Introduction
Lung cancer (LC) is the leading cause of
cancer-related mortality worldwide, with a mortality rate of 18.7%
(1) and a 5-year survival rate of
~16.6%. Lung adenocarcinoma (LUAD) is the most frequent
histological type, accounting for ~40% of LC cases. Although
multiple therapies, such as surgical resection, chemotherapy,
radiotherapy and molecular targeted therapy have been developed in
the past decades to treat LUAD, the overall survival time of
patients with LUAD has not markedly improved, primarily due to the
lack of useful molecular biomarkers (2). It remains a matter of debate whether
chemoimmunotherapy or immune checkpoint inhibitor monotherapy
should be used as first-line treatment for patients with advanced
non-small cell LC (NSCLC) and have high expression levels of
programmed cell death-ligand 1 (PD-L1) (3). To the best of our knowledge, the lack
of prognostic markers for cancer has always limited the choice of
first-line therapies (4,5). Similarly, the currently available
targeted drugs such as gefitinib, erlotinib, osimertinib, afatinib
and crizotinib, have limited clinical efficacy in the treatment of
LC due to their off-target effects or drug resistance issues
(6). Hu et al (7) demonstrated that high Toll-like
receptor 7 expression serves as a robust clinical feature
predictive of patient prognosis, immunotherapy response and
candidate drug efficacy, providing deeper insights for LUAD
treatment. In addition, Zhang and Lin (8) identified the TDP-43 co-expressed gene
risk score, a prognostic model based on the expression of kinesin
family member 20A, WD repeat domain 4, proline rich 11 and glia
maturation factor γ, as a reliable biomarker for predicting
prognosis and treatment response in LUAD, offering valuable
insights for guiding clinical strategies. Therefore, a
comprehensive understanding of how LUAD occurs and progresses is
essential to improve the diagnosis and prognosis of patients with
LUAD in the future.
The methylation of RNA molecules to
N6-methyladenosine (m6A) is a universal modification found in all
eukaryotes, but its biological relevance remains to be elucidated.
Modification of m6A affects mRNA splicing, export, translation and
stability (9). Previous studies
have reported that m6A is involved in RNA modulation as ‘writers’
[such as methyltransferase-like (METTL)3/METTL14 complex for
methylation] (10), ‘erasers’
[including fat mass and obesity-associated gene (FTO) and AlkB
homolog 5 for demethylation] (11),
and ‘readers’ [such as YT521-B homology domain-containing family
proteins (YTHDFs) that dictate the functional outcomes of m6A
modifications] (12), which
collectively modulate RNA metabolism (13). Several enzymes, including METTL3
(14) and heterogeneous nuclear
ribonucleoprotein A2/B1 (15),
participate in the m6A system. Previous studies have reported that
modifications to m6A contribute to the progression of a number of
diseases, such as obesity (16),
cancer (17) and embryonic
development (18). However, to the
best of our knowledge, comprehensive analysis of the expression of
m6A RNA methylation regulators in LC and particularly in LUAD, is
largely lacking. The diagnostic and prognostic value of such
regulators remains to be explored.
The present study aimed to profile the mRNA
expression patterns of m6A-related genes in LUAD using data
obtained from The Cancer Genome Atlas (TCGA) and the University of
California, Santa Cruz (UCSC) Xena databases. A survival analysis
was performed to assess the prognostic value of m6A-related genes
in patients with LUAD and correlations between m6A genes and the
expression of immunomodulatory factors in LUAD were investigated.
The current study aimed to comprehensively analyze the expression
and prognostic significance of m6A methylation regulators in LUAD
using data from TCGA and UCSC Xena databases.
Materials and methods
Tissue samples
Samples of paracancerous and LUAD tissues used for
immunohistochemistry were obtained from 10 patients admitted to the
Cancer Center of Guangzhou Twelfth People's Hospital (Guangzhou,
China) from February 2019 to May 2021. The cohort comprised 6 men
and 4 women, with a median age of 57 years (range, 31–76 years).
Clinical data for the patients are shown in Table SI. All patients provided their
written informed consent for study participation.
Datasets
Gene expression profiles and clinical information
for a cohort of 585 patients with LUAD from The Cancer Genome Atlas
(TCGA-LUAD) were downloaded to serve as the training set. Notably,
all 585 samples belong to this single TCGA-LUAD cohort, with data
obtained from two distinct sources: TCGA database (http://portal.gdc.cancer.gov/), which provided
284 tumor and 37 adjacent normal samples, and the UCSC Xena
database (https://xena.ucsc.edu/), which provided
242 tumor and 22 adjacent normal samples. The dataset used in the
analysis is provided in Table SII.
In addition, gene expression datasets [accession nos. GSE135222
(19) and GSE126044 (20)] were downloaded from the Gene
Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) series to analyze
the expression levels of m6A-related genes.
Identification of differentially
expressed immune-related genes and differentially expressed
pyroptosis-related genes in LUAD
Based on the method previously described (21), the ‘DESeq2’ R package (v 3.5.1;
http://bioconductor.org/packages/release/bioc/html/DESeq2.html)
was used to identify genes that were differentially expressed
between the 526 LUAD and 59 adjacent non-tumor samples, with a
threshold of log2 FC >1 and P<0.05 was considered to indicate
a statistically significant difference.
Construction of a risk model and
nomogram based on prognostic markers
The Tumor IMmune Estimation Resource (TIMER)
database (http://timer.cistrome.org/) was used
to evaluate the correlation between m6A regulators and the levels
of immune cell infiltration [including cancer-associated
fibroblasts, bone marrow dendritic cells (BMDCs), CD4+ T
cells, neutrophils, regulatory T cells (Tregs), CD8+ T
cells and macrophages]. The Tumor and Immune System Interaction
Database (TISIDB) was used to assess the correlation between the
expression of m6A regulators and immunoregulators [including
immunosuppressant, immunostimulator and major histocompatibility
complex (MHC) molecules].
Genetic alteration of m6A regulators
in LUAD
cBio Cancer Genomics Portal (cBioPortal; http://www.cbioportal.org/), an open-access website
that explores, visualizes and analyzes multidimensional cancer
genomics data, was used to analyze the genetic alterations of m6A
regulators in LUAD.
Mutation analysis of m6A genes in
LUAD
Mutation annotation format files were analyzed using
the R package ‘Maftools’ (v 2.25.10; Bioconductor; http://bioconductor.org/packages/devel/bioc/html/maftools.html)
to summarize, visualize and interpret somatic mutations in m6A
regulator genes across the LUAD cohort.
Survival analysis
The correlation between m6A regulator aberrations
and the survival time of patients with cancer was determined using
the cBioPortal database. The correlation between the overall
survival time and the expression of m6A regulators was evaluated
using Kaplan-Meier curves.
T cell exhaustion correlation
analysis
First, the correlation between m6A gene expression
and biomarkers of T cell exhaustion [TNF, IL-2, IFN-γ and cytotoxic
T lymphocyte (CTL)] was analyzed using the R software package
‘psych’ (version 2.4.3; http://www.rdocumentation.org/packages/psych/versions/2.4.3),
based on research datasets in a previous study (22).
Next, the expression levels of m6A genes in patients
with LUAD treated with anti-programmed cell death
protein-1(PD-1)/PD-L1 were analyzed the GSE135222 and GSE126044
datasets. In these datasets, patients with LUAD were grouped
according to whether they had responded to anti-PD-1/PD-L1
treatment [19 non-responders and 8 responders in GSE135222
(23); 11 non-responders and 5
responders in GSE126044 (20)].
Furthermore, analyzing the correlation between the expression
levels of m6A-related genes and immune cell marker genes was
analyzed using Spearman's rank correlation to clarify the role of
m6A-related genes in predicting immune infiltration responses.
Immunohistochemistry (IHC)
Samples of LUAD and paracancerous tissue were
collected at Cancer Center of Guangzhou Twelfth People's Hospital.
Immunohistochemical procedures were performed on the tissues to
assess gene expression according to the methods reported in a
previous study (24). In brief, the
tissues were fixed in 10% formalin, embedded in paraffin, and cut
into 4-µm sections. Subsequently, the sections were dewaxed using
gradient ethanol and subjected to antigen retrieval in citrate
buffer (cat. no. G1202; Wuhan Servicebio Technology Co., Ltd.) at
95°C for 20 min. The slides were then treated with 3%
H2O2 at 25°C for 30 min to block endogenous
peroxidase activity, followed by blocking with 3% bovine serum
albumin (cat. no. GC305006; Wuhan Servicebio Technology Co., Ltd.)
at room temperature for 30 min. Next, the sections were incubated
with anti-heterogeneous nuclear ribonucleoprotein C (HNRNPC; cat.
no. ab75822; dilution 1:800), anti-insulin-like growth factor 2
mRNA binding protein (IGF2BP)1 (cat. no. ab184305; dilution
1:4,000), anti-IGF2BP3 (cat. no. ab179807; dilution 1:1,000),
anti-PD-L1 (cat. no. ab205921; dilution 1:1,000), anti-CD4 (cat.
no. ab133616; dilution 1:500), anti-CD8 (cat. no. ab217344;
dilution 1:2,000) and anti-Forkhead box P3 (FOXP3; cat. no.
ab20034; dilution 1:500), respectively, followed by incubation with
goat anti-rabbit IgG (HRP; cat. no. ab7090; dilution 1:1,000) (all
from Abcam) and DAB chromogen. The tissue sections were then
counterstained with hematoxylin, observed and captured under a
light microscope.
Statistical analysis
Statistical analyses were performed using SPSS
(version 26.0; IBM Corp.) and GraphPad Prism (version 10.1.2;
Dotmatics). Differential expression analysis of m6A methylation
regulators between tumor and paired adjacent normal tissues was
assessed using the paired Student's t-test for parametric data or
the Wilcoxon signed-rank test for non-parametric data, with 59
paired tumor and normal tissue samples. Univariate Cox proportional
hazards regression analysis was used to evaluate the association
between individual m6A regulator expression levels and overall
survival time. Kaplan-Meier survival curves were generated and
compared using the log-rank test. The correlation between T-cell
exhaustion markers and other variables of interest was assessed
using Spearman's rank correlation coefficient (ρ), with statistical
significance determined by the corresponding P-value. Differences
in gene expression across ordinal clinical variables [tumor, lymph
node and metastasis (TNM) stages) were assessed using one-way ANOVA
with Tukey's post hoc test for multiple comparisons. Mutation
analysis of m6A genes was performed. Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analyses were performed on genes identified through Pearson's
correlation analysis (r>0.5, P<0.001) of m6A-related genes
from TCGA expression matrix that were co-expressed with HNRNPC,
IGF2BP1 and IGF2BP3. P<0.05 was considered to indicate a
statistically significant difference for all analyses, unless
otherwise indicated.
Results
Tumor mutation load analysis
To evaluate the mutation load in LUAD, the tumor
mutation load in 33 TCGA tumor types was compared. The present
study data demonstrated that LUAD had a high mutation load, which
was lower compared with the mutation loads of bladder urothelial
carcinoma, lung squamous cell carcinoma and skin cutaneous melanoma
(Fig. 1).
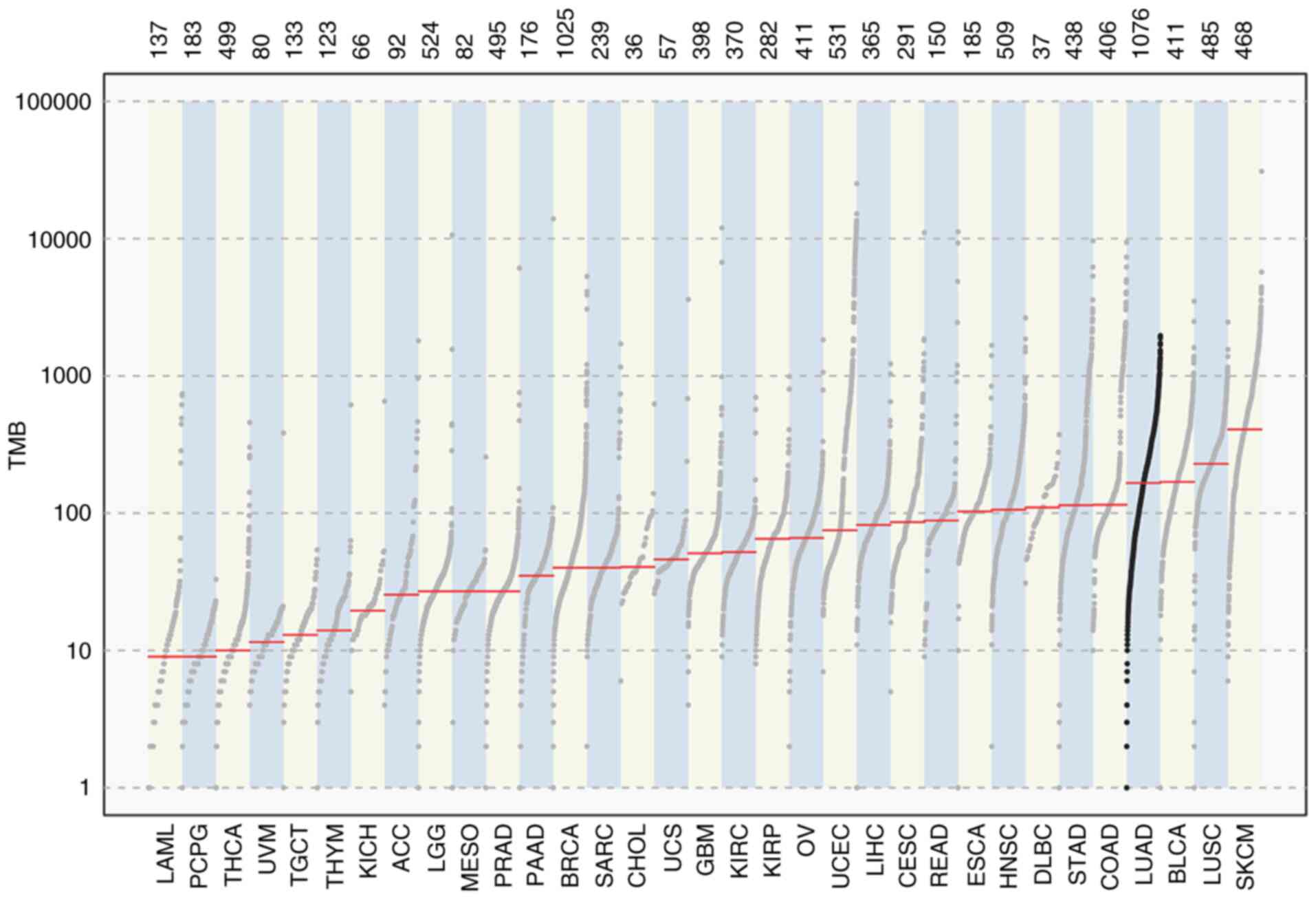 | Figure 1.Mutation load in LUAD. TMB, tumor
mutation burden; LAML, acute myeloid leukemia; PCPG,
pheochromocytoma and paraganglioma; THCA, thyroid carcinoma; UVM,
uveal melanoma; TGCT, testicular germ cell tumors; THYM, thymoma;
KICH, kidney chromophobe; ACC, adrenocortical carcinoma; LGG, brain
lower grade glioma; MESO, mesothelioma; PRAD, prostate
adenocarcinoma; PAAD, pancreatic adenocarcinoma; BRCA, breast
invasive carcinoma; SARC, sarcoma; CHOL, cholangiocarcinoma; UCS,
uterine carcinosarcoma; GBM, glioblastoma; KIRC, kidney renal clear
cell carcinoma; KIRP, kidney renal papillary cell carcinoma; OV,
ovarian serous cystadenocarcinoma; UCEC, uterine corpus endometrial
carcinoma; LIHC, liver hepatocellular carcinoma; CESC, cervical
squamous cell carcinoma and endocervical adenocarcinoma; READ,
rectum adenocarcinoma; ESCA, esophageal carcinoma; HNSC, head and
neck squamous cell carcinoma; DLBC, lymphoid neoplasm diffuse large
B-cell lymphoma; STAD, stomach adenocarcinoma; COAD, colon
adenocarcinoma; LUAD, lung adenocarcinoma; BLCA, bladder urothelial
carcinoma; LUSC, lung squamous cell carcinoma; SKCM, skin cutaneous
melanoma. |
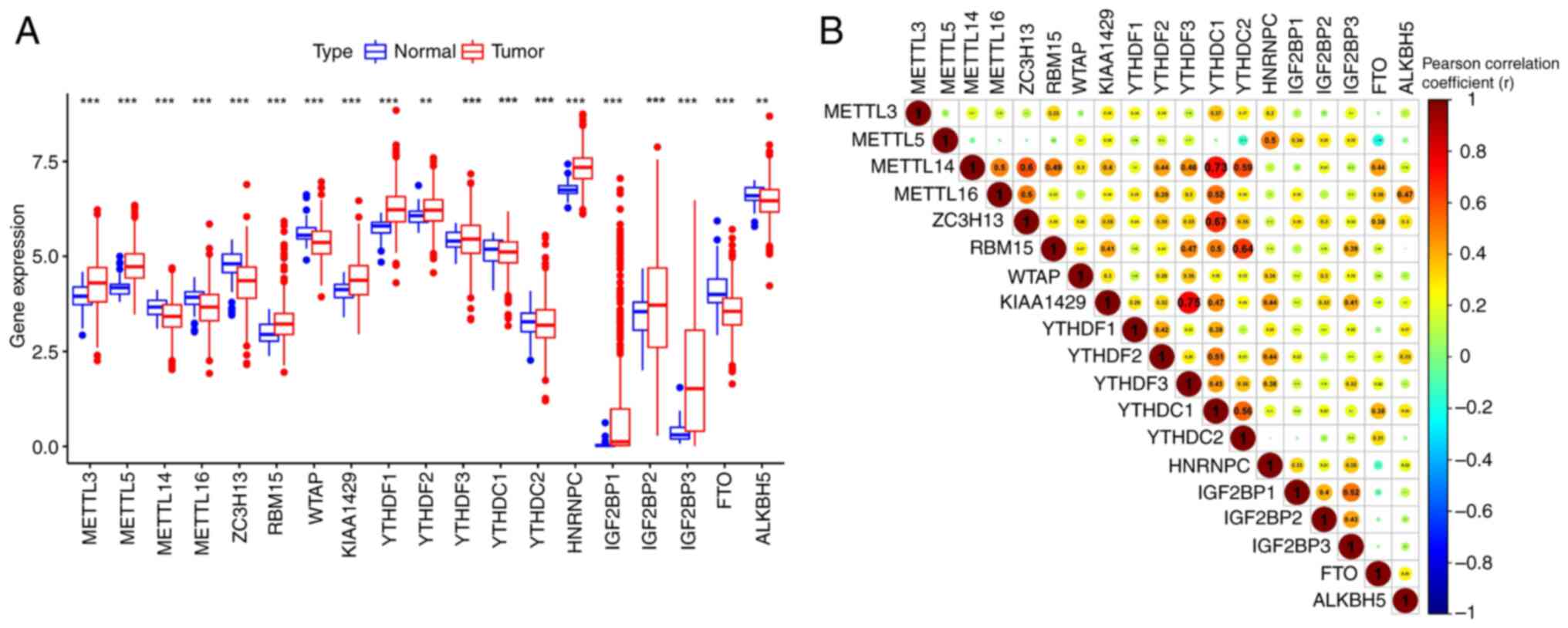
Differential expression levels of m6A
RNA regulators in normal and tumor samples
The differential expression levels of m6A genes in
the tumor and control groups and the correlation patterns were
analyzed. Excluding YTHDF3, YTHDC protein (YTHDC)1, YTHDC2 and
IGF2BP2, the expression levels of all m6A genes significantly
differed between the tumor and control groups. (P<0.01; Fig. 2A).
A correlation analysis indicated a moderate positive
correlation between m6A genes, among which, YTHDF3 had the
strongest correlation with vir like m6A methyltransferase
associated (KIAA1429) and YTHDC1 a slightly weaker correlation with
METTL14 and Zinc finger CCCH domain-containing protein 13 (ZC3H13).
Furthermore, FTO was negatively correlated with METTL5 (r=0.19;
Fig. 2B).
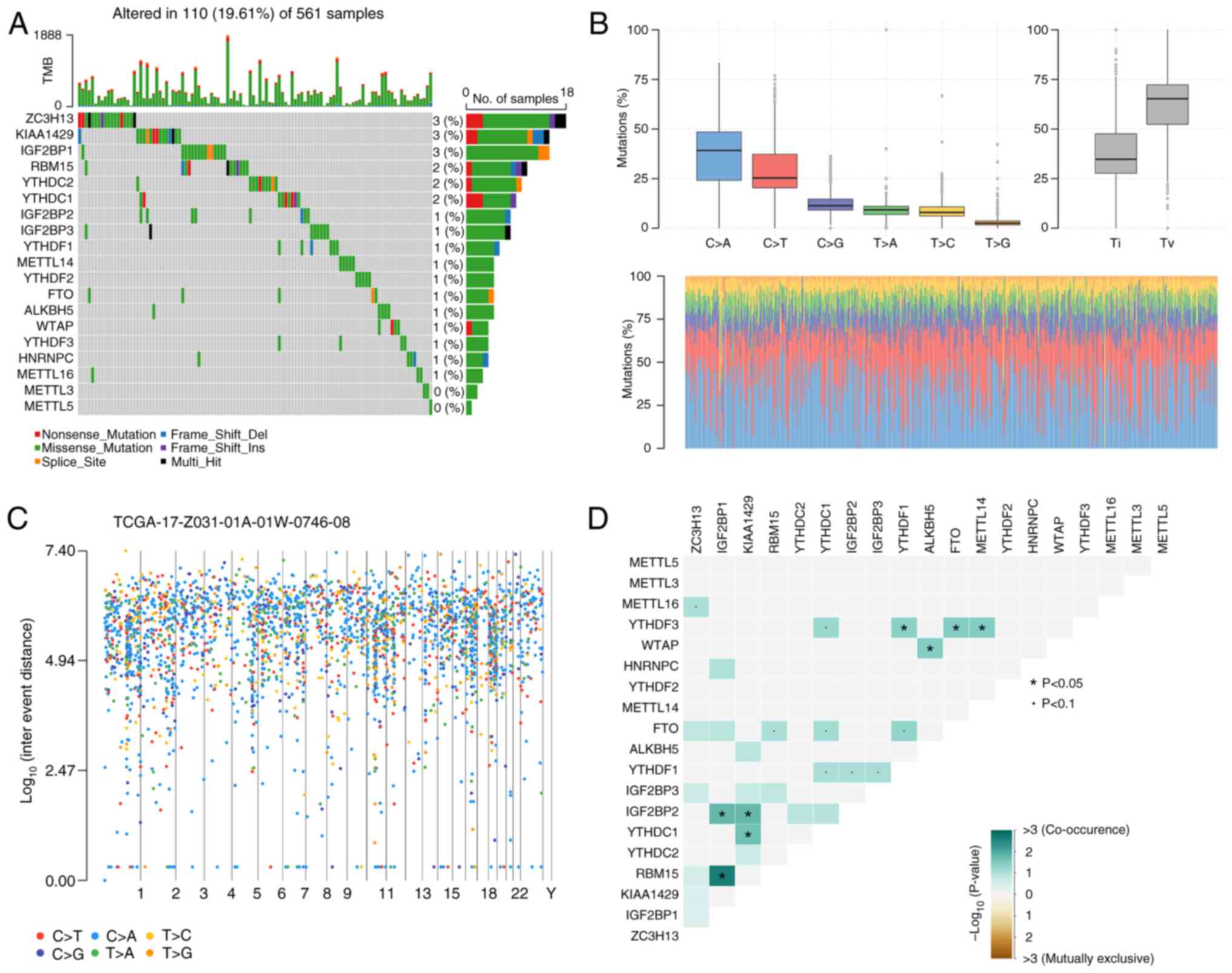
Mutation analysis of m6A genes in
LUAD
To identify somatic mutations in patients with LUAD,
mutation data were analyzed using the R software package
‘Maftools’. Results indicated that 110 of 561 patients with LUAD
had mutated m6A-related genes. As shown in Fig. 3A, the mutated m6A-related genes
included KIAA1429 (3%), ZC3H13 (3%), IGF2BP1 (3%), YTHDC1 (2%),
YTHDC2 (2%), RNA binding motif protein 15 (RBM15; 2%), IGF2BP2
(1%), IGF2BP3 (1%), HNRNPC (1%), YTHDF1 (1%), METTL14 (1%), YTHDF3
(1%), METTL14 (1%), FTO (1%), Wilms' tumor 1-associating protein
(1%), bacterial alkane hydroxylase homolog 5, RNA demethylase (1%)
and METTL116 (1%).
Furthermore, the somatic interactions between m6A
genes and the status of single nucleotide polymorphisms (SNPs) and
highly mutated genomic regions were further explored. C>A and
C>T were the two main mutation types. The proportion of each
sample is shown in a stacked histogram (Fig. 3B). The rainfall plot demonstrated
high mutation genomic regions based on different SNP mutation types
(Fig. 3C). As shown in Fig. 3D, IGF2BP1 had significant
coexpression frequencies with RBM15 and IGF2BP2, and KIAA1429 had
significant coexpression frequencies with IGF2BP2 and YTHDC1
(P<0.05).
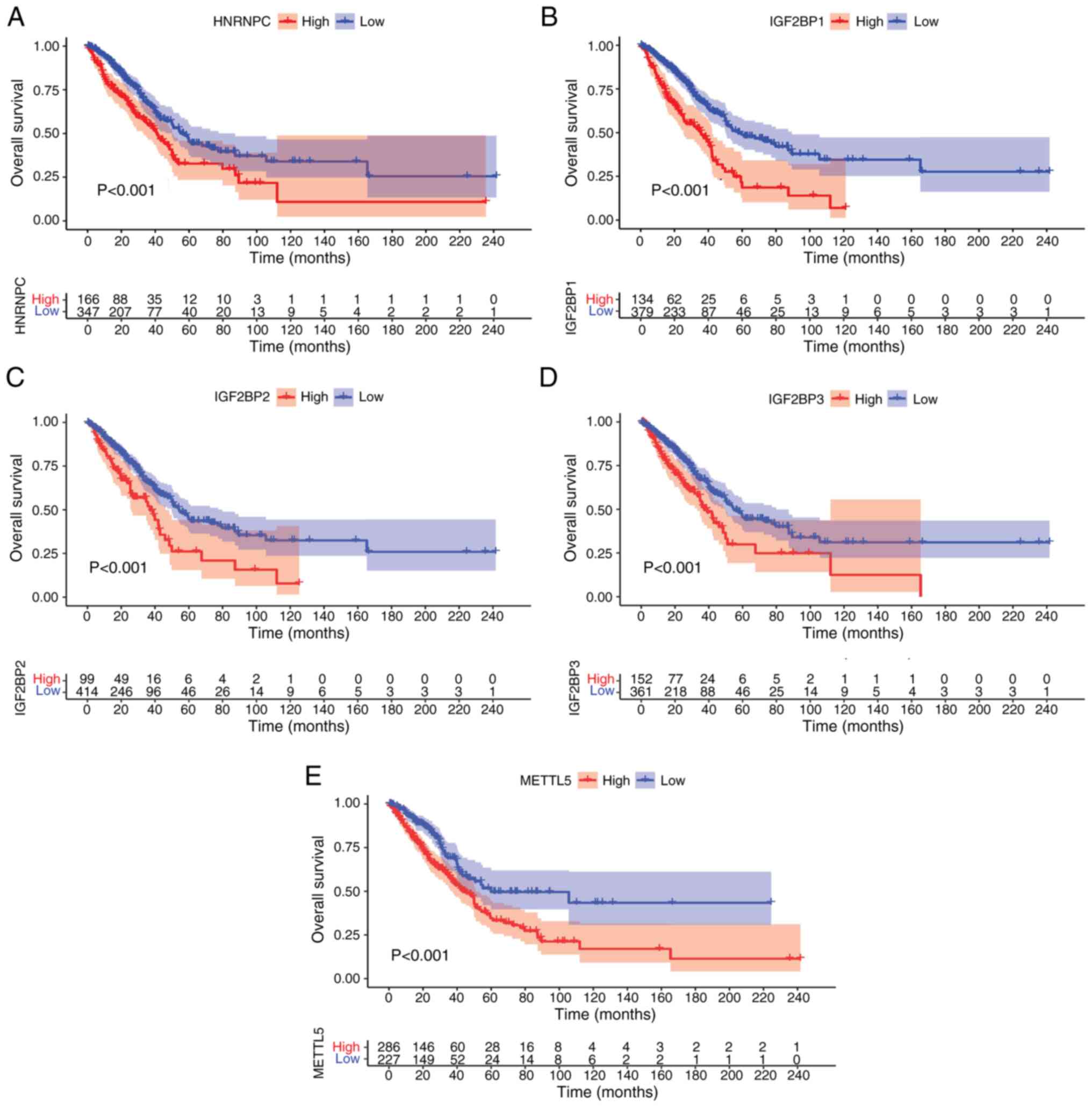
Survival analysis for the m6A gene in
LUAD
To further evaluate the prognostic value of m6A RNA
methylation regulators in LUAD, the relationship between their
expression levels and overall patient survival in TCGA database was
determined by a univariate Cox regression and Kaplan-Meier
analysis. The Cox regression results revealed that five genes were
associated with LUAD prognosis. High expression levels of HNRNPC,
IGF2BP1, IGF2BP2, IGF2BP3 and METTL5 were with significantly
associated with a poor prognosis in LUAD (P<0.001; Fig. 4A-E).
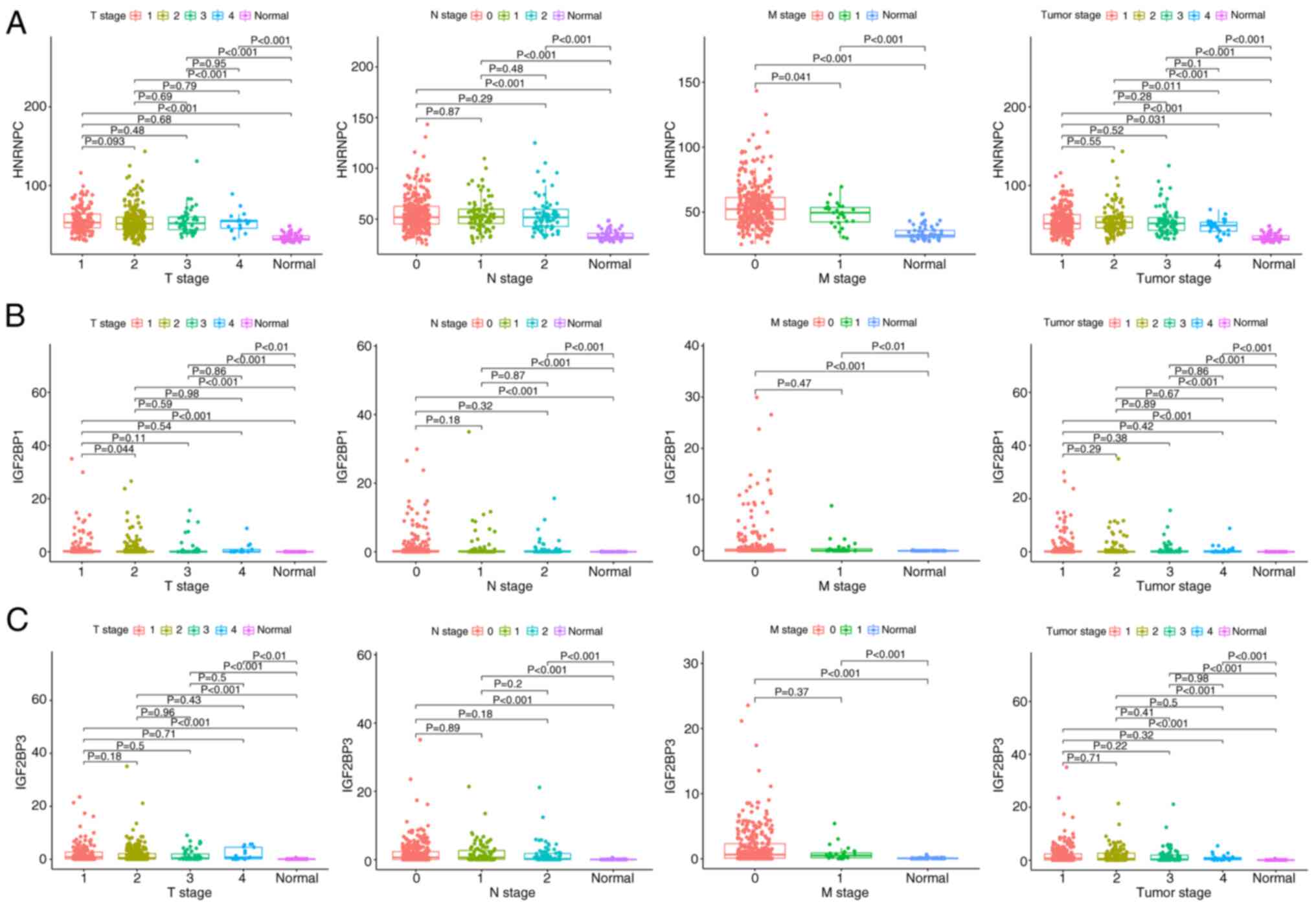
Construction of a nomogram for
LUAD
The associations between HNRNPC, IGF2BP1 and IGF2BP3
expression and clinical parameters, including tumor and TNM stages
(25), were evaluated. The present
study results indicated that there was a significant difference in
HNRNPC expression between the normal group and the four stages
(P<0.001; Fig. 5A). In addition,
a statistical difference was observed between stage 1 and 4
(P=0.031). There were significant differences between the normal
group and the four stages in the T stage (all P<0.001), and
there was a significant difference between the normal group and the
two stages in the M stage (both P<0.001). Significant
differences were also observed between the normal group and the
three stages in the N stage (all P<0.001).
For IGF2BP1 expression, results indicated that there
was a statistically significant difference between the normal group
and the four stages in stage (P<0.001; Fig. 5B). Significant differences were also
found between the normal group and the four stages in the T stage
(P<0.01). Furthermore, a significant difference was observed
between T stages 1 and 2 (P=0.044). There were significant
differences between the normal group and M0 and M1 stages in the M
stage (P<0.001 and P<0.01, respectively). Significant
differences were also found between the normal group and the three
stages in the N stage (P<0.001).
For IGF2BP3 expression, results indicated that there
was a statistically significant difference between the normal group
and the four stages (P<0.001; Fig.
5C). Significant differences were observed between the normal
group and the four stages in the T stage (all P<0.001). There
was also a significant difference between the normal group and M0
and M1 stages in the M stage (both P<0.001). Significant
differences were also found between the normal group and the three
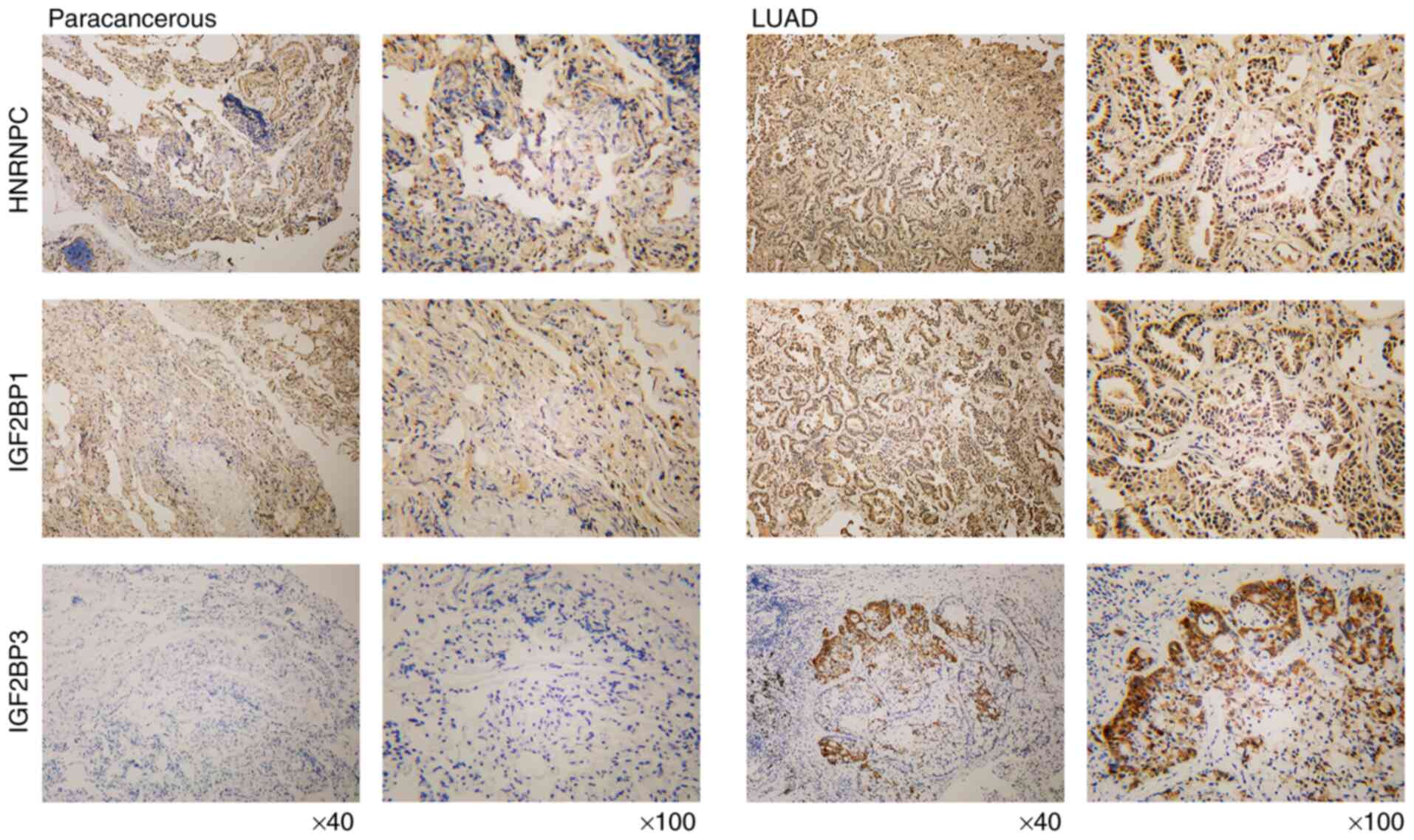
stages in the N stage (P<0.001 for all). Next, the expression
levels of HNRNPC, IGF2BP1 and IGF2BP3 were detected in LC tissues
by IHC. The results demonstrated that these three genes were all
upregulated in LUAD when compared with paracancerous tissues
(Fig. 6).
Correlation analysis between m6A gene
expression and immune infiltration in LUAD
The TIMER database was used to evaluate the
correlation between m6A genes and immune cell infiltration level in
LUAD. Results revealed that BMDCs (ρ=0.494;
P=8.96×10−32) and Tregs (ρ=−0.181;
P=5.02×10−5) were significantly positively and
negatively correlated with HNRNPC, respectively (Fig. 7A). Cancer-associated fibroblasts
(ρ=0.155; P=5.02×10−4), BMDCs (ρ=0.374;
P=6.84×10−18) and macrophages (ρ=0.155;
P=5.54×10−4) were significantly positively correlated
with IGF2BP1 (Fig. 7B). IGF2BP3
indicated significant positive correlations with cancer-associated
fibroblasts (ρ=0.208; P=3.09×10−6), BMDCs (ρ=0.389;
P=2.65×10−19), neutrophils (ρ=0.16;
P=3.56×10−4), CD8+ T cells (ρ=0.224;
P=4.66×10−7), and macrophages (ρ=0.239;
P=7.39×10−8); however, there was a negative correlation
with CD4+ T cells (ρ=−0.094; P=3.74×10−2)
(Fig. 7C). Furthermore, IHC
staining was performed to assess the expression levels of PD-L1,
CD4, CD8 and FOXP3 in clinical samples. Results indicated that the
expression levels of PD-L1, CD4, CD8 and FOXP3 were elevated in
LUAD cancer tissues (Fig. 7D).
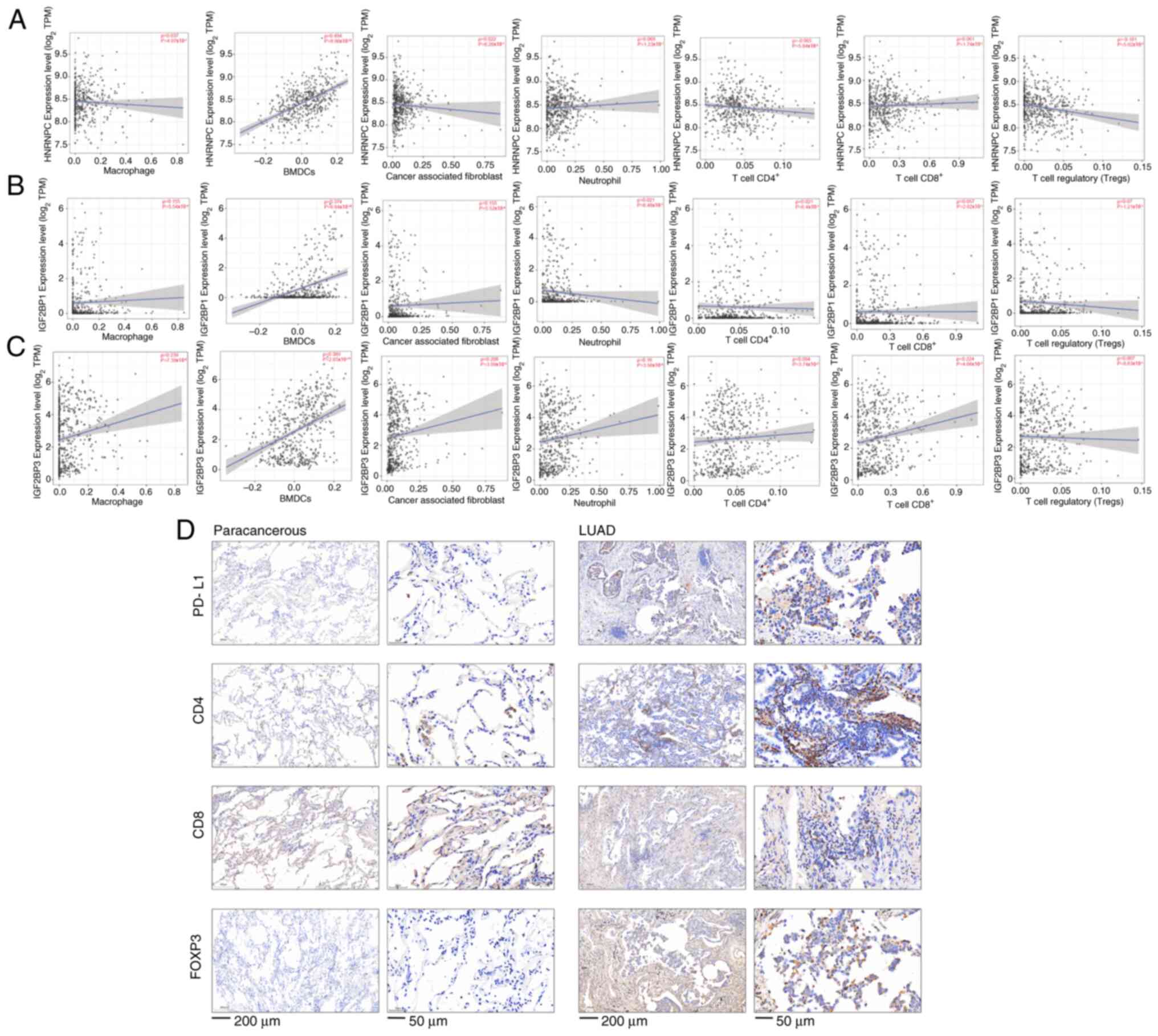 | Figure 7.Correlation analysis between m6A gene
expression and immune infiltration in LUAD. (A) Bone marrow
dendritic cells and regulatory T cells were significantly
positively and negatively correlated with HNRNPC, respectively. (B)
Cancer-associated fibroblasts, BMDCs and macrophages were
significantly positively correlated with IGF2BP1. (C) IGF2BP3 was
positively correlated with cancer-associated fibroblasts, BMDCs,
neutrophils, CD8+ T cells, and macrophages, but
negatively correlated with CD4+ T cells. (D)
Immunohistochemical detection of PD-L1, CD4, CD8 and FOXP3
expression in LUAD (scale bar, 200 and 50 µm) and paracancerous
tissues (scale bar, 200 and 50 µm). m6A, N6-methyladenosine; LUAD,
lung adenocarcinoma; HNRNPC, heterogeneous nuclear
ribonucleoprotein C; IGF2BP, insulin-like growth factor 2 mRNA
binding protein; BMDCs, bone marrow dendritic cells; PD-L1,
programmed cell death-ligand 1; FOXP3, Forkhead box P3; TPM,
transcripts per million. |
Correlation between m6A gene and T
cell exhaustion in LUAD
First, the correlation between m6A gene expression
and biomarkers of T cell exhaustion (TNF, IL-2, IFN-γ and CTL). The
results indicated that HNRNPC was negatively correlated with TNF
expression (P<0.05), IL-2 expression (P<0.001) and CTL
expression (P<0.05), IGF2BP3 was positively correlated with
IFN-γ expression (P<0.01), while IGF2BP1 (P>0.05) was not
correlated with any of the biomarkers (Fig. 8A). Next, the expression levels of
m6A genes in patients treated with anti-PD-1/PD-L1 were analyzed
based on GEO datasets (dataset nos. GSE135222 and GSE126044).
However, the expression levels of HNRNPC (P=0.163; P=0.583),
IGF2BP1 (P=0.69; P=0.743) and IGF2BP3 (P=0.621; P=0.583) were not
significantly different between the non-response group and response
group (Fig. 8B and C).
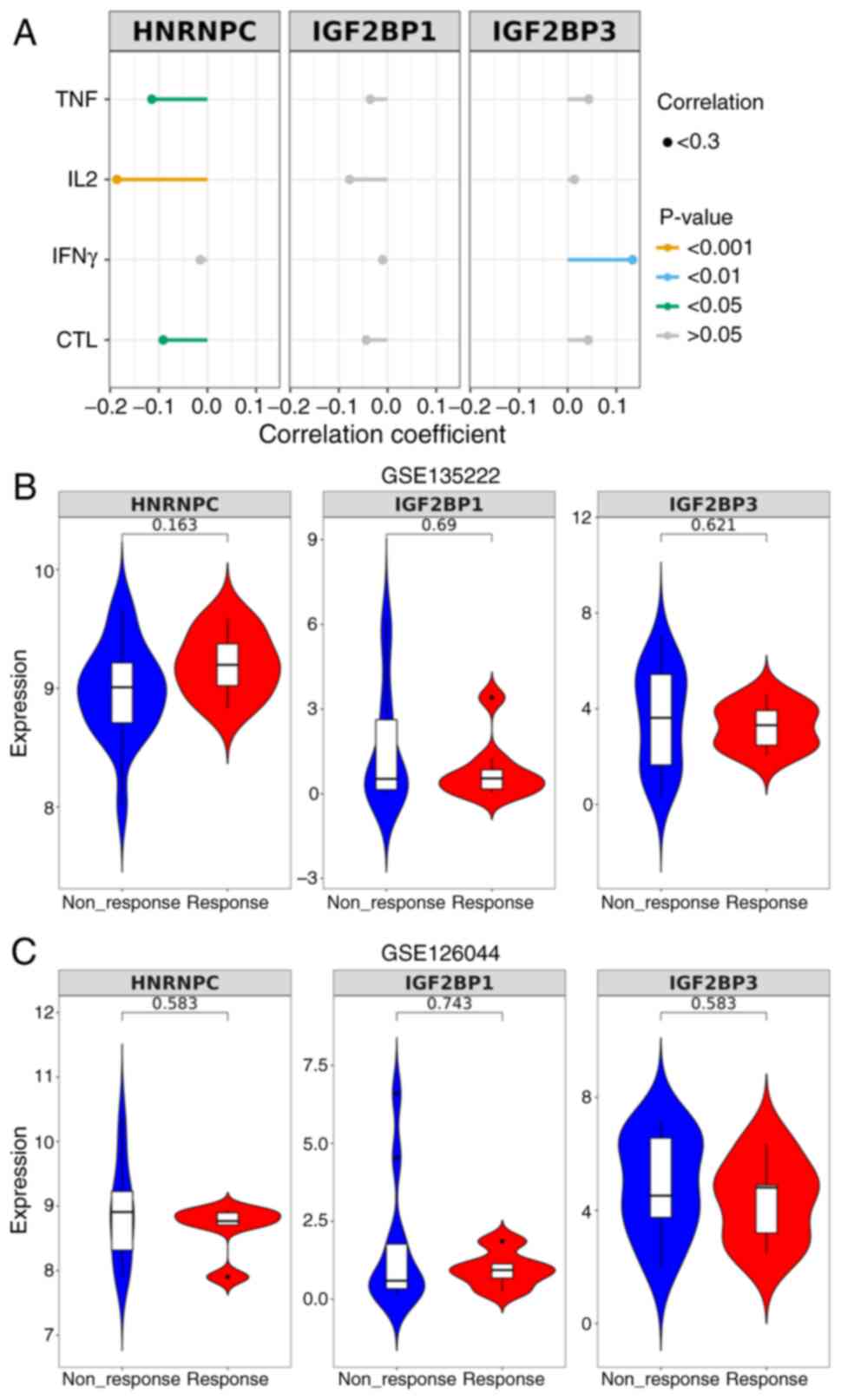 | Figure 8.Correlation between m6A gene
expression and T cell exhaustion in LUAD. (A) The correlation
between m6A gene expression and biomarkers of T cell exhaustion
(TNF, IL-2, IFN-γ and CTL) was analyzed based on previous research
datasets (PMID, 37284314 and 37284314). (B) Expression levels of
m6A genes in patients treated with anti-PD-1/PD-L1 based on a GEO
dataset (dataset no. GSE135222). (C) Expression levels of m6A genes
in patients treated with anti-PD-1/PD-L1 based on a GEO dataset
(dataset no. GSE126044). GEO, Gene Expression Omnibus; m6A,
N6-methyladenosine; LUAD, lung adenocarcinoma; HNRNPC,
heterogeneous nuclear ribonucleoprotein C; IGF2BP, insulin-like
growth factor 2 mRNA binding protein; PD-L1, programmed cell
death-ligand 1; PD-1, programmed cell death protein-1; PMID, PubMed
identifier; CTL, cytotoxic T lymphocytes. |
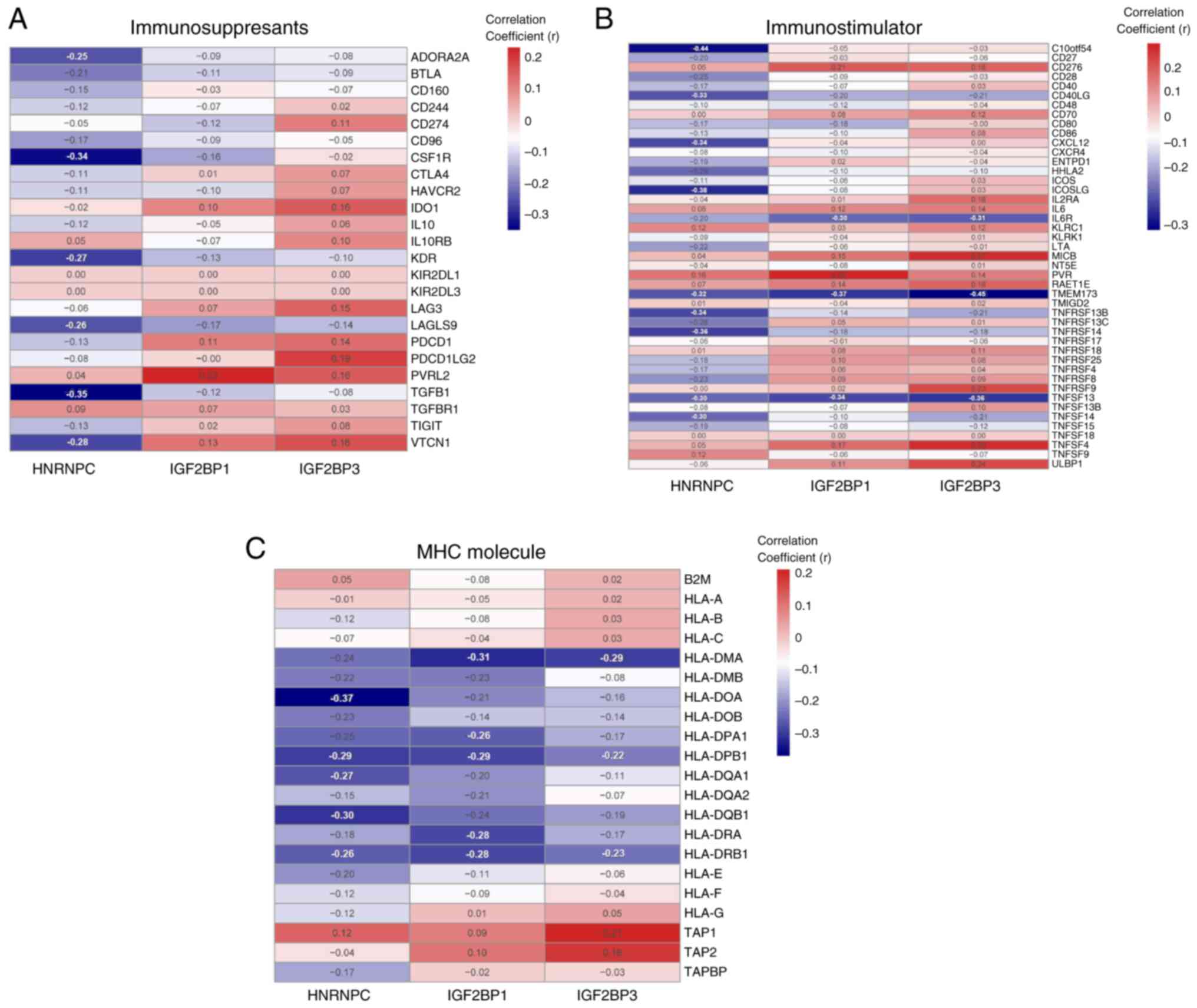
Correlation between m6A genes and the
expression of immunomodulatory factors in LUAD
The TISIDB database was used to analyze the
correlation between m6A genes and the expression of
immunomodulators. Furthermore, to explore the effect of m6A
regulatory factors on tumor immune response, the correlation
between m6A regulatory factors and the expression of
immunoregulatory factors was assessed. The present results
demonstrated that HNRNPC and IGF2BP1 were negatively correlated
with immunosuppressants (Fig. 9A),
immunostimulators (Fig. 9B) and MHC
molecules (Fig. 9C), whereas
IGF2BP3 was negatively correlated with immunostimulants and MHC
molecules and positively correlated with immunosuppressants.
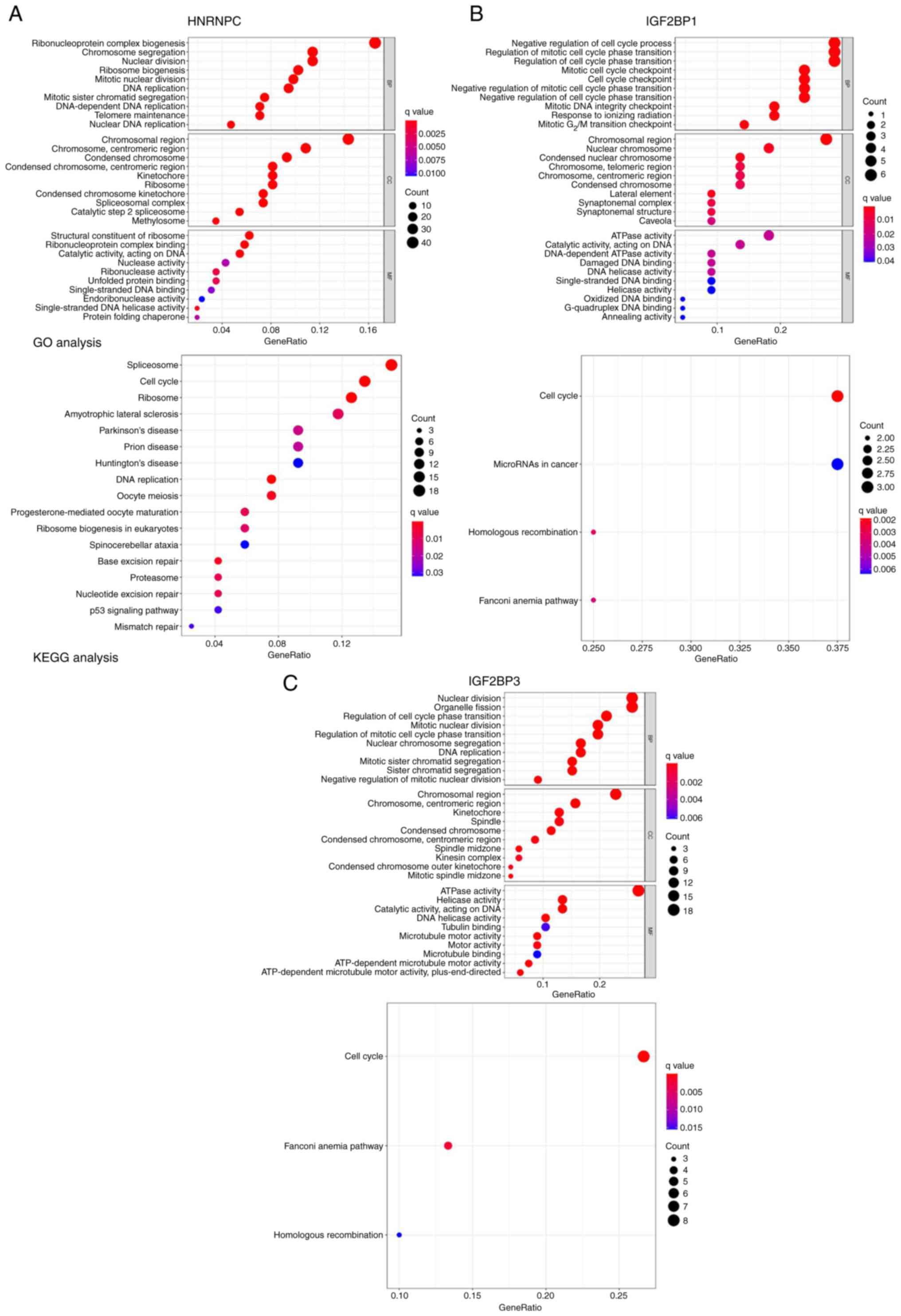
Enrichment analysis of the hub gene
and its related gene pathways
To identify the genes co-expressed with HNRNPC,
IGF2BP1 and IGF2BP3, a Pearson's correlation analysis of m6A genes
from TCGA expression matrix was performed. r>0.5 and P<0.001
were used for screening in the subsequent analysis. A total of 276
genes exhibiting co-expression with HNRNPC were identified, along
with 22 for IGF2BP1 and 80 for IGF2BP3. To investigate the
downstream pathways of hub m6A regulators in LUAD, GO and KEGG
(26–28) analyses were performed using
coexpression genes of the three m6A regulators. The present study
demonstrated that HNRNPC was mainly enriched in the
‘ribonucleoprotein complex biogenesis’ pathway (Fig. 10A), IGF2BP1 in the ‘mitotic cell
cycle checkpoint’ pathway (Fig.
10B) and IGF2BP3 in the ‘nuclear division’ pathway (Fig. 10C).
Discussion
Globally, LUAD is one of the leading causes of
cancer-related mortality (29).
Numerous studies have confirmed that genomic and epigenetic changes
can facilitate tumor occurrence and progression (30), such as DNA methylation (31). For example, modifications of m6A,
one of the most modified mRNAs, are considered to affect tumor
proliferation, invasion and metastasis. Zhou et al (32) demonstrated that m6A alteration was
associated with a pathologic stage in clear cell renal cancer. A
previous study by Lin et al (33) revealed that METTL3, a modified form
of M6A, stimulated the growth and mobility of gastric cancer
cells.
The present study assessed whether m6A-related genes
could serve as novel biomarkers for LUAD. An examination of TCGA
database revealed that the expression levels of certain
m6A-associated genes, including YTHDF3, YTHDC1, YTHDC2 and IGF2BP2,
was significantly different between the tumor and control groups
(P<0.001). Pearson correlation analysis showed that YTHDF3 had
the strongest correlation with KIAA1429 (r=0.75, P<0.001). Luo
et al (34) reported that
YTHDF3 could upregulate the transcription stability of PD-L1 mRNA
and accelerate NSCLC immune evasion by targeting CD8+ T
lymphocytes. Yuan et al (35) suggested YTHDC1 as a tumor
progression suppressor in LC and a ferroptosis regulator that
functions by modulating ferroptosis suppressor protein 1 mRNA
stability. YTHDC2 was found to be suppressed (36). In the present study, further
investigation of the somatic interactions between m6A genes and the
status of SNPs and highly mutated genomic regions revealed that
IGF2BP1, RBM15, IGF2BP2, IGF2BP1 and KIAA1429 had significant
co-expression frequencies. Furthermore, the present study results
indicated that high expression levels of HNRNPC, IGF2BP1, IGF2BP2,
IGF2BP3 and METTL5 were associated with a poor prognosis in LUAD.
Similarly, another previous study reported that IGF2BP2 promoted
angiogenesis and metastasis in LUAD via exosome-mediated transfer
to endothelial cells, where it stabilized FMS-related receptor
tyrosine kinase 4 mRNA via m6A modification and activated the
PI3K-AKT pathway (37). High
expression levels of METTL5 were associated with a poor prognosis
in LUAD and the METTL5-associated prognostic signature served as an
independent biomarker with immune implications (38).
HNRNPC expression was compared in different clinical
parameters, including tumor stage and the TNM stages. The results
demonstrated a statistically significant difference between stage 1
and 4 and between the normal group and the four stages in the T, M
and N stages. Furthermore, a correlation was observed between m6A
genes and immune cell infiltration levels in LUAD. BMDCs and Tregs
indicated significant positive and negative correlations with
HNRNPC, respectively. The present study indicated that HNRNPC
promotes NSCLC progression and metastasis by stabilizing
m6A-modified transcription factor activating enhancer binding
protein-2α mRNA (39). A
correlation analysis between m6A genes and the expression of
immunomodulators also demonstrated that HNRNPC and IGF2BP1 were
negatively correlated with immunosuppressants, immunostimulators
and MHC molecules. Hu et al (40) also reported that IGF2BP1 upregulates
budding uninhibited by benzimidazoles 1 mitotic checkpoint
serine/threonine kinase B expression via m6A modification to drive
malignant progression, stemness and immune resistance in NSCLC stem
cells. The GO and KEGG enrichment analyses in the present study
demonstrated that HNRNPC was mainly enriched in the
‘ribonucleoprotein complex biogenesis’ pathway, IGF2BP1 in the
‘mitotic cell cycle checkpoint’ pathway and IGF2BP3 in the ‘nuclear
division’ pathway. A previous study by Fujiwara et al
(41) suggested that IGF2BP3 drives
malignancy in early-stage LUAD by controlling microRNA structural
diversity. The present study identified m6A RNA methylation
regulators (HNRNPC, IGF2BP1 and IGF2BP3) as novel prognostic
markers and immune modulators in LUAD and therefore provides a
bridge between epigenetic regulation and tumor immunology. We
hypothesize that small-molecule inhibitors targeting m6A ‘writers’
(for example, METTL3) or ‘readers’ (for example, IGF2BPs) might
enter clinical trials within the next 5 years.
In the current study, m6A-related genes were found
to be partially expressed in LUAD and may serve as potential
diagnostic and prognostic indicators. There are three categories of
enzymes involved in RNA modification, including ‘writers’,
‘erasers’ and ‘readers’, which modify RNA via the m6A modification
mechanism (17,42). Proteins that act as m6A ‘readers’
recognize m6A modifications by binding specific domains and this
binding leads to RNA splicing, mRNA decay and translation
regulation. (43,44). While it is assumed that m6A-related
molecules manipulate the progression and deterioration of human
cancer types via those mechanisms, current understanding of the
mechanistic relationship between m6A and human cancer remains
limited. The findings of the present study suggested an association
between m6A-related genes and immune infiltration; however, the
correlation lacks experimental validation, a limitation
attributable to the retrospective nature and exclusive use of
public genomic datasets in the current work, which prevents the
confirmation of causal relationships between m6A-related genes and
immune infiltration. Therefore, investigating how m6A contributes
to tumor progression is warranted in future research and can
potentially provide novel insights into cancer therapy and drug
development.
In conclusion, the present study identified HNRNPC,
IGF2BP1 and IGF2BP3 as novel immune-related prognostic m6A
regulators in LUAD, which holds notable potential in improving
patient risk stratification and guiding therapies targeting m6A
pathways. However, their precise mechanisms in modulating LUAD
immunity and progression remain unclear and it is necessary to
focus on functional validation in models in future research. The
field may evolve by integrating multi-omics approaches within the
tumor microenvironment and potentially advance the development of
targeted inhibitors against these specific regulators, paving the
way for novel LUAD treatments in the future.
Future studies with larger clinical cohorts are
warranted to experimentally validate the predicted correlations
between m6A-related genes and immune markers (for example, PD-L1,
CD8+ T cells) identified in the present bioinformatics
analysis.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by Guangzhou Science and Technology
Planning Project (grant no. 2023A03J0491), High-level Hospital
Construction Project (grant no. DFJH201801), Guangdong Provincial
People's Hospital Young Talent Project (grant no. GDPPHYTP201902),
GDPH Scientific Research Funds for Leading Medical Talents and
Distinguished Young Scholars in Guangdong Province (grant no.
KJ012019449), Guangdong Basic and Applied Basic Research Foundation
(grant no. 2019B1515130002), and National Science Foundation of
China (grant no. 81872510).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JY and JC conceptualized and devised the methodology
for the present study. XC, YL and ZC conducted the formal analysis
and investigation. XC and JY prepared the original draft of the
manuscript. JC obtained funding and reviewed and edited the
manuscript. JY and XC obtained resources. JY and JC confirm the
authenticity of the data in the present study. All authors have
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
All procedures performed in the present study
involving human participants were in accordance with the ethical
standards of the institutional and/or national research committee
and with the Declaration of Helsinki. The present study was
approved by the ethical committee of Guangzhou Twelfth People's
Hospital (approval no. 2021065; Guangzhou, China). Informed consent
was obtained from all individual participants included in the
present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Laversanne M and Sung H: Global
cancer statistics 2022: GLOBOCAN estimates of incidence and
mortality worldwide for 36 cancers in 185 countries. CA Cancer J
Clin. 74:229–263. 2024.PubMed/NCBI
|
|
2
|
Chen D, Wang R, Yu C, Cao F, Zhang X, Yan
F, Chen L, Zhu H, Yu Z and Feng J: FOX-A1 contributes to
acquisition of chemoresistance in human lung adenocarcinoma via
transactivation of SOX5. EBioMedicine. 44:150–161. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rizzo A: Identifying optimal first-line
treatment for advanced non-small cell lung carcinoma with high
PD-L1 expression: A matter of debate. Br J Cancer. 127:1381–1382.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sahin TK, Ayasun R, Rizzo A and Guven DC:
Prognostic value of neutrophil-to-eosinophil ratio (NER) in cancer:
A systematic review and meta-analysis. Cancers (Basel).
16:36892024. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bas O, Sahin TK, Karahan L, Rizzo A and
Guven DC: Prognostic significance of the cachexia index (CXI) in
patients with cancer: A systematic review and meta-analysis. Clin
Nutr ESPEN. 68:240–247. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lamberti G, Andrini E, Sisi M, Rizzo A,
Parisi C, Di Federico A, Gelsomino F and Ardizzoni A: Beyond EGFR,
ALK and ROS1: Current evidence and future perspectives on newly
targetable oncogenic drivers in lung adenocarcinoma. Crit Rev Oncol
Hematol. 156:1031192020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu F, Hu C, He Y, Sun Y, Han C, Zhang X,
Yu L, Shi D, Sun Y, Zhang J, et al: TLR7: A key prognostic
biomarker and immunotherapeutic target in lung adenocarcinoma.
Biomedicines. 13:1512025. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang H and Lin J: Comprehensive analysis
of co-expressed genes with TDP-43: Prognostic and therapeutic
potential in lung adenocarcinoma. J Cancer Res Clin Oncol.
150:442024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kwok CT, Marshall AD, Rasko JE and Wong
JJ: Genetic alterations of m(6)A regulators predict poorer survival
in acute myeloid leukemia. J Hematol Oncol. 10:392017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Adamopoulos PG, Athanasopoulou K, Daneva
GN and Scorilas A: The repertoire of RNA modifications orchestrates
a plethora of cellular responses. Int J Mol Sci. 24:23872023.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Balacco DL, Hewitt BJ, Bardhan A, Shriane
LM, Hunjan M, Hickerson R, Heagerty AHM and Chapple IL: Unlocking
the potential: m6A-RNA methylation in severe epidermolysis bullosa
simplex. Biosci Rep. 45:429–438. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bai R, Sun M, Chen Y, Zhuo S, Song G, Wang
T and Zhang Z: H19 recruited N 6-methyladenosine (m 6 A) reader
YTHDF1 to promote SCARB1 translation and facilitate angiogenesis in
gastric cancer. Chin Med J. 136:1719–1731. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen J, Zhang YC, Huang C, Shen H, Sun B,
Cheng X, Zhang YJ, Yang YG, Shu Q, Yang Y and Li X: m6A
regulates neurogenesis and neuronal development by modulating
histone methyltransferase Ezh2. Genomics Proteomics Bioinformatics.
17:154–168. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Athanasopoulou K and Adamopoulos PG: New
insights into the dynamics of m6A epitranscriptome: Hybrid-seq
identifies novel mRNAs of the m6A writers METTL3/14. Epigenomics.
16:1159–1174. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Alarcón CR, Goodarzi H, Lee H, Liu X,
Tavazoie S and Tavazoie SF: HNRNPA2B1 is a mediator of
m(6)A-dependent nuclear RNA processing events. Cell. 162:1299–1308.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rong ZX, Li Z, He JJ, Liu LY, Ren XX, Gao
J, Mu Y, Guan YD, Duan YM, Zhang XP, et al: Downregulation of fat
mass and obesity associated (FTO) promotes the progression of
intrahepatic cholangiocarcinoma. Front Oncol. 9:3692019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen XY, Zhang J and Zhu JS: The role of
m6A RNA methylation in human cancer. Mol Cancer.
18:1032019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Y, Li Y, Yue M, Wang J, Kumar S,
Wechsler-Reya RJ, Zhang Z, Ogawa Y, Kellis M, Duester G and Zhao
JC: N(6)-methyladenosine RNA modification regulates embryonic
neural stem cell self-renewal through histone modifications. Nat
Neurosci. 21:195–206. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jung H, Kim HS, Kim JY, Sun JM, Ahn JS,
Ahn MJ, Park K, Esteller M, Lee SH and Choi JK: DNA methylation
loss promotes immune evasion of tumours with high mutation and copy
number load. Nat Commun. 10:42782019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cho JW, Hong MH, Ha SJ, Kim YJ, Cho BC,
Lee I and Kim HR: Genome-wide identification of differentially
methylated promoters and enhancers associated with response to
anti-PD-1 therapy in non-small cell lung cancer. Exp Mol Med.
52:1550–1563. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chenard S, Jackson C, Vidotto T, Chen L,
Hardy C, Jamaspishvilli T, Berman D, Siemens DR and Koti M: Sexual
Dimorphism in outcomes of non-muscle-invasive bladder cancer: A
role of CD163+ macrophages, B cells, and PD-L1 immune checkpoint.
Eur Urol Open Sci. 29:50–58. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yuan K, Zhao S, Ye B, Wang Q, Liu Y, Zhang
P, Xie J, Chi H, Chen Y, Cheng C and Liu J: A novel T-cell
exhaustion-related feature can accurately predict the prognosis of
OC patients. Front Pharmacol. 14:11927772023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim JY, Choi JK and Jung H: Genome-wide
methylation patterns predict clinical benefit of immunotherapy in
lung cancer. Clin Epigenetics. 12:1192020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu R, Lee YJ, Kim CH, Min GH, Kim YB, Park
JW, Kim DH, Kim JH and Yim H: Invasive FoxM1 phosphorylated by PLK1
induces the polarization of tumor-associated macrophages to promote
immune escape and metastasis, amplified by IFITM1. J Exp Clin
Cancer Res. 42:3022023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Olawaiye AB, Baker TP, Washington MK and
Mutch DG: The new (Version 9) American Joint Committee on Cancer
tumor, node, metastasis staging for cervical cancer. CA Cancer J
Clin. 71:287–298. 2021.PubMed/NCBI
|
|
26
|
Ogata H, Goto S, Sato K, Fujibuchi W, Bono
H and Kanehisa M: KEGG: Kyoto encyclopedia of genes and genomes.
Nucleic Acids Res. 27:29–34. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kanehisa M: Toward understanding the
origin and evolution of cellular organisms. Protein Sci.
28:1947–1951. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kanehisa M, Furumichi M, Sato Y, Kawashima
M and Ishiguro-Watanabe M: KEGG for taxonomy-based analysis of
pathways and genomes. Nucleic Acids Res. 51:D587–D592. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang Y, Yuan Y, Li Y, Zhang P, Chen P and
Sun S: An inverse interaction between HOXA11 and HOXA11-AS is
associated with cisplatin resistance in lung adenocarcinoma.
Epigenetics. 14:949–960. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kanda M, Sugimoto H and Kodera Y: Genetic
and epigenetic aspects of initiation and progression of
hepatocellular carcinoma. World J Gastroenterol. 21:10584–10597.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu M, Chen X, Lin K, Zeng K, Liu X, Pan B,
Xu X, Xu T, Hu X, Sun L, et al: The long noncoding RNA SNHG1
regulates colorectal cancer cell growth through interactions with
EZH2 and miR-154-5p. Mol Cancer. 17:1412018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou J, Wang J, Hong B, Ma K, Xie H, Li L,
Zhang K, Zhou B, Cai L and Gong K: Gene signatures and prognostic
values of m6A regulators in clear cell renal cell carcinoma-a
retrospective study using TCGA database. Aging (Albany NY).
11:1633–1647. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin S, Liu J, Jiang W, Wang P, Sun C, Wang
X, Chen Y and Wang H: METTL3 promotes the proliferation and
mobility of gastric cancer cells. Open Med (Wars). 14:25–31. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Luo Y, Zeng C, Ouyang Z, Zhu W, Wang J,
Chen Z, Xiao C, Wu G, Li L, Qian Y, et al: YTH domain family
protein 3 accelerates non-small cell lung cancer immune evasion
through targeting CD8(+) T lymphocytes. Cell Death Discov.
10:3202024. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yuan S, Xi S, Weng H, Guo MM, Zhang JH, Yu
ZP, Zhang H, Yu Z, Xing Z, Liu MY, et al: YTHDC1 as a tumor
progression suppressor through modulating FSP1-dependent
ferroptosis suppression in lung cancer. Cell Death Differ.
30:2477–2490. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang X, Hu Y, Li X, Zhu C and Chen F:
YTHDC2-mediated m6A mRNA modification of Id3 suppresses cisplatin
resistance in non-small cell lung cancer. J Thorac Dis.
15:1247–1257. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fang H, Sun Q, Zhou J, Zhang H, Song Q,
Zhang H, Yu G, Guo Y, Huang C, Mou Y, et al: m(6)A methylation
reader IGF2BP2 activates endothelial cells to promote angiogenesis
and metastasis of lung adenocarcinoma. Mol Cancer. 22:992023.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun S, Fei K, Zhang G, Wang J, Yang Y, Guo
W, Yang Z, Wang J, Xue Q, Gao Y and He J: Construction and
comprehensive analyses of a METTL5-associated prognostic signature
with immune implication in lung adenocarcinomas. Front Genet.
11:6171742020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liao M, Li C, Yang R, Li J, Wu K, Zhang J,
Zhu Q, Shi Y and Zhang X: HNRNPC promotes progression of non-small
cell lung cancer by maintaining TFAP2A mRNA stability. Cancer Cell
Int. 25:852025. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hu S, Yan X, Bian W and Ni B: The m6A
reader IGF2BP1 manipulates BUB1B expression to affect malignant
behaviors, stem cell properties, and immune resistance of
non-small-cell lung cancer stem cells. Cytotechnology. 75:517–532.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fujiwara Y, Takahashi RU, Saito M,
Umakoshi M, Shimada Y, Koyama K, Yatabe Y, Watanabe SI, Koyota S,
Minamiya Y, et al: Oncofetal IGF2BP3-mediated control of microRNA
structural diversity in the malignancy of early-stage lung
adenocarcinoma. Proc Natl Acad Sci USA. 121:e24070161212024.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Song H, Feng X, Zhang H, Luo Y, Huang J,
Lin M, Jin J, Ding X, Wu S, Huang H, et al: METTL3 and ALKBH5
oppositely regulate m6A modification of TFEB mRNA, which
dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes.
Autophagy. 15:1419–1437. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lan Q, Liu PY, Haase J, Bell JL,
Hüttelmaier S and Liu T: The critical role of RNA m6A
methylation in cancer. Cancer Res. 79:1285–1292. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Deng X, Su R, Weng H, Huang H, Li Z and
Chen J: RNA N6-methyladenosine modification in cancers:
current status and perspectives. Cell Res. 28:507–517. 2018.
View Article : Google Scholar : PubMed/NCBI
|