
to AR promoters by the transcription factor Sp1 in LNCAP cell lines
to promote tumor cell proliferation (39). PRMT7, a member of the same family as
PRMT5 and the only representative of the type III group, can
monomethylate arginine. To the best of our knowledge, studies on
the mechanism of action of PRMT7 in cancer are currently limited.
Only a part of its mechanism of action in breast cancer has been
identified and these previous findings suggested that it may be an
important target for breast cancer treatment (27,40,41).
However, due to its regulatory role in cancer stem cells (42,43),
PRMT7 may also serve a key epigenetic regulatory role in several
cancer types, including PCa. Rodrigo-Faus et al (44) proposed that in metastatic
castration-resistant PCa cells, PRMT7 methylates various
transcription factors (such as forkhead box protein K1) to
reprogram the expression of several adhesion molecules, which lead
to the loss of adhesiveness in the primary tumor and increases its
migratory ability. However, the experiments of the present study
suggested a different conclusion. The present study suggested that
PRMT7 was carcinogenic in PCa and may serve a role in cell cycle
regulation of cancer cells. This was consistent with the results of
other PRMT7 studies in renal cell carcinoma (24) and human non-small-cell lung cancer
cells (45), which suggested that
PRMT7 is likely a strong tumor-associated epigenetic factor.
Based on a previous study by Vieira et al
(32), the present study explored
the specific molecular mechanism of PRMT7 in PCa to gain a more
comprehensive understanding of the activity of PRMT7 and provide
novel strategies for the clinical treatment and diagnosis of PCa.
Tissues from 159 patients with PCa were collected and the
correlation between PRMT7 expression and age, Gleason score, PSA
and TNM stage were analyzed using tissue chip technology. PRMT7
expression significantly associated with PSA levels in patients
with PCa. After analyzing the histochemical results of the patient
samples, the present study demonstrated that PRMT7 was abnormally
expressed in cancer tissues, which suggests that it may be
associated with the occurrence and progression of PCa. RT-qPCR
results of paired PCa clinical specimens suggested that PRMT7
expression was also increased in PCa tissues. Similarly, PRMT7
expression was significantly increased in PCa cells compared with
that in prostate epithelial cells.
Subsequently, siRNAs were used to knock down PRMT7
expression in PC3 and DU145 cells. PRMT7 promoted PCa cell
proliferation, migration and invasion and affected the cell cycle
and apoptosis. These results demonstrated that PRMT7 is an oncogene
that contributes to the malignant phenotype of PCa.
Furthermore, the present study explored the specific
regulatory mechanisms by which PRMT7 exerts oncogenic effects in
PCa. Bioinformatics analysis and transcriptome sequencing were used
to search for PRMT7-related differentially expressed genes in PCa
and predict the potential downstream cancer-promoting signaling
molecules. The present study determined that PRMT7 may be involved
in cell cycle control pathways, particularly the
YY1/TP53/CCND2/CDK6/RB1 signaling pathway.
p53 is a stress-activated transcription factor that
regulates the expression of target genes involved in DNA repair,
cell cycle arrest, and apoptosis to maintain genomic stability and
suppress tumorigenesis (46).
Mutations in the p53 gene have been reported in 50% of all human
malignancies, including breast, colon, lung, liver, prostate,
bladder and skin cancer (47). p53
orchestrates cell cycle checkpoints (G1/S, G2/M arrest),
senescence, and metabolic adaptation to preserve genomic integrity.
As a tumor suppressor, it is indispensable for eliminating damaged
normal cells, while its dysfunction in cancer promotes genomic
instability, therapy resistance, and metastasis (46,48–52).
However, the results of the present study indicated that PRMT7
depletion did not significantly affect TP53 protein expression
levels. PC3 cells harbor homozygous deletion of TP53, which
explains the lack of detectable changes. This confirms that the
pro-proliferative effects of PRMT7 in PC3 cells are independent of
canonical p53 pathways. However, PRMT7 may still engage alternative
survival pathways (for example, MDM2 and AKT) in p53-null cells to
bypass p53-mediated cell cycle checkpoints.
YY1 is a member of the GLI-Kruppel zinc finger
protein family that has two opposing abilities: Inhibiting and
activating gene transcription (53). PRMT7 interacts with YY1, which leads
to breast cancer cell migration and invasion (26). In the present study, YY1 exhibited a
small yet statistically significant decrease in expression upon
PRMT7 knockdown. PRMT7 may methylate YY1 to stabilize its protein
or enhance its transcriptional activity. Loss of PRMT7-dependent
methylation could impair the ability of YY1 to activate downstream
targets, including cell cycle regulators such as CCND2/CDK6
(54–56). In indirect regulation, YY1
downregulation might reflect secondary effects of PRMT7 knockdown,
such as altered chromatin accessibility at YY1-binding loci or
dysregulation of upstream signaling pathways (56,57).
This mild reduction suggests YY1 is not the primary effector of
PRMT7 in PC3 cells, but its partial loss may synergize with other
pathways to amplify CCND2/CDK6 suppression.
CCND2 is a member of the cyclin family that encodes
cyclin D2. It regulates cancer cell processes through the cell
cycle. Cyclin D2 reduces the inhibitory effect of miR-615 on the
proliferation, migration and invasion of PCa cells (58). The expression levels of E2F
transcription factor 2 and CCND2 was downregulated by let-7a; the
latter may inhibit PCa growth in an in vivo PCa xenograft
model (59). CCND2 promotes PCa
development and CDK6 and RB1 are well-known cell cycle regulators;
several studies have reported that targeting them can affect the
cell cycle of PCa cells and inhibit their proliferation (60–63).
Analysis of the western blotting experiments in the present study
demonstrated that CCND2 and CDK6 levels were markedly downregulated
following PRMT7 depletion. PRMT7-mediated symmetric dimethylation
of histone H4R3 (H4R3me2s) is key for maintaining an active
chromatin state at the CCND2 promoter. PRMT7 knockdown may induce
heterochromatin formation, which represses transcription. PRMT7
might methylate RNA-binding proteins to stabilize CCND2 mRNA; its
absence could accelerate mRNA degradation. PRMT7 may methylate
histones (for example, H3R2me2s) at the CDK6 promoter to facilitate
transcriptional elongation by RNA Polymerase II. Furthermore, YY1
has been reported to bind the CDK6 promoter (64). The concurrent downregulation of YY1
and CDK6 expression suggests a PRMT7-YY1-CDK6 regulatory axis. In
the present study, total RB1 protein levels demonstrated no
significant alteration after PRMT7 knockdown. RB1 function is
primarily governed by phosphorylation status compared with total
protein abundance. The observed CCND2/CDK6 downregulation likely
reduces RB1 phosphorylation, activating its growth-suppressive
function without altering total RB1 levels. PRMT7 may methylate RB1
to modulate its interaction with E2F or chromatin remodelers.
However, such modifications might not affect total RB1 stability,
which necessitates phospho-specific assays. Therefore, these
results suggested that PRMT7 likely serves a role in TP53-related
cell cycle pathways, ultimately contributing to PCa
progression.
Previous studies on blocking AR resistance have
indicated the role of AR-related cell cycle pathways in PCa
(65–67). The binding of androgens to their
receptors induces cell cycle progression by directly affecting the
transcription-regulated expression of cell cycle regulatory
proteins (68). This involves
ligand-activated AR signaling regulation of the cell cycle, which
causes androgen-deprived ADT-sensitive cells to exit the cell cycle
and stall in the G0 phase (69–71).
As CDK complexes control transition within the cell cycle and
maintain progression from the G1 phase to mitosis
(72–74), AR can promote tumor cell
proliferation by affecting the activity of CDKs and CDK inhibitors.
After androgen blocking, the expression of major D-type cyclin is
inhibited in ADT-reactive PCa cells, which results in cell cycle
arrest and the inhibition of cell proliferation (71,75).
The growth factor-mediated accumulation of D-type cyclins in cells
further induces CDK4/6 activity and maintains the cell cycle
(74), which initiates the
phosphorylation/inactivation of the RB tumor suppressor and
blocking the RB-mediated negative regulation of cell cycle
transition and DNA replication (76). These results suggested that further
research on cell cycle changes in PCa could help to elucidate the
possible mechanism of androgen-blocking drug resistance and
tumorigenesis.
The present study had several limitations. For
instance, the sample collection period was relatively short (~6
months). Although an adequate sample size was achieved from this
high-volume tertiary hospital (Affiliated Hospital of Jiangnan
University; Wuxi, China), the potential for temporal bias requires
consideration. Furthermore, the present study primarily relied on
in vitro experiments. Future research should validate these
findings through multicenter cohorts and further investigate the
molecular mechanisms underlying PRMT7-mediated cell cycle
regulation. Ideally, a larger and directly matched control group
would enhance the reliability of the findings. Furthermore, future
collaborations with other institutions aim to obtain a more
comprehensive dataset, thereby improving the external validity of
the present study results. Although the present study revealed that
PRMT7 modulates cell cycle progression in PCa cells through a
specific signaling axis, the mechanisms underlying cell cycle
G0 arrest and the downstream phosphorylation of
associated factors remains to be elucidated. Relying on
bioinformatics prediction methods, specific research directions
were identified to study the mechanism by which PRMT7 promotes PCa.
These findings suggested a functional involvement of PRMT7 in
modulating PCa cell cycle progression, but more rigorous
mechanistic investigations are required to fully delineate its
underlying molecular actions. However, due to the lack of further
evidence, the present study was unable to confirm the precise
mechanism by which PRMT7 affected cell cycle-related factors such
as YY1, TP53, CCND2, CDK6 and RB1 in PCa. As members of the same
enzyme family, PRMT7 and other PRMTs may exhibit synergistic or
antagonistic interactions in vivo. Notably, studies have
reported that PRMT7-mediated monomethylation of histone H4 at
Arg-17 can directly modulate PRMT5-catalyzed symmetric
dimethylation at Arg-3 within the same histone protein (77,78).
Therefore, investigating the functional role of PRMT7 in PCa
progression must account for the coordinated or competitive
activities of other PRMT family members, particularly in regulating
cell cycle checkpoints through key effectors such as RB1
phosphorylation and CDK6 activation. These mechanisms require
further investigation in future studies. Future experiments will
include exploring the specific mechanism of PRMT7 in the PCa cell
cycle and identifying effective molecularly targeted drugs
associated with PRMT7 to provide research directions for the
precise treatment of PCa.
The present study confirmed that PRMT7 was involved
in the proliferation and invasion of PCa cells and potentially
promoted the malignant progression through the
YY1/TP53/CCND2/CDK6/RB1 cell cycle signaling axis.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the following
entities: The National Natural Science Foundation (grant no.
81802576), Wuxi Commission of Health and Family Planning (grant
nos. T202102, Z202011 and M202330) and Talent plan of Taihu Lake in
Wuxi (Double Hundred Medical Youth Professionals Program) from
Health Committee of Wuxi (grant nos. BJ2020061 and BJ2023051).
Furthermore, the Clinical Trial of Affiliated Hospital of Jiangnan
University (grant nos. LCYJ202227 and LCYJ202323), Research Topic
of Jiangsu Health Commission (grant no. Z2022047) and Jiangsu
Province 7th phase ‘333’ high-level talents [grant no.
(2024)3-2425] also provided funding support for the present
study.
Availability of data and materials
The data generated in the present study may be found
in the Gene Expression Omnibus under accession number (GSE302155)
or at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE302155.
The remaining data generated in the present study may be requested
from the corresponding author.
Authors' contributions
HW, HZ, JG, NF and DY conceived and designed the
present study; HW, RW, JN and YM performed the experiments and
acquired the data. SW, YQ, QQ and LZ contributed to data analysis.
HW and YM confirm the authenticity of all the raw data. All authors
were involved in drafting and revising the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Affiliated Hospital of Jiangnan University (approval
no. LS2023099; Wuxi, China) and written informed consent was
obtained from all patients prior to sample collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Giaquinto AN and Jemal A:
Cancer statistics, 2024. CA Cancer J Clin. 74:12–49.
2024.PubMed/NCBI
|
|
2
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
3
|
Sugiura M, Sato H, Kanesaka M, Imamura Y,
Sakamoto S, Ichikawa T and Kaneda A: Epigenetic modifications in
prostate cancer. Int J Urol. 28:140–149. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu J, Dong L, Zhu Y, Dong B, Sha J, Zhu
HH, Pan J and Xue W: Prostate cancer treatment-China's perspective.
Cancer Lett. 550:2159272022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu X, Yu C, Bi Y and Zhang ZJ: Trends and
age-period-cohort effect on incidence and mortality of prostate
cancer from 1990 to 2017 in China. Public Health. 172:70–80. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang F, Wang C, Xia H, Lin Y, Zhang D, Yin
P and Yao S: Burden of prostate cancer in China, 1990–2019:
Findings from the 2019 global burden of disease study. Front
Endocrinol (Lausanne). 13:8536232022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Han B, Zheng R, Zeng H, Wang S, Sun K,
Chen R, Li L, Wei W and He J: Cancer incidence and mortality in
China, 2022. J Natl Cancer Cent. 4:47–53. 2024.PubMed/NCBI
|
|
8
|
Cao W, Chen HD, Yu YW, Li N and Chen WQ:
Changing profiles of cancer burden worldwide and in China: A
secondary analysis of the global cancer statistics 2020. Chin Med J
(Engl). 134:783–791. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al: Global surveillance of trends in cancer survival
2000–14 (CONCORD-3): Analysis of individual records for 37 513 025
patients diagnosed with one of 18 cancers from 322 population-based
registries in 71 countries. Lancet. 391:1023–1075. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pound CR, Partin AW, Eisenberger MA, Chan
DW, Pearson JD and Walsh PC: Natural history of progression after
PSA elevation following radical prostatectomy. JAMA. 281:1591–1597.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lobo J, Barros-Silva D, Henrique R and
Jeronimo C: The emerging role of epitranscriptomics in cancer:
Focus on urological tumors. Genes (Basel). 9:5522018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rebello RJ, Oing C, Knudsen KE, Loeb S,
Johnson DC, Reiter RE, Gillessen S, Van der Kwast T and Bristow RG:
Prostate cancer. Nat Rev Dis Primers. 7:92021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Barsouk A, Padala SA, Vakiti A, Mohammed
A, Saginala K, Thandra KC, Rawla P and Barsouk A: Epidemiology,
staging and management of prostate cancer. Med Sci (Basel).
8:282020.PubMed/NCBI
|
|
14
|
Feldman BJ and Feldman D: The development
of androgen-independent prostate cancer. Nat Rev Cancer. 1:34–45.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hirst M and Marra MA: Epigenetics and
human disease. Int J Biochem Cell Biol. 41:136–146. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Miremadi A, Oestergaard MZ, Pharoah PD and
Caldas C: Cancer genetics of epigenetic genes. Hum Mol Genet.
16:R28–R49. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fisk JC, Sayegh J, Zurita-Lopez C, Menon
S, Presnyak V, Clarke SG and Read LK: A type III protein arginine
methyltransferase from the protozoan parasite Trypanosoma brucei. J
Biol Chem. 284:11590–11600. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Feng Y, Hadjikyriacou A and Clarke SG:
Substrate specificity of human protein arginine methyltransferase 7
(PRMT7): The importance of acidic residues in the double E loop. J
Biol Chem. 289:32604–32616. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zurita-Lopez CI, Sandberg T, Kelly R and
Clarke SG: Human protein arginine methyltransferase 7 (PRMT7) is a
type III enzyme forming omega-NG-monomethylated arginine residues.
J Biol Chem. 287:7859–7870. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Debler EW, Jain K, Warmack RA, Feng Y,
Clarke SG, Blobel G and Stavropoulos P: A glutamate/aspartate
switch controls product specificity in a protein arginine
methyltransferase. Proc Natl Acad Sci USA. 113:2068–2073. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bedford MT and Richard S: Arginine
methylation an emerging regulator of protein function. Mol Cell.
18:263–272. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bedford MT and Clarke SG: Protein arginine
methylation in mammals: Who, what, and why. Mol Cell. 33:1–13.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang Y and Bedford MT: Protein arginine
methyltransferases and cancer. Nat Rev Cancer. 13:37–50. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu F, Wan L, Zou H, Pan Z, Zhou W and Lu
X: PRMT7 promotes the growth of renal cell carcinoma through
modulating the beta-catenin/C-MYC axis. Int J Biochem Cell Biol.
120:1056862020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Baldwin RM, Haghandish N, Daneshmand M,
Amin S, Paris G, Falls TJ, Bell JC, Islam S and Côté J: Protein
arginine methyltransferase 7 promotes breast cancer cell invasion
through the induction of MMP9 expression. Oncotarget. 6:3013–3032.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yao R, Jiang H, Ma Y, Wang L, Wang L, Du
J, Hou P, Gao Y, Zhao L, Wang G, et al: PRMT7 induces
epithelial-to-mesenchymal transition and promotes metastasis in
breast cancer. Cancer Res. 74:5656–5667. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma W, Sun X, Zhang S, Chen Z and Yu J:
Circ_0039960 regulates growth and Warburg effect of breast cancer
cells via modulating miR-1178/PRMT7 axis. Mol Cell Probes.
64:1018292022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fuhrmann J and Thompson PR: Protein
arginine methylation and citrullination in epigenetic regulation.
ACS Chem Biol. 11:654–668. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fuhrmann J, Clancy KW and Thompson PR:
Chemical biology of protein arginine modifications in epigenetic
regulation. Chem Rev. 115:5413–5461. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yao B, Zhu S, Wei X, Chen MK, Feng Y, Li
Z, Xu X, Zhang Y, Wang Y, Zhou J, et al: The circSPON2/miR-331-3p
axis regulates PRMT5, an epigenetic regulator of CAMK2N1
transcription and prostate cancer progression. Mol Cancer.
21:1192022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Beketova E, Fang S, Owens JL, Liu S, Chen
X, Zhang Q, Asberry AM, Deng X, Malola J, Huang J, et al: Protein
arginine methyltransferase 5 promotes pICln-dependent androgen
receptor transcription in castration-resistant prostate cancer.
Cancer Res. 80:4904–4917. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vieira FQ, Costa-Pinheiro P,
Ramalho-Carvalho J, Pereira A, Menezes FD, Antunes L, Carneiro I,
Oliveira J, Henrique R and Jerónimo C: Deregulated expression of
selected histone methylases and demethylases in prostate carcinoma.
Endocr Relat Cancer. 21:51–61. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Epstein JI, Egevad L, Amin MB, Delahunt B,
Srigley JR and Humphrey PA: The 2014 international society of
urological pathology (ISUP) consensus conference on gleason grading
of prostatic carcinoma: Definition of grading patterns and proposal
for a new grading system. Am J Surg Pathol. 40:244–252. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xiao WJ, Zhu Y, Zhu Y, Dai B and Ye DW:
Evaluation of clinical staging of the American Joint Committee on
Cancer (eighth edition) for prostate cancer. World J Urol.
36:769–774. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Feng Y, Sun C, Zhang L, Wan H, Zhou H,
Chen Y, Zhu L, Xia G and Mi Y: Upregulation of COPB2 promotes
prostate cancer proliferation and invasion through the MAPK/TGF-β
signaling pathway. Front Oncol. 12:8653172022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mi YY, Ji Y, Zhang L, Sun CY, Wei BB, Yang
DJ, Wan HY, Qi XW, Wu S and Zhu LJ: A first-in-class HBO1 inhibitor
WM-3835 inhibits castration-resistant prostate cancer cell growth
in vitro and in vivo. Cell Death Dis. 14:672023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jaiswal B, Agarwal A and Gupta A: Lysine
acetyltransferases and their role in AR signaling and prostate
cancer. Front Endocrinol (Lausanne). 13:8865942022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cocchiola R, Romaniello D, Grillo C,
Altieri F, Liberti M, Magliocca FM, Chichiarelli S, Marrocco I,
Borgoni G, Perugia G and Eufemi M: Analysis of STAT3
post-translational modifications (PTMs) in human prostate cancer
with different Gleason Score. Oncotarget. 8:42560–42570. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Deng X, Shao G, Zhang HT, Li C, Zhang D,
Cheng L, Elzey BD, Pili R, Ratliff TL, Huang J and Hu CD: Protein
arginine methyltransferase 5 functions as an epigenetic activator
of the androgen receptor to promote prostate cancer cell growth.
Oncogene. 36:1223–1231. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu L, Zhang X, Ding H, Liu X, Cao D, Liu
Y, Liu J, Lin C, Zhang N, Wang G, et al: Arginine and lysine
methylation of MRPS23 promotes breast cancer metastasis through
regulating OXPHOS. Oncogene. 40:3548–3563. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu Y, Li L, Liu X, Wang Y, Liu L, Peng L,
Liu J, Zhang L, Wang G, Li H, et al: Arginine methylation of SHANK2
by PRMT7 promotes human breast cancer metastasis through activating
endosomal FAK signalling. Elife. 9:e576172020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nicot C: PRMT7: A survive-or-die switch in
cancer stem cells. Mol Cancer. 21:1272022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu C, Zou W, Nie D, Li S, Duan C, Zhou M,
Lai P, Yang S, Ji S, Li Y, et al: Loss of PRMT7 reprograms glycine
metabolism to selectively eradicate leukemia stem cells in CML.
Cell Metab. 34:818–35.e7. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rodrigo-Faus M, Vincelle-Nieto A, Vidal N,
Puente J, Saiz-Pardo M, Lopez-Garcia A, Mendiburu-Eliçabe M, Palao
N, Baquero C, Linzoain-Agos P, et al: CRISPR/Cas9 screenings
unearth protein arginine methyltransferase 7 as a novel essential
gene in prostate cancer metastasis. Cancer Lett. 588:2167762024.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cheng D, He Z, Zheng L, Xie D, Dong S and
Zhang P: PRMT7 contributes to the metastasis phenotype in human
non-small-cell lung cancer cells possibly through the interaction
with HSPA5 and EEF2. Onco Targets Ther. 11:4869–4876. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen J: The Cell-cycle arrest and
apoptotic functions of p53 in tumor initiation and progression.
Cold Spring Harb Perspect Med. 6:a0261042016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Marei HE, Althani A, Afifi N, Hasan A,
Caceci T, Pozzoli G, Morrione A, Giordano A and Cenciarelli C: p53
signaling in cancer progression and therapy. Cancer Cell Int.
21:7032021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hashimoto N, Nagano H and Tanaka T: The
role of tumor suppressor p53 in metabolism and energy regulation,
and its implication in cancer and lifestyle-related diseases.
Endocr J. 66:485–496. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu J, Zhang C, Hu W and Feng Z: Tumor
suppressor p53 and metabolism. J Mol Cell Biol. 11:284–292. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang H, Xu J, Long Y, Maimaitijiang A, Su
Z, Li W and Li J: Unraveling the guardian: P53′s multifaceted role
in the DNA Damage response and tumor treatment strategies. Int J
Mol Sci. 25:129282024. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mirzayans R, Andrais B, Kumar P and Murray
D: Significance of Wild-type p53 signaling in suppressing apoptosis
in response to chemical genotoxic agents: Impact on chemotherapy
outcome. Int J Mol Sci. 18:9282017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Madan E, Gogna R, Bhatt M, Pati U,
Kuppusamy P and Mahdi AA: Regulation of glucose metabolism by p53:
Emerging new roles for the tumor suppressor. Oncotarget. 2:948–957.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang W, Li D and Sui G: YY1 is an inducer
of cancer metastasis. Crit Rev Oncog. 22:1–11. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li Z, Wang F, Tian X, Long J, Ling B,
Zhang W, Xu J and Liang A: HCK maintains the self-renewal of
leukaemia stem cells via CDK6 in AML. J Exp Clin Cancer Res.
40:2102021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Buontempo F, McCubrey JA, Orsini E,
Ruzzene M, Cappellini A, Lonetti A, Evangelisti C, Chiarini F,
Evangelisti C, Barata JT and Martelli AM: Therapeutic targeting of
CK2 in acute and chronic leukemias. Leukemia. 32:1–10. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Rizkallah R, Alexander KE, Kassardjian A,
Lüscher B and Hurt MM: The transcription factor YY1 is a substrate
for Polo-like kinase 1 at the G2/M transition of the cell cycle.
PLoS One. 6:e159282011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Dong X, Guo R, Ji T, Zhang J, Xu J, Li Y,
Sheng Y, Wang Y, Fang K, Wen Y, et al: YY1 safeguard
multidimensional epigenetic landscape associated with extended
pluripotency. Nucleic Acids Res. 50:12019–12038. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Huang F, Zhao H, Du Z and Jiang H: miR-615
inhibits prostate cancer cell proliferation and invasion by
directly targeting Cyclin D2. Oncol Res. 27:293–299. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Dong Q, Meng P, Wang T, Qin W, Qin W, Wang
F, Yuan J, Chen Z, Yang A and Wang H: MicroRNA let-7a inhibits
proliferation of human prostate cancer cells in vitro and in vivo
by targeting E2F2 and CCND2. PLoS One. 5:e101472010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chen X, Wu Y, Wang X, Xu C, Wang L, Jian
J, Wu D and Wu G: CDK6 is upregulated and may be a potential
therapeutic target in enzalutamide-resistant castration-resistant
prostate cancer. Eur J Med Res. 27:1052022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yu Z, Zhan C, Du H and Zhang L, Liang C
and Zhang L: Baicalin suppresses the cell cycle progression and
proliferation of prostate cancer cells through the CDK6/FOXM1 axis.
Mol Cell Biochem. 469:169–178. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chakraborty G, Armenia J, Mazzu YZ,
Nandakumar S, Stopsack KH, Atiq MO, Komura K, Jehane L, Hirani R,
Chadalavada K, et al: Significance of BRCA2 and RB1 Co-loss in
aggressive prostate cancer progression. Clin Cancer Res.
26:2047–2064. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ku SY, Rosario S, Wang Y, Mu P, Seshadri
M, Goodrich ZW, Goodrich MM, Labbé DP, Gomez EC, Wang J, et al: Rb1
and Trp53 cooperate to suppress prostate cancer lineage plasticity,
metastasis, and antiandrogen resistance. Science. 355:78–83. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Li Y, Wang F, Xu J, Ye F, Shen Y, Zhou J,
Lu W, Wan X, Ma D and Xie X: Progressive miRNA expression profiles
in cervical carcinogenesis and identification of HPV-related target
genes for miR-29. J Pathol. 224:484–495. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
McNair C, Urbanucci A, Comstock CE,
Augello MA, Goodwin JF, Launchbury R, Zhao SG, Schiewer MJ, Ertel
A, Karnes J, et al: Cell cycle-coupled expansion of AR activity
promotes cancer progression. Oncogene. 36:1655–1668. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Oner M, Lin E, Chen MC, Hsu FN, Shazzad
Hossain Prince GM, Chiu KY, Teng CJ, Yang TY, Wang HY, Yue CH, et
al: Future aspects of CDK5 in prostate cancer: From pathogenesis to
therapeutic implications. Int J Mol Sci. 20:38812019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zolochevska O and Figueiredo ML: Cell
cycle regulator cdk2ap1 inhibits prostate cancer cell growth and
modifies androgen-responsive pathway function. Prostate.
69:1586–1597. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Schiewer MJ, Augello MA and Knudsen KE:
The AR dependent cell cycle: Mechanisms and cancer relevance. Mol
Cell Endocrinol. 352:34–45. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Agus DB, Cordon-Cardo C, Fox W, Drobnjak
M, Koff A, Golde DW and Scher HI: Prostate cancer cell cycle
regulators: Response to androgen withdrawal and development of
androgen independence. J Natl Cancer Inst. 91:1869–1876. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Huggins C and Hodges CV: Studies on
prostatic cancer: I. The effect of castration, of estrogen and of
androgen injection on serum phosphatases in metastatic carcinoma of
the prostate. 1941. J Urol. 168:9–12. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Knudsen KE, Arden KC and Cavenee WK:
Multiple G1 regulatory elements control the androgen-dependent
proliferation of prostatic carcinoma cells. J Biol Chem.
273:20213–20222. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lee YM and Sicinski P: Targeting cyclins
and cyclin-dependent kinases in cancer: Lessons from mice, hopes
for therapeutic applications in human. Cell Cycle. 5:2110–2114.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Malumbres M and Barbacid M: Cell cycle
kinases in cancer. Curr Opin Genet Dev. 17:60–65. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Sherr CJ and Roberts JM: Living with or
without cyclins and cyclin-dependent kinases. Genes Dev.
18:2699–2711. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Xu Y, Chen SY, Ross KN and Balk SP:
Androgens induce prostate cancer cell proliferation through
mammalian target of rapamycin activation and post-Transcriptional
increases in cyclin D proteins. Cancer Res. 66:7783–7792. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Knudsen ES and Knudsen KE: Tailoring to
RB: Tumour suppressor status and therapeutic response. Nat Rev
Cancer. 8:714–724. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Jain K, Jin CY and Clarke SG: Epigenetic
control via allosteric regulation of mammalian protein arginine
methyltransferases. Proc Natl Acad Sci USA. 114:10101–10106. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Jain K and Clarke SG: PRMT7 as a unique
member of the protein arginine methyltransferase family: A review.
Arch Biochem Biophys. 665:36–45. 2019. View Article : Google Scholar : PubMed/NCBI
|