Introduction
According to GLOBOCAN statistics, lung cancer is the
most frequently diagnosed cancer globally and a major contributor
to cancer-related deaths worldwide, accounting for ~12.4% of all
new cancer cases and 18.7% of all cancer deaths in 2022 (1). Non-small cell lung cancer (NSCLC),
primarily consisting of lung adenocarcinoma (LUAD) and lung
squamous cell carcinoma (LUSC), is the predominant form of lung
cancer (2). In previous years,
substantial strides have been made in the treatment of lung cancer,
including advancements in screening, diagnosis and minimally
invasive treatments. Additionally, progress in radiotherapy,
including stereotactic ablative radiotherapy, has become an
effective approach for treating lung cancer (3–5).
Meanwhile, the emergence of new targeted therapies and
immunotherapies has markedly improved the survival rate of patients
with NSCLC (6,7). Patients with NSCLC who are eligible
for targeted therapy and immunotherapy now have a longer survival
time, with a 5-year survival rate ranging from 15.0 to 62.5%,
depending on the biomarkers (8).
This underscores the importance of identifying new molecular
markers and therapeutic targets for NSCLC.
Leucine-rich repeat-containing G protein-coupled
receptor 4 (LGR4/GPCR48) belongs to the GPCR family and can
regulate developmental pathways through typical G protein signaling
(9). LGR4 is also involved in cell
proliferation and organ development. For example, LGR4 deficiency
decreases the migration and proliferation of eyelid epidermal
keratinocytes, and it also disrupts postnatal intestinal crypt
development, leading to defective epithelial proliferation and
abnormal Paneth cell differentiation (10). Moreover, numerous studies have
reported that LGR4 promotes tumor progression, such as in
colorectal (11), breast (12) and prostate (13) cancer. A recent study indicates that
the R-spondin (RSPO)-LGR4/5-E3 ubiquitin-protein ligase ZNRF3
(ZNRF3)/E3 ubiquitin-protein ligase RNF43 (RNF43) signaling complex
critically regulates Wnt/β-catenin signaling in hepatic biology
(14). Dysregulation of the
RSPO-LGR4/5-ZNRF3/RNF43 complex, commonly caused by RSPO
overexpression or loss-of-function mutations in ZNRF3/RNF43,
constitutes a major oncogenic driver in hepatocellular carcinoma
that induces constitutive Wnt/β-catenin activation (15). However, another study indicates that
LGR4 potentiates breast cancer metastasis via a Wnt-independent
mechanism, wherein it directly interacts with epidermal growth
factor receptor (EGFR) and suppresses E3 ubiquitin-protein ligase
CBL-mediated ubiquitination, thereby impeding EGFR degradation and
augmenting EGFR signaling activation (16). Nevertheless, the function of LGR4 in
NSCLC requires further investigation.
The present study aimed to investigate the role of
LGR4 in NSCLC and potential mechanisms underlying its involvement
in tumor progression. Understanding the contribution of LGR4 may
provide insights into its significance in NSCLC pathophysiology and
its potential as a therapeutic target.
Materials and methods
Analysis of The Cancer Genome Atlas
(TCGA) data
NSCLC mRNA sequencing data were retrieved from TCGA,
comprising 1,043 tumor samples and 110 normal samples from
TCGA-LUAD and TCGA-LUSC (cancer.gov/tcga). The data were analyzed
using R software (version 4.3.3; R Foundation) to determine LGR4
gene expression levels in NSCLC. Patients were divided into high
and low LGR4 expression groups based on the median expression.
Kaplan-Meier survival analysis was performed using the log-rank
test.
Tissue samples from patients
To evaluate the prognostic relevance of LGR4
expression in NSCLC, two independent tissue microarrays (TMA1 and
TMA2) were utilized to validate its association with clinical
outcomes. TMA1 (cat. no. ZL-lug1201) included 60 pairs of NSCLC
tumors and paracancerous tissue, sourced from Superbiotek, for
comparing LGR4 expression. TMA2 comprised samples from 140 patients
with NSCLC and 10 normal controls (28 female, 112 males) who
underwent surgery at the Department of Thoracic Surgery, Zhongshan
Hospital, Fudan University (Shanghai, China) between January and
December 2005. The normal control lung tissue samples were resected
from patients undergoing surgery for benign pulmonary diseases
(such as pulmonary bulla) and confirmed tumor-free by
histopathological examination. Clinical follow-ups were conducted
until July 2013 and stages Ia-IIIa, according to the American Joint
Committee on Cancer and Union for International Cancer Control
criteria (17).
Immunohistochemical (IHC)
staining
The TMA of both groups was stained
immunohistochemically with rabbit anti-LGR4 (Proteintech Group,
Inc.; cat. no. 20150-1-AP; 1:400). Tissue sections were baked at
59°C for 60 min and then immersed sequentially in xylene and an
ethanol series for 10 min each. After hydration and washing in
distilled water for 10 min, the sections were exposed to 3%
H2O2 for 10 min. Antigen retrieval was
performed by diluting the Antigen Retrieval Solution (Weiao; cat.
no. WH1034; 1:1 and heating it in a pressure cooker to boiling
(~100°C). Tissue sections were steamed for 2 min 30 sec, then
cooled to room temperature and washed twice with 1X PBS (Weiao,
China; cat. no. WB6020) for 15 min each. The sections were blocked
with 5% BSA (Weiao, China; cat. no. WH2051) at room temperature for
35 min, then incubated overnight with the primary antibody (rabbit
anti-LGR4; Proteintech, Wuhan, China; cat. no. 20150-1-AP; 1:400)
at 4°C. After four washes with PBS (15 min each), the sections were
incubated with the secondary antibody (HRP-conjugated goat
anti-rabbit IgG; ImmunoWay, China; cat. no. RS0002; 1:100) at 37°C
for 35 min, followed by further washing. The color reaction was
developed according to the manufacturer's instructions, followed by
hematoxylin counterstaining at room temperature for 1 min and a 10
min wash. Finally, dehydration of the sections was carried out
through an ethanol gradient and xylene, followed by mounting with a
sealing agent.
The average optical density (AOD) values from TMA1
were analyzed using a paired Student's t-test with GraphPad Prism
software (Dotmatics; version 10.1.2). For TMA2, AOD values were
divided into high- and low-expression groups based on the median
cutoff. Clinicopathological characteristics are summarized in
Table I, and univariate and
multivariate Cox regression analyses were performed to evaluate
whether LGR4 expression serves as an independent prognostic factor
in NSCLC. Kaplan-Meier survival analysis was then conducted to
assess the prognostic significance of LGR4 expression during the
clinical follow-up period.
 | Table I.Clinicopathological characteristics of
126 patients with non-small cell lung cancer. |
Table I.
Clinicopathological characteristics of
126 patients with non-small cell lung cancer.
| Characteristic | Total (n=126) | High LGR4
expression (n=63) | Low LGR4 expression
(n=63) | P-value |
|---|
| Mean age,
years | 59.8±10.2 | 59.1±10.6 | 60.5±9.8 | 0.455 |
| Sex, n (%) |
|
|
| 0.826 |
|
Male | 100 (79.4) | 51 (81.0) | 49 (77.8) |
|
|
Female | 26 (20.6) | 12 (19.0) | 14 (22.2) |
|
| Histological type,
n (%) |
|
|
| 0.017 |
|
Adenocarcinoma | 48 (38.1) | 17 (27.0) | 31 (49.2) |
|
|
Squamous cell carcinoma | 78 (61.9) | 46 (73.0) | 32 (50.8) |
|
| Tumor stage, n
(%) |
|
|
| 0.304 |
| I | 48 (38.1) | 20 (31.7) | 28 (44.4) |
|
| II | 40 (31.7) | 21 (33.3) | 19 (30.2) |
|
|
III | 38 (30.2) | 22 (34.9) | 16 (25.4) |
|
| Ki-67 (%) |
|
|
| 0.434 |
|
Negative | 89 (70.6) | 42 (66.7) | 47 (74.6) |
|
|
Positive | 37 (29.4) | 21 (33.3) | 16 (25.4) |
|
| Tumor location, n
(%) |
|
|
| 0.284 |
|
Left | 59 (46.8) | 26 (41.3) | 33 (52.4) |
|
|
Right | 67 (53.2) | 37 (58.7) | 30 (47.6) |
|
Cell and cell line culture
Human healthy lung epithelial cells (BEAS-2B; cat.
no. TCH-C132), and NSCLC cell lines A549 (cat. no. TCH-C116) and
H226 (cat. no. TCH-C279) were obtained from Haixing Biosciences and
cultured in DMEM (cat. no. BL305A, Biosharp) supplemented with 10%
fetal bovine serum (FBS; cat. no. SLB-13011-8611; Zhejiang Tianhang
Biotechnology Co., Ltd.) and 1% penicillin-streptomycin at
37°C.
Small interfering RNA (siRNA)
transfection
Cells were transfected with one of three siRNAs
targeting LGR4 from the ‘siRNA 3-in-1 package’ (Ruibo Bio,
Shanghai, China), and with a non-targeting scrambled control si-NC.
The sequences (5′-3′) were as follows: si-LGR4 #1: forward
GAAAGAACUCAAAGUUCUAAC, reverse UAGAACUUUGAGUUCUUUCAA. si-LGR4#2:
forward GGUAGUUCUGCAUCUUCAUAA, reverse AUGAAGAUGCAGAACUACCAG.
si-LGR4#3: forward GCUGCGGCGGACUGCUGAAGG, reverse
UUCAGCAGUCCGCCGCAGCGG si-NC: forward UUCUUCGAAGGUGUCACGUTT, reverse
ACGUGACACCUUCGAAGAATT. A549 and H226 cells were seeded
3×105 into 6-well plates, each with 2 ml of complete
medium as aforementioned. This allowed the cells to reach ~50%
confluence by the time of transfection. Cells were transfected
using Lipo 8000™ Transfection Reagent (cat. no. C0533
Beyotime Institute of Biotechnology, China). Cells were transfected
with siRNAs at a final concentration of 50 nM at 37°C
(CLM-170B-8-CN, ESCO, Singapore). Transfection efficiency was
evaluated by reverse transcription-quantitative PCR (RT-qPCR) at 48
h post-transfection.
RT-qPCR
Total RNA was isolated from the NSCLC cell lines and
BEAS-2B cells using the FastPure® Cell/Tissue Total RNA
Isolation Kit V2 (Vazyme Biotech Co., Ltd.), following the protocol
provided by the manufacturer. cDNA synthesis was performed using
the HiScript III 1st Strand cDNA Synthesis Kit (+gDNA wiper)
(Vazyme Biotech Co., Ltd.), according to the manufacturer's
instructions. Any genomic DNA present was removed and then the
extracted RNA was reverse-transcribed into cDNA. Next,
gene-specific primers were designed for the target genes using
SnapGene software (version 7.2.1, GSL Biotech LLC), ensuring that
the primers only amplified the desired gene fragment and avoided
spanning splice junctions. Primer specificity was validated using
the NCBI Primer-BLAST tool (version 2.53.0,
ncbi.nlm.nih.gov/tools/primer-blast/) to ensure primers would not
amplify non-target sequences. Finally, the primers were synthesized
through Agena Bioscience, Inc., and the sequences of the primers
are provided in Table II.
 | Table II.Primer sequence for reverse
transcription-quantitative PCR. |
Table II.
Primer sequence for reverse
transcription-quantitative PCR.
| Gene name and
primer direction | Primer sequence
(5′-3′) |
|---|
| LGR4 forward |
ACTCAAAGTTCTAACGCTCCAG |
| LGR4 reverse |
AAAGCACTCAGCCCTCGAATG |
| GAPDH forward |
GGAGCGAGATCCCTCCAAAAT |
| GAPDH reverse |
GGCTGTTGTCATACTTCTCATGG |
RT-qPCR was conducted with the Taq Pro Universal
SYBR quantitative PCR (qPCR) Master Mix (Vazyme Biotech Co., Ltd.)
under the following thermal cycling conditions: An initial
denaturation at 95°C for 30 sec followed by 40 cycles consisting of
denaturation at 95°C for 5 sec and extension at 60°C for 60 sec.
Gene expression levels were determined using the 2−ΔΔCq
method (18), with GAPDH serving as
the endogenous control.
Western blot assay
Western blotting was used to evaluate the protein
expression of LGR4. Proteins were extracted from A549 and H226
cells using RIPA lysis buffer (Beyotime Institute of Biotechnology)
with protease and phosphatase inhibitors, quantified by BCA assay.
10% Cell-Free Acrylamide System (CFAS) separation gels were
prepared by combining CFAS PAGE Separation Gel A (cat. no. PE004-A,
Zhonghui Hecai, China) and Separation Gel B (cat. no. PE004-B,
Zhonghui Hecai, China) at a 1:1 ratio, following the manufacturer's
instructions (10% CFAS PAGE Rapid Gel Preparation Kit, PE004,
Zhonghui Hecai, China), and thoroughly homogenized before use.
Proteins were transferred onto PVDF membranes using 1× rapid
transfer buffer and blocked with 5% non-fat dried milk in 1× TBST
(0.1% Tween-20, Biosharp, China) at room temperature for 2 h.
Membranes were incubated with rabbit polyclonal primary antibodies
against LGR4 (cat. no. ER63609; HuaAn Biotech, China; 1:400) and
GAPDH (cat. no. R1210-1; HuaAn Biotech, China; 1:400) overnight at
4°C, washed with 1× TBST (0.1% Tween-20) 5×7 min, then incubated
with HRP-linked goat anti-rabbit IgG (cat. no. A0208; Beyotime
Biotechnology, China; 1:10,000) at room temperature for 80 min,
followed by the same TBST washing cycles 5×7 min. Protein signals
were detected using Super ECL Chemiluminescent Substrate (cat. no.
BL520A, Biosharp, China). Images were analyzed using Tanon Image
software (https://tanon.cnreagent.com/).
Cell Counting Kit-8 (CCK-8) assay
To examine the effect of LGR4 on the proliferative
capacity of NSCLC cells, cell proliferation in A549 and H226 cells
was measured at 0, 24, 48 and 72 h after transfection with si-LGR4
using the CCK-8 assay. A total of 2,500 cells (A549 and H226) were
seeded in 100 µl of complete medium into each well of a 96-well
plate. Then 10 µl enhanced CCK-8 reagent (cat. no. C0041; Beyotime
Institute of Biotechnology) was dispensed into each well. Cells
without the culture medium and CCK-8 solution served as the blank
control. After incubating both the experimental and control groups
in a cell incubator for 2 h, absorbance was measured at 450 nm.
Flow cytometry apoptosis assay
Following transfection, A549 and H226 cells were
digested with 0.25% trypsin (without EDTA and phenol red; cat. no.
T1350; Beijing Solarbio, China) for 3 min at room temperature until
the cells detached. The detached cells were centrifuged at 1,000 ×
g for 5 min at room temperature. Thereafter, the supernatant was
discarded and the cells were collected. After washing twice with
PBS, the cells were stained with Annexin V-FITC and propidium
iodide using the Cell Apoptosis Detection Kit (cat. no. C1062M,
Beyotime Biotechnology, China), and incubated on ice in the dark
for 20 min. Finally, apoptosis was then analyzed using BD
Accuri™ C6 software (version 1.0.264.21, BD Biosciences)
on the FACS Accuri C6 flow cytometer (BD Biosciences, USA).
Transwell assay
A549 and H226 cells were cultured in DMEM/F12 (1:1)
medium supplemented with 10% FBS (cat. no. SLB-13011-8611,
Sijiqing, China) and 1% penicillin-streptomycin (cat. no. BL505A,
Biosharp, China). Transwell inserts with an 8-µm pore size, 24-well
format (cat. no. 3422, Corning, USA) were used for both migration
and invasion assays. For invasion assays, the upper surface of the
inserts was precoated with Matrigel (cat. no. 082704; Mogengel) at
37°C for 30 min; uncoated inserts were used for migration assays. A
total of ~5×10⁵ cells were seeded into the upper chambers
containing serum-free DMEM/F12, while 500 µl of medium supplemented
with 10% FBS was added to the lower chambers as a chemoattractant.
After 48 h of incubation at 37°C with 5% CO₂, non-migrated or
non-invaded cells remaining on the upper membrane surfaces were
removed with a cotton swab. Cells on the lower membrane surfaces
were fixed with methanol for 30 min and stained with 0.1% crystal
violet (cat. no. BL802A, Biosharp, China) for 20 min at room
temperature. Migrated or invaded cells were quantified in five
random fields under an inverted light microscope (×400; Model
CKx53, Olympus, Japan).
Gene Set Enrichment Analysis
(GSEA)
To explore the possible oncogenic mechanisms of LGR4
in NSCLC, The LGR4 expression matrix was extracted from TCGA NSCLC
dataset (https://www.cancer.gov/tcga),
comprising 1,043 tumor samples, using R software (version 4.3.3; R
Foundation), and subsequently converted into gct and cls files for
GSEA. Thereafter, GSEA 4.3.3 software (Broad Institute) was
employed to analyze the signaling pathways activated in tumor
samples exhibiting high LGR4 expression. High and low expression
groups were defined using the median LGR4 expression as the cut-off
(4.7589). Specifically, the KEGG canonical pathway gene set
(c2.cp.kegg.v2023.1.Hs.symbols.gmt,
ftp://ftp.broadinstitute.org/pub/gsea/msigdb/human/gene_sets/c2.cp.kegg.v2023.1.Hs.symbols.gmt)
and the HALLMARK gene set (h.all.v2025.1.Hs.symbols.gmt,
ftp://ftp.broadinstitute.org/pub/gsea/msigdb/human/gene_sets/h.all.v2025.1.Hs.symbols.gmt)
were used. Pathways with |normalized enrichment score|>1,
P<0.05 and FDR <0.05 were regarded as significantly
enriched.
Statistical analysis
Intergroup All quantitative data are presented as
the mean ± SD from at least three independent experiments.
Two-group comparisons were performed using paired or unpaired
Student's t-tests. Multi-group comparisons were analyzed by one-way
ANOVA with Dunnett's post hoc test. Time-course experiments, such
as CCK8 assays, were evaluated by two-way ANOVA with Šídák's
multiple comparisons test. Survival probability estimations were
performed with Kaplan-Meier methodology and log-rank testing.
Prognostic factor screening used uni- and multivariate Cox
proportional hazards models. Statistical analyses were performed
with GraphPad Prism (version 10.1.2; Dotmatics) and R software
(version 4.3.3; R Foundation). P<0.05 was considered to indicate
a statistically significant difference. All analyses were repeated
at least three times.
Results
LGR4 expression is upregulated in
NSCLC
The present investigation into the role of LGR4 in
NSCLC began with an evaluation of its expression using data
obtained from the TCGA database. The analysis revealed that LGR4
expression was significantly higher in NSCLC samples than in
healthy lung tissue (P<0.001; Fig.
1A). Kaplan-Meier survival curves were plotted to determine the
prognostic impact of LGR4. The findings indicated that patients
with elevated LGR4 expression had shorter overall survival times
compared with those with low expression (P=0.013; Fig. 1B). To further confirm these results,
the expression of LGR4 in NSCLC cell lines was assessed by western
blotting and RT-qPCR. Both the protein and mRNA protein levels of
LGR4 were significantly elevated in NSCLC compared with those in
healthy tissues (Fig. 1C and D),
with differences considered statistically significant (both
P<0.05). Significant downregulation of LGR4 was also
consistently observed across all three siRNA knockdown groups via
qPCR analysis (all P<0.05; Fig.
1E). Among the three siRNAs targeting LGR4, si-LGR4 #1 and #2
showed comparable knockdown efficiency in preliminary experiments.
To maintain consistency in subsequent functional assays, si-LGR4 #2
was used for all following experiments.
Expression of LGR4 in NSCLC
tissues
IHC staining of two independent TMAs revealed
predominant cytoplasmic localization of LGR4 within tumor cells.
Analysis of TMA1 demonstrated elevated LGR4 expression in both LUAD
and LUSC specimens compared with matched adjacent paracancerous
tissues (Fig. 2A and B). Consistent
with this finding, IHC evaluation of TMA2 displayed increased LGR4
levels in malignant tissues compared with those in healthy lung
tissues (Fig. 2C). Matched-pair
analysis of 60 tumor/paracancerous samples from TMA1 confirmed
significantly higher LGR4 expression in the cancerous component vs.
the relevant paracancerous counterpart (P<0.0001; Fig. 2D). Moreover, LGR4 expression was
significantly increased in tumor specimens compared with healthy
lung tissue (n=10) (P<0.0001; Fig.
2E).
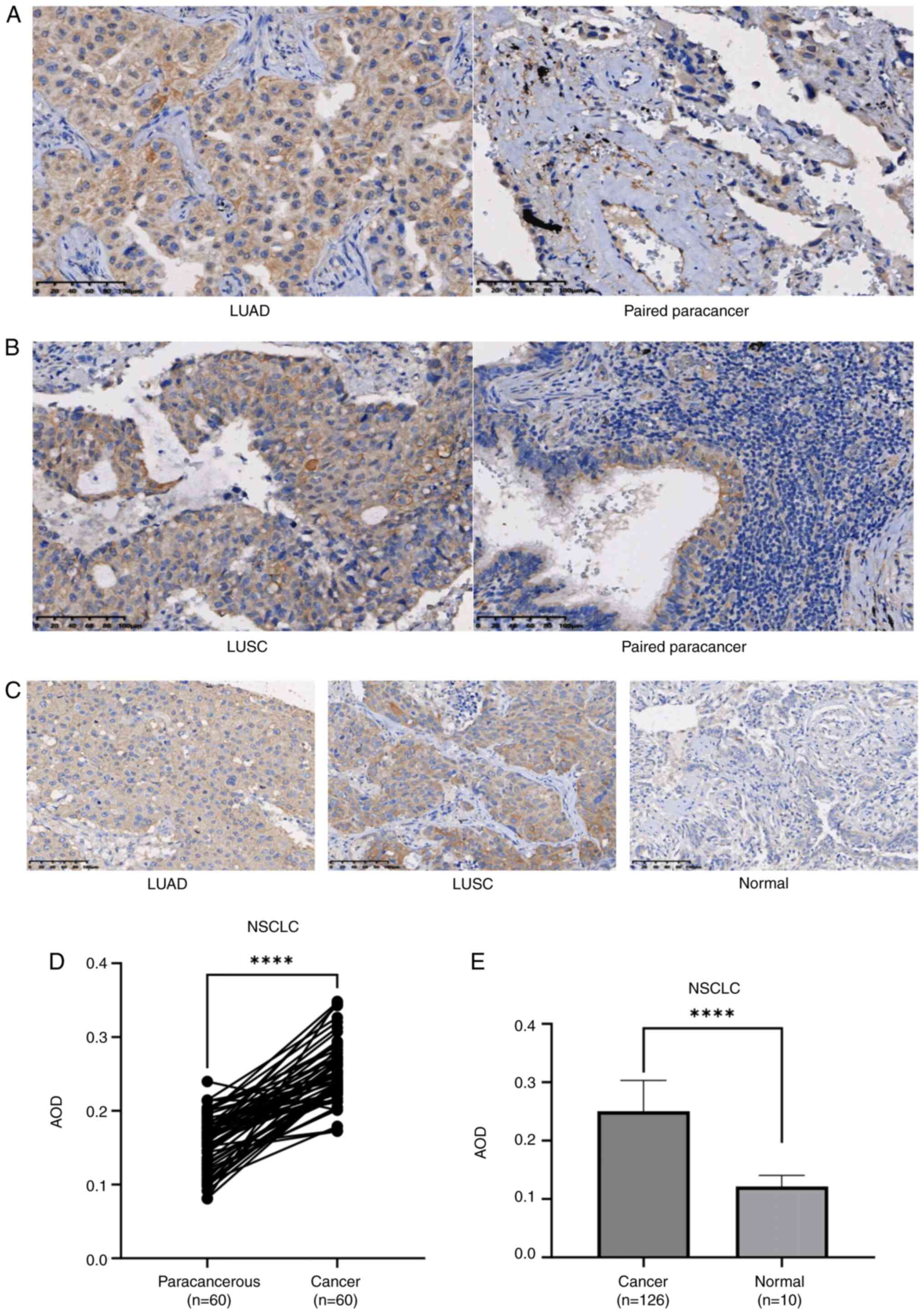 | Figure 2.LGR4 is overexpressed in tumor
tissues. (A) Lung adenocarcinoma tissue (×200 magnification) and
adjacent non-tumor tissue (×200 magnification). (B) LUSC tissue
(×200 magnification) and adjacent non-tumor tissue (×200
magnification). (C) LUAD tissue (×200 magnification), LUSC tissue
(×200 magnification) and healthy tissue (×200 magnification). Scale
bar, 100 µm. (D) Comparison of LGR4 expression in TMA1 (paired
Student's t-test). (E) LGR4 expression in TMA2 (unpaired Student's
t-test). ****P<0.0001. LGR4, leucine-rich repeat-containing
G-protein coupled receptor 4; LUSC, lung squamous cell carcinoma;
LUAD, lung adenocarcinoma; TMA1, tissue microarray 1; TMA2, tissue
microarray 2; NSCLC, non-small cell lung cancer; HR, hazard ratio;
AOD, average optical density. |
Association of LGR4 expression with
clinicopathological characteristics
IHC staining of TMA2 resulted in tissue core
detachment in some cases. Therefore, patients lacking evaluable
staining due to detachment were excluded, leaving 126 patients for
analysis. The baseline clinicopathological characteristics are
detailed in Table I. Patients were
stratified into high- and low-expression groups based on the median
AOD value of LGR4 (0.2512). As presented in Table I, LGR4 expression demonstrated a
significant association with histological tumor type (P=0.017;
Table I). However, no significant
associations were observed between LGR4 expression levels and
patient age (P=0.455), sex (P=0.826), TNM tumor stage P=0.304),
Ki-67 expression status (P=0.434) or tumor location (P=0.284).
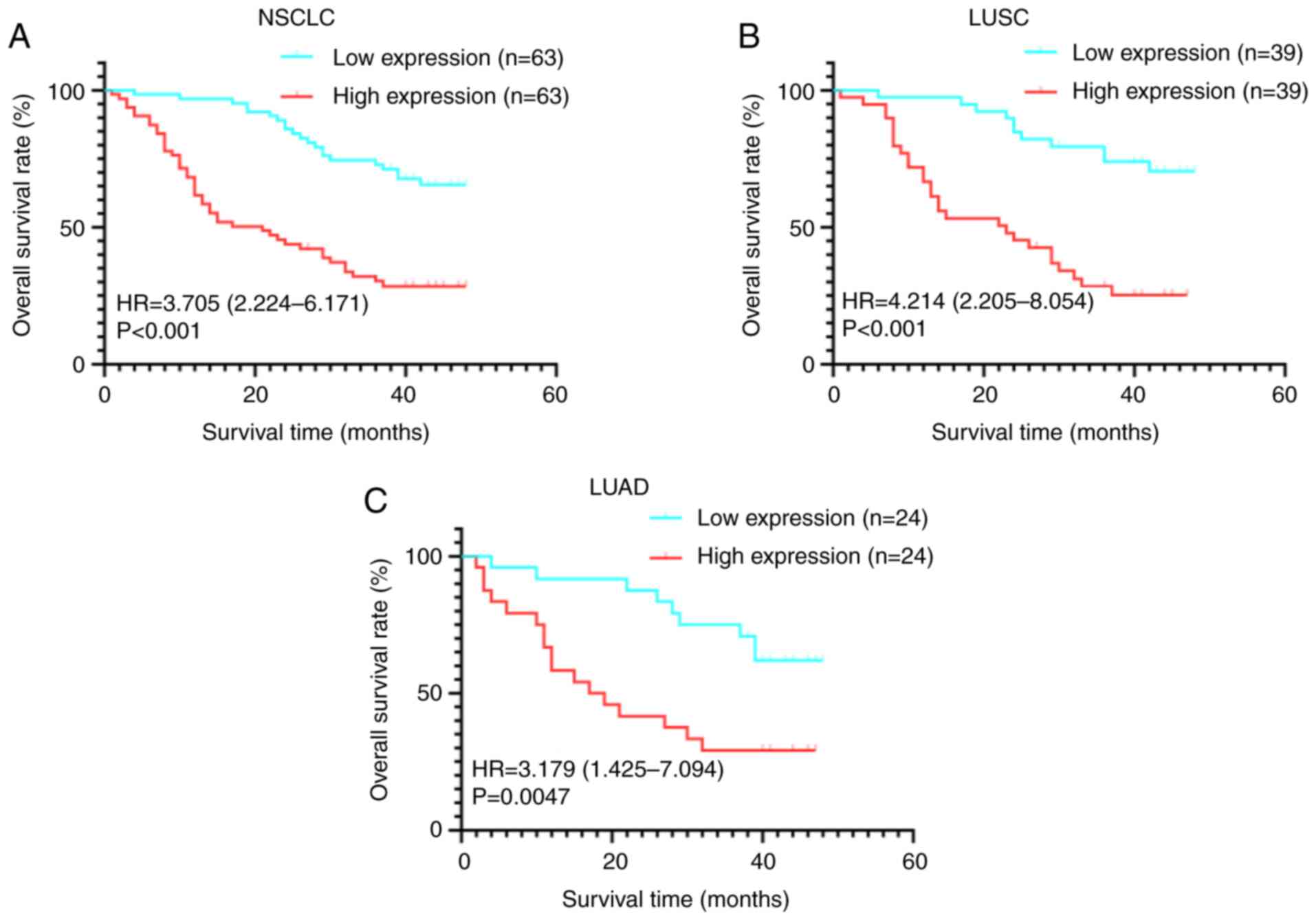
Prognostic value of LGR4 in NSCLC
Analysis of long-term follow-up data from TMA2
demonstrated that patients with high LGR4 expression exhibited
significantly shorter overall survival times compared with those
with low LGR4 expression (Fig. 3A,
P<0.001). Furthermore, this adverse association was consistently
observed in both LUAD and LUSC subtypes, where high LGR4 expression
was associated with poorer survival outcomes relative to low
expression groups (both P<0.05; Fig.
3B and C). Subsequent univariate and multivariate Cox
proportional hazards regression analyses confirmed elevated LGR4
expression as an independent predictor of poor prognosis in
patients with NSCLC (P<0.001; Table III). Ultimately, both high LGR4
levels and advanced tumor stage (stage II and III; both P<0.05)
were identified as independent adverse prognostic factors.
 | Table III.Univariate and multivariate
analysis. |
Table III.
Univariate and multivariate
analysis.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Characteristic | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age | 0.98
(0.96–1.01) | 0.321 | 0.99
(0.97–1.03) | 0.833 |
| Sex (Male) | 1.14
(0.62–2.09) | 0.681 | 1.16
(0.62–2.15) | 0.648 |
| Ki-67
(Positive) | 1.41
(0.84–2.35) | 0.193 | 1.75
(0.98–3.14) | 0.061 |
| LGR4 | 3.55
(2.10–5.99) | <0.001 | 4.31
(2.43–7.63) | <0.001 |
| Histological
type |
|
|
|
|
|
Squamous cell carcinoma | 1.00
(Reference) | NA | 1.00
(Reference) | NA |
|
Adenocarcinoma | 1.10
(0.67–1.81) | 0.702 | 2.22
(1.21–4.09) | 0.011 |
| Tumor stage |
|
|
|
|
| I | 1.00
(Reference) | NA | 1.00
(Reference) | NA |
| II | 2.23
(1.20–4.16) | 0.012 | 2.30
(1.20–4.40) | 0.012 |
|
III | 2.08
(1.10–3.95) | 0.024 | 1.96
(1.01–3.83) | 0.049 |
| Tumor location |
|
|
|
|
|
Left | 1.00
(Reference) | NA | 1.00
(Reference) | NA |
|
Right | 1.37
(0.84–2.26) | 0.209 | 1.01
(0.58–1.74) | 0.988 |
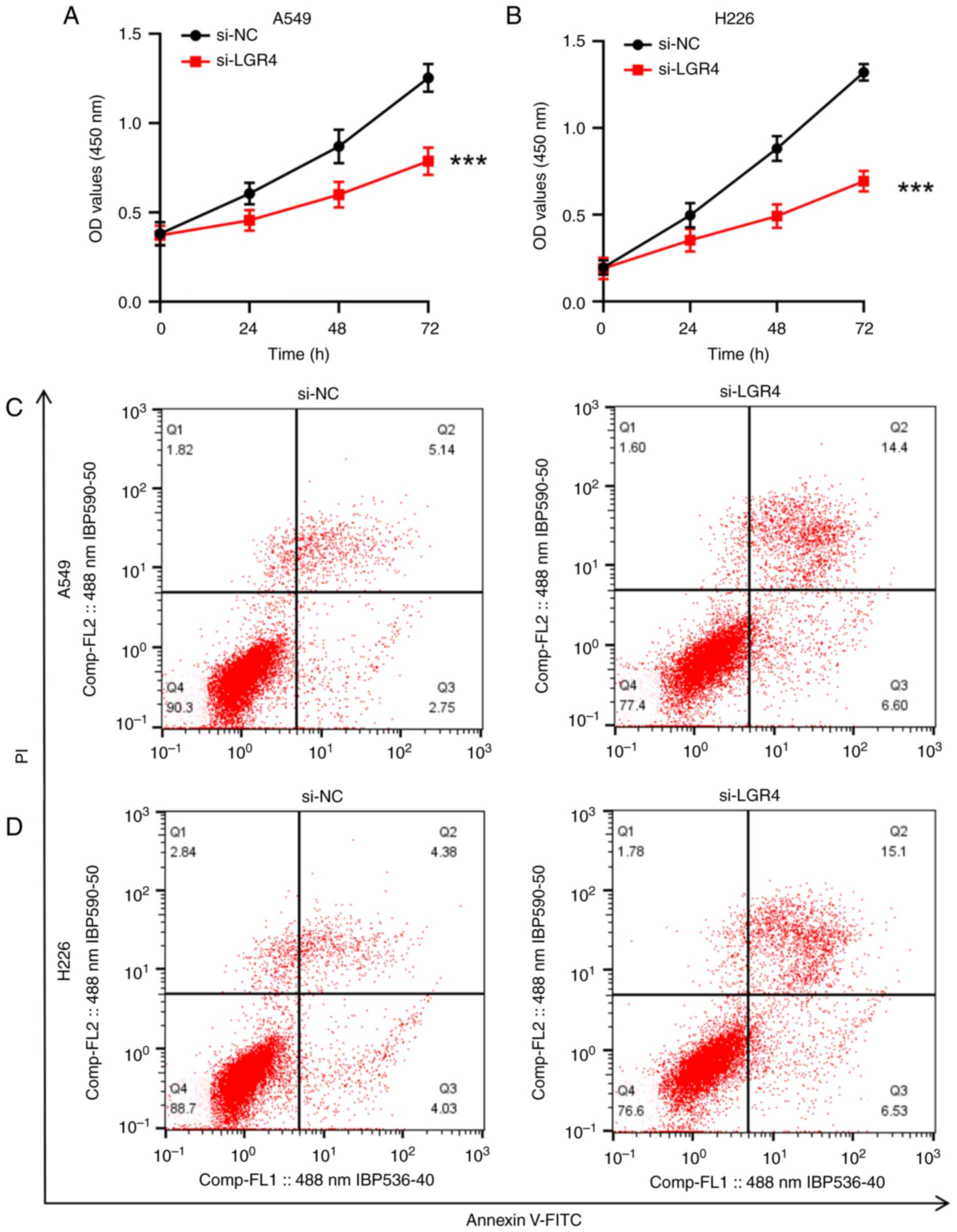
Knockdown of LGR4 inhibits the
proliferation of A549 and H226 cells
To investigate whether LGR4 knockdown could inhibit
the proliferation of NSCLC cells, a CCK-8 assay was conducted. The
results revealed a significant reduction in the proliferation of
both A549 and H226 cells at 24, 48 and 72 h following si-LGR4
transfection, (P<0.001; Fig. 4A and
B).
Knockdown of LGR4 promotes apoptosis
in A549 and H226 cells
Flow cytometry analysis indicated that silencing
LGR4 considerably increased the apoptotic rate in both A549 and
H226 cells relative to the si-NC control group, suggesting that
LGR4 depletion promotes apoptosis in these NSCLC cell lines (both
P<0.001; Fig. 4C and D).
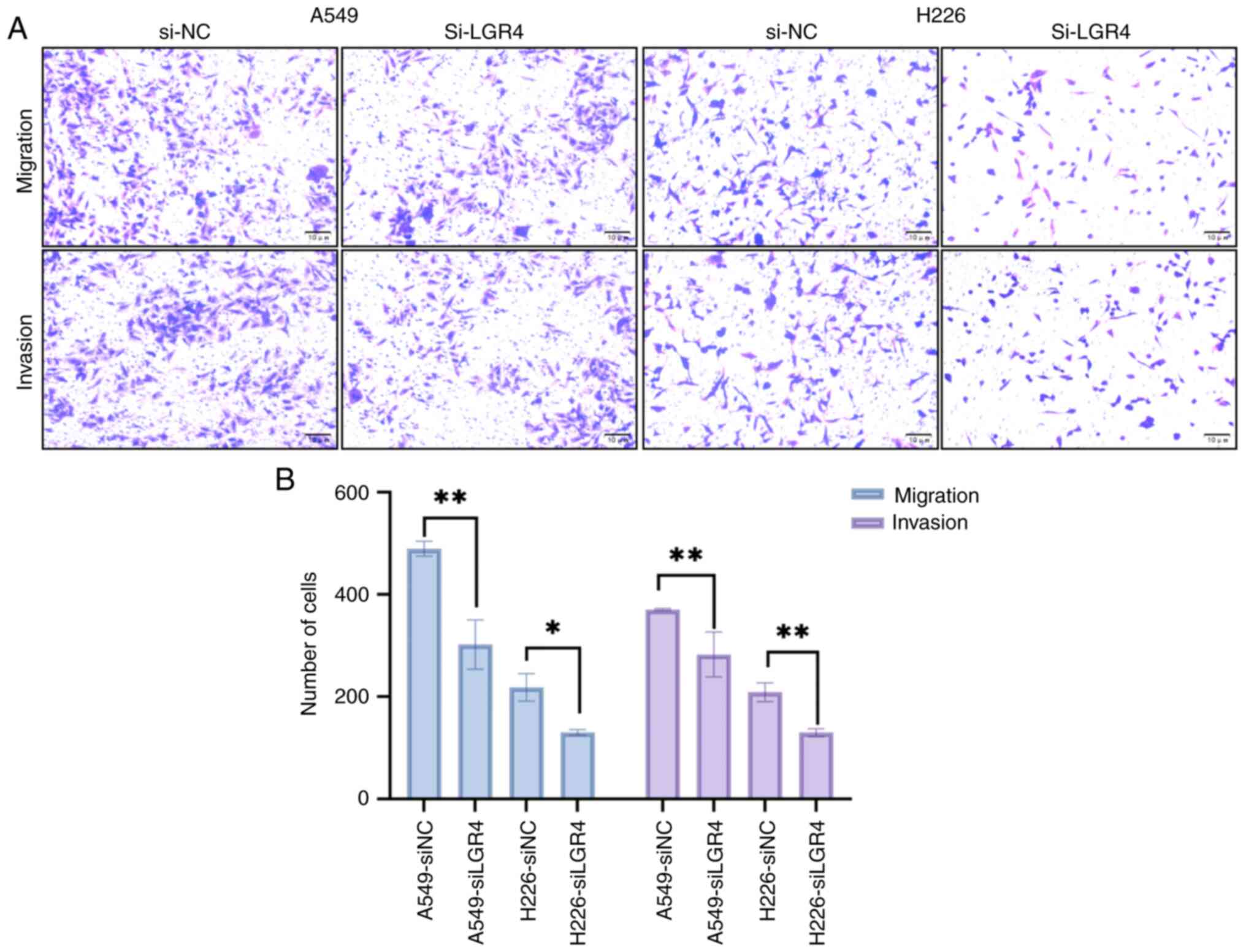
Knockdown of LGR4 inhibits invasion
and migration of A549 and H226 cells
To determine if LGR4 contributes to the invasive and
migratory behavior of NSCLC cells, the effect of LGR4 silencing on
the invasion and migration abilities of A549 and H226 cells was
evaluated using a Transwell assay. The results demonstrated that
transfection with si-LGR4 significantly decreased both the invasion
and migration abilities of A549 and H226 cells, indicating that
LGR4 depletion inhibits these key cellular processes (all
P<0.05; Fig. 5A and B).
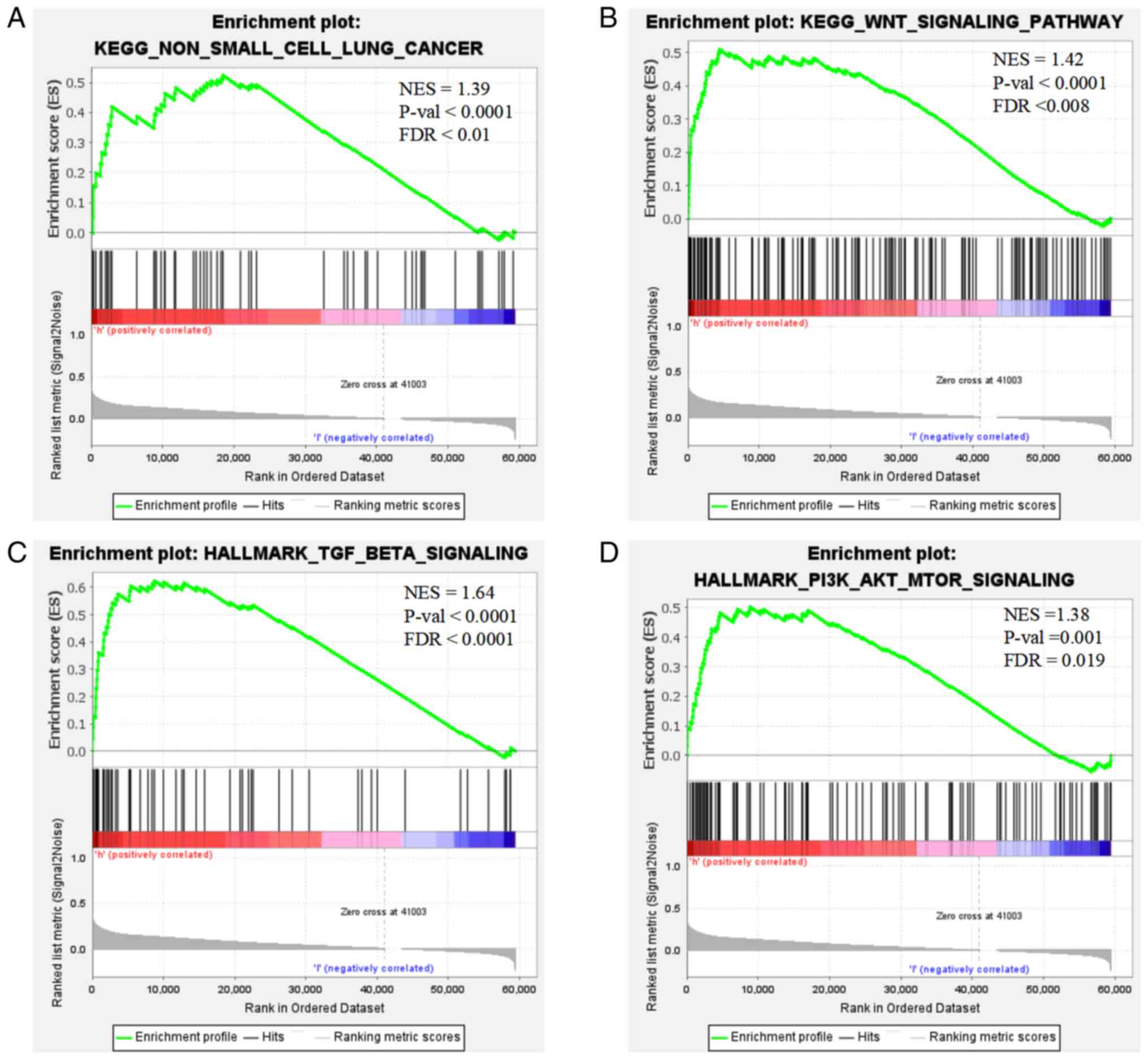
Enrichment analysis of LGR4
GSEA demonstrated that elevated LGR4 expression in
tumor samples was significantly enriched in the KEGG) ‘non-small
cell lung cancer’ pathway (Fig. 6A)
and was associated with activation of ‘Wnt signaling pathway’,
‘PI3K/AKT/mTOR signaling’ and ‘TGF-β signaling’ (Fig. 6B-D).
Discussion
GPCRs constitute the most extensive class of cell
surface receptors that mediate signal transduction, and are notable
drivers of tumor progression and metastasis (19). As a member of the GPCR family, LGR4
exerts various regulatory effects in multiple cell types and acts
on numerous targets. In the liver, it controls key gluconeogenic
enzymes (such as phosphoenolpyruvate carboxykinase) via
Wnt/β-catenin, affecting glucose output (20). In osteoblasts, LGR4 regulates
glycolysis, and its loss reduces bone formation and strength
(21). Abnormal LGR4 signaling is
highly influential in cancer and other disease, with
loss-of-function mutations associated with osteoporosis,
electrolyte imbalance, and reduced body weight, and
gain-of-function mutations linked to increased bone density,
insulin resistance, and obesity (22). A growing body of evidence indicates
that LGR4 is integral to both the development and progression of
various cancer types. For instance, Steffen et al (23) observed that the transcriptional and
translational levels of LGR4 and LGR6 are upregulated in gastric
cancer, with LGR4 expression significantly associated with lymph
node metastasis. Additionally, Cui et al (24) found that the expression of LGR4,
LGR5 and LGR6 is elevated in gastrointestinal cancer, with LGR5
being enriched in cancer stem cells. Research has demonstrated that
RSPO2 and receptor activator of nuclear factor κ-B ligand co-opt
LGR4 as a shared receptor to synergistically over-activate
osteoclastogenesis, thereby disrupting bone homeostasis and
establishing a premetastatic niche conducive to tumor colonization,
which ultimately drives breast cancer bone metastasis. Targeting
LGR4 may represent a novel therapeutic strategy to intercept this
process (25). Upregulation of
microRNA (miR)-449b significantly suppresses the growth and
invasive capacity of NSCLC by modulating LGR4 (26). The association between LGR4
expression and the development of NSCLC remains to be
elucidated.
In the present study, LGR4 expression was initially
confirmed in NSCLC tissues using immunohistochemistry. A marked
elevation of LGR4 was detected in NSCLC tissues when benchmarked
against normal controls and histologically healthy adjacent areas.
Notably, relatively high LGR4 expression was observed in some
non-cancerous tissues. This may be attributed to the physiological
roles of LGR4 in healthy tissue homeostasis and regeneration. In
addition, the adjacent non-tumor tissues may be influenced by
tumor-associated factors such as inflammation or hypoxia, which
could upregulate LGR4 expression. Alternatively, individual
variation and tissue heterogeneity might also contribute to this
observation. Furthermore, higher LGR4 expression was linked to a
poor prognosis in patients with NSCLC, consistent with the
bioinformatics analysis in the present study. Univariate and
multivariate analyses established high LGR4 expression as an
independent predictor of poor prognosis in NSCLC. The functional
role of LGR4 was then investigated in two NSCLC cell lines.
Relative to healthy lung epithelial cells, LGR4 expression was
significantly elevated in the A549 lung adenocarcinoma cell line
and the H226 squamous cell carcinoma cell line. In addition, LGR4
knockdown increased apoptosis and decreased proliferation, invasion
and migration abilities in both A549 and H226 cells.
The present enrichment analysis revealed that high
LGR4 expression levels may be associated with the activation of
major signaling pathways, including the Wnt, PI3K/AKT/mTOR and
TGF-β signaling pathways. Moreover, the significant enrichment of
LGR4 within the KEGG ‘non-small cell lung cancer’ pathway indicates
that LGR4 may be a potential key driver in NSCLC progression.
Numerous studies have reported that LGR4 and its homologs LGR5 and
LGR6 influence cellular signaling primarily through the Wnt pathway
via their interaction with the ligand R-spondin (27–30).
Furthermore, research has shown that LGR4 can regulate the
expression of TGF-β1, thereby triggering the TGF-β1/Smad signaling
pathway and driving multiple myeloma progression (31). Additionally, according to a study by
Liang et al (32),
overexpression of LGR4 upregulates mTOR and the key effector AKT in
the PI3K/AKTt pathway. Building on these findings, it is proposed
that LGR4 may promote NSCLC progression through the activation of
these pathways. These results from previous studies imply that LGR4
could be strongly associated with the initiation and progression of
NSCLC.
In conclusion, LGR4 may represent a promising
therapeutic approach for patients with NSCLC. However, the present
study is confined to in vitro experiments, and animal
studies have yet to be conducted. In the present research,
bioinformatics analysis was utilized to predict the potential
signaling pathways that LGR4 may activate in NSCLC. To further
validate the reliability of LGR4 as a diagnostic biomarker, our
future studies will focus on exploring the upstream regulatory
molecules of LGR4 and conducting additional experiments for further
confirmation.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Changzhou High-Level
Medical Talents Training Project (grant no. 2022CZBJ069), the
Changzhou Sci&Tech Program (grant no. CZ20220025), the ‘333
Project’ of Jiangsu Province (grant no. BRA2020157) and the 333
High-Level Talent Training Project (grant no. 2022-2).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XD was responsible for the conceptualization,
methodology, investigation, formal analysis, writing the original
draft, and reviewing and editing the manuscript. XL provided the
software, analyzed and curated the data (which involved ollection,
organization, cleaning, and preservation of research data,
including the archiving of experimental results, preparation of
data tables, and processing and management of images/raw data), and
provided experimental supervision. YS and SZ interpreted data. ML
and ZG performed the experiments. KY conceived and designed the
study, supervised the project, and revised the manuscript for
important scientific content. All authors have read and approved
the final manuscript. XD and KY confirm the authenticity of all the
raw data.
Ethics approval and consent to
participate
The Research Ethics Committee of The Third
Affiliated Hospital of Nanjing Medical University (Changzhou,
China) reviewed and approved the study [approval no. (2023)
KY422-01]. All participating patients and their families
voluntarily provided written informed consent. All tissue samples
were anonymized following ethical and legal guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48.
2023.PubMed/NCBI
|
|
3
|
National Lung Screening Trial Research
Team, . Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD,
Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM and Sicks JD:
Reduced lung-cancer mortality with low-dose computed tomographic
screening. N Engl J Med. 365:395–409. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Timmerman R, Paulus R, Galvin J, Michalski
J, Straube W, Bradley J, Fakiris A, Bezjak A, Videtic G, Johnstone
D, et al: Stereotactic body radiation therapy for inoperable early
stage lung cancer. JAMA. 303:1070–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun B, Brooks ED, Komaki R, Liao Z, Jeter
M, McAleer M, Balter PA, Welsh JD, O'Reilly M, Gomez D, et al:
Long-term outcomes of salvage stereotactic ablative radiotherapy
for isolated lung recurrence of non-small cell lung cancer: A phase
II clinical trial. J Thorac Oncol. 12:983–992. 2017. View Article : Google Scholar
|
|
6
|
Reck M, Rodríguez-Abreu D, Robinson AG,
Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe
S, et al: Pembrolizumab versus chemotherapy for PD-L1-positive
non-small-cell lung cancer. N Engl J Med. 375:1823–1833. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shaw AT, Kim DW, Nakagawa K, Seto T, Crinó
L, Ahn MJ, De Pas T, Besse B, Solomon BJ, Blackhall F, et al:
Crizotinib versus chemotherapy in advanced ALK-positive lung
cancer. N Engl J Med. 368:2385–2394. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Riely GJ, Wood DE, Ettinger DS, Aisner DL,
Akerley W, Bauman JR, Bharat A, Bruno DS, Chang JY, Chirieac LR, et
al: Non-Small cell lung cancer, version 4.2024, NCCN clinical
practice guidelines in oncology. J Natl Compr Canc Netw.
22:249–274. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Filipowska J, Kondegowda NG, Leon-Rivera
N, Dhawan S and Vasavada RC: LGR4, a G protein-coupled receptor
with a systemic role: From development to metabolic regulation.
Front Endocrinol (Lausanne). 13:8670012022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ordaz-Ramos A, Rosales-Gallegos VH,
Melendez-Zajgla J, Maldonado V and Vazquez-Santillan K: The role of
LGR4 (GPR48) in normal and cancer processes. Int J Mol Sci.
22:46902021. View Article : Google Scholar
|
|
11
|
Zheng H, Liu J, Cheng Q, Zhang Q, Zhang Y,
Jiang L, Huang Y, Li W, Zhao Y, Chen G, et al: Targeted activation
of ferroptosis in colorectal cancer via LGR4 targeting overcomes
acquired drug resistance. Nat Cancer. 5:572–589. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yue Z, Yuan Z, Zeng L, Wang Y, Lai L, Li
J, Sun P, Xue X, Qi J, Yang Z, et al: LGR4 modulates breast cancer
initiation, metastasis, and cancer stem cells. FASEB J.
32:2422–2437. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liang F, Zhang H, Cheng D, Gao H, Wang J,
Yue J, Zhang N, Wang J, Wang Z and Zhao B: Ablation of LGR4
signaling enhances radiation sensitivity of prostate cancer cells.
Life Sci. 265:1187372021. View Article : Google Scholar
|
|
14
|
Annunziato S, Sun T and Tchorz JS: The
RSPO-LGR4/5-ZNRF3/RNF43 module in liver homeostasis, regeneration,
and disease. Hepatology. 76:888–899. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Planas-Paz L, Orsini V, Boulter L,
Calabrese D, Pikiolek M, Nigsch F, Xie Y, Roma G, Donovan A, Marti
P, et al: The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver
zonation and size. Nat Cell Biol. 18:467–479. 2016. View Article : Google Scholar
|
|
16
|
Yue F, Jiang W, Ku AT, Young AIJ, Zhang W,
Souto EP, Gao Y, Yu Z, Wang Y, Creighton CJ, et al: A
Wnt-independent LGR4-egfr signaling axis in cancer metastasis.
Cancer Res. 81:4441–4454. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: The eighth edition AJCC cancer staging manual:
Continuing to build a bridge from a population-based to a more
‘personalized’ approach to cancer staging. CA Cancer J Clin.
67:93–99. 2017.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dorsam RT and Gutkind JS:
G-protein-coupled receptors and cancer. Nat Rev Cancer. 7:79–94.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fang Q, Ye L, Han L, Yao S, Cheng Q, Wei
X, Zhang Y, Huang J, Ning G, Wang J, et al: LGR4 is a key regulator
of hepatic gluconeogenesis. Free Radic Biol Med. 229:183–194. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang YY, Zhou YM, Xu JZ, Sun LH, Tao B,
Wang WQ, Wang JQ, Zhao HY and Liu JM: Lgr4 promotes aerobic
glycolysis and differentiation in osteoblasts via the canonical
Wnt/β-catenin pathway. J Bone Miner Res. 36:1605–1620. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang L, Wang J, Gong X, Fan Q, Yang X, Cui
Y, Gao X, Li L, Sun X, Li Y and Wang Y: Emerging roles for LGR4 in
organ development, energy metabolism and carcinogenesis. Front
Genet. 12:7288272022. View Article : Google Scholar
|
|
23
|
Steffen JS, Simon E, Warneke V, Balschun
K, Ebert M and Röcken C: LGR4 and LGR6 are differentially expressed
and of putative tumor biological significance in gastric carcinoma.
Virchows Arch. 461:355–365. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cui J, Toh Y, Park S, Yu W, Tu J, Wu L, Li
L, Jacob J, Pan S, Carmon KS and Liu QJ: Drug conjugates of
antagonistic R-spondin 4 mutant for simultaneous targeting of
leucine-rich repeat-containing g protein-coupled receptors 4/5/6
for cancer treatment. J Med Chem. 64:12572–12581. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yue Z, Niu X, Yuan Z, Qin Q, Jiang W, He
L, Gao J, Ding Y, Liu Y, Xu Z, et al: RSPO2 and RANKL signal
through LGR4 to regulate osteoclastic premetastatic niche formation
and bone metastasis. J Clin Invest. 132:e1445792022. View Article : Google Scholar
|
|
26
|
Yang D, Li JS, Xu QY, Xia T and Xia JH:
Inhibitory effect of MiR-449b on cancer cell growth and invasion
through LGR4 in non-small-cell lung carcinoma. Curr Med Sci.
38:582–589. 2018. View Article : Google Scholar
|
|
27
|
de Lau W, Barker N, Low TY, Koo BK, Li VS,
Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M,
et al: Lgr5 homologues associate with Wnt receptors and mediate
R-spondin signalling. Nature. 476:293–297. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Luo J, Yang Z, Ma Y, Yue Z, Lin H, Qu G,
Huang J, Dai W, Li C, Zheng C, et al: LGR4 is a receptor for RANKL
and negatively regulates osteoclast differentiation and bone
resorption. Nat Med. 22:539–546. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
de Lau W, Peng WC, Gros P and Clevers H:
The R-spondin/Lgr5/Rnf43 module: Regulator of Wnt signal strength.
Genes Dev. 28:305–316. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Han XH, Jin YR, Tan L, Kosciuk T, Lee JS
and Yoon JK: Regulation of the follistatin gene by RSPO-LGR4
signaling via activation of the WNT/β-catenin pathway in skeletal
myogenesis. Mol Cell Biol. 34:752–764. 2014. View Article : Google Scholar
|
|
31
|
Yi Z, Ma T, Liu J, Tie W, Li Y, Bai J, Li
L and Zhang L: LGR4 promotes tumorigenesis by activating
TGF-β1/Smad signaling pathway in multiple myeloma. Cell Signal.
110:1108142023. View Article : Google Scholar
|
|
32
|
Liang F, Yue J, Wang J, Zhang L, Fan R,
Zhang H and Zhang Q: GPCR48/LGR4 promotes tumorigenesis of prostate
cancer via PI3K/Akt signaling pathway. Med Oncol. 32:492015.
View Article : Google Scholar
|















![LGR4 is highly expressed in NSCLC. (A)
Unpaired analysis. TCGA [NSCLC (n=1,043) vs. healthy tissues
(n=110); unpaired Student's t-test]. (B) High LGR4 expression is
associated with a poor prognosis in patients with NSCLC (data
fromTCGA). (C) Western blot analysis showed the expression level of
LGR4 protein in NSCLC cell lines. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001. (D) RT-qPCR analysis showed the
expression level of LGR4 mRNA in NSCLC cell lines (one-way ANOVA
followed by Dunnett's multiple comparison test). (E) RT-qPCR
analysis showed that si-LGR4 transfection significantly reduced
LGR4 expression in A549 and H226 cells compared with the si-NC
group (one-way ANOVA followed by Dunnett's multiple comparison
test). LGR4, leucine-rich repeat-containing G-protein coupled
receptor 4; NSCLC, non-small cell lung cancer; TCGA, The Cancer
Genome Atlas; RT-qPCR, reverse transcription-quantitative PCR;
si-NC, small-interfering RNA negative control.](/article_images/ol/30/6/ol-30-06-15304-g00.jpg)