Introduction
Gastric cancer (GC) ranks among the most prevalent
malignancies worldwide, with a notable clinical burden. According
to GLOBOCAN 2022, GC accounts for ~1 million new cases annually,
and remains the fourth leading cause of cancer-related mortality,
characterized by a poor 5-year survival rate of <30% in advanced
stages (1). For patients with GC
with an adequate performance status and organ function, those who
receive combination chemotherapy have a median overall survival
(OS) of ~1 year, compared with 3–4 months for those treated solely
with supportive care (2). In recent
years, immunotherapy has markedly expanded treatment options for
solid tumors, and chemo-immunotherapy holds particular promise for
advanced GC; however, GC prognosis remains poor. Over half of
patients are diagnosed with advanced/metastatic stage disease, with
a median OS of ~1 year (3).
The pathogenesis of GC involves multifactorial
etiologies, including a genetic predisposition, Helicobacter
pylori (H. pylori) infection, inflammatory stimulation
and environmental factors (4–7). Early
diagnosis of GC is challenging due to nonspecific symptoms, leading
to delayed therapeutic interventions and poor prognosis (8). In addition, chemoresistance notably
undermines treatment efficacy, as standard regimens often fail to
achieve durable responses (9,10).
This resistance is attributed to dynamic molecular adaptations in
tumor cells, such as drug efflux mechanisms and apoptosis evasion
(11). Furthermore, GC progression
is driven by an intricate regulatory network involving aberrant
signaling pathways, epigenetic modifications and tumor
microenvironment (TME) interactions (12–15).
Despite advances in understanding GC biology, key gaps persist
(16). The molecular mechanisms
underlying tumorigenesis, metastasis and chemoresistance remain
incompletely elucidated, particularly regarding context-dependent
roles of non-coding RNAs (17).
Addressing these knowledge gaps is critical for developing targeted
therapies and improving clinical outcomes.
Previous studies have increasingly focused on the
role of long non-coding (lnc)RNA in the progression of several
cancers, including GC. Among them, maternally expressed gene 3
(MEG3) has emerged as a critical player in tumor suppression. MEG3
is known for its ability to regulate several cellular processes
such as proliferation, apoptosis and migration, which are essential
for cancer progression (18–20).
Therefore, understanding the role of MEG3 in GC could provide novel
insights into its pathogenesis and open new avenues for early
diagnosis and targeted therapies.
Studies have reported that MEG3 is frequently
downregulated in GC tissues, compared with in normal gastric
tissues, suggesting its potential role as a tumor suppressor
(21–23). Moreover, in vitro studies
have reported that reintroducing MEG3 into GC cell lines leads to
increased apoptosis and decreased cell viability, migration and
invasion (24,25). This is achieved through the
modulation of epithelial-to-mesenchymal transition (EMT), a process
critical for cancer metastasis (26). MEG3 has been reported to enhance the
expression of epithelial markers such as E-cadherin, whilst
suppressing mesenchymal markers such as vimentin and fibronectin,
effectively inhibiting the EMT process (27). Furthermore, the interaction between
MEG3 and numerous signaling pathways has been a focus of research.
MEG3 has been reported to interact with microRNAs (miRNAs/miRs),
particularly miR-21, which is known to promote tumorigenesis in
several types of cancer. By acting as a competing endogenous
(ce)RNA, MEG3 can sequester miR-21, thereby reducing its oncogenic
effects (28). This regulatory axis
highlights the intricate network of interactions among non-coding
RNAs in cancer biology and reinforces the importance of lncRNAs
such as MEG3 in modulating tumor behavior.
In addition to its role in cellular processes, MEG3
expression has been associated with the presence of H.
pylori, a notable risk factor for GC. Studies have reported
that H. pylori infection is associated with altered
expression levels of MEG3 and other lncRNAs, suggesting that
microbial factors may influence the lncRNA landscape in GC
(29,30). Understanding how H. pylori
and other environmental factors interact with lncRNAs such as MEG3
could provide valuable insights into the multifactorial nature of
GC development. Furthermore, the therapeutic implications of
targeting MEG3 in GC are profound. Given its role in inhibiting
tumor growth and metastasis, strategies aimed at restoring MEG3
expression or mimicking its function could serve as potential
therapeutic approaches. For instance, the use of small molecules or
gene editing technologies to enhance MEG3 levels in GC cells may
reverse the malignant phenotype and sensitize tumors to
conventional therapies (31).
Therefore, the role of lncRNA MEG3 in GC is a
burgeoning area of research that holds promise for both
understanding the mechanisms of the disease and developing novel
therapeutic strategies. The evidence supporting the
tumor-suppressive functions of MEG3, its interactions with key
signaling pathways and its potential as a biomarker underscore the
importance of further investigations into this lncRNA. As research
continues to unravel the complexities of GC biology, MEG3 may
emerge as a pivotal target for innovative treatment modalities
aimed at combating this formidable malignancy.
MEG3 structure and function
MEG3 gene structure
MEG3 is a lncRNA located within the imprinted δ-like
non-canonical Notch ligand 1-MEG3 locus on chromosome 14q32.3
(32). This gene spans ~35 kb and
consists of 10 exons, which are crucial for its functional
integrity (33). The MEG3 gene is
characterized by its imprinted expression, meaning it is expressed
from the maternal allele, whilst the paternal allele is silenced
(34). This unique genetic
regulation is notable, as it contributes to the functional
diversity of lncRNAs in several biological processes. The
transcription of MEG3 results in a 1.6 kb lncRNA that does not code
for proteins but serves essential roles in regulating gene
expression through numerous mechanisms, including through its
interaction with miRNAs and proteins (35,36).
Notably, MEG3 has been reported to form secondary structures,
including pseudoknots, which are vital for its interaction with the
p53 pathway, further emphasizing the importance of its structural
features in mediating biological functions (37).
Major biological functions of
MEG3
MEG3 has emerged as a critical player in several
biological processes, particularly in tumor suppression. Its
primary functions include regulating cell proliferation, apoptosis
and migration, particularly in the context of cancer (38). MEG3 exerts its tumor-suppressive
effects by modulating key signaling pathways, such as the p53
pathway, where it enhances p53 activity, leading to increased
apoptosis in cancer cells (39). In
addition, MEG3 functions as a molecular sponge for multiple
oncogenic miRNAs, including miR-21, which is known to promote
oncogenesis (40). By sequestering
miR-21, MEG3 indirectly upregulates tumor suppressor genes and
inhibits pathways that contribute to cancer progression (41). Studies have reported that MEG3
downregulation is associated with poor prognosis in several types
of cancer, including hepatocellular carcinoma, GC and colorectal
cancer, indicating its potential as a diagnostic and therapeutic
target (42–44). In addition, beyond its role in
cancer, MEG3 has been implicated in other physiological processes,
such as fibrosis and metabolic regulation. It has been reported to
influence the expression of genes involved in EMT, a critical
process in cancer metastasis, and to regulate insulin signaling
pathways, highlighting its multifaceted role in both cancer biology
and metabolic diseases (45,46).
Expression characteristics of MEG3 in
cells
MEG3 exhibits distinct expression patterns across
numerous tissues and cell types, reflecting its functional
diversity. In healthy tissues, MEG3 is generally expressed at
higher levels, particularly in the brain, liver and skeletal muscle
(47). However, its expression is
often markedly reduced in several cancer types, where it is
associated with tumor progression and metastasis (48,49).
This downregulation is frequently attributed to epigenetic
modifications, such as the hypermethylation of the MEG3 promoter.
Specifically, the downregulation of MEG3 is driven by abnormal CpG
methylation in its promoter region. Among these CpG sites, linear
changes in MEG3 expression show a negative correlation with the
methylation levels of two specific CpG sites: MEG3_4_CpG_9 and
MEG3_5_CpG_2 (50,51). The methylation process of MEG3
promoters is mainly mediated by DNA methyltransferases (DNMTs)
including three major DNMTs: DNMT1, DNMT3a and DNMT3b in mammals.
In addition, MEG3 expression is influenced by several physiological
conditions (52,53). For instance, studies have reported
that MEG3 levels can be modulated in response to inflammatory
stimuli and metabolic changes, indicating its potential role as a
biomarker for disease states (54,55).
In the context of obesity and insulin resistance, MEG3 expression
has been reported to be negatively associated with the severity of
these conditions, suggesting its involvement in metabolic
regulation (56). Furthermore, the
dynamic association between MEG3 and nuclear speckles, and its
transient expression in response to transcriptional activity,
highlight its regulatory role in gene expression within the
cellular context, suggesting that MEG3 may serve as a sensor for
cellular stress and a regulator of gene expression in response to
environmental cues (57).
MEG3 expression changes in GC
Comparison of expression in normal and
cancerous tissues
The expression levels of the lncRNA MEG3 have been
reported to markedly differ between normal gastric and GC tissues
(58). In normal gastric cells,
MEG3 is typically expressed at higher levels, functioning as a
tumor suppressor that regulates several cellular processes,
including apoptosis and cell proliferation (59). However, studies have consistently
reported that MEG3 expression is markedly decreased in GC cells, as
compared with their normal counterparts (25,60)
For instance, a study indicated that MEG3 transfection in GC cells
led to an increased expression of E-cadherin, a key epithelial
marker, whilst simultaneously inhibiting the expression of
mesenchymal markers, such as vimentin and fibronectin. This
suggests that MEG3 serves a crucial role in suppressing EMT
(24). Furthermore, the
differential expression of MEG3 has been reported to be associated
with several clinical parameters in patients with GC. Lower levels
of MEG3 have been associated with advanced disease stages and worse
prognostic outcomes, indicating its potential as a biomarker for
disease progression (60). The
downregulation of MEG3 in cancerous tissues not only highlights its
role in GC pathogenesis but also underscores its potential as a
therapeutic target (61).
Association between MEG3 expression
and clinical pathological features
The association between MEG3 expression and clinical
pathological features in GC has been a focal point in recent
studies (62,63). Numerous studies have reported that
the expression levels of MEG3 are notably associated with several
clinicopathological characteristics, including tumor size, lymph
node metastasis and overall patient survival (61,64).
For instance, lower MEG3 expression levels have been associated
with larger tumor sizes and a higher incidence of lymph node
metastasis, suggesting that MEG3 may serve a critical role in tumor
aggressiveness (65). These
findings indicate that MEG3 could serve as a valuable prognostic
marker in GC, aiding in the stratification of patients based on
their risk of disease progression. Furthermore, the expression of
MEG3 has been associated with the OS rates of patients with GC.
Studies have reported that patients with higher levels of MEG3
expression tend to have improved survival outcomes, as compared
with those with lower expression levels (66). This association suggests that MEG3
may not only be involved in the tumorigenic processes, but it also
serves as a potential therapeutic target.
Impact of MEG3 on the biological behavior of
GC cells
Cell proliferation
lncRNA MEG3 has emerged as a critical regulator of
cell proliferation in several types of cancer, including GC
(67). Studies have reported that
MEG3 functions as a tumor suppressor, with its downregulation
associated with increased cell proliferation and malignancy
(68,69). This reduction in MEG3 has been
associated with the activation of oncogenic pathways that promote
cell cycle progression and proliferation (70). For instance, MEG3 has been reported
to inhibit the expression of cyclin D1 and other cell cycle
regulators, thereby slowing down the transition from the G1 phase
to the S phase of the cell cycle (71). In addition, the overexpression of
MEG3 in GC cell lines has been reported to markedly reduce cell
growth and proliferation rates, suggesting that MEG3 acts to
restrain the proliferative capacity of these cells (72). Furthermore, the interaction between
MEG3 and several miRNAs has been implicated in its regulatory role,
as MEG3 can act as a sponge for miRNAs that promote proliferation,
thus further inhibiting cancer cell growth (73).
Cell apoptosis
The role of MEG3 in promoting apoptosis in GC cells
has been extensively investigated, and it has been established that
it is critically involved in the regulation of programmed cell
death (58). MEG3 has been reported
to enhance the sensitivity of GC cells to apoptotic stimuli,
effectively increasing the rate of apoptosis in these cells
(74). This is particularly
important given that numerous types of GC exhibit resistance to
apoptosis, contributing to tumor progression and treatment failure
(75,76). Mechanistically, MEG3 facilitates
apoptosis through several pathways. It has been reported to
upregulate pro-apoptotic proteins such as Bax and downregulate
anti-apoptotic proteins such as Bcl-2, thereby tipping the balance
in favor of cell death (77,78).
In addition, MEG3 can activate the caspase cascade, a series of
proteolytic enzymes critical for the execution of apoptosis. For
instance, the increased expression of MEG3 has been associated with
the enhanced activation of caspase-3 and caspase-9, leading to the
induction of apoptosis in GC cells (79,80).
Beyond direct protein regulation, MEG3 further potentiates its
pro-apoptotic effects by interacting with miRNAs. Studies have
indicated that MEG3 can interact with miRNAs that regulate
apoptosis, further enhancing its pro-apoptotic effects. For
example, MEG3 has been reported to sequester miR-21, a known
anti-apoptotic miRNA, thereby promoting apoptosis in GC cells
(21). The ability of MEG3 to
induce apoptosis not only highlights its potential as a tumor
suppressor, but also suggests that therapeutic strategies aimed at
increasing MEG3 expression or mimicking its function could enhance
the efficacy of existing treatments for GC.
Cell migration and invasion
MEG3 also serves a central regulatory role in
regulating the migration and invasion of GC cells, processes that
are decisive for metastasis. The downregulation of MEG3 has been
associated with increased migration and invasion rates of GC cells,
which is a hallmark of aggressive tumor behavior (72). Moreover, studies have reported that
MEG3 overexpression is associated with a marked reduction in the
migration and invasion rates of GC cells, indicating its function
as a suppressor of these malignant behaviors (21,24).
The mechanisms through which MEG3 exerts its effects on cell
migration and invasion involve the modulation of EMT, a process
that is critical for cancer cell dissemination (81). MEG3 has been reported to inhibit EMT
by promoting the expression of epithelial markers, such as
E-cadherin, whilst suppressing mesenchymal markers such as
N-cadherin and vimentin (82). This
shift in cellular phenotype is crucial for maintaining the adhesive
properties of cancer cells and reducing their invasive potential
(83). Furthermore, MEG3 has been
reported to interact with several signaling pathways that regulate
cell motility, including the Rho GTPase family, which is known to
control cytoskeletal dynamics and cell movement (84). By affecting these pathways, MEG3 can
alter the migratory behavior of GC cells, thereby reducing their
ability to invade surrounding tissues and cause distant metastasis
(72). Given the importance of
migration and invasion in the progression of GC, targeting MEG3 or
its regulatory networks may provide a novel therapeutic strategy to
inhibit metastasis and improve patient prognosis.
MEG3 signaling pathways and mechanisms
Relationship between MEG3 and the p53
signaling pathway
The p53 signaling pathway is a crucial regulator of
the cell cycle, apoptosis and genomic stability, often referred to
as the ‘guardian of the genome’. MEG3 has been reported to interact
with this pathway in multiple cancer types and research has
indicated that MEG3 can enhance the expression of p53, thereby
promoting apoptosis in cancer cells (85). For instance, in neuroblastoma, MEG3
overexpression led to increased p53 levels, which in turn activated
pro-apoptotic factors and inhibited cell proliferation (86). This interaction suggests that MEG3
may function as a tumor suppressor by stabilizing p53 and enhancing
its tumor-suppressive activities (87). In addition to directly regulating
p53 expression, MEG3 also indirectly influences p53 activity
through interactions with miRNAs. MEG3 has been identified as a
ceRNA that can sponge several miRNAs, including miR-21, which is
known to inhibit p53 expression. By sequestering miR-21, MEG3
indirectly promotes p53 activity, leading to enhanced apoptosis and
reduced tumor growth (88). This
regulatory mechanism highlights the potential of MEG3 in modulating
the p53 pathway and its implications for cancer therapy,
particularly in tumors where p53 is often mutated or dysfunctional.
In addition to its role in apoptosis, the interaction between MEG3
and p53 also extends to cellular senescence and DNA damage
response. Studies have reported that MEG3 can influence the
expression of genes involved in these processes, thereby
contributing to the maintenance of genomic integrity (89,90).
The ability of MEG3 to modulate p53 activity positions it as a
critical player in cancer progression and treatment resistance,
making it a promising target for therapeutic intervention (Fig. 1).
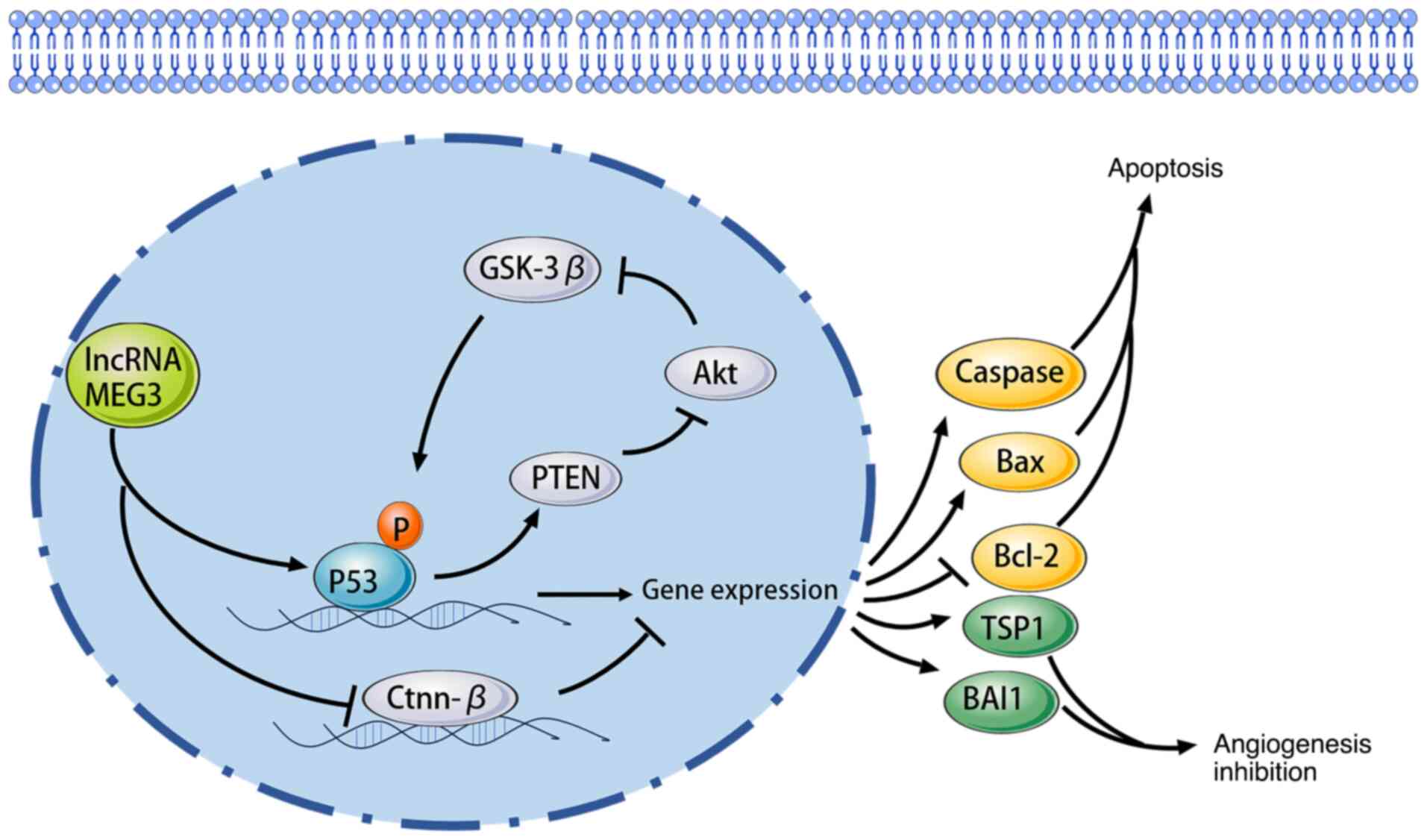 | Figure 1.Relationship between MEG3 and the p53
signaling pathway. MEG3, maternally expressed gene 3; p53, protein
53; lncRNA, long non-coding RNA; GSK-3β, glycogen synthase
kinase-3; Akt, protein kinase B; PTEN, phosphatase and tensin
homologue deleted on chromosome ten; Ctnn-β, catenin beta; Caspase,
cysteinyl aspartate specific proteinase; Bcl-2, B-cell lymphoma-2;
Bax, Bcl-2 associated X; TSP1,thrombin sensitive protein 1; BAI1,
brain-specific angiogenesis inhibitor 1. |
Role of MEG3 in miRNA regulation
MEG3 serves a pivotal role in the regulation of
miRNAs, which are small non-coding RNAs that post-transcriptionally
regulate gene expression. By acting as a ceRNA, MEG3 can bind to
specific miRNAs, thereby preventing them from interacting with
their target mRNAs. This function has been particularly noted with
miR-21, which is often overexpressed in several types of cancer and
is associated with poor prognosis (91). The ability of MEG3 to sponge miR-21
results in the downregulation of its target genes, including those
involved in apoptosis and cell cycle regulation (92). In GC, for example, MEG3 has been
reported to inhibit the expression of miR-21, leading to the
upregulation of pro-apoptotic proteins and the downregulation of
anti-apoptotic factors, thus promoting apoptosis in cancer cells
(21). This regulatory axis
underscores the importance of MEG3 in maintaining cellular
homeostasis and preventing tumorigenesis. Furthermore, the
interaction of MEG3 with other miRNAs, such as miR-548d-3p and
miR-376a, has been documented, indicating its broader role in
modulating several signaling pathways associated with cancer
progression (93,94). By regulating these miRNAs, MEG3 can
influence key cellular processes, including proliferation,
migration and invasion, thereby contributing to the overall TME and
its dynamics. The therapeutic potential of targeting the MEG3-miRNA
axis is notable, as restoring MEG3 levels in tumors with low
expression could enhance the efficacy of existing therapies by
re-establishing normal apoptotic signaling and inhibiting oncogenic
pathways (95).
Impact of MEG3 on the TME
The TME exerts a pivotal influence on cancer
progression and metastasis, influencing tumor behavior and response
to therapy. Emerging evidence demonstrates that MEG3 markedly
impacts the TME by modulating the behavior of several cell types
within it, including immune cells, fibroblasts and endothelial
cells. For instance, MEG3 can promote the polarization of
macrophages towards the M1 phenotype, which is associated with
antitumor immunity, whilst inhibiting the M2 phenotype that
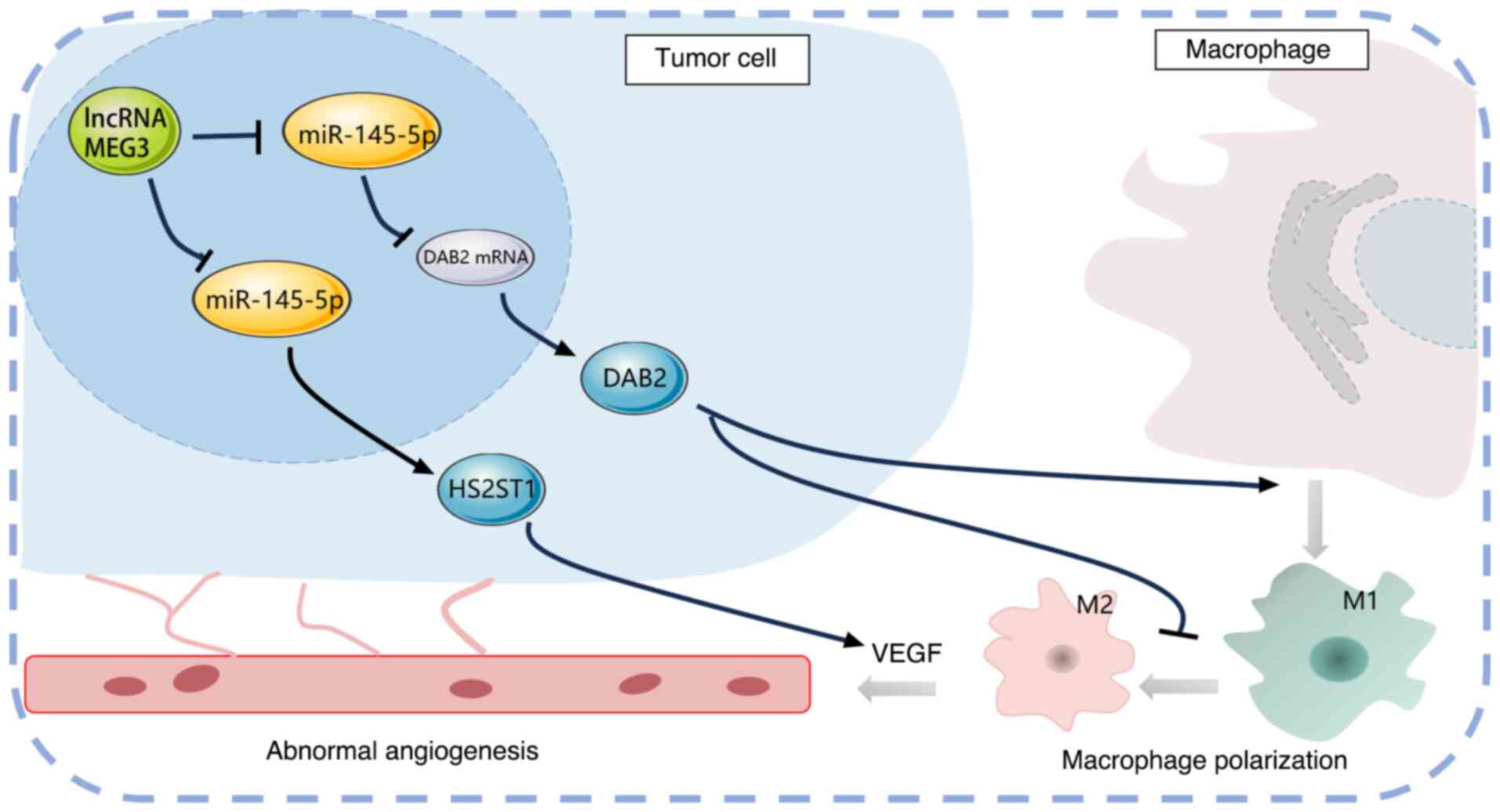
supports tumor growth and metastasis (96). In addition, MEG3 has been reported
to inhibit M2 macrophage polarization, thereby reducing tumor
growth and metastasis (97). This
effect is mediated through its interaction with miR-145-5p, which
regulates the expression of disabled-2, a protein involved in
macrophage polarization. By modulating macrophage phenotypes, MEG3
can alter the inflammatory landscape of the TME, potentially
leading to improved therapeutic outcomes. Furthermore, the role of
MEG3 in regulating angiogenesis is notable. For example, MEG3 has
been reported to inhibit angiogenesis by reducing the levels of
miR-421, which promotes endothelial cell migration and tube
formation (98). These findings
imply that MEG3 not only influences immune cell behavior, but also
impacts the vascular components of the TME, further underscoring
its multifaceted role in cancer biology. The ability of MEG3 to
shape the TME underscores its promising potential as a therapeutic
target. By restoring or enhancing MEG3 expression, it may be
possible to reprogram the TME to favor antitumor responses, and
lead to an improved drug delivery and enhanced efficacy of existing
therapies (21). This comprehensive
understanding of the signaling pathways and mechanisms of MEG3
provides valuable insights into its potential as a therapeutic
target in cancer treatment (Fig.
2).
MEG3 as a potential biomarker for GC
Diagnostic value of MEG3
The lncRNA MEG3 has been established as a promising
biomarker in the diagnosis and prognosis of GC. Numerous studies
have reported that MEG3 is downregulated in GC tissues and cell
lines compared with normal gastric tissues and cell lines,
indicating its potential as a diagnostic biomarker (25,54,55).
For instance, research has shown that the expression of the lncRNA
MEG3 was markedly lower in GC cells than in normal gastric cells,
and its transfection led to an increased expression of E-cadherin,
a marker associated with epithelial cells, whilst inhibiting the
expression of mesenchymal markers, such as vimentin and fibronectin
(24). This suggests that MEG3
serves a crucial role in EMT, a process that is pivotal in cancer
progression and metastasis. Furthermore, a previous study
demonstrated the diagnostic utility of MEG3 using receiver
operating characteristic curve analyses, which indicated that MEG3
levels could effectively discriminate between tumor and non-tumor
tissues. The area under the curve of MEG3 was 0.8736, and using the
cut-off value of 0.0014, the sensitivity and specificity of MEG3
were 79 and 86%, respectively, indicating the strong diagnostic
performance of GC (32). In
addition, another study reported that the A allele at the rs7158663
19 loci of MEG3 was a risk factor for GC (odds ratio, 1.41; 95%
confidence interval, 1.14–1.74; P=0.002) (99). This association underscores the
potential of MEG3 not only as a diagnostic marker but also as a
prognostic indicator for patient outcomes. The role of MEG3 in GC
is further supported by its involvement in several molecular
pathways that regulate cell proliferation, apoptosis and migration.
Studies have reported that MEG3 can inhibit tumor growth by
modulating the expression of key oncogenic pathways, such as the
PI3K/Akt and Wnt/β-catenin signaling pathways (21). The downregulation of MEG3 has been
associated with the increased expression of oncogenic microRNAs,
such as miR-21, which promotes tumor growth and metastasis
(28). Thus, the restoration of
MEG3 expression in GC cells has been proposed as a therapeutic
strategy that could reverse the malignant phenotype and improve
patient prognosis (72).
Furthermore, the potential of MEG3 as a biomarker extends beyond
tissue expression levels. Its presence in the serum has been
investigated, with studies suggesting that circulating MEG3 could
serve as a non-invasive biomarker for GC diagnosis (100,101). The ability to detect MEG3 in serum
samples adds a layer of practicality to its application in clinical
settings, allowing for early detection and monitoring of disease
progression. This is particularly important given the asymptomatic
nature of early-stage GC, which can lead to late diagnosis and poor
outcomes. The diagnostic value of MEG3 in GC is supported by its
downregulation in tumor tissues, association with clinical
parameters, involvement in critical signaling pathways, and
potential as a circulating biomarker. These findings collectively
highlight the potential of MEG3 as a valuable tool for the early
detection and management of GC, warranting further exploration in
clinical trials and translational research.
Feasibility of MEG3 as a therapeutic
target
The feasibility of targeting MEG3 for therapeutic
interventions in GC is increasingly supported by research that
underscores its role as a tumor suppressor. MEG3 has been reported
to exert notable antitumor effects through several mechanisms,
making it an attractive candidate for targeted therapies. For
instance, studies have indicated that MEG3 can inhibit cell
proliferation, migration and invasion in GC cells by modulating the
expression of several key genes involved in these processes
(24,60). Specifically, MEG3 has been reported
to inhibit the expression of anti-apoptotic proteins, such as
Bcl-2, whilst promoting pro-apoptotic proteins, such as caspase-3
and caspase-9, thereby enhancing apoptosis in GC cells (24). The therapeutic potential of MEG3 is
further highlighted by its ability to regulate the EMT process,
which is crucial for cancer metastasis (102). By inhibiting EMT, MEG3 can
potentially reduce the invasive capabilities of GC cells, thereby
limiting the spread of the disease (103). This regulatory function aligns
with the growing interest in targeting EMT as a therapeutic
strategy for several types of cancer, including GC. Furthermore,
the restoration of MEG3 expression in cancer cells has been
reported to reverse EMT and restore the epithelial phenotype,
suggesting that MEG3 could serve as a therapeutic target to inhibit
cancer progression (104). In
addition to its direct effects on tumor cells, the interactions of
MEG3 with other non-coding RNAs and signaling pathways further
enhance its therapeutic feasibility. For example, MEG3 has been
identified as a ceRNA that can sponge miRNAs, such as miR-21, which
are known to promote oncogenic processes (29). By sequestering these miRNAs, MEG3
can counteract their effects, providing a novel mechanism through
which MEG3 exerts its tumor-suppressive functions (105). This interplay between MEG3 and
miRNAs presents an opportunity for developing RNA-based therapies
that could enhance the expression of MEG3 or mimic its function in
GC cells. Furthermore, the potential for using MEG3 as a
therapeutic target is supported by its association with patient
outcomes (106–108). Studies have reported that patients
with low MEG3 expression levels tend to have worse prognoses,
reinforcing the notion that MEG3 serves a critical role in tumor
suppression (102,103,109). This association suggests that
therapies aimed at restoring or enhancing MEG3 function could
improve patient survival and quality of life. Furthermore, the
feasibility of targeting MEG3 as a therapeutic intervention in GC
is supported by its role as a tumor suppressor, its ability to
inhibit critical cancer processes, such as EMT, and its
interactions with other regulatory RNAs. The development of
strategies to enhance MEG3 expression or mimic its function holds
promise for improving therapeutic outcomes in patients with GC.
Further research into the mechanisms underlying the effects of MEG3
and its potential applications in clinical settings is warranted to
fully realize its therapeutic potential.
Conclusions and limitations
The exploration of lncRNAs has notably expanded the
understanding of the molecular mechanisms underlying numerous types
of cancer, particularly GC. Among these lncRNAs, MEG3 has emerged
as a critical player, demonstrating a complex relationship with the
biological characteristics of GC. The present review highlights the
multifaceted roles of MEG3 in the initiation and progression of GC,
suggesting that its expression levels are intricately associated
with the behavior of the tumor and patient outcomes.
The development of research surrounding MEG3
underscores the need for a balanced approach when interpreting
diverse findings across studies. As the understanding deepens, it
is crucial to integrate data from molecular biology, clinical
observations and bioinformatics to construct a comprehensive
picture of the function of MEG3. However, the dual nature of MEG3
as both a tumor suppressor and a potential contributor to
tumorigenesis presents a challenge: Whilst certain studies have
elucidated its role in inhibiting GC cell proliferation and
promoting apoptosis, others have pointed to contexts where MEG3 may
be involved in facilitating tumor growth. This dichotomy emphasizes
the necessity of contextualizing research findings within specific
biological and environmental frameworks.
Furthermore, the impact of MEG3 on GC not only
pertains to its mechanistic insights but also extends to its
potential as a biomarker for early diagnosis and a target for
therapeutic interventions. The association between MEG3 expression
and several clinicopathological features of GC suggests that it
could serve as a prognostic indicator, aiding in risk
stratification and personalized treatment plans. However, for MEG3
to transition from bench to bedside, future research must
rigorously validate its clinical utility across diverse patient
populations and genetic backgrounds.
Additionally, when considering the clinical
application of MEG3, it is essential to address the challenges and
limitations inherent in translating basic research into therapeutic
strategies. The heterogeneity of GC, characterized by its diverse
molecular subtypes and varying responses to treatment, necessitates
a nuanced understanding of how MEG3 interacts with other molecular
pathways. Collaborative efforts that bring together oncologists,
molecular biologists and bioinformaticians are critical to
deciphering these complex interactions and refining the potential
therapeutic applications of MEG3.
Moreover, the evolving landscape of cancer
treatment, particularly with the advent of precision medicine,
provides a fertile ground for investigating the role of MEG3 in
targeted therapies. As the intricate networks in which MEG3 is
involved continue to be elucidated, there lies an opportunity to
develop innovative approaches that leverage the properties of this
lncRNA to enhance treatment efficacy and minimize adverse effects.
The integration of MEG3 modulation into existing treatment
regimens, such as chemotherapy or immunotherapy, could yield
notable benefits, warranting further exploration.
In conclusion, whilst MEG3 presents promising
avenues for both research and clinical application in GC, a
concerted effort is needed to harmonize the diverse findings and
perspectives within the field. Future studies should strive for a
multidisciplinary approach, focusing on elucidating the precise
roles of MEG3 in GC biology and its implications for patient
management. By fostering collaboration and innovation, MEG3 could
become a cornerstone in the future of GC diagnosis and therapy,
ultimately improving outcomes for patients affected by this
challenging disease.
Despite the comprehensive synthesis of current
knowledge on MEG3 in GC, this review had several limitations that
should be acknowledged. Most included studies relied on in
vitro cell line models and animal xenograft experiments, with
relatively few well-powered clinical trials or prospective cohort
studies. This discrepancy limited the direct translation of
findings to human GC patients, as cell lines and animal models may
not fully recapitulate the complex TME and genetic heterogeneity of
clinical tumors. In addition, the molecular mechanisms underlying
MEG3′s regulatory role remain partially elusive. While key
signaling pathways, such as p53 and PI3K/Akt are discussed in
existing literature, the crosstalk between MEG3 and other
non-coding RNAs or epigenetic modifiers is insufficiently explored,
leaving gaps in understanding its multifaceted functions. These
limitations highlight the need for more uniform, large-scale
clinical and mechanistic studies to advance MEG3-related research
in GC.
Acknowledgements
Not applicable.
Funding
The present work was supported by the Open Project of Key
Laboratory of Xi'an Medicine, Ministry of Education, Anhui
University of Chinese Medicine (grant nos. 2024×ayx09 and
2024×ayx10), Scientific Research Project of Anhui Higher Education
Institutions (grant nos. 2022AH050450 and 2023AH050742) and Talent
Support Program of Anhui University of Chinese Medicine (grant nos.
2022rcyb002 and 2022rcyb007).
Availability of data and materials
Not applicable.
Authors' contributions
BJ designed and conceived the study, wrote the
manuscript and acquired funding. HC, PY and XW performed the
literature analysis and visualization. SL and JW revised the
manuscript. All authors read and approved the final manuscript.
Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GC
|
gastric cancer
|
|
lncRNA
|
long non-coding RNA
|
|
MEG3
|
maternally expressed gene 3
|
|
EMT
|
epithelial-to-mesenchymal
transition
|
|
ceRNA
|
competing endogenous RNA
|
|
miRNA
|
microRNA
|
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
2
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rihawi K, Ricci AD, Rizzo A, Brocchi S,
Marasco G, Pastore LV, Llimpe FLR, Golfieri R and Renzulli M:
Tumor-associated macrophages and inflammatory microenvironment in
gastric cancer: Novel translational implications. Int J Mol Sci.
22:38052021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Y, Liu C, Peng H, Zhang J and Feng
Q: IL1 receptor antagonist gene IL1-RN variable number of tandem
repeats polymorphism and cancer risk: A literature review and
meta-analysis. PLoS One. 7:e460172012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He J, Hu W, Ouyang Q, Zhang S, He L, Chen
W, Li X and Hu C: Helicobacter pylori infection induces stem
cell-like properties in Correa cascade of gastric cancer. Cancer
Lett. 542:2157642022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mommersteeg MC, Simovic I, Yu B, van
Nieuwenburg SAV, Bruno IMJ, Doukas M, Kuipers EJ, Spaander MCW,
Peppelenbosch MP, Castaño-Rodríguez N and Fuhler GM: Autophagy
mediates ER stress and inflammation in Helicobacter pylori-related
gastric cancer. Gut Microbes. 14:20152382022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Molaei F, Forghanifard MM, Fahim Y and
Abbaszadegan MR: Molecular signaling in tumorigenesis of gastric
cancer. Iran Biomed J. 22:217–230. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang J, Du L and Chen X: Adenosine
signaling: Optimal target for gastric cancer immunotherapy. Front
Immunol. 13:10278382022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Q, Cao T, Zhang X, Hui J, Wang C,
Zhang W, Wang P, Zhou Y and Han S: ATXN2-mediated PI3K/AKT
activation confers gastric cancer chemoresistance and attenuates
CD8+ Tcell cytotoxicity. J Immunol Res. 2022:68632402022.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu K, Yuan S, Wang C and Zhu H:
Resistance to immune checkpoint inhibitors in gastric cancer. Front
Pharmacol. 14:12853432023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Khan I, Kamal A and Akhtar S: Diabetes
driven oncogenesis and anticancer potential of repurposed
antidiabetic drug: A systemic review. Cell Biochem Biophys.
82:1907–1929. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li J, Ye D, Shen P, Liu X, Zhou P, Zhu G,
Xu Y, Fu Y, Li X, Sun J, et al: Mir-20a-5p induced WTX deficiency
promotes gastric cancer progressions through regulating PI3K/AKT
signaling pathway. J Exp Clin Cancer Res. 39:2122020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu Z, Jiang X, Qin L, Deng H, Wang J, Ren
W, Li H, Zhao L, Liu H and Jiao Z: A novel UBE2T inhibitor
suppresses Wnt/β-catenin signaling hyperactivation and gastric
cancer progression by blocking RACK1 ubiquitination. Oncogene.
40:1027–1042. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zeng W, Zhu J, Zeng D, Guo J, Huang G,
Zeng Y, Wang L, Bin J, Liao Y, Shi M and Liao W: Epigenetic
modification-associated molecular classification of gastric cancer.
Lab Invest. 103:1001702023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen D, Xiong L, Zhang L, Yu H, Xu Y, Wang
M, Jiang X and Xiong Z: CSF1R is a prognostic biomarker and
correlated with immune cell infiltration in the gastric cancer
microenvironment. Pharmgenomics Pers Med. 14:445–457.
2021.PubMed/NCBI
|
|
16
|
van Schooten TS, Derks S, Jiménez-Martí E,
Carneiro F, Figueiredo C, Ruiz E, Alsina M, Molero C, Garrido M,
Riquelme A, et al: The LEGACy study: A European and Latin American
consortium to identify risk factors and molecular phenotypes in
gastric cancer to improve prevention strategies and personalized
clinical decision making globally. BMC Cancer. 22:6462022.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peña-Flores JA, Enríquez-Espinoza D,
Muela-Campos D, Álvarez-Ramírez A, Sáenz A, Barraza-Gómez AA, Bravo
K, Estrada-Macías ME and González-Alvarado K: Functional relevance
of the long intergenic non-coding RNA regulator of reprogramming
(Linc-ROR) in cancer proliferation, metastasis, and drug
resistance. Noncoding RNA. 9:122023.PubMed/NCBI
|
|
18
|
Liu W, Luo M, Zou L, Liu X, Wang R, Tao H,
Wu D, Zhang W, Luo Q and Zhao Y: uNK cell-derived TGF-β1 regulates
the long noncoding RNA MEG3 to control vascular smooth muscle cell
migration and apoptosis in spiral artery remodeling. J Cell
Biochem. 120:15997–16007. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ye HH, Yang SH and Zhang Y: MEG3 damages
fetal endothelial function induced by gestational diabetes mellitus
via AKT pathway. Eur Rev Med Pharmacol Sci. 22:8553–8560.
2018.PubMed/NCBI
|
|
20
|
Wang X, Li X and Wang Z: lncRNA MEG3
inhibits pituitary tumor development by participating in cell
proliferation, apoptosis and EMT processes. Oncol Rep. 45:402021.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Wang Y, Jiang Y, Bai H and Wen Y:
The meaningful function of the emerging clinical targets-lncRNA
MEG3 in gastric cancer. Curr Pharm Des. 29:2204–2212. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo W, Dong Z, Liu S, Qiao Y, Kuang G, Guo
Y, Shen S and Liang J: Promoter hypermethylation-mediated
downregulation of miR-770 and its host gene MEG3, a long non-coding
RNA, in the development of gastric cardia adenocarcinoma. Mol
Carcinog. 56:1924–1934. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li CY, Liang GY, Yao WZ, Sui J, Shen X,
Zhang YQ, Ma SM, Ye YC, Zhang ZY, Zhang WH, et al: Identification
and functional characterization of long non-coding RNAs in human
gastric cancer. Oncol Lett. 15:8805–8815. 2018.PubMed/NCBI
|
|
24
|
Jiao J and Zhang S: Long non-coding RNA
MEG-3 suppresses gastric carcinoma cell growth, invasion and
migration via EMT regulation. Mol Med Rep. 20:2685–2693.
2019.PubMed/NCBI
|
|
25
|
Najafi D, Siri G, Sadri M, Yazdani O,
Esbati R, Karimi P, Keshavarz A, Mehmandar-Oskuie A and Ilktac M:
Combination MEG3 lncRNA and Ciprofloxacin dramatically decreases
cell migration and viability as well as induces apoptosis in GC
cells in vitro. Biotechnol Appl Biochem. 71:809–816. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lüönd F, Sugiyama N, Bill R, Bornes L,
Hager C, Tang F, Santacroce N, Beisel C, Ivanek R, Bürglin T, et
al: Distinct contributions of partial and full EMT to breast cancer
malignancy. Dev Cell. 56:3203–3221. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Leung DHL, Phon BWS, Sivalingam M,
Radhakrishnan AK and Kamarudin MNA: Regulation of EMT markers,
extracellular matrix, and associated signalling pathways by long
non-coding RNAs in glioblastoma mesenchymal transition: A scoping
review. Biology (Basel). 12:8182023.PubMed/NCBI
|
|
28
|
Giordo R, Ahmadi FAM, Husaini NA,
Al-Nuaimi NRAM, Ahmad SMS, Pintus G and Zayed H: microRNA 21 and
long non-coding RNAs interplays underlie cancer pathophysiology: A
narrative review. Noncoding RNA Res. 9:831–852. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Amini F, Khalaj-Kondori M, Moqadami A and
Rajabi A: Expression of HOTAIR and MEG3 are negatively associated
with H. pylori positive status in gastric cancer patients. Genes
Cancer. 13:1–8. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Khosroshahi NS, Akbarzadeh S, Abbaslou EM,
Rajabi A, Abdi A, Saber A, Pakmanesh AR and Safaralizadeh R:
Upregulation of lncRNA ANRASSF1 is associated with the lymph node
metastasis and H. pylori infection in gastric cancer. Biomark Med.
19:571–576. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rashwan HH, Taher AM, Hassan HA, Awaji AA,
Kiriacos CJ, Assal RA and Youness RA: Harnessing the supremacy of
MEG3 LncRNA to defeat gastrointestinal malignancies. Pathol Res
Pract. 256:1552232024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu J, Wang X, Zhu C and Wang K: A review
of current evidence about lncRNA MEG3: A tumor suppressor in
multiple cancers. Front Cell Dev Biol. 10:9976332022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Laurentino S, Beygo J, Nordhoff V, Kliesch
S, Wistuba J, Borgmann J, Buiting K, Horsthemke B and Gromoll J:
Epigenetic germline mosaicism in infertile men. Hum Mol Genet.
24:1295–1304. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dirks RAM, van Mierlo G, Kerstens HHD,
Bernardo AS, Kobolák J, Bock I, Maruotti J, Pedersen RA, Dinnyés A,
Huynen MA, et al: Allele-specific RNA-seq expression profiling of
imprinted genes in mouse isogenic pluripotent states. Epigenetics
Chromatin. 12:142019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu JL, Meng FM and Li HJ: High expression
of lncRNA MEG3 participates in non-small cell lung cancer by
regulating microRNA-7-5p. Eur Rev Med Pharmacol Sci. 22:5938–5945.
2018.PubMed/NCBI
|
|
36
|
Qiu YY, Wu Y, Lin MJ, Bian T, Xiao YL and
Qin C: LncRNA-MEG3 functions as a competing endogenous RNA to
regulate Treg/Th17 balance in patients with asthma by targeting
microRNA-17/ RORγt. Biomed Pharmacother. 111:386–394. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Uroda T, Anastasakou E, Rossi A, Teulon
JM, Pellequer JL, Annibale P, Pessey O, Inga A, Chillón I and
Marcia M: Conserved Pseudoknots in lncRNA MEG3 are essential for
stimulation of the p53 pathway. Mol Cell. 75:982–995. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang M, Wang F and Zhang X: miRNA-627
inhibits cell proliferation and cell migration, promotes cell
apoptosis in prostate cancer cells through upregulating MAP3K1,
PTPRK and SRA1. Int J Clin Exp Pathol. 11:255–261. 2018.PubMed/NCBI
|
|
39
|
Zhang Y, Wu J, Jing H, Huang G, Sun Z and
Xu S: Long noncoding RNA MEG3 inhibits breast cancer growth via
upregulating endoplasmic reticulum stress and activating NF-κB and
p53. J Cell Biochem. 120:6789–6797. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sannigrahi MK, Sharma R, Panda NK and
Khullar M: Role of non-coding RNAs in head and neck squamous cell
carcinoma: A narrative review. Oral Dis. 24:1417–1427. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhu M, Wang X, Gu Y, Wang F, Li L and Qiu
X: MEG3 overexpression inhibits the tumorigenesis of breast cancer
by downregulating miR-21 through the PI3K/Akt pathway. Arch Biochem
Biophys. 661:22–30. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhan QY, Xie LX and Wang C: Promoting
critical care system and capacity building in pulmonary and
critical care medicine subspecialties. Zhonghua Yi Xue Za Zhi.
103:3149–3151. 2023.(In Chinese). PubMed/NCBI
|
|
43
|
Zamani M, Sadeghizadeh M, Behmanesh M and
Najafi F: Dendrosomal curcumin increases expression of the long
non-coding RNA gene MEG3 via up-regulation of epi-miRs in
hepatocellular cancer. Phytomedicine. 22:961–967. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH
and Guo RH: Decreased expression of long noncoding RNA MEG3 affects
cell proliferation and predicts a poor prognosis in patients with
colorectal cancer. Tumour Biol. 36:4851–4859. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wu W, Zhou S, Fei G and Wang R: The role
of long noncoding RNA MEG3 in fibrosis diseases. Postgrad Med J.
100:529–538. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Dunn-Davies H, Dudnakova T, Nogara A,
Rodor J, Thomas AC, Parish E, Gautier P, Meynert A, Ulitsky I,
Madeddu P, et al: Control of endothelial cell function and
arteriogenesis by MEG3:EZH2 epigenetic regulation of integrin
expression. Mol Ther Nucleic Acids. 35:1021732024. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu W, Huang L, Zhang C and Liu Z: lncRNA
MEG3 is downregulated in ankylosing spondylitis and associated with
disease activity, hospitalization time and disease duration. Exp
Ther Med. 17:291–297. 2019.PubMed/NCBI
|
|
48
|
Zheng Q, Lin Z, Xu J, Lu Y, Meng Q, Wang
C, Yang Y, Xin X, Li X, Pu H, et al: Long noncoding RNA MEG3
suppresses liver cancer cells growth through inhibiting β-catenin
by activating PKM2 and inactivating PTEN. Cell Death Dis.
9:2532018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yang Y, Tian Z, He L, Meng H, Xie X, Yang
Z, Wang X, Zhao Y and Huang C: RhoGDIβ inhibition via
miR-200c/AUF1/SOX2/miR-137 axis contributed to lncRNA MEG3
downregulation-mediated malignant transformation of human bronchial
epithelial cells. Mol Carcinog. 63:977–990. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cheng X, Ali MS, Moran M, Viana MP,
Schlichte SL, Zimmerman MC, Khalimonchuk O, Feinberg MW and Sun X:
Long non-coding RNA Meg3 deficiency impairs glucose homeostasis and
insulin signaling by inducing cellular senescence of hepatic
endothelium in obesity. Redox Biol. 40:1018632021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yao H, Duan M, Lin L, Wu C, Fu X, Wang H,
Guo L, Chen W, Huang L, Liu D, et al: TET2 and MEG3 promoter
methylation is associated with acute myeloid leukemia in a Hainan
population. Oncotarget. 8:18337–18347. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yan J, Guo X, Xia J, Shan T, Gu C, Liang
Z, Zhao W and Jin S: MiR-148a regulates MEG3 in gastric cancer by
targeting DNA methyltransferase 1. Med Oncol. 31:8792014.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pan T, Ding H, Jin L, Zhang S, Wu D, Pan
W, Dong M, Ma X and Chen Z: DNMT1-mediated demethylation of lncRNA
MEG3 promoter suppressed breast cancer progression by repressing
Notch1 signaling pathway. Cell Cycle. 21:2323–2337. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
McMinn J, Wei M, Schupf N, Cusmai J,
Johnson EB, Smith AC, Weksberg R, Thaker HM and Tycko B: Unbalanced
placental expression of imprinted genes in human intrauterine
growth restriction. Placenta. 27:540–549. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
El Said NH, Abdrabou W, Mahmood SR, Venit
T, Idaghdour Y and Percipalle P: Nuclear actin-dependent Meg3
expression suppresses metabolic genes by affecting the chromatin
architecture at sites of elevated H3K27 acetylation levels. Nucleic
Acids Res. 53:gkaf2802025. View Article : Google Scholar
|
|
56
|
Luo Y, Wang H, Wang L, Wu W, Zhao J, Li X,
Xiong R, Ding X, Yuan D and Yuan C: LncRNA MEG3: Targeting the
molecular mechanisms and pathogenic causes of metabolic diseases.
Curr Med Chem. 31:6140–6153. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hasenson SE, Alkalay E, Atrash MK,
Boocholez A, Gershbaum J, Hochberg-Laufer H and Shav-Tal Y: The
association of MEG3 lncRNA with nuclear speckles in living cells.
Cells. 11:19422022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ding L, Tian Y, Wang L, Bi M, Teng D and
Hong S: Hypermethylated long noncoding RNA MEG3 promotes the
progression of gastric cancer. Aging (Albany NY). 11:8139–8155.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhou X, Ji G, Ke X, Gu H, Jin W and Zhang
G: MiR-141 inhibits gastric cancer proliferation by interacting
with long noncoding RNA MEG3 and down-regulating E2F3 expression.
Dig Dis Sci. 60:3271–3282. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Peng W, Si S, Zhang Q, Li C, Zhao F, Wang
F, Yu J and Ma R: Long non-coding RNA MEG3 functions as a competing
endogenous RNA to regulate gastric cancer progression. J Exp Clin
Cancer Res. 34:792015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Cui HB, Ge HE, Wang YS and Bai XY:
MiR-208a enhances cell proliferation and invasion of gastric cancer
by targeting SFRP1 and negatively regulating MEG3. Int J Biochem
Cell Biol. 102:31–39. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Guo LL, Song CH, Wang P, Dai LP, Zhang JY
and Wang KJ: Competing endogenous RNA networks and gastric cancer.
World J Gastroenterol. 21:11680–11687. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li C, Liang G, Yao W, Sui J, Shen X, Zhang
Y, Ma S, Ye Y, Zhang Z, Zhang W, et al: Differential expression
profiles of long non-coding RNAs reveal potential biomarkers for
identification of human gastric cancer. Oncol Rep. 35:1529–1540.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Soghala S, Harsiny K, Momeni P, Hatami M,
Oskooei VK, Hussen BM, Taheri M and Ghafouri-Fard S:
Down-regulation of LINC-ROR, HOXA-AS2 and MEG3 in gastric cancer.
Heliyon. 8:e111552022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sun M, Xia R, Jin F, Xu T, Liu Z, De W and
Liu X: Downregulated long noncoding RNA MEG3 is associated with
poor prognosis and promotes cell proliferation in gastric cancer.
Tumour Biol. 35:1065–1073. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Gao S, Zhao ZY, Wu R, Zhang Y and Zhang
ZY: Prognostic value of long noncoding RNAs in gastric cancer: A
meta-analysis. Onco Targets Ther. 11:4877–4891. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wei GH and Wang X: lncRNA MEG3 inhibit
proliferation and metastasis of gastric cancer via p53 signaling
pathway. Eur Rev Med Pharmacol Sci. 21:3850–3856. 2017.PubMed/NCBI
|
|
68
|
Li Y, Lou S, Zhang J, Zhao S and Lou G:
m6A methylation-mediated regulation of LncRNA MEG3 suppresses
ovarian cancer progression through miR-885-5p and the VASH1
pathway. J Transl Med. 22:1132024. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ye M, Lu H, Tang W, Jing T, Chen S, Wei M,
Zhang J, Wang J, Ma J, Ma D and Dong K: Downregulation of MEG3
promotes neuroblastoma development through FOXO1-mediated autophagy
and mTOR-mediated epithelial-mesenchymal transition. Int J Biol
Sci. 16:3050–3061. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
An C, Wang I, Li X, Xia R and Deng F: Long
non-coding RNA in prostate cancer. Am J Clin Exp Urol. 10:170–179.
2022.PubMed/NCBI
|
|
71
|
Ramezani M, Shamsabadi FT and Shahbazi M:
Harnessing the TP53INP1/TP53I3 axis for inhibition of colorectal
cancer cell proliferation through MEG3 and Linc-ROR Co-expression.
Heliyon. 10:e340752024. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Dan J, Wang J, Wang Y, Zhu M, Yang X, Peng
Z, Jiang H and Chen L: LncRNA-MEG3 inhibits proliferation and
metastasis by regulating miRNA-21 in gastric cancer. Biomed
Pharmacother. 99:931–938. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Terashima M, Tange S, Ishimura A and
Suzuki T: MEG3 long noncoding RNA contributes to the epigenetic
regulation of epithelial-mesenchymal transition in lung cancer cell
lines. J Biol Chem. 292:82–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Li T, Zhang X, Cheng L, Li C, Wu Z, Luo Y,
Zhou K, Li Y, Zhao Q and Huang Y: Modulation of lncRNA H19 enhances
resveratrol-inhibited cancer cell proliferation and migration by
regulating endoplasmic reticulum stress. J Cell Mol Med.
26:2205–2217. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kuai X, Jia L, Yang T, Huang X, Zhao W,
Zhang M, Chen Y, Zhu J, Feng Z and Tang Q: Trop2 promotes multidrug
resistance by regulating Notch1 signaling pathway in gastric cancer
cells. Med Sci Monit. 26:e9195662020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ma Y, Wei X and Wu Z: HNF-4α promotes
multidrug resistance of gastric cancer cells through the modulation
of cell apoptosis. Oncol Lett. 14:6477–6484. 2017.PubMed/NCBI
|
|
77
|
Zhu YX, Yao J, Liu C, Hu HT, Li XM, Ge HM,
Zhou YF, Shan K, Jiang Q and Yan B: Long non-coding RNA MEG3
silencing protects against light-induced retinal degeneration.
Biochem Biophys Res Commun. 496:1236–1242. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Tu Y, Xie L, Chen L, Yuan Y, Qin B, Wang
K, Zhu Q, Ji N, Zhu M and Guan H: Long non-coding RNA MEG3 promotes
cataractogenesis by upregulating TP53INP1 expression in age-related
cataract. Exp Eye Res. 199:1081852020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang X, Wu N, Wang J and Li Z: LncRNA
MEG3 inhibits cell proliferation and induces apoptosis in laryngeal
cancer via miR-23a/APAF-1 axis. J Cell Mol Med. 23:6708–6719. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Wu D, Ma Z, Ma D and Li Q: Long non-coding
RNA maternally expressed gene 3 affects cell proliferation,
apoptosis and migration by targeting the microRNA-9-5p/midkine axis
and activating the phosphoinositide-dependent kinase/AKT pathway in
hepatocellular carcinoma. Oncol Lett. 21:3452021. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Mitra R, Chen X, Greenawalt EJ, Maulik U,
Jiang W, Zhao Z and Eischen CM: Decoding critical long non-coding
RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat
Commun. 8:16042017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang R, Zou L and Yang X: microRNA-210/
Long non-coding RNA MEG3 axis inhibits trophoblast cell migration
and invasion by suppressing EMT process. Placenta. 109:64–71. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Liu H, Chen C, Zeng J, Zhao Z and Hu Q:
MicroRNA-210-3p is transcriptionally upregulated by hypoxia
induction and thus promoting EMT and chemoresistance in glioma
cells. PLoS One. 16:e02535222021. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Wang Z, Xia P, Hu J, Huang Y, Zhang F, Li
L, Wang E, Guo Q and Ye Z: LncRNA MEG3 alleviates diabetic
cognitive impairments by reducing mitochondrial-derived apoptosis
through promotion of FUNDC1-related mitophagy via Rac1-ROS axis.
ACS Chem Neurosci. 12:2280–2307. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Elimam H, Zaki MB, Abd-Elmawla MA, Darwish
HA, Hatawsh A, Aborehab NM, Mageed SSA, Moussa R, Mohammed OA,
Abdel-Reheim MA and Doghish AS: Natural products and long
non-coding RNAs in prostate cancer: Insights into etiology and
treatment resistance. Naunyn Schmiedebergs Arch Pharmacol.
18:2025.
|
|
86
|
Bai Z, Hu K, Yu J, Shen Y and Chen C:
Macrophage migration inhibitory factor protects bone marrow
mesenchymal stem cells from hypoxia/ischemia-induced apoptosis by
regulating lncRNA MEG3. J Zhejiang Univ Sci B. 23:989–1001. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J,
Wei L, Jin Y, Fu H, Wu Y and Zheng X: Long Noncoding RNA MEG3
interacts with p53 protein and regulates partial p53 target genes
in hepatoma cells. PLoS One. 10:e01397902015. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Ali MS, Cheng X, Moran M, Haemmig S,
Naldrett MJ, Alvarez S, Feinberg MW and Sun X: LncRNA Meg3 protects
endothelial function by regulating the DNA damage response. Nucleic
Acids Res. 47:1505–1522. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Zheng H, Li BH, Liu C, Jia L and Liu FT:
Comprehensive analysis of lncRNA-mediated ceRNA crosstalk and
identification of prognostic biomarkers in Wilms' tumor. Biomed Res
Int. 2020:49516922020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Bauer NC, Yang A, Wang X, Zhou Y,
Klibanski A and Soberman RJ: A cross-nearest neighbor/Monte Carlo
algorithm for single-molecule localization microscopy defines
interactions between p53, Mdm2, and MEG3. J Biol Chem.
296:1005402021. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Luo Q, Cui M, Deng Q and Liu J:
Comprehensive analysis of differentially expressed profiles and
reconstruction of a competing endogenous RNA network in papillary
renal cell carcinoma. Mol Med Rep. 19:4685–4696. 2019.PubMed/NCBI
|
|
92
|
Kumar S, Williams D, Sur S, Wang JY and Jo
H: Role of flow-sensitive microRNAs and long noncoding RNAs in
vascular dysfunction and atherosclerosis. Vascul Pharmacol.
114:76–92. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Tan J, Xiang L and Xu G: LncRNA MEG3
suppresses migration and promotes apoptosis by sponging miR-548d-3p
to modulate JAK-STAT pathway in oral squamous cell carcinoma. IUBMB
Life. 71:882–890. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Li Y, Zhang L, Zhao Y, Peng H, Zhang N and
Bai W: MEG3 sponges miRNA-376a and YBX1 to regulate angiogenesis in
ovarian cancer endothelial cells. Heliyon. 9:e132042023. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhang C, Qin Y, Tang Y, Gu M, Li Z and Xu
H: MEG3 in hematologic malignancies: from the role of disease
biomarker to therapeutic target. Pharmacogenet Genomics.
34:209–216. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Wei Q, Liu G, Huang Z, Huang Y, Huang L,
Huang Z, Wu X, Wei H and Pu J: LncRNA MEG3 inhibits tumor
progression by modulating macrophage phenotypic polarization via
miR-145-5p/DAB2 axis in hepatocellular carcinoma. J Hepatocell
Carcinoma. 10:1019–1035. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Lin J, Huang J, Tan C, Wu S, Lu X and Pu
J: LncRNA MEG3 suppresses hepatocellular carcinoma by stimulating
macrophage M1 polarization and modulating immune system via
inhibiting CSF-1 in vivo/vitro studies. Int J Biol Macromol.
281:1364592024. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Huang CY, Chou ST, Hsu YM, Chao WJ, Wu GH,
Hsiao JR, Wang HD and Shiah SG: MEG3-mediated oral
squamous-cell-carcinoma-derived exosomal miR-421 activates
angiogenesis by targeting HS2ST1 in vascular endothelial cells. Int
J Mol Sci. 25:75762024. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Kong X, Yang S, Liu C, Tang H, Chen Y,
Zhang X, Zhou Y and Liang G: Relationship between MEG3 gene
polymorphism and risk of gastric cancer in Chinese population with
high incidence of gastric cancer. Biosci Rep. 40:BSR202003052020.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Liu H, Ye D, Chen A, Tan D, Zhang W, Jiang
W, Wang M and Zhang X: A pilot study of new promising non-coding
RNA diagnostic biomarkers for early-stage colorectal cancers. Clin
Chem Lab Med. 57:1073–1083. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Ghaedi H, Mozaffari MAN, Salehi Z, Ghasemi
H, Zadian SS, Alipoor S, Hadianpour S and Alipoor B: Co-expression
profiling of plasma miRNAs and long noncoding RNAs in gastric
cancer patients. Gene. 687:135–142. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Li MK, Liu LX, Zhang WY, Zhan HL, Chen RP,
Feng JL and Wu LF: Long non-coding RNA MEG3 suppresses
epithelial-to-mesenchymal transition by inhibiting the
PSAT1-dependent GSK-3β/Snail signaling pathway in esophageal
squamous cell carcinoma. Oncol Rep. 44:2130–2142. 2020.PubMed/NCBI
|
|
103
|
Li J, Jiang X, Li C, Liu Y, Kang P, Zhong
X and Cui Y: LncRNA-MEG3 inhibits cell proliferation and invasion
by modulating Bmi1/RNF2 in cholangiocarcinoma. J Cell Physiol.
234:22947–22959. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Ji Y, Feng G, Hou Y, Yu Y, Wang R and Yuan
H: Long noncoding RNA MEG3 decreases the growth of head and neck
squamous cell carcinoma by regulating the expression of miR-421 and
E-cadherin. Cancer Med. 9:3954–3963. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Ning J, Zhang L, Guo H, Zhou S, Sun X and
Bao W: LncRNA MEG3 inhibits the development of nasopharyngeal
carcinoma by sponging miR-543 targeting KLF4. Transl Cancer Res.
9:958–971. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Jin L, Cai Q, Wang S, Wang S, Mondal T,
Wang J and Quan Z: Long noncoding RNA MEG3 regulates LATS2 by
promoting the ubiquitination of EZH2 and inhibits proliferation and
invasion in gallbladder cancer. Cell Death Dis. 9:10172018.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Liu L, Liu Y, Zhang S, Zhang J, Meng Y,
Liu D, Gu L, Zhang Y, Xu L, Zhang Z, et al: Celastrol promotes
apoptosis of breast cancer MDA-MB-231 cells by targeting HSDL2.
Acupuncture and Herbal Medicine. 4:92–101. 2024. View Article : Google Scholar
|
|
108
|
Huang D, Tang Z, Pu X, Gao F and Chong L:
A novel cabazitaxel liposomes modified with ginsenoside Rk1 for
cancer targeted therapy. Acupuncture and Herbal Medicine.
4:113–121. 2024. View Article : Google Scholar
|
|
109
|
Zhang L, Zhao F, Li W, Song G, Kasim V and
Wu S: The biological roles and molecular mechanisms of long
non-coding RNA MEG3 in the hallmarks of cancer. Cancers (Basel).
14:60322022. View Article : Google Scholar : PubMed/NCBI
|