Introduction
Metabolism serves as the fundamental basis of
cellular life, providing energy for biological processes, enabling
the biosynthesis of macromolecules and maintaining redox
homeostasis (1). The cellular
metabolic network, comprising the four core pathways, amino acid,
glucose, lipid and nucleotide metabolism, forms the structural and
functional foundation of cellular biochemistry (2). Amino acid metabolism occupies a
particularly pivotal position within this network due to its
notable diversity in molecular species and metabolic routes,
enabling its critical involvement in multiple cellular processes
(3–8). Emerging evidence has revealed that
deficiencies in essential amino acids, including tryptophan,
tyrosine, branched-chain amino acids (such as isoleucine, leucine
and valine), methionine, phenylalanine and threonine, can disrupt
lipid homeostasis, with potential implications for
neurodevelopmental disorders (9).
In diabetic murine models, depletion of serine and glycine has been
demonstrated to exacerbate neuropathy progression (10). A mechanistic study has further shown
that essential amino acid deficiency in hepatocytes leads to
functional impairment of ubiquitin protein ligase E3 component
n-recognin 1, causing aberrant ubiquitination and subsequent
degradation of the lipid droplet-stabilizing protein, perilipin 2.
This molecular cascade ultimately suppresses hepatic lipid
catabolism and promotes fatty liver pathogenesis (11).
To sustain their rapid proliferation, tumor cells
must overcome three fundamental metabolic demands: i) Rapid energy
production; ii) biosynthesis of macromolecules; and iii)
maintenance of redox homeostasis (1). Amino acids serve as essential
providers of both carbon and nitrogen sources, playing pivotal
roles in meeting these metabolic challenges. Consequently, amino
acid restriction therapies have emerged as a promising therapeutic
strategy against tumors (12–17).
Notably, the sulfur-containing amino acid, cysteine, assumes
particular importance in tumor metabolism, serving as a critical
precursor for glutathione (GSH) synthesis and providing cellular
protection against ferroptosis (18,19).
Ferroptosis is an iron-dependent form of regulated cell death
characterized by three key metabolic features: i) The glutathione
peroxidase 4/GSH antioxidant system (dependent on cysteine
availability); ii) Fe2+-mediated Fenton reactions; and
iii) acyl-CoA synthetase long chain family member 4
(ACSL4)-catalyzed biosynthesis of polyunsaturated fatty
acid-containing phospholipids (PUFA-PLs) (20). Notably, cellular cysteine can be
acquired through two distinct pathways: Exogenous uptake via the
solute carrier family 7 member 11 (SLC7A11) transporter or
endogenous synthesis from methionine via the methionine
adenosyltransferase 2A (MAT2A)-initiated transsulfuration pathway.
Both SLC7A11 and MAT2A are frequently upregulated in tumors, making
them attractive therapeutic targets (21–25).
Notably, ACSL4 plays a contradictory role in the occurrence and
development of tumors. On the one hand, ACSL4 can provide
biomacromolecules for tumor proliferation by regulating the
synthesis of PUFA-PLs (26). On the
other hand, an increase in PUFA-PLs levels also increases the
sensitivity of tumor cells to ferroptosis (27). Beyond their canonical roles in
protein biosynthesis and cellular metabolism, amino acids serve as
pivotal allosteric modulators that fine-tune metabolic flux through
the direct regulation of rate-limiting enzymes (28). While the crosstalk between amino
acid metabolism and glucose homeostasis has been extensively
characterized, particularly through the dynamic interconversion of
non-essential amino acid networks, the unidirectional nature of
amino acid-lipid metabolic interactions has resulted in
comparatively limited understanding of the amino acid-mediated
regulation of lipid metabolism.
The present study aimed to systematically
investigate the metabolic consequences of essential amino acid
deprivation, with particular emphasis on delineating the
mechanistic role of methionine in regulating intracellular lipid
homeostasis. To assess alterations in lipid metabolism, flow
cytometry coupled with fluorescent lipid probes were employed to
evaluate intracellular lipid accumulation and cellular fatty acid
uptake efficiency following essential amino acid deprivation.
Comprehensive analyses were performed to elucidate the underlying
molecular mechanisms of methionine-mediated lipid metabolic
regulation. Western blotting and reverse transcription-quantitative
PCR (RT-qPCR) were conducted to examine methionine
restriction-induced modulation of ACSL4 expression, a critical
regulator of fatty acid metabolism (27). Furthermore, the feedback regulation
within the methionine-MAT2A-S-adenosylmethionine (SAM) metabolic
axis was systematically investigated through combined western
blotting and RT-qPCR analyses, with a specific focus on the
SAM-dependent regulation of MAT2A protein stability and its
consequent impact on lipid metabolic homeostasis.
Materials and methods
Cell culture
Mouse embryonic fibroblasts (MEFs) (CRL-2991; ATCC)
and the HT1080, DU145 and H1299 cell lines were obtained from the
American Tissue Culture Collection (ATCC) and cultured following
ATCC recommendations. These cells were also subjected to STR
identification and mycoplasma testing. The cells were sustained in
Dulbecco's modified Eagle's medium (DMEM) with high glucose, sodium
pyruvate (1 mM), glutamine (2 mM), penicillin (100 U/ml),
streptomycin (0.1 mg/ml) and 10% (v/v) fetal bovine serum (FBS;
MeisenCTCC; Zhejiang Meisen Cell Technology Co., Ltd.) at 37°C and
5% CO2. In addition, the cultivation conditions for
methionine deficiency are based on DMEM (cat. no. D0422;
MilliporeSigma) with high glucose, sodium pyruvate (1 mM),
glutamine (2 mM), penicillin (100 U/ml), streptomycin (0.1 mg/ml)
and 10% (v/v) fetal bovine serum (FBS) (MeisenCTCC) at 37°C and 5%
CO2. Alternatively, S-adenosylmethionine (SAM; 0.4 mM),
S-adenosylhomocysteine (SAH; 0.4 mM), homocysteine (Hcy; 0.4 mM) or
adenosine dialdehyde (ADOX; 0.4 mM) were added separately to
methionine deficient culture medium to treat cells. Cells were also
treated with a constructed culture medium lacking essential amino
acids (isoleucine, 0.4 mM; isoleucine, 0.4 mM; lysine, 0.4 mM;
methionine, 0.4 mM; phenylalanine, 0.4 mM; threonine, 0.4 mM;
tryptophan, 0.4 mM; valine, 0.4 mM; histidine, 0.4 mM; and
arginine, 0.4 mM).
Reagents
The reagents used included propidium iodide (PI;
cat. no. P4170; MilliporeSigma), BODIPY581/591 C11 (cat. no. D3861;
Thermo Fisher Scientific, Inc.), BODIPY (cat. no. HY-D1614;
MedChemExpress), BODIPY™ FL C12 (cat. no. D3822; Thermo
Fisher Scientific, Inc.), SAM (cat. no. A7007; MilliporeSigma),
S-Adenosylhomocysteine (SAH; cat. no. A9384; MilliporeSigma),
homocysteine (cat. no. H4628; MilliporeSigma), anti-MAT2A (cat. no.
A19272; ABclonal Biotech Co., Ltd.), anti-ACSL4 (cat. no. A20414;
ABclonal Biotech Co., Ltd.) and DMEM with high glucose (cat. no.
D0422-100 ml; MilliporeSigma). In addition, all the components of
the D777 culture medium (with 4,500 mg/l glucose, L-glutamine and
sodium pyruvate, without sodium bicarbonate, powder, suitable for
cell culture) were purchased to construct an amino acid-deficient
culture medium. The medium mainly included methionine (cat. no.
M0960000), leucine (cat. no. L8000), isoleucine (cat. no. I2752),
lysine (cat. no. L5501), phenylalanine (cat. no. P17008), tyrosine
(cat. no. T1145), tryptophan (cat. no. T0254), valine (cat. no.
V0500), histidine (cat. no. H8125) and arginine (cat. no. A5006),
all from MilliporeSigma.
Fatty acid uptake and intracellular
lipid detection
Fatty acid uptake was measured by flow cytometry.
Briefly, HT1080 and H1299 cells were plated at a density of
5×104 cells per well in a 24-well dish and cultured for
12 h in DMEM supplemented with 10% FBS, 1% penicillin and
streptomycin at 37°C with 5% CO2. Subsequently, 2 µM
BODIPY-C12 was added to the cell culture medium and incubated for
10 h alongside the indicated treatment. Following this, the cells
were trypsinized, washed with PBS containing 5% FBS and resuspended
in 0.3 ml of PBS with 5% FBS for flow cytometry analysis (Attune
NxT; Thermo Fisher Scientific, Inc.). Acquired data were analyzed
using FlowJo version 10 software (FlowJo LLC). A minimum of 10,000
cells were analyzed per condition. Intracellular lipids were also
measured by flow cytometry (Attune NxT.). Acquired data were
analyzed using FlowJo version 10 software. The protocol was the
same as that aforementioned except 1 µg/ml BODIPY was added to the
cell culture medium and incubated for 30 min after the indicated
treatment.
Lipid reactive oxygen species (ROS)
detection
Lipid ROS were analyzed by flow cytometry. Briefly,
cells were plated at a density of 1×105 cells per well
in a 24-well dish and cultured overnight in DMEM. Subsequently, 4
µM BODIPY581/591 C11 was added to the cell culture medium and
incubated for 30 min after the indicated treatment. Excess
BODIPY581/591 C11 was then removed by washing the cells with PBS
twice. Labeled cells were trypsinized and resuspended in PBS with
5% FBS. Oxidation of BODIPY581/591 C11 resulted in a shift of the
fluorescence emission peak from 590 to 510 nm proportional to the
lipid ROS generation, which was detected using a flow cytometer
(Attune NxT). Acquired data were analyzed using FlowJo version 10
software.
Cell death detection
Cell death was analyzed by flow cytometry. Briefly,
cells were plated at a density of 2×105 cells per well
in a 12-well dish for 12 h in fresh medium. After the indicated
treatment, 1 µg/ml PI was added to the cell culture medium and
incubated for 30 min. Then, cells were trypsinized, washed with PBS
containing 5% FBS and resuspended in 0.3 ml of PBS with 5% FBS for
flow cytometry (Attune NxT). Acquired data were analyzed using
FlowJo software.
Apoptosis detection
Apoptosis was analyzed by flow cytometry. Briefly,
cells were plated in 12-well culture plates at a density of
1×105 cells per well and cultured in DMEM for 12 h.
Cells were then trypsinized, washed with PBS containing 5% FBS and
resuspended in 0.3 ml of PBS. Subsequently, PI and Annexin V-FITC
were added to the cells and incubated for 30 min, and cell
apoptosis was detected by flow cytometry (Attune NxT). Acquired
data were analyzed using FlowJo.
Transfections
Transfections were conducted using
Lipofectamine™ 2000 transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. HT1080 cells seeded in a 6-well dish were transfected
with 40 ng of the following small interfering RNAs (Suzhou
GenePharma Co., Ltd.): NC (scrambled), sense:
5′-UUCUCCGAACGUGUCACGUUTT-3′, antisense:
5′-AACGUGACACGUUCGGAGAATT-3′; MAT2A-1, sense:
5′-GGAUCGAGGUGCUGUGCUUTT-3′, antisense: 5′AAGCACAGCACCUCGAUCCTT-3′;
MAT2A-2, sense: 5′-GGGAUGCCCUAAAGGAGAATT-3′, antisense:
5′-UUCUCCUUUAGGGCAUCCCTT-3′ and MAT2A-3, sense:
5′-GCCUAUGGCCACUUUGGUATT-3′, antisense:
5′-UACCAAAAGUGGCCAUAGGCTT-3′. After transfection, the cells were
placed in a 37°C cell culture incubator and the medium was changed
after 8 h. After 48 h, the cells were lysed and the interference
efficiency was measured using western blotting.
Western blotting
Cell lysate preparation, SDS-PAGE and
electrophoretic transference were accomplished as previously
described (19). Briefly, the
protein sample was quantified using a NanoPhotometer®
N30 Touch and each lane of the 8% SDS gel was loaded with 10 µg
protein sample. Membranes were blocked with 5% non-fat dried milk
in TBS (pH 7.2) containing 0.1% Tween 20 (TBST), incubated with the
appropriate primary antibodies in 5% non-fat dried milk in TBST
overnight at 4°C. Then, the membrane was incubated with the
appropriate secondary antibodies in 5% non-fat dried milk in TBST
at room temperature for 50 min. The following primary antibodies
were used: Anti-MAT2A, anti-ACSL4 and β-actin (cat. no. HC201-01;
TransGen Biotech Co., Ltd.). The following secondary antibodies
were used: HRP-conjugated goat anti-rabbit (cat. no. AS014;
ABclonal Biotech Co., Ltd.) and HRP-conjugated goat anti-mouse
(cat. no. AS003; ABclonal Biotech Co., Ltd.). Chemiluminescence was
detected using the ChampChemi 610 (SinSage Technology Co., Ltd.),
and the software used for densitometry was ImageJ (National
Institutes of Health).
RT-qPCR
Total RNA was extracted from cells using TRNzol
(cat. no. DP424; Tiangen Biotech Co., Ltd.) and reverse transcribed
with the PrimeScript™ RT reagent Kit with gDNA Eraser
(cat. no. RR047A; Takara Bio, Inc.) according to the manufacturer's
protocol. The resulting cDNA was then subjected to qPCR using the
TB Green Premix Ex Taq™ (Tli RNase H Plus) (cat. no.
RR820A; Takara Bio, Inc.). The detection and analysis were
accomplished as previously described (19). The sequences of the primers used for
qPCR were as follows: ACSL4 (human) forward,
5′-CATCCCTGGAGCAGATACTCT-3′ and reverse,
5′-TCACTTAGGATTTCCCTGGTCC-3′; MAT2A (human) forward,
5′-ACCAGAAAGTGGTTCGTGAAG-3′ and reverse,
5′-CAAGGCTACCAGCACGTTACA-3′; MAT2B (human) forward,
5′-TTCACTGGTCTGGCAATGAAC-3′ and reverse,
5′-AGGGCTGTCAGTAATAGGTCTT-3′; MAT2A (mouse) forward,
5′-GCTTCCACGAGGCGTTCAT-3′ and reverse,
5′-AGCATCACTGATTTGGTCACAA-3′; MAT2B (mouse) forward,
5′-AGGGAACCTTTCACTGGTCTG-3′ and reverse
5′-ATTTGGAGCAATCGAGCTGAG-3′; β-actin (human) forward,
5′-GTTGTCGACGACGAGCG-3′ and reverse, 5′-GCACAGAGCCTCGCCTT-3′; and
β-actin (mouse) forward, 5′-GGCTGTATTCCCCTCCATCG-3′ and reverse,
5′-CCAGTTGGTAACAATGCCATGT-3′.
Public database analyses
ACLS4 expression in cancer
The expression levels of ACSL4 in tumor and
normal tissues were analyzed using the Expression DIY module on the
GEPIA 2 website (http://gepia2.cancer-pku.cn/#index). The analysis
parameter settings were as follows: Log2 FC cut-off (1); q-value cut-off (0.01); log scale
(yes); jitter size (0.4); and matched normal data (match TCGA
normal and GTEx data).
Overall survival
The survival analysis module on the GEPIA 2 website
was used to analyze the relationship between ACSL4 expression and
patient survival. The analysis parameter settings were as follows:
Methods (overall survival); group cut-off (median); cut-off high
(%) (50); cut-off low (%) (50); hazard ratio (yes); 95% confidence
interval (yes); and axis units (months).
Statistical analysis
GraphPad Prism (v.10.4.0; Dotmatics) software was
used to calculate the P-values using two-sided unpaired t-test or
one-way ANOVA followed by Tukey's Honestly Significant Difference.
All data presented in the figures represent the mean ± SD.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Methionine restriction downregulates
cellular lipid levels
To systematically evaluate the role of essential
amino acids in lipid metabolism regulation, an essential amino
acid-deficient culture system was established by excluding leucine,
isoleucine, lysine, methionine, phenylalanine, tyrosine,
tryptophan, valine, histidine and arginine from the medium. HT1080
cells cultured in this modified medium were subsequently analyzed
for intracellular lipid accumulation using BODIPY staining.
Quantitative analysis revealed that methionine deprivation
specifically caused a significant reduction in cellular lipid
content (Fig. 1A). Next, the
functional consequences of essential amino acid deficiency on fatty
acid uptake capacity were examined using BODIPY-C12 fluorescence
assays. Notably, methionine-deficient conditions produced a notable
impairment in fatty acid internalization among all tested amino
acid deprivations (Fig. 1B).
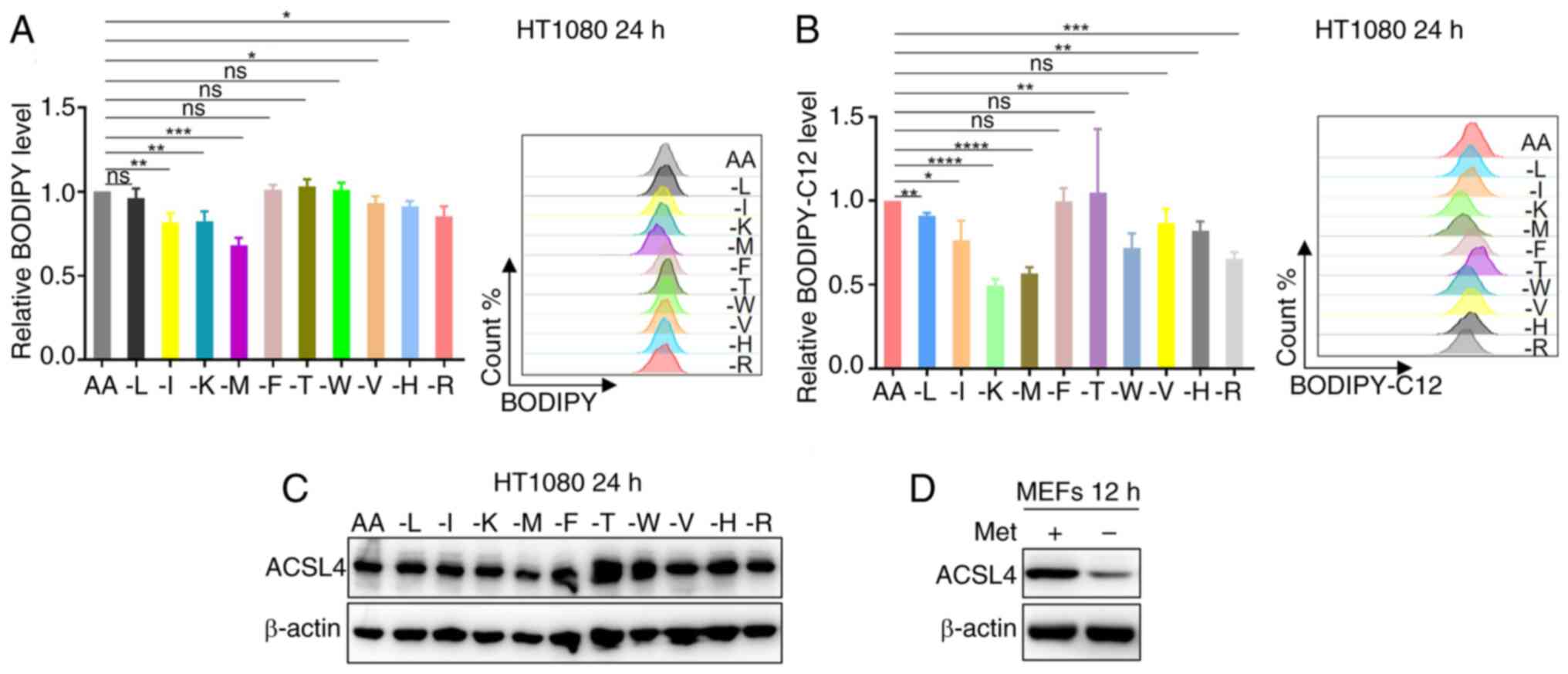 | Figure 1.Methionine restriction downregulates
cellular lipid levels. (A) HT1080 cells were cultured without
leucine, isoleucine, lysine, methionine, phenylalanine, tyrosine,
tryptophan, valine, histidine or arginine as indicated for 24 h,
and the intracellular lipids were determined by BODIPY staining
coupled with flow cytometry (n=3). (B) HT1080 cells were cultured
without leucine, isoleucine, lysine, methionine, phenylalanine,
tyrosine, tryptophan, valine, histidine, or arginine as indicated
for 24 h, and fatty acid uptake capacity was determined by
BODIPY-C12 staining coupled with flow cytometry (n=3). (C) Western
blot analysis of ACSL4 expression in HT1080 cells treated with
medium without leucine, isoleucine, lysine, methionine,
phenylalanine, tyrosine, tryptophan, valine, histidine or arginine
as indicated for 24 h. (D) Western blot analysis of ACSL4
expression in MEFs cells treated with medium without methionine for
12 h. Data are presented as the mean ± SD with P-values determined
by one-way ANOVA followed by Tukey's Honestly Significant
Difference. *P<0.05, **P<0.01, ***P<0.001,
****P<0.0001. AA, all amino acids; L, isoleucine; I, isoleucine;
K, lysine; M, methionine; F, phenylalanine; T, threonine; W,
tryptophan; V, valine; H, histidine; R, arginine; ACSL4, acyl-CoA
synthetase long chain family member 4; MEFs, mouse embryonic
fibroblasts; Met, methionine. |
Given the established role of ACSL4 in facilitating
fatty acid uptake through its dual function of converting fatty
acids to acyl-CoA derivatives and inhibiting fatty acid efflux,
ACSL4 protein expression in HT1080 cells under essential amino acid
deprivation conditions was first examined. Western blot analysis
revealed that methionine deficiency reduced the ACSL4 protein
levels (Fig. 1C). To validate this
finding across different cellular contexts, the investigation was
extended to MEFs under methionine-restricted conditions. Consistent
with the initial observation, methionine deprivation significantly
decreased ACSL4 expression in MEFs cells (Fig. 1D). Taken together, the results
suggest that methionine deficiency attenuates cellular lipid
accumulation and the fatty acid uptake capacity, potentially
through downregulation of ACSL4 protein expression, revealing a
novel mechanistic link between methionine availability and lipid
metabolic regulation.
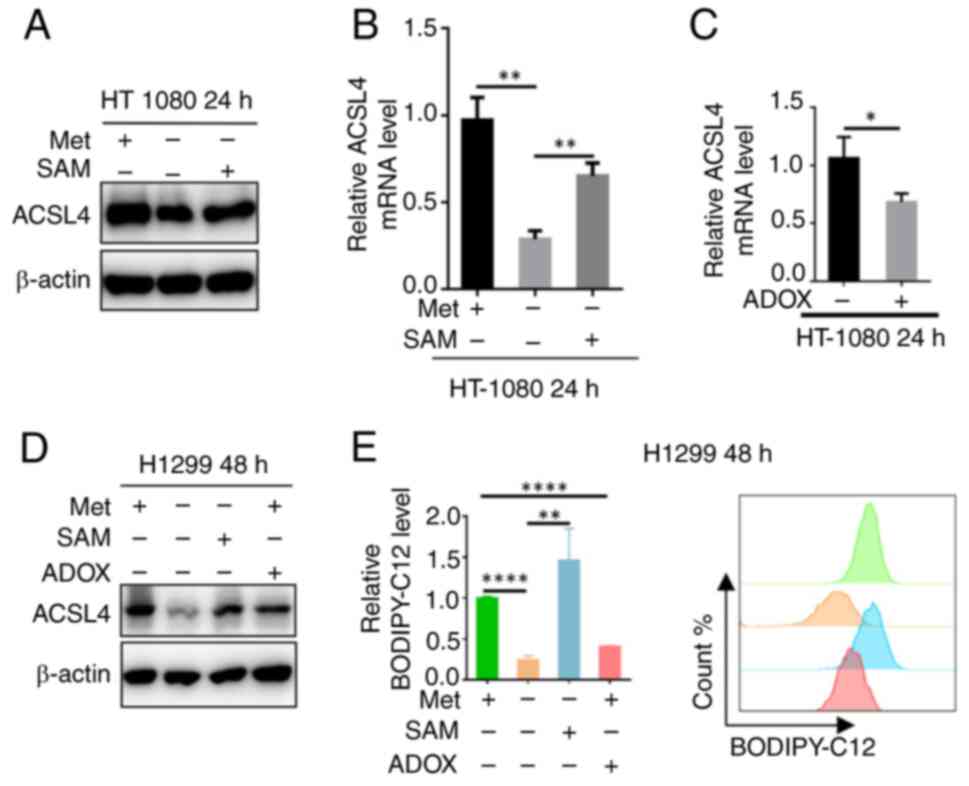
Methionine regulates ACSL4 through
SAM
As the sulfur-containing amino acid methionine
functions as a vital cellular methyl donor primarily through its
metabolic derivative SAM, it was next determined whether methionine
regulates ACSL4 expression and fatty acid uptake capacity via
SAM-dependent mechanisms. To test this hypothesis, methionine
deprivation experiments in HT1080 cells were performed followed by
SAM supplementation. Western blot analysis demonstrated that SAM
administration rescued the methionine deficiency-induced reduction
in ACSL4 protein levels (Fig. 2A).
Complementary RT-qPCR analysis further revealed that SAM
supplementation similarly restored the suppressed ACSL4 mRNA
expression under methionine-depleted conditions (Fig. 2B). To elucidate whether SAM
regulates ACSL4 expression through methylation-dependent
mechanisms, HT1080 cells were treated with the methylation
inhibitor, adenosine dialdehyde (ADOX), and the ACSL4
transcript levels were analyzed. RT-qPCR analysis revealed that
ADOX treatment significantly suppressed ACSL4 mRNA
expression (Fig. 2C), indicating
methylation-dependent regulation. This regulatory mechanism in
H1299 cells was further validated, as western blot analysis
confirmed that the methionine-SAM axis controls ACSL4 protein
abundance possibly through methylation modifications (Fig. 2D). Functional assays demonstrated
that this methylation-dependent regulation directly impacts
cellular fatty acid uptake capacity (Fig. 2E). Collectively, these results
establish that SAM-mediated methylation may represent a critical
epigenetic mechanism through which methionine regulates
ACSL4 expression and function in lipid metabolism.
MAT2A can be dynamically regulated by
the level of SAM
As the key enzyme catalyzing methionine-to-SAM
conversion, MAT2A occupies a central position in the methionine-SAM
metabolic axis (24). To
investigate whether MAT2A serves as a critical regulator of SAM
homeostasis and lipid metabolism, MAT2A protein expression in
HT1080 cells under essential amino acid deprivation was first
examined. Western blot analysis revealed that the MAT2A levels were
specifically upregulated by methionine deficiency (Fig. 3A). Using a dose-response approach,
it was demonstrated that MAT2A expression exhibits sensitivity to
extracellular methionine concentrations in DU145 cells (Fig. 3B). Notably, SAM supplementation
reversed the observed methionine deprivation-induced MAT2A
upregulation (Fig. 3C), suggesting
a feedback regulatory mechanism. Further mechanistic studies in
H1299 cells showed that MAT2A protein levels remained unchanged
following ADOX-mediated methylation inhibition, compared with the
methionine deprivation only group (Fig.
3D). Consistent with this result, transcriptional analysis
across multiple cell lines (MEFs, HT1080 and H1299) revealed that,
while the methionine-SAM axis regulates MAT2A mRNA
expression, this control occurs independently of the
methyltransferase activity of SAM (Fig.
3E). Notably, the regulatory subunit, MAT2B, was unaffected by
these metabolic perturbations (Fig.
3F). These results establish that MAT2A is subject to
SAM-mediated feedback regulation through a novel
methylation-independent mechanism, highlighting the sophisticated
control of methionine metabolism.
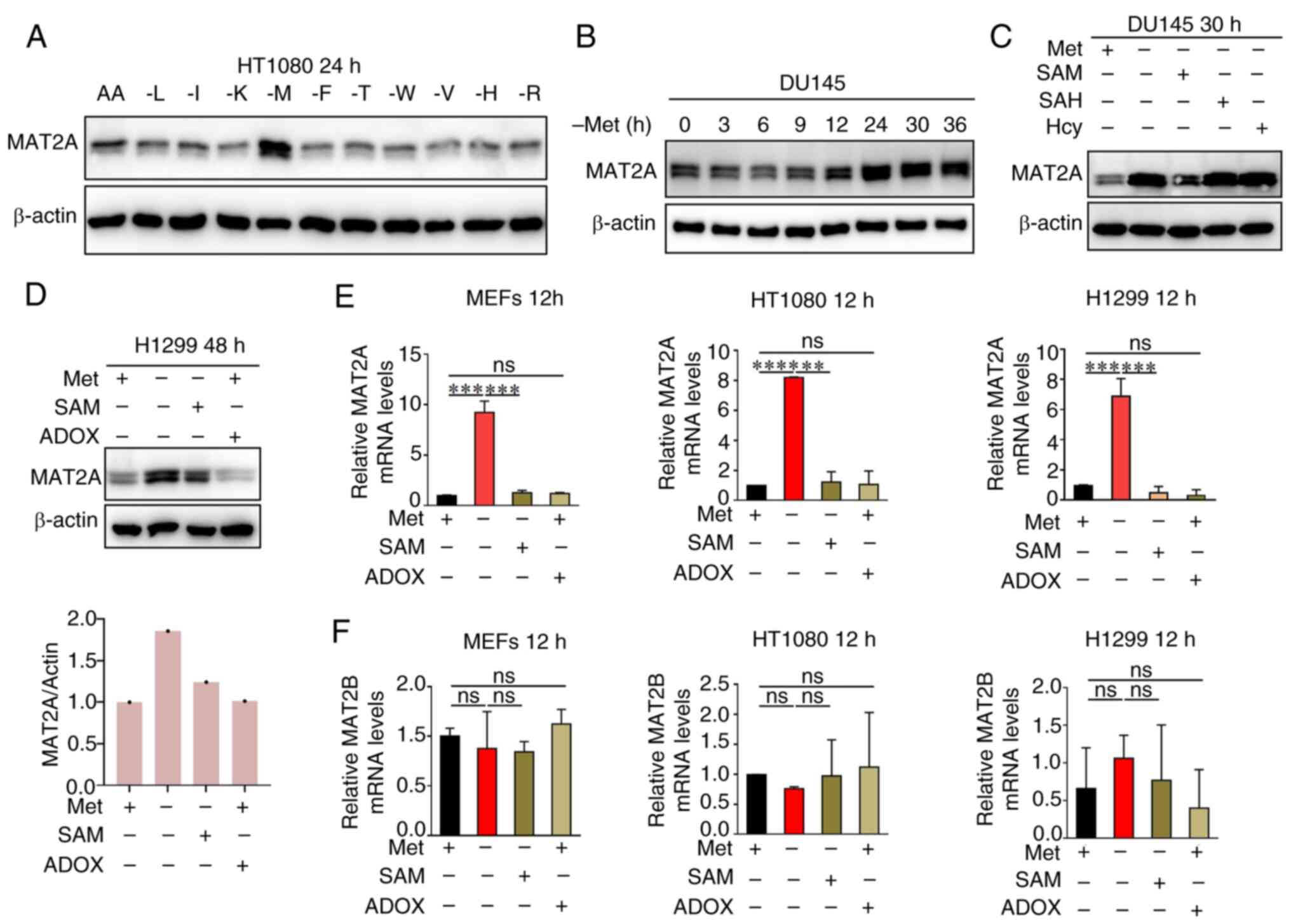 | Figure 3.MAT2A can be dynamically regulated by
the level of SAM. (A) Western blot analysis of MAT2A expression in
HT1080 cells treated with medium without leucine, isoleucine,
lysine, methionine, phenylalanine, tyrosine, tryptophan, valine,
histidine, or arginine as indicated for 24 h. (B) Western blot
analysis of MAT2A expression in DU145 cells treated with medium
without methionine as indicated for 0, 3, 6, 9, 12, 24, 30 and 36
h. (C) Western blot analysis of MAT2A expression in DU145 cells
cultured ± methionine, ± SAM, ± SAH or ± Hcy as indicated for 30 h.
(D) Western blot analysis of MAT2A expression in H1299 cells
cultured ± methionine, ± SAM or ± ADOX, as indicated for 48 h. The
(E) MAT2A and (F) MAT2B mRNA levels in MEFs and
HT1080 and H1299 cells cultured ± methionine, ± SAM or ± ADOX as
indicated for 12 h, were analyzed using reverse
transcription-quantitative PCR (n=3). Data are presented as mean ±
SD with P-values determined by one-way ANOVA followed by Tukey's
Honestly Significant Difference. **P<0.01, ***P<0.001,
****P<0.0001. ns, non-significant; AA, all amino acids; L,
isoleucine; I, isoleucine; K, lysine; M, methionine; F,
phenylalanine; T, threonine; W, tryptophan; V, valine; H,
histidine; R, arginine; MEFs, mouse embryonic fibroblasts; SAM,
S-adenosylmethionine; ADOX, adenosine dialdehyde; SAH,
S-adenosylhomocysteine; Hcy, homocysteine; MAT2A/B, methionine
adenosyltransferase 2A/B; Met, methionine. |
Knocking down MAT2A reduces
intracellular lipid levels
To systematically validate the regulatory role of
methionine in lipid metabolism, complementary experimental
approaches were performed using HT1080 cells. First, methionine
deprivation (using commercial methionine deficient medium) led to
significant reductions in both intracellular lipid accumulation
(BODIPY staining) and fatty acid uptake capacity (BODIPY-C12 assay)
(Fig. 4A and B). To directly assess
the involvement of MAT2A in these metabolic processes, RNA
interference-mediated knockdown was employed. MAT2A downregulation
similarly reduced the intracellular lipid content (Fig. 4C and D) and impaired fatty acid
internalization (Fig. 4E),
phenocopying the effects of methionine deprivation. These
consistent findings across orthogonal experimental approaches
establish MAT2A as a key metabolic regulator and potential
therapeutic target for modulating cellular lipid homeostasis.
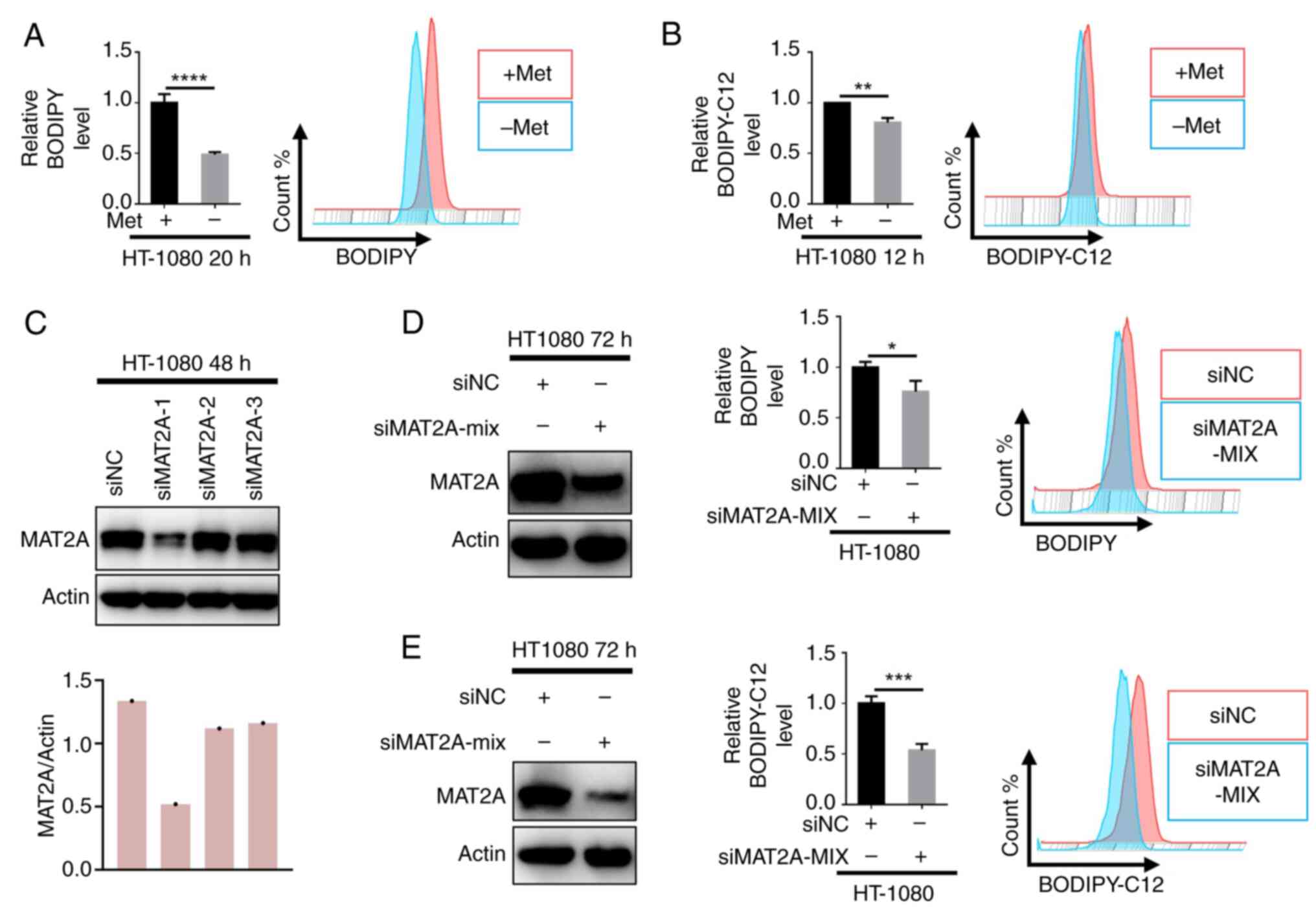 | Figure 4.Knocking down MAT2A expression
reduces the intracellular lipid levels. (A) HT1080 cells were
cultured ± methionine as indicated for 20 h, and the intracellular
lipids were determined by BODIPY staining coupled with flow
cytometry (n=3). (B) HT1080 cells were cultured ± methionine as
indicated for 12 h, and fatty acid uptake capacity was determined
by BODIPY-C12 staining coupled with flow cytometry (n=3). (C)
Western blot analysis of MAT2A expression in HT1080 cells
transfected with siMAT2A or siNC. (D) The intracellular lipids in
HT1080 cells transfected with siMAT2A or siNC were determined by
BODIPY staining coupled with flow cytometry (n=3). (E) Fatty acid
uptake capacity in HT1080 cells transfected with siMAT2A or siNC
was determined by BODIPY-C12 staining coupled with flow cytometry
(n=3). Data are presented as mean ± SD, with P-values determined by
one-way ANOVA followed by Tukey's Honestly Significant Difference.
*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. MAT2A,
methionine adenosyltransferase 2A; si, small interfering (RNA); NC,
negative control; Met, methionine. |
Methionine restriction suppresses
tumor cell proliferation by inhibiting ACLS4
Lipids serve as fundamental structural components of
cellular membranes and play a critical role in supporting the rapid
proliferation of tumor cells. The aforementioned results
demonstrated that methionine functions as a positive regulator of
intracellular lipid homeostasis. Analysis of The Cancer Genome
Atlas datasets revealed elevated ACSL4 expression in
multiple malignancies, including esophageal carcinoma, liver
hepatocellular carcinoma, pancreatic adenocarcinoma and stomach
adenocarcinoma (Fig. 5A). Notably,
high ACSL4 expression was significantly associated with
poorer overall survival in patients with cancer, including
cholangiocarcinoma, esophageal carcinoma and colon adenocarcinoma
(Fig. 5B), suggesting its clinical
relevance as a potential prognostic marker. Functional studies in
HT1080 cells showed that methionine deprivation simultaneously
suppressed ACSL4 expression (Fig. 5C) and impaired HT1080 cell
proliferation (Fig. 5D), while not
inducing ferroptosis (Fig. 5E and
F); however, it has a certain inducing effect on apoptosis
(Fig. 5G). Taken together, these
results establish methionine as an essential metabolic requirement
for tumor cell proliferation, operating through ACSL4-mediated
lipid metabolic pathways.
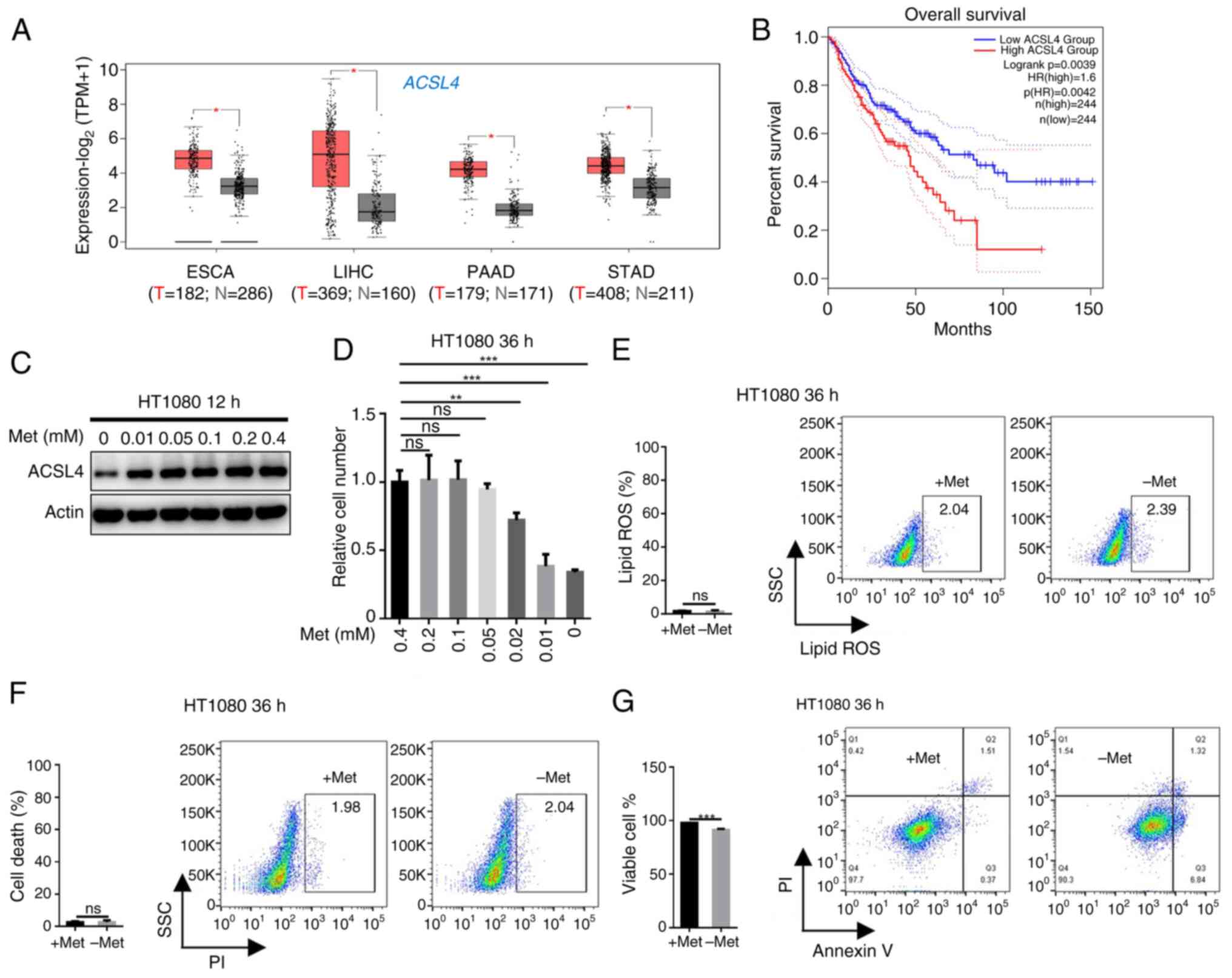 | Figure 5.Methionine restriction suppresses
tumor cell proliferation by inhibiting ACLS4. (A) Analysis of
ACSL4 expression in tumor and adjacent tissues based on TCGA
database. (B) Analysis of the correlation between ACSL4 expression
and overall survival based on TCGA database through GEPIA2
(http://gepia2.cancer-pku.cn/). (C)
Western blot analysis of ACSL4 expression in HT1080 cells treated
with medium without methionine as indicated for 12 h. (D) The
number of HT1080 cells after methionine deprivation as indicated
for 36 h were counted using a microscope and cell counter (n=3).
(E) HT1080 cells were cultured ± methionine as indicated for 36 h
and the lipid ROS level was determined by BODIPY-C11 staining
coupled with flow cytometry (n=3). (F) HT1080 cells were cultured ±
methionine as indicated for 36 h and cell death was determined by
PI staining coupled with flow cytometry (n=3). (G) HT1080 cells
were cultured ± methionine as indicated for 36 h and cell apoptosis
was determined by PI staining coupled with flow cytometry (n=3).
Data are presented as mean ± SD, with P-values determined by
one-way ANOVA followed by Tukey's Honestly Significant Difference.
*P<0.05, **P<0.01, ***P<0.001. ns, non-significant; TCGA,
The Cancer Genome Atlas; TPM, transcripts per million; ESCA,
esophageal carcinoma; LIHC, liver hepatocellular carcinoma; PAAD,
pancreatic adenocarcinoma; STAD, stomach adenocarcinoma; T, tumor;
N, normal; HR, hazard ratio; ACSL4, acyl-CoA synthetase long chain
family member 4; SSC, side Scatter; ROS, reactive oxygen species;
PI, propidium iodide; Met, methionine. |
Discussion
Amino acid metabolism serves as a critical metabolic
hub supporting the biosynthetic and energetic demands of rapidly
proliferating tumor cells. Among the essential amino acids,
methionine, a sulfur-containing metabolite with pleiotropic
functions, has emerged as a promising therapeutic target in
oncology (14,29). The present study identified a unique
role for methionine in governing lipid homeostasis, demonstrating
its superior capacity to modulate intracellular lipid accumulation
and fatty acid uptake compared with other essential amino acids.
Mechanistically, it was established that methionine exerts this
control through SAM-dependent regulation of ACSL4, a master
regulator of fatty acid metabolism (27). Downregulation of MAT2A, the
rate-limiting enzyme in methionine-to-SAM conversion (24), phenocopied methionine deprivation by
reducing lipid stores and impairing fatty acid internalization.
Notably, these metabolic perturbations translated to significant
anti-proliferative effects in tumor cells, highlighting the
therapeutic potential of targeting methionine metabolism.
Furthermore, methionine restriction has emerged as a promising
therapeutic strategy in oncology, primarily through its depletion
of SAM, the universal methyl donor critical for numerous cellular
processes (30–32). The present study extends this
paradigm by revealing an additional mechanism whereby methionine
restriction impairs tumor proliferation through ACSL4
downregulation-mediated inhibition of fatty acid uptake. The
therapeutic potential of targeting methionine metabolism is further
underscored by the frequent upregulation of MAT2A across diverse
malignancies (33,34). In view of this, MAT2A has become an
important target for cancer treatment, and inhibitors developed
around it are constantly emerging (24,35,36).
Notably, the MAT2A inhibitor, AG-270, has entered phase I clinical
trials as a potential therapeutic agent for malignant tumors
(24). Furthermore, ACSL4, a
crucial enzyme in lipid metabolism regulation, has been
demonstrated to facilitate metastatic extravasation in ovarian
cancer through its enhancement of membrane fluidity and cellular
invasiveness (37). Consequently,
targeted disruption of the methionine-MAT2A-ACSL4 axis represents a
potential therapeutic strategy to simultaneously suppress tumor
proliferation and metastasis.
MAT2A is essential for maintaining intracellular SAM
homeostasis. Emerging evidence indicates that the regulatory
subunit, MAT2B, mediates SAM-dependent feedback inhibition of MAT2A
activity (38–41). In addition, the present study
extended these findings by demonstrating that: i) Methionine
deprivation upregulates MAT2A expression; ii) SAM supplementation
suppresses this induction; and iii) this regulatory mechanism
operates independently of the methyltransferase activity of SAM. We
hypothesize that mTORC1 may serve as a key mediator in
SAM-dependent regulation of MAT2A expression. This speculation is
supported by existing evidence demonstrating that mTORC1 can
modulate MAT2A expression through its downstream effector, MYC
(42). Furthermore, a study has
suggested that SAM may exert feedback control on MAT2A through the
SAM sensor, SAMTOR, which interfaces with both the mTORC1 and MYC
signaling pathways (43).
Therefore, the feedback regulation of MAT2A by SAM contributes to
the steady-state of methionine-MAT2A-SAM.
ACSL4, a key regulator of lipid metabolism,
catalyzes the ATP-dependent conversion of fatty acids to acyl-CoA
esters. Extensive studies have established that ACSL4 governs
cellular fatty acid uptake capacity by maintaining the
intracellular acyl-CoA pool (44,45).
The present study revealed that methionine deprivation decreased
ACSL4 protein abundance, leading to impaired fatty acid
internalization. In future research, the construction of isogenic
ACSL4-knockout cell lines will be critical for characterizing
ACSL4-dependent lipid metabolic pathways and for determining
whether methionine restriction can further modulate intracellular
lipid dynamics in ACSL4-deficient contexts. Notably, this lipid
metabolic perturbation can be rescued by SAM supplementation,
suggesting SAM-dependent regulation of ACSL4. However, the
molecular mechanisms underlying SAM-mediated control of ACSL4
expression and activity require further investigation. Previous
evidence provides important mechanistic insights into this
regulatory process. A previous study demonstrated that SAM
modulates mTORC1 activity through its sensor protein, SAMTOR
(43). As a central
nutrient-sensing hub, mTORC1 plays a pivotal role in coordinating
cellular anabolism and metabolic reprogramming. Notably, mTORC1 has
been shown to directly regulate ACSL4 expression (46). Based on these established
relationships, we propose a model wherein SAM regulates ACSL4
levels through SAMTOR-mediated modulation of mTORC1 signaling
activity.
As the primary product of the methionine-MAT2A-SAM
metabolic axis, SAM serves as the universal methyl donor for
diverse biological methylation reactions, including protein, DNA,
RNA and small molecule modifications (47). Given this extensive substrate
diversity, we hypothesize that the regulatory mechanisms by which
the methionine-MAT2A-SAM axis influences ACSL4 and lipid metabolism
remain incompletely characterized. Furthermore, the additional role
of SAM as a precursor for polyamine biosynthesis (48) suggests potential cross-regulation
between polyamine metabolism and ACSL4-mediated lipid pathways.
Collectively, these observations highlight the multifaceted
regulatory capacity of the methionine-MAT2A-SAM axis in cellular
metabolism.
The present study has certain limitations. Notably,
the BODIPY probe used in the present study functions as a
broad-spectrum lipid stain, lacking molecular specificity.
Consequently, while this method effectively detects overall lipid
uptake, it does not discriminate between distinct lipid species.
This represents a limitation of the current approach, as it
precludes detailed characterization of the specific lipid subtypes
involved in the observed processes.
In conclusion, the present study elucidated a novel
regulatory mechanism by which methionine governs cellular lipid
homeostasis through modulation of fatty acid uptake. These findings
establish the methionine-MAT2A-SAM axis as a central metabolic
regulator and reveal its therapeutic potential for targeted cancer
interventions.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Zhejiang Sci-Tech
University Research Start-up Fund (grant no. 24042191-Y).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
CX conceived the project, designed the research and
led the entire experiment, from design to execution. JM assisted
with the experiments. CX analyzed the data and wrote the
manuscript. CX and JM confirm the authenticity of all the raw data.
All authors have read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cairns RA, Harris IS and Mak TW:
Regulation of cancer cell metabolism. Nat Rev Cancer. 11:85–95.
2011. View
Article : Google Scholar
|
|
2
|
Mossmann D, Park S and Hall MN: mTOR
signalling and cellular metabolism are mutual determinants in
cancer. Nat Rev Cancer. 18:744–757. 2018. View Article : Google Scholar
|
|
3
|
Wang W and Zou W: Amino acids and their
transporters in T cell immunity and cancer therapy. Mol Cell.
80:384–395. 2020. View Article : Google Scholar
|
|
4
|
Yoneshiro T, Wang Q, Tajima K, Matsushita
M, Maki H, Igarashi K, Dai Z, White PJ, McGarrah RW, Ilkayeva OR,
et al: BCAA catabolism in brown fat controls energy homeostasis
through SLC25A44. Nature. 572:614–619. 2019. View Article : Google Scholar
|
|
5
|
Gao X, Locasale JW and Reid MA: Serine and
methionine metabolism: Vulnerabilities in lethal prostate cancer.
Cancer Cell. 35:339–341. 2019. View Article : Google Scholar
|
|
6
|
Kelly B and Pearce EL: Amino assets: How
amino acids support immunity. Cell Metab. 32:154–175. 202
View Article : Google Scholar
|
|
7
|
Labuschagne CF, van den Broek NJ, Mackay
GM, Vousden KH and Maddocks OD: Serine, but not glycine, supports
one-carbon metabolism and proliferation of cancer cells. Cell Rep.
7:1248–1258. 2014. View Article : Google Scholar
|
|
8
|
Longchamp A, Mirabella T, Arduini A,
MacArthur MR, Das A, Treviño-Villarreal JH, Hine C, Ben-Sahra I,
Knudsen NH, Brace LE, et al: Amino acid restriction triggers
angiogenesis via GCN2/ATF4 regulation of VEGF and H(2)S production.
Cell. 173:117–129.e14. 2018. View Article : Google Scholar
|
|
9
|
Knaus LS, Basilico B, Malzl D, Gerykova
Bujalkova M, Smogavec M, Schwarz LA, Gorkiewicz S, Amberg N, Pauler
FM, Knittl-Frank C, et al: Large neutral amino acid levels tune
perinatal neuronal excitability and survival. Cell.
186:1950–1967.e25. 2023. View Article : Google Scholar
|
|
10
|
Handzlik MK, Gengatharan JM, Frizzi KE,
McGregor GH, Martino C, Rahman G, Gonzalez A, Moreno AM, Green CR,
Guernsey LS, et al: Insulin-regulated serine and lipid metabolism
drive peripheral neuropathy. Nature. 614:118–124. 2023. View Article : Google Scholar
|
|
11
|
Zhang Y, Lin S, Peng J, Liang X, Yang Q,
Bai X, Li Y, Li J, Dong W, Wang Y, et al: Amelioration of hepatic
steatosis by dietary essential amino acid-induced ubiquitination.
Mol Cell. 82:1528–1542.e10. 2022. View Article : Google Scholar
|
|
12
|
Schulte ML, Fu A, Zhao P, Li J, Geng L,
Smith ST, Kondo J, Coffey RJ, Johnson MO, Rathmell JC, et al:
Pharmacological blockade of ASCT2-dependent glutamine transport
leads to antitumor efficacy in preclinical models. Nat Med.
24:194–202. 2018. View
Article : Google Scholar
|
|
13
|
Muthusamy T, Cordes T, Handzlik MK, You L,
Lim EW, Gengatharan J, Pinto AFM, Badur MG, Kolar MJ, Wallace M, et
al: Serine restriction alters sphingolipid diversity to constrain
tumour growth. Nature. 586:790–795. 2020. View Article : Google Scholar
|
|
14
|
Gao X, Sanderson SM, Dai Z, Reid MA,
Cooper DE, Lu M, Richie JP Jr, Ciccarella A, Calcagnotto A, Mikhael
PG, et al: Dietary methionine influences therapy in mouse cancer
models and alters human metabolism. Nature. 572:397–401. 2019.
View Article : Google Scholar
|
|
15
|
Mossmann D, Müller C, Park S, Ryback B,
Colombi M, Ritter N, Weißenberger D, Dazert E, Coto-Llerena M,
Nuciforo S, et al: Arginine reprograms metabolism in liver cancer
via RBM39. Cell. 186:5068–5083.e23. 2023. View Article : Google Scholar
|
|
16
|
Krall AS, Mullen PJ, Surjono F, Momcilovic
M, Schmid EW, Halbrook CJ, Thambundit A, Mittelman SD, Lyssiotis
CA, Shackelford DB, et al: Asparagine couples mitochondrial
respiration to ATF4 activity and tumor growth. Cell Metab.
33:1013–1026.e6. 2021. View Article : Google Scholar
|
|
17
|
Missiaen R, Anderson NM, Kim LC, Nance B,
Burrows M, Skuli N, Carens M, Riscal R, Steensels A, Li F and Simon
MC: GCN2 inhibition sensitizes arginine-deprived hepatocellular
carcinoma cells to senolytic treatment. Cell Metab.
34:1151–1167.e7. 2022. View Article : Google Scholar
|
|
18
|
Badgley MA, Kremer DM, Maurer HC,
DelGiorno KE, Lee HJ, Purohit V, Sagalovskiy IR, Ma A, Kapilian J,
Firl CEM, et al: Cysteine depletion induces pancreatic tumor
ferroptosis in mice. Science. 368:85–89. 2020. View Article : Google Scholar
|
|
19
|
Xue Y, Lu F, Chang Z, Li J, Gao Y, Zhou J,
Luo Y, Lai Y, Cao S, Li X, et al: Intermittent dietary methionine
deprivation facilitates tumoral ferroptosis and synergizes with
checkpoint blockade. Nat Commun. 14:47582023. View Article : Google Scholar
|
|
20
|
Zheng J and Conrad M: The metabolic
underpinnings of ferroptosis. Cell Metab. 32:920–937. 2020.
View Article : Google Scholar
|
|
21
|
Zhang W, Li Q, Zhang Y, Wang Z, Yuan S,
Zhang X, Zhao M, Zhuang W and Li B: Multiple myeloma with high
expression of SLC7A11 is sensitive to erastin-induced ferroptosis.
Apoptosis. 29:412–423. 2024. View Article : Google Scholar
|
|
22
|
Koppula P, Zhuang L and Gan B: Cystine
transporter SLC7A11/xCT in cancer: Ferroptosis, nutrient
dependency, and cancer therapy. Protein Cell. 12:599–620. 2021.
View Article : Google Scholar
|
|
23
|
Dixon SJ, Patel DN, Welsch M, Skouta R,
Lee ED, Hayano M, Thomas AG, Gleason CE, Tatonetti NP, Slusher BS
and Stockwell BR: Pharmacological inhibition of cystine-glutamate
exchange induces endoplasmic reticulum stress and ferroptosis.
Elife. 3:e025232014. View Article : Google Scholar
|
|
24
|
Gounder M, Johnson M, Heist RS, Shapiro
GI, Postel-Vinay S, Wilson FH, Garralda E, Wulf G, Almon C, Nabhan
S, et al: MAT2A inhibitor AG-270/S095033 in patients with advanced
malignancies: A phase I trial. Nat Commun. 16:4232025. View Article : Google Scholar
|
|
25
|
Cacciatore A, Shinde D, Musumeci C,
Sandrini G, Guarrera L, Albino D, Civenni G, Storelli E, Mosole S,
Federici E, et al: Epigenome-wide impact of MAT2A sustains the
androgen-indifferent state and confers synthetic vulnerability in
ERG fusion-positive prostate cancer. Nat Commun. 15:66722024.
View Article : Google Scholar
|
|
26
|
Hou PP, Zheng CM, Wu SH, Liu XX, Xiang GX,
Cai WY, Chen G and Lou YL: Extracellular vesicle-packaged ACSL4
induces hepatocyte senescence to promote hepatocellular carcinoma
progression. Cancer Res. 84:3953–3966. 2024. View Article : Google Scholar
|
|
27
|
Doll S, Proneth B, Tyurina YY, Panzilius
E, Kobayashi S, Ingold I, Irmler M, Beckers J, Aichler M, Walch A,
et al: ACSL4 dictates ferroptosis sensitivity by shaping cellular
lipid composition. Nat Chem Biol. 13:91–98. 2017. View Article : Google Scholar
|
|
28
|
Chaneton B, Hillmann P, Zheng L, Martin
ACL, Maddocks ODK, Chokkathukalam A, Coyle JE, Jankevics A, Holding
FP, Vousden KH, et al: Serine is a natural ligand and allosteric
activator of pyruvate kinase M2. Nature. 491:458–462. 2012.
View Article : Google Scholar
|
|
29
|
Cunningham A, Erdem A, Alshamleh I,
Geugien M, Pruis M, Pereira-Martins DA, van den Heuvel FAJ,
Wierenga ATJ, Ten Berge H, Dennebos R, et al: Dietary methionine
starvation impairs acute myeloid leukemia progression. Blood.
140:2037–2052. 2022. View Article : Google Scholar
|
|
30
|
Wang QL, Chen Z, Lu X, Lin H, Feng H, Weng
N, Chen L, Liu M, Long L, Huang L, et al: Methionine metabolism
dictates PCSK9 expression and antitumor potency of PD-1 blockade in
MSS colorectal cancer. Adv Sci (Weinh). 12:e25016232025. View Article : Google Scholar
|
|
31
|
Hong XL, Huang CK, Qian H, Ding CH, Liu F,
Hong HY, Liu SQ, Wu SH, Zhang X and Xie WF: Positive feedback
between arginine methylation of YAP and methionine transporter
SLC43A2 drives anticancer drug resistance. Nat Commun. 16:872025.
View Article : Google Scholar
|
|
32
|
Zhang X, Zhao Z, Wang X, Zhang S, Zhao Z,
Feng W, Xu L, Nie J, Li H, Liu J, et al: Deprivation of methionine
inhibits osteosarcoma growth and metastasis via C1orf112-mediated
regulation of mitochondrial functions. Cell Death Dis. 15:3492024.
View Article : Google Scholar
|
|
33
|
Huang Z, Chen P and Liu Y: RBM15-mediated
the m6A modification of MAT2A promotes osteosarcoma cell
proliferation, metastasis and suppresses ferroptosis. Mol Cell
Biochem. 480:2923–2933. 2025. View Article : Google Scholar
|
|
34
|
Xia S, Liang Y, Shen Y, Zhong W and Ma Y:
MAT2A inhibits the ferroptosis in osteosarcoma progression
regulated by miR-26b-5p. J Bone Oncol. 41:1004902023. View Article : Google Scholar
|
|
35
|
Yang S, Gu X, Chen L and Zhu W: Discovery
of novel spirocyclic MAT2A inhibitors demonstrating high in vivo
efficacy in MTAP-Null xenograft models. J Med Chem. 68:3480–3494.
2025. View Article : Google Scholar
|
|
36
|
Zhou Y, Wang L, Ren R, Zhang J, Huan X,
Yang P, Miao ZH, Xiong B, Wang Y and Liu T: Structure-based
discovery of a series of novel MAT2a inhibitors. ACS Med Chem Lett.
16:646–650. 2025. View Article : Google Scholar
|
|
37
|
Wang Y, Hu M, Cao J, Wang F, Han JR, Wu
TW, Li L, Yu J, Fan Y, Xie G, et al: ACSL4 and polyunsaturated
lipids support metastatic extravasation and colonization. Cell.
188:412–429.e27. 2025. View Article : Google Scholar
|
|
38
|
Shafqat N, Muniz JR, Pilka ES,
Papagrigoriou E, von Delft F, Oppermann U and Yue WW: Insight into
S-adenosylmethionine biosynthesis from the crystal structures of
the human methionine adenosyltransferase catalytic and regulatory
subunits. Biochem J. 452:27–36. 2013. View Article : Google Scholar
|
|
39
|
LeGros HL Jr, Halim AB, Geller AM and Kotb
M: Cloning, expression, and functional characterization of the beta
regulatory subunit of human methionine adenosyltransferase (MAT
II). J Biol Chem. 275:2359–2366. 2000. View Article : Google Scholar
|
|
40
|
LeGros L, Halim AB, Chamberlin ME, Geller
A and Kotb M: Regulation of the human MAT2B gene encoding the
regulatory beta subunit of methionine adenosyltransferase, MAT II.
J Biol Chem. 276:24918–24924. 2001. View Article : Google Scholar
|
|
41
|
Li Z, Wang F, Liang B, Su Y, Sun S, Xia S,
Shao J, Zhang Z, Hong M, Zhang F and Zheng S: Methionine metabolism
in chronic liver diseases: An update on molecular mechanism and
therapeutic implication. Signal Transduct Target Ther. 5:2802020.
View Article : Google Scholar
|
|
42
|
Villa E, Sahu U, O'Hara BP, Ali ES, Helmin
KA, Asara JM, Gao P, Singer BD and Ben-Sahra I: mTORC1 stimulates
cell growth through SAM synthesis and m6A mRNA-dependent
control of protein synthesis. Mol Cell. 81:2076–2093.e9. 2021.
View Article : Google Scholar
|
|
43
|
Gu X, Orozco JM, Saxton RA, Condon KJ, Liu
GY, Krawczyk PA, Scaria SM, Harper JW, Gygi SP and Sabatini DM:
SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway.
Science. 358:813–818. 2017. View Article : Google Scholar
|
|
44
|
Ibrahim A, Yucel N, Kim B and Arany Z:
Local mitochondrial ATP production regulates endothelial fatty acid
uptake and transport. Cell Metab. 32:309–319.e7. 2020. View Article : Google Scholar
|
|
45
|
Milger K, Herrmann T, Becker C, Gotthardt
D, Zickwolf J, Ehehalt R, Watkins PA, Stremmel W and Füllekrug J:
Cellular uptake of fatty acids driven by the ER-localized acyl-CoA
synthetase FATP4. J Cell Sci. 119:4678–4688. 2006. View Article : Google Scholar
|
|
46
|
Huang B, Nie G, Dai X, Cui T, Pu W and
Zhang C: Environmentally relevant levels of Cd and Mo coexposure
induces ferroptosis and excess ferritinophagy through AMPK/mTOR
axis in duck myocardium. Environ Toxicol. 39:4196–4206. 2024.
View Article : Google Scholar
|
|
47
|
Ouyang Y, Wu Q, Li J and Sun S and Sun S:
S-adenosylmethionine: A metabolite critical to the regulation of
autophagy. Cell Prolif. 53:e128912020. View Article : Google Scholar
|
|
48
|
Fernández-Ramos D, Lopitz-Otsoa F, Lu SC
and Mato JM: S-adenosylmethionine: A multifaceted regulator in
cancer pathogenesis and therapy. Cancers (Basel). 17:5352025.
View Article : Google Scholar
|