Introduction
Lung cancer, a predominant contributor to global
cancer mortality (1), manifests in
~2,206,771 new cases each year, with lung adenocarcinoma (LUAD)
representing a substantial 39% of these cases (2). By 2020, LUAD had become the most
prevalent subtype of lung cancer worldwide, presenting an enduring
and formidable challenge to global public health efforts (3). While surgical resection remains
effective for early-stage LUAD, the prognostic assessment of
postoperative recurrence and the therapeutic options for patients
with advanced-stage LUAD remain notably limited (4). Despite recent advancements in
immunotherapy, the pronounced heterogeneity of LUAD tumors and the
intricate complexity of their microenvironments contribute to
unpredictable treatment responses (5), 5-year overall survival rates of
<20% and persistent challenge of acquired resistance (6). Consequently, the identification of
novel therapeutic targets and innovative strategies is imperative
to optimize the management of patients with LUAD (7).
The endoplasmic reticulum (ER), often regarded as
the cellular epicenter of protein synthesis, folding and
post-translational modification, orchestrates essential
intracellular processes (8,9). Upon exposure to extrinsic or intrinsic
stressors, the functional integrity of the ER is compromised,
precipitating ER stress (ERS), a phenomenon pervasive across
multiple tumor types, including LUAD (10,11).
ERS orchestrates the regulation of numerous malignant phenotypes in
cancer cells and exerts a profound effect on the functionality and
behavior of immune cells (12).
Notably, under the persistent activation of ERS signaling cascades,
tumor-infiltrating leukocytes not only initiate the classical
unfolded protein response (UPR), but also distinctively modulate
the transcriptional and metabolic programming of immune cells, thus
reshaping the tumor immune microenvironment (13). Therefore, strategic targeting of ERS
and its related UPR pathways holds promise in unveiling novel
therapeutic avenues to enhance the efficacy of immune checkpoint
inhibitors (ICIs) and adoptive T-cell therapies (14). Additionally, cancer cells
experiencing ERS can secrete a cascade of signaling molecules that
recruit or reprogram myeloid cells within the tumor
microenvironment (TME), thereby promoting immune modulation and
potentially impeding tumor progression (15). As research advances, ERS is
increasingly recognized as a promising therapeutic target in
oncology, although its precise mechanisms and clinical
applicability remain to be thoroughly elucidated.
Considering the critical role of genetic factors in
the pathogenesis of LUAD, genome-wide association studies (GWAS)
have emerged as robust tools for identifying LUAD-associated
genetic variations (16). The aim
of the present study was to comprehensively explore the expression
profiles of ERS-related genes (ERSGs) in LUAD and their association
with the immune microenvironment by integrating RNA sequencing
(RNA-seq) data from platforms such as Gene Expression Omnibus (GEO)
and The Cancer Genome Atlas (TCGA) through sophisticated
bioinformatics approaches. By developing an ERS-related predictive
model, the aim was to provide a fresh perspective for prognostic
evaluation in patients with LUAD. The current study concentrated on
brain-derived neurotrophic factor (BDNF), a potentially pivotal
gene, employing summary-data-based Mendelian Randomization (SMR)
and colocalization analyses to prioritize its relevance in LUAD,
followed by experimental validation of its expression and function,
thereby contributing to the scientific foundation for the
development of precision therapeutic strategies for LUAD (Fig. 1).
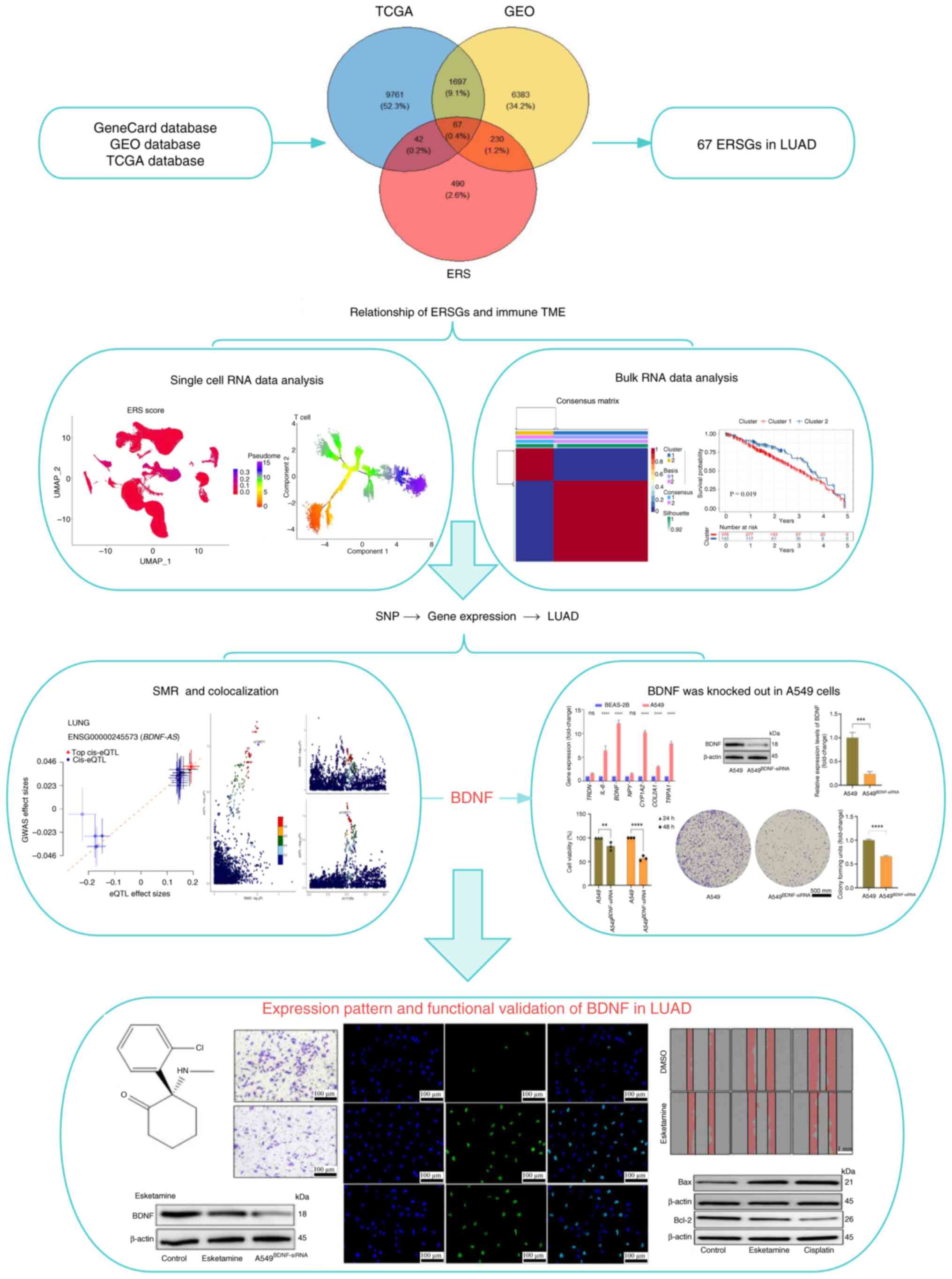 | Figure 1.The main workflow of this study. A
total of 67 ERSGs were identified by combining three databases and
the ERSGs were analyzed and determined using single-cell and bulk
RNA data. The causal relationship between BDNF and LUAD was
analyzed by SMR and colocalization. The difference of BNDF
expression and its effect on A549 cells were verified. Molecular
docking and in vitro experiments verified the effectiveness
of Esketamine in inhibiting A549 cells via downregulated BDNF
level. **P<0.01, ***P<0.005, ****P<0.001. ERSGs,
endoplasmic reticulum stress related genes; BDNF, brain-derived
neurotrophic factor; LUAD, lung adenocarcinoma; SMR,
summary-data-based Mendelian randomization; ERS, endoplasmic
reticulum stress; TCGA, The Cancer Genome Atlas; GEO, Gene
Expression Omnibus; SNP, single nucleotide polymorphism. |
Materials and methods
Data resources
In the present study, the gene set associated with
ERS was systematically curated from the Gene Cards database
(https://www.genecards.org/), employing
‘Endoplasmic Reticulum Stress’ as the defining keyword (17). Subsequently, the GSE139032 dataset,
which encompasses RNA expression profiles from 77 matched pairs of
LUAD and normal lung tissues, was retrieved from the GEO database
(https://www.ncbi.nlm.nih.gov/geo/)
(18). Additionally, TCGA database,
accessed via the UCSC Xena platform (https://xena.ucsc.edu/), was used to acquire a more
extensive dataset of LUAD tumor expression profiles along with
corresponding clinical information, comprising 576 tumor samples
and clinical data from 641 patients (19). Moreover, a single-cell mRNA dataset,
GSE131907 (comprising 208,506 cells), along with an independent
LUAD validation dataset, GSE31210 (containing 226 samples), were
procured from the GEO database (20,21).
Furthermore, GWAS summary statistics from the IEU database
(ieu-a-984; http://gwas.mrcieu.ac.uk/datasets/ieu-a-984/),
encompassing 11,245 LUAD cases and 54,619 controls, were
incorporated into the analysis.
Transcriptional profiling and pathway
exploration
To characterize transcriptional variations between
LUAD and normal pulmonary specimens, dimensionality reduction was
first performed through principal component analysis (PCA) on
RNA-seq data from TCGA cohort. The DESeq2 algorithm in R
(https://bioconductor.org/packages/DESeq2; version,
1.44.0) was subsequently employed for the rigorous identification
of differentially expressed genes (DEGs), applying stringent
thresholds of Benjamin-Hochberg adjusted P<0.05 and |log2 fold
change|>0.58. Functional annotation studies including Gene
Ontology (GO) terms categorization and Kyoto Encyclopedia of Genes
and Genomes (KEGG) pathway enrichment were systematically conducted
using the cluster Profiler toolkit (https://bioconductor.org/packages/clusterProfiler;
version, 4.12.0). Prognostically relevant ERSGs were screened
through survival-associated filtration via univariate Cox
proportional hazards modeling (P<0.05). Molecular interaction
networks of these critical ERSGs were subsequently reconstructed
through STRING platform (https://string-db.org; version, 12.0) interrogation,
maintaining a minimum interaction confidence score >0.4
(Tables SI and SII).
Single-cell transcriptomic profiling
and functional dynamics
To ensure analytical robustness in processing the
GSE131907 RNA-seq dataset, multistep quality assurance procedures
were implemented. Cellular quality control thresholds excluded
outliers with transcriptome coverage <200 or >2,500 unique
gene counts, along with cells demonstrating mitochondrial read
contribution >10% (mitochondrial read fraction >10%).
Normalized expression matrices underwent nonlinear dimensionality
reduction and unsupervised clustering via the Seurat toolkit
(https://satijalab.org/seurat/; version
4.0), employing variance stabilization transformation and
graph-based clustering algorithms. Cellular subpopulations were
partitioned at a Leiden algorithm resolution parameter of 0.5,
visualized through uniform manifold approximation projection across
the first 20 principal components. Iterative marker validation
refined 11 algorithmically derived clusters into eight biologically
coherent lineages using lineage-specific canonical markers from the
Cell Marker repository. ERS activation patterns were quantified at
single-cell resolution using the Add Module Score framework
(https://satijalab.org/seurat/reference/addmodulescore;
version, Seurat v4), incorporating a curated ERSG signature.
Functional phenotyping included cytotoxicity potential estimation
via granzyme-perforin axis expression and T-cell exhaustion
profiling using programmed cell death protein 1 (PD-1/PDCD1),
cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) and
lymphocyte-activation gene 3 weighted indices, implemented through
the Seurat AddModuleScore function (v4.0). Intercellular signaling
topologies were reconstructed via Cell Chat (version 1.6.0;
http://bioconductor.org/packages/CellChat) with
ligand-receptor interaction probability thresholds >0.25 and
co-expression pattern validation. Developmental trajectories were
inferred through pseudotemporal ordering algorithms in Monocle3
(version 1.3; http://cole-trapnell-lab.github.io/monocle3/),
incorporating branch point analysis and gene expression momentum
modeling to resolve lineage bifurcation events (Table SIII, Table SIV, Table V).
Molecular taxonomy and TME
deconvolution
To resolve the intrinsic molecular stratification of
LUAD, a consensus clustering framework was implemented through
non-negative matrix decomposition (NMF; version 0.23.0; http://pypi.org/project/nmf/) on TCGA transcriptomic
profiles, optimizing factorization rank via cophenetic coefficient
stability assessment (22).
Survival disparities among molecular subtypes were interrogated
through multivariate survival modeling (survival version 3.2;
http://cran.r-project.org/web/packages/survival/index.html)
incorporating log-rank testing and stratified survival probability
distributions, with hazard ratios (HR) and 95% confidence intervals
(CI) computed for subtype-specific prognostic outcomes (23). Pathway perturbation landscapes were
mapped via bootstrapped Gene Set Enrichment Analysis (GSEA; 1,000
permutations; FDR <0.1) using Hallmark gene sets, quantifying
enrichment of oncogenic signature pathways such as mTORC1 signaling
and epithelial-mesenchymal transition, and immune regulatory
modules (24). The TME was
computationally deconvoluted through: i) Stromal-immune
quantification, ESTIMATE algorithm (estimate version 1.1;
http://bioconductor.org/packages/estimate/)
implementation calculating four-dimensional microenvironment
indices, tumor purity, immune infiltration score, stromal
activation index and composite TME complexity metric, with batch
correction for technical covariates; ii) immune phenotyping,
relative composition: CIBERSORTx (version 1.06; http://cibersortx.stanford.edu) constrained
regression model with 22-leukocyte signature matrix, applying
P<0.05 confidence threshold for lymphocyte subset fraction
estimation. Absolute quantification: MCP-counter (version 2.2.0;
http://bioconductor.org/packages/MCPcounter) digital
cytometry-based quantitation of eight cytotoxic populations
[CD8+ T cells, natural killer (NK) cells] and two
stromal lineages (fibroblasts and endothelial cells), normalized to
transcripts per million (TPM). Immunotherapeutic vulnerability
prediction employed the TIDE framework (version 2.0.3; http://tide.dfci.harvard.edu) incorporating: i)
Tumor-associated immunosuppression markers (CTLA-4, PDCD1); ii)
exclusion signature scores (WNT/TCFβ activation); and iii)
dysfunctional T-cell infiltration patterns. A predefined TIDE
threshold (>75th percentile) identified immunotherapy-refractory
cases, validated through sensitivity analysis against alternative
predictors (TIS score, IFN-γ response signature) (Table SVI, Table SVII, Table SVIII, Table SIX, Table SX, Table SXI).
Construction of prognostic features
using machine learning
To establish robust predictive biomarkers, a
consensus machine learning architecture integrating
survival-optimized feature engineering was designed (25). The analytical workflow comprised
(26): i) Nonlinear feature
prioritization: √p, node size, 15 to quantify variable impact on
event-time distributions. Prognostically influential transcripts
were selected via variable hunting mode (var. used=‘all’) with
minimal depth thresholding; ii) High-Dimensional Regularization
(λα): The multi-algorithmic integration generated a parsimonious
transcriptional signature where: Predictors: Log2-transformed
expression values (TPM normalized); Outcomes: Right-censored
survival tuples (status indicator δ ϵ {0,1}, event time T).
Weighting: LASSO-derived β coefficients scaled by RSF variable
importance metrics. Individual risk stratification employed a
composite scoring algorithm where ω i represented stabilized
coefficients from nested cross-validation (10-fold outer; 5-fold
inner loops) and ε the baseline hazard offset. Predictive
performance validation incorporated time-dependent discrimination
analysis (time-ROC version 0.4; http://cran.r-project.org/web/packages/timeROC/index.html)
with trapezoidal area under the curve integration.
Bootstrap-corrected concordance index (C-index) estimation.
Decision curve analysis quantifying clinical net benefit (Table SXII, Table XIII, Table XIV).
SMR and colocalization analysis
To investigate the potential causal or pleiotropic
relationships between gene expression and LUAD, the current study
implemented SMR and colocalization analyses (27). In the SMR analysis, single
nucleotide polymorphisms were employed as instrumental variables,
with a particular emphasis on blood and colon-specific
cis-expression quantitative trait loci (eQTL). Gene expression
levels, derived from summary eQTL data, functioned as the exposure
factor, while LUAD, based on summary GWAS data, constituted the
outcome variable. SMR version 1.03 (https://yanglab.westlake.edu.cn/software/smr/) was
used to execute the analysis under default settings, with the
objective of identifying genetic factors that may modulate gene
expression and, in turn, influence LUAD risk. To enhance
specificity, the Meta-analysis of cis-eQTL in associated samples
method was employed prior to SMR to integrate Genotype-Tissue
Expression lung tissue and related cis-eQTL data, thereby
mitigating the effect of sample overlap. A stringent P-value
threshold was applied in the SMR analysis to screen for top eQTLs,
with relevant variants being searched within a 1 Mb region
surrounding the target gene. Statistical significance was
determined by Bonferroni-corrected P-values. Additionally, the
Heterogeneity in Dependent Instruments (HEIDI) test was conducted
to exclude potential effects of linkage disequilibrium, where a
P_HEIDI >0.05 was indicative of passing the heterogeneity test.
To interrogate potential pleiotropic effects at shared genomic
loci, Bayesian causal variant colocalization through the coloc
framework (version 5.2.1; http://cran.r-project.org/web/packages/coloc/index.html)
was implemented employing a hierarchical probabilistic model. The
analysis integrated harmonized GWAS summary statistics (MAF >1%;
imputation quality score >0.8) with cis-eQTL data (window, ±500
kb from TSS), calculating approximate Bayes factors through
adaptive quadrature integration (28).
Structural bioinformatics and
ligand-receptor profiling
Three-dimensional conformations of BDNF and
therapeutic ligands were acquired from the PubChem Compound
repository (29). A comprehensive
preprocessing protocol was implemented, including: Solvent excision
via topological analysis; Protonation state optimization at
physiological pH 7.4; Partial charge assignment using
Gasteiger-Marsili formalism and Energy minimization (AMBER ff14SB
force field; 5,000 steepest descent steps). Molecular recognition
dynamics were simulated through semi-empirical free energy
calculations in Auto Dock Tools (version 1.2.2; http://autodock.scripps.edu/resources/adt) employing:
Conformational constraint docking methodology (ligand flexibility
≤5° rotational tolerance); Grid parameterization centered on the
tyrosine kinase receptor B binding domain of BDNF and Lamarckian
genetic algorithm (250 runs; population size 300) with cluster RMSD
cutoff, 2.0. Binding thermodynamics were quantified via molecular
mechanics Poisson-Boltzmann surface area calculations, with
intermolecular stability benchmarks defined as: Moderate affinity:
Gibbs free energy (ΔG) ≤-20.9 kJ/mol (−5.0 kcal/mol); high-affinity
molecular recognition: ΔG ≤-29.3 kJ/mol (−7.0 kcal/mol). Visual
validation of π-π stacking, hydrogen bonding networks and
hydrophobic complementarity was performed in UCSF Chimera (version
1.16; http://www.cgl.ucsf.edu/chimera/) with subsequent
binding pose validation through 50 ns molecular dynamics
simulations (version 3.0) (30).
Cell model establishment
The bronchial epithelial BEAS-2B cells and the
non-small cell lung cancer (NSCLC)-derived A549 cells were
commercially acquired from certified biobanking institutions
(Procell Life Science & Technology Co., Ltd.). Cell populations
were propagated in high-glucose DMEM/F12 hybrid medium containing
heat-inactivated fetal bovine serum (FBS; Thermo Fisher Scientific,
Inc.) under standardized antibiotic prophylaxis. Continuous culture
maintenance occurred in tri-gas incubators with precisely regulated
parameters of 37±0.2°C, 5.0% CO2 and 95% relative
humidity. All cell stocks underwent mycoplasma screening prior to
experimental use, with passage numbers restricted to ≤15
generations to preserve phenotypic stability.
Cell treatment and transfection
In light of the molecular docking results
demonstrating optimal binding between Esketamine and BDNF, A549
cells were divided into control, Esketamine (16.25 µM, 24 h) and
cisplatin (5 µM, 24 h) treatment groups to assess its inhibitory
effects. For the small interfering RNA (siRNA) transfection
experiments, a separate group of A549 cells was transfected with
BDNF-targeting siRNA (siBDNF) or negative control siRNA (NC siRNA)
(Thermo Fisher Scientific, Inc.) to achieve specific protein
knockdown or serve as a transfection control, respectively. The
transfection was performed using the Lipofectamine™ 3000
Transfection Reagent (Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. Briefly, 50 nM of either siRNA was
mixed with the reagent in opti-MEM medium (Thermo Fisher
Scientific, Inc.) and applied to the cells. The transfection
complex was incubated with the cells at 37°C for 6 h, after which
the medium was replaced with fresh complete growth medium.
Subsequent experimentation (including Esketamine or cisplatin
treatment) was performed 24 h after the initiation of transfection
to allow for sufficient gene knockdown. The siRNA sequences used
were as follows: siBDNF sense 5′-GCAUGGCAUUUGACACUUU-3′ and
antisense 5′-AAGUGUCAAAUGCCAUGCUG-3′; NC siRNA sense
5′-UUCUCCGAACGUGUCACGU-3′ and anti-sense
5′-ACGUGACACGUUCGGAGAA-3′.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA isolation was performed with TRizol-based
purification (Beyotime Institute of Biotechnology; cat. no. R0016)
followed by RT using Prime Script RT Master Mix (Takara Bio Inc.)
according to the manufacturer's instructions. Quantitative analysis
of target transcripts was conducted on a CFX96 Real-Time system
(Bio-Rad Laboratories, Inc.) employing SYBR Green chemistry (cat.
no. D7268S; Beyotime Institute of Biotechnology). Expression data
normalization was carried out using GAPDH (forward primer:
5′-GAAGGTGAAGGTCGGAGTC-3′; reverse primer:
5′-GAAGATGGTGATGGGATTTC-3′) as the endogenous reference gene, with
baseline correction and threshold cycle determination performed by
instrument software. The standard thermal cycling conditions used
for the qPCR were: Initial denaturation at 95°C for 2 min;
amplification (40 cycles): Denaturation at 95°C for 15 sec and
annealing/extension at 60°C for 1 min; melting curve analysis:
60–95°C. Primer validation included melt curve analysis and
efficiency testing (95–105%), with sequence details provided in
Table SI. Relative quantification
between experimental conditions was computed through comparative
2-ΔΔCq methodology (31).
Cell proliferation
Post-transfection cell proliferation dynamics were
evaluated through dual complementary approaches. Metabolic activity
quantification employed Cell Counting Kit-8 (CCK-8) reagent (cat.
no. C0038; Beyotime Institute of Biotechnology) with optical
density measurements at 450 nm using a microplate reader. For
long-term proliferation analysis, transfected cells (800–1,000
viable cells/well) were cultured in 6-well plates under standard
conditions for 14 days. Resultant colonies were fixed with 100%
methanol at room temperature for 15 min, stained with 0.5% crystal
violet (cat. no. P0099; Beyotime Institute of Biotechnology) at
room temperature for 15 min and subjected to automated particle
analysis using ImageJ (National Institutes of Health; version 6.0)
with size exclusion criteria (>50 cells/cluster). Clonogenic
survival rates were calculated relative to untransfected
controls.
Migration analysis (Transwell
assay)
Cell migration was assessed using 8 µm-pore
polycarbonate membranes coated with matrix basement membrane
extract (Matrigel; BD Biosciences) incubated at 37°C for 1 h. A549
suspensions (1×105 cells/ml) in serum-free medium were
loaded into upper chambers, while lower chambers contained 10%
FBS-supplemented medium as chemoattractant. After 24 h of
incubation, transmigrated cells were fixed with 4% paraformaldehyde
at room temperature for 20 min and stained with 0.1% crystal violet
at room temperature for 15 min. A total of five random microscopic
fields (magnification, ×200) per insert were captured using an
inverted phase-contrast microscope (Nikon Eclipse Ti; Nikon
Corporation), with cell counts performed by two independent
investigators blinded to experimental conditions.
Wound healing assay
Confluent A549 monolayers in 35 mm culture dishes
were mechanically wounded using 200 µl pipette tips. Debris removal
was achieved through PBS washing before introducing fresh medium
containing 2% FBS. Cells were treated with Esketamine at a
concentration of 16.25 µM. For comparative purposes, parallel
experiments were performed using cisplatin at a concentration of 5
µM. Time-lapse imaging was conducted at 0, 24 and 48 h
post-wounding using an Incu Cyte S3 live-cell imaging system
(Sartorius AG). Wound closure kinetics were quantified through
image analysis software (Image Pro Plus; version 6.0; Media
Cybernetics, Inc.) using edge detection algorithms, with migration
rates expressed as percentage wound area reduction relative to
baseline measurements.
Apoptosis detection (TUNEL assay)
A549 cell apoptosis was quantified using
fluorometric TUNEL assay (cat. no. C1086; Beyotime Institute of
Biotechnology). Fixed cells were permeabilized with ice-cold 0.3%
Triton X-100/PBS, then incubated with reaction mixture containing
TdT enzyme and FITC-dUTP for 1 h at 37°C protected from light.
Nuclear counterstaining employed DAPI (1 µg/ml) with mounting in
anti-fade medium. Fluorescence signals were captured using a
confocal microscope (Leica TCS SP8; Leica Microsystems, Inc.) with
standardized exposure settings. Apoptotic indices were calculated
as (TUNEL+ nuclei/DAPI + nuclei) ×100%, with positive controls
treated with DNase I and negative controls omitting TdT enzyme.
Western blotting
Total protein was extracted from these groups using
RIPA lysis buffer (Beijing Solarbio Science & Technology Co.,
Ltd.; cat. no. A0020). Protein concentration was determined using a
Protein Quantification Kit (BCA Assay; Abbkine Scientific Co.,
Ltd.; cat. no. KTD3001) according to the manufacturer's
instructions. Briefly, a standard curve was generated using BSA,
and the absorbance of samples was measured at 562 nm after
incubation at 37°C for 25 min. Equal amounts of protein (30 µg per
lane) were separated by electrophoresis on 10% SDS-polyacrylamide
gels (prepared using SDS-PAGE Gel Preparation Kit; Beyotime
Institute of Biotechnology; cat. no. P0012A) and then transferred
onto polyvinylidene difluoride membranes (Immobilon®-P;
Merck KGaA; cat. no. ISEQ00010). The membranes were blocked with 5%
bovine serum albumin (Beijing Solarbio Science & Technology
Co., Ltd.; cat. no. A8020) in TBST (20 mM Tris-HCl, 150 mM NaCl,
0.1% Tween-20, pH 7.5) for 2 h at 25°C. Subsequently, the membranes
were incubated overnight at 4°C with the following primary
antibodies: Rabbit anti-BDNF (1:1,000; cat. no. GB300035), rabbit
anti-Bax (1:1,500; cat. no. GB114122), rabbit anti-Bcl-2 (1:800;
cat. no. GB154380) and rabbit anti-β-actin (1:4,000; cat. no.
GB15003), all from Beijing Solarbio Science & Technology Co.,
Ltd. After washing, the membranes were incubated with
HRP-conjugated secondary antibodies: Goat anti-rabbit IgG
(1:50,000; Abbkine Scientific Co., Ltd.; cat. no. A21020) for 1 h
at 25°C. Protein bands were visualized using an enhanced
chemiluminescence detection reagent (SuperKine™ West Femto Maximum
Sensitivity Substrate; Abbkine Scientific Co., Ltd.; cat. no.
BMU102) and captured on a chemiluminescence imaging system.
Densitometric analysis of the bands was performed using ImageJ
software (version 1.53t; National Institutes of Health).
Statistical analysis
For comparative analyses of non-normally distributed
datasets (Shapiro-Wilk test; P>0.05), non-parametric Wilcoxon
rank-sum tests were applied using the stats package in R version
4.3.0 (https://www.r-project.org/). If the
groups were independent, experimental datasets were analyzed via
unpaired two-tailed Student's t-tests. If the measurements were
paired/matched, experimental datasets were analyzed via paired
two-tailed Student's t-tests, with continuous variables expressed
as mean ± standard deviation. Computational workflows were executed
in RStudio incorporating the tidyverse ecosystem (https://www.tidyverse.org/; version 2.0.0), while
differential expression patterns were visualized through ggplot2
(version 3.4.2; http://cran.r-project.org/web/packages/ggplot2/index.html)
and GraphPad Prism (version 9.0; Dotmatics; ANOVA module). For
one-way ANOVA, Tukey's post hoc test was used. Statistical
significance thresholds followed Benjamini-Hochberg correction for
multiple comparisons. P<0.05 was considered to indicate a
statistically significant difference. All the experiments were
repeated three times (n=3).
Results
Identification of ERSGs in LUAD
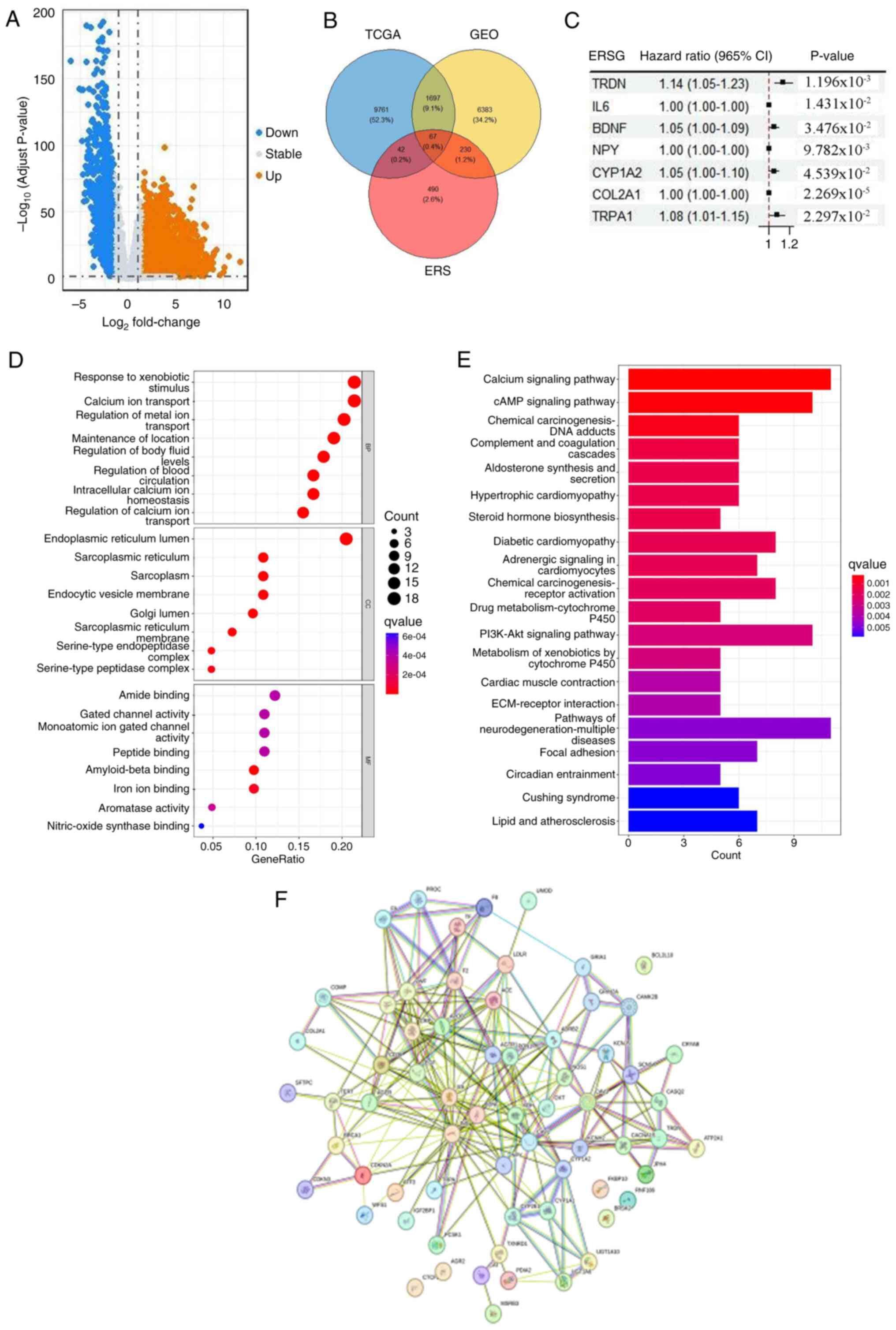
The present study systematically investigated the
molecular characteristics of ERSGs in LUAD. By conducting a
comprehensive analysis of TCGA-LUAD cohort, 11,576 genes were
identified with markedly differential expression between LUAD and
normal lung tissues (Fig. 2A). To
bolster the robustness and generalizability of the findings, the
expression dataset GSE139032 from the GEO database, comprising 77
LUAD samples and 77 normal lung tissue controls, was additionally
analyzed. Through the integration of data from TCGA and GEO, and by
cross-referencing with the ERSG list from the Gene Cards database,
67 critical ERSGs were identified in LUAD, which constituted the
primary focus of the present study (Fig. 2B). Prognostic screening via Cox
proportional hazards regression (univariate analysis with Benjamini
correction) delineated seven risk-associated ERSGs, predominantly
encoding proto-oncogene products, with Kaplan-Meier validation
revealing distinct survival stratification (log-rank P<0.01;
Fig. 2C). Subsequently, GO
enrichment analysis provided deeper insights into the specific
roles of ERSGs in various biological processes, such as the
regulation of responses to external stimuli and the transport of
calcium and metal ions (Fig. 2D).
Additionally, KEGG pathway enrichment profiling (hypergeometric
test, FDR <0.05) identified topologically organized pathways
including calcium homeostasis regulation, cAMP-PKA signaling and
PI3K-Akt-mTOR cascade as core mechanisms linked to ERSGs, as
visualized through the cluster Profiler package (version 4.8.1;
Fig. 2E). Protein interactome
reconstruction using STRING version 12.0 (confidence score >0.7)
and Cytoscape Cyto Hubba plugin identified critical network hubs
(betweenness centrality >0.3), including BDNF (degree, 28), IL6
(degree, 25) and INS (degree, 22), forming a functional module
enriched in inflammatory response and growth factor binding
(Fig. 2F).
Single-cell RNA-seq uncovers the
intrinsic link between ERS-score and the TME
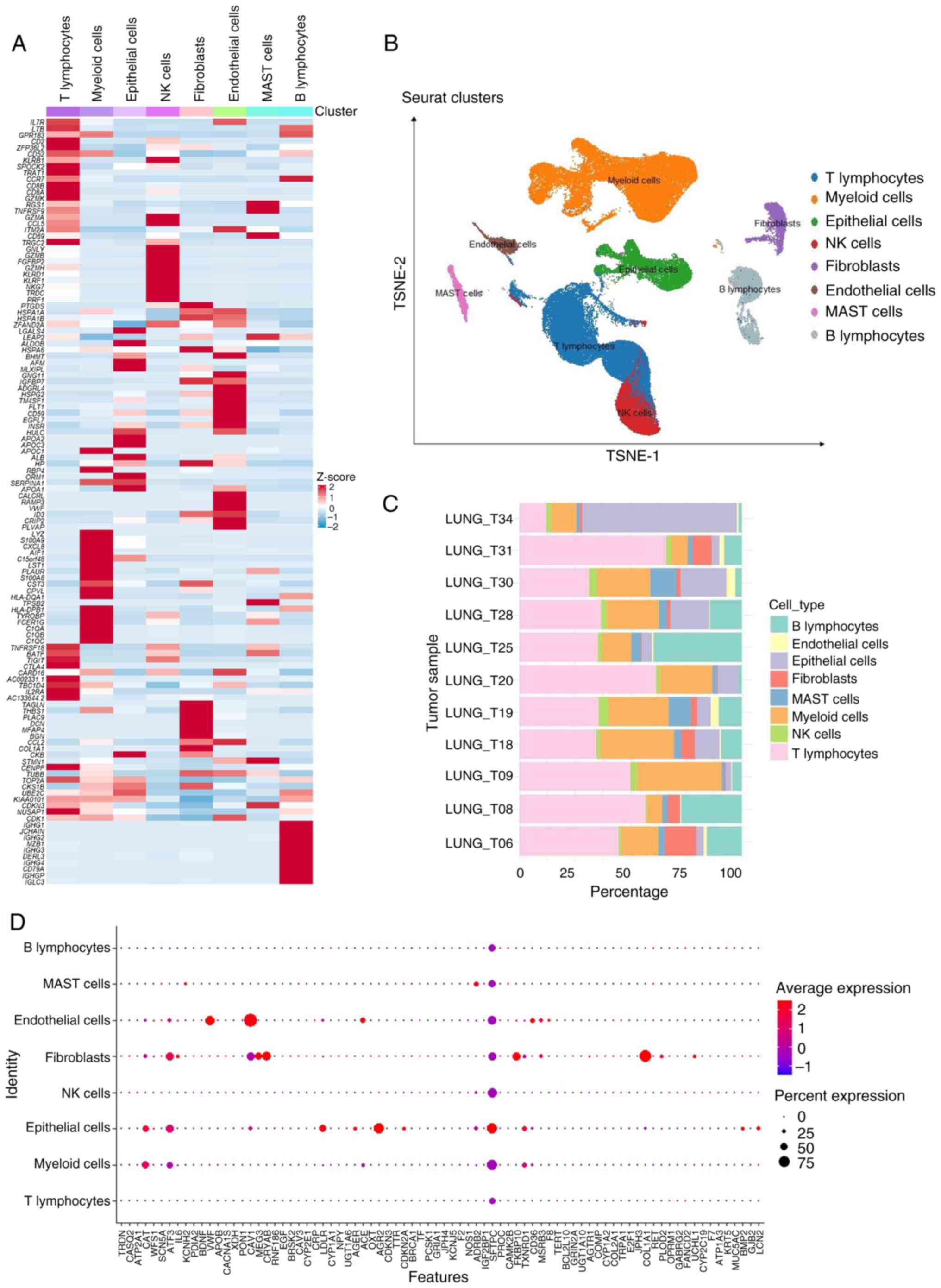
To investigate the specific mechanisms of ERSGs
within the TME, the current study extracted high-quality
single-cell mRNA expression profiles from the publicly available
GSE131907 dataset. Following stringent cellular quality control and
tSNE clustering analysis, 208,506 cells were isolated from 11 tumor
samples and classified into eight biologically distinct cell
clusters (Fig. 3A-C). By
integrating cluster marker genes with original cell type
annotations, eight distinct cell types were definitively
identified, including T lymphocytes (50.2%), NK cells (3.51%), B
lymphocytes (15.6%), endothelial cells (1.56%), epithelial cells
(3.05%), fibroblasts (6.47%), mast cells (2.81%) and myeloid cells
(16.8%), thereby establishing a solid foundation for subsequent
analyses. Further analysis revealed diverse expression patterns of
the 67 ERSGs across cell types, with notably higher expression in
fibroblasts and epithelial cells (Fig.
3D), implicating these cell types in the ERSG-mediated
pathological processes of LUAD. Importantly, the elevated
expression of genes such as BDNF, XDH, OXT, IGF2BP1 and CAMK2B
highlights their potential significance in regulating the TME. A
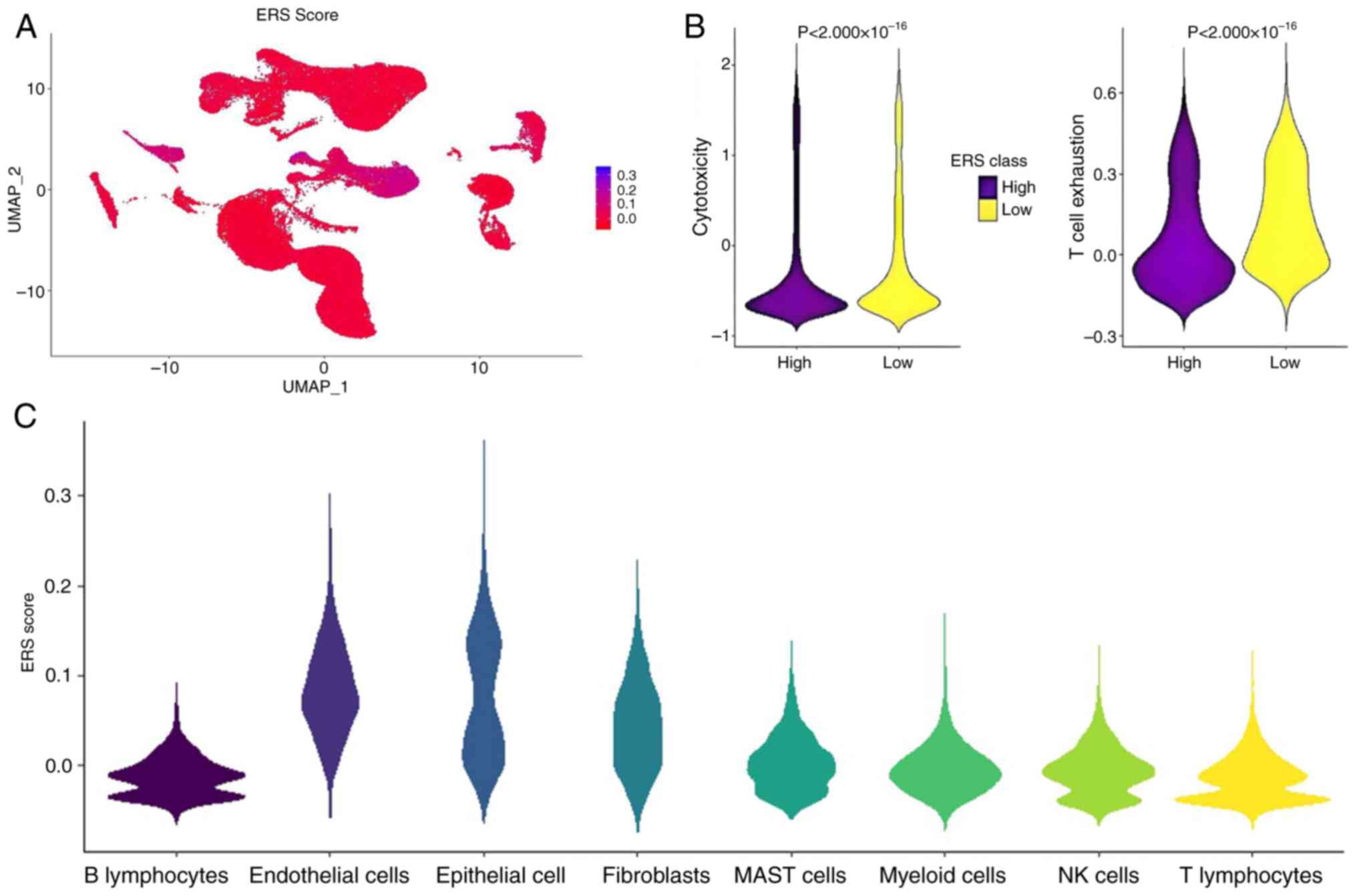
composite ERS activity index was computed using Seurat's Add Module
Score (n=50 control genes; scale=TRUE) with kernel density
estimation revealing bimodal distribution in epithelial cells
(Fig. 4A and C). Tertile-based
stratification (cutoff, 33rd percentile) identified high-ERS
subpopulations exhibiting enhanced cytotoxic lymphocyte
infiltration and attenuated T-cell exhaustion compared to low-ERS
counterparts (Mann-Whitney U test; Fig.
4B).
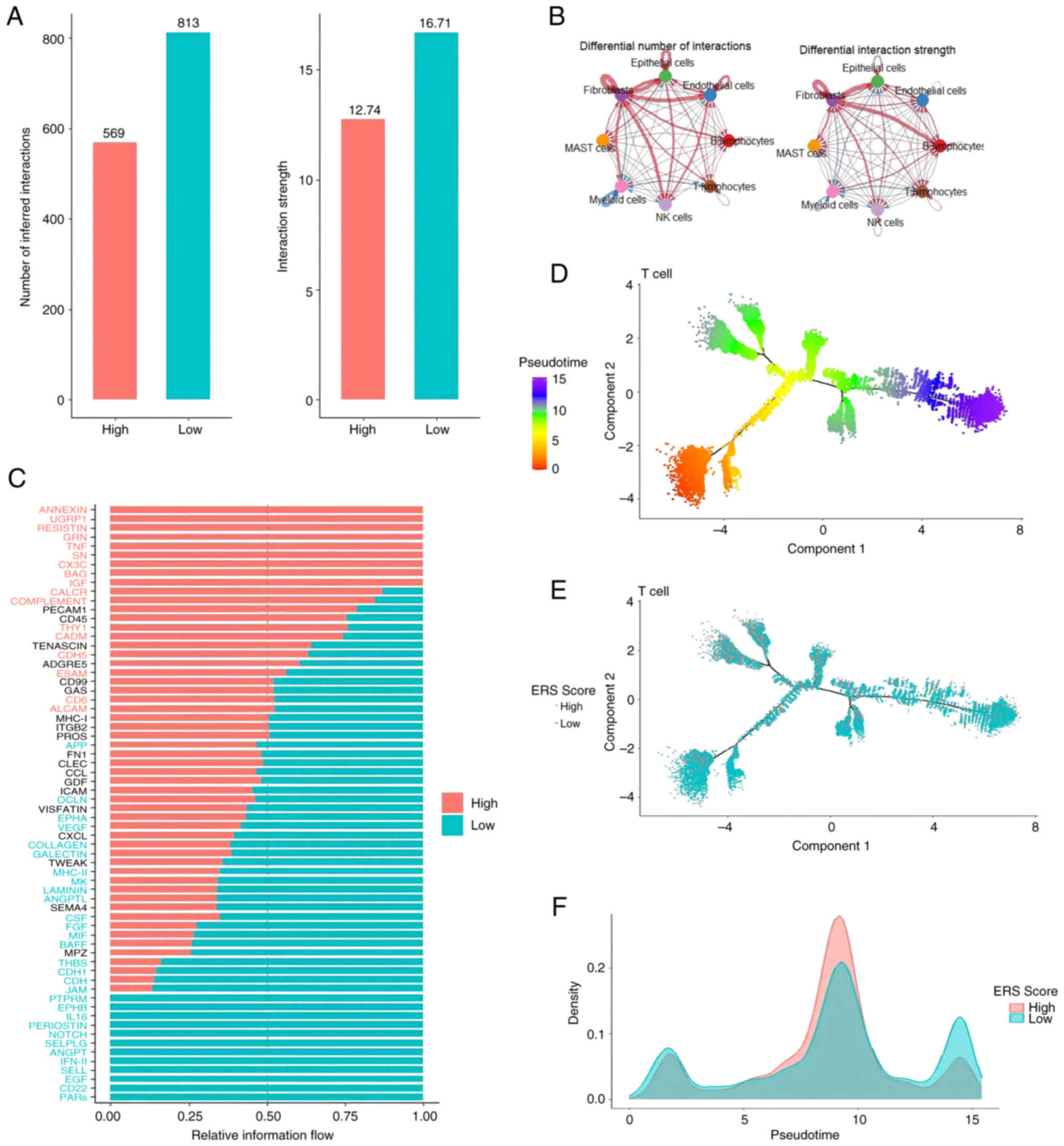
Further investigation into cell-cell interaction
differences between the two groups revealed more frequent and
stable communication within the low ERS-score group, particularly
among B lymphocytes, endothelial cells and epithelial cells
(Fig. 5A and B), potentially
reflecting a more coordinated immune response under low ERS
conditions. Moreover, signaling pathway analysis revealed
significant enrichment of the PARs pathway in the low ERS-score
group (P<0.05) and pathways involving ANNEXIN, UGRP1 and
RESISTIN in the high ERS-score group (P<0.05), offering new
insights into the specific regulatory mechanisms of ERS within the
TME (Fig. 5C). Furthermore, pseudo
time trajectory analysis of T-cell subsets revealed a significant
association between ERS-score and T-cell maturation status; early T
cells had lower ERS-scores, while late T cells exhibited higher
ERS-scores (P<0.001; Fig. 5D-F).
This finding suggested that T cells with lower ERS-scores may be at
a more mature stage, thereby executing their immune functions with
greater efficacy.
To analyze the role of ERSGs in the
molecular subtypes of LUAD based on bulk RNA-seq data
To investigate the potential roles of ERSGs in the
molecular subtypes of LUAD, the present study used TCGA-LUAD cohort
and applied the NMF algorithm to conduct a comprehensive molecular
subtyping of LUAD based on the expression profiles of 67 ERSGs. The
consensus matrix heatmap displayed distinct boundaries at
clustering number k=1, confirming the effectiveness of clustering
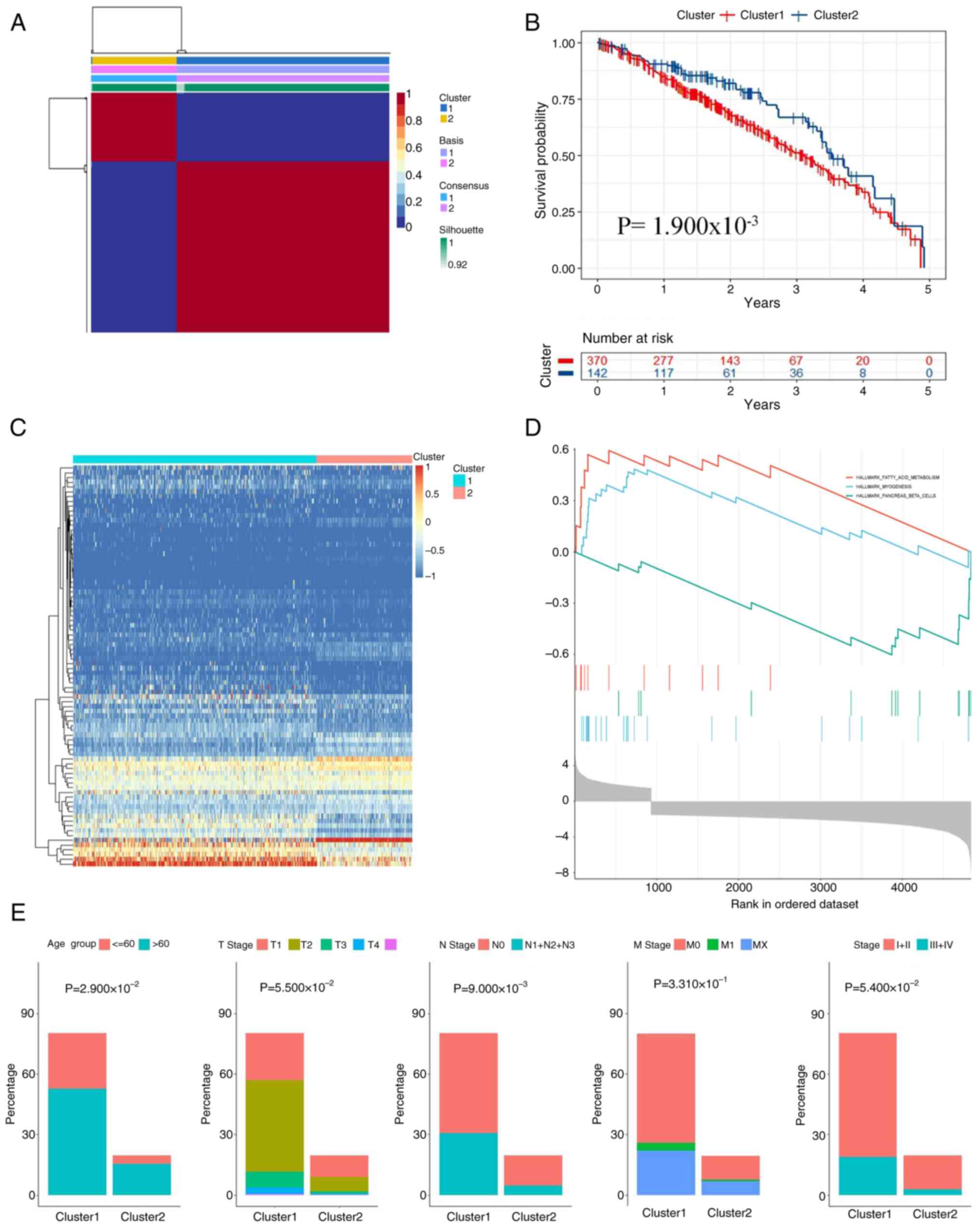
and ensuring the stability and reliability of the results (Fig. 6A). Consequently, LUAD cases were
stratified into two molecular clusters: i) Cluster A containing 877
individuals; and ii) cluster B including 364 subjects. Survival
evaluation demonstrated that Cluster A cases showed markedly
improved prognostic profiles relative to Cluster B counterparts
(log-rank, P=0.018; Fig. 6B).
Further analysis of ERSG expression patterns in the two subtypes
revealed a heatmap that clearly illustrated a slight downregulation
of multiple ERSGs in Subtype 1 (Fig.
6C), suggesting that these genes may have distinct roles in
different subtypes. GSEA highlighted the enrichment of tumor and
immune-related pathways and activities in Subtype 1, particularly
the activation of fatty acid metabolism, myogenesis and pancreatic
β-cell pathways, offering potential clues for the development of
subtype-specific therapeutic targets (Fig. 6D). Additionally, an in-depth
analysis of clinical features identified significant differences in
age and primary tumor stage between the two subtypes (age, P=0.029;
N Stage, P=0.009; Fig. 6E), further
highlighting the importance of molecular subtyping in precision
medicine.
To comprehensively dissect the differences in the
immune microenvironment between the two subtypes, the present study
employed various algorithms, including CIBERSORT, MCP-Counter and
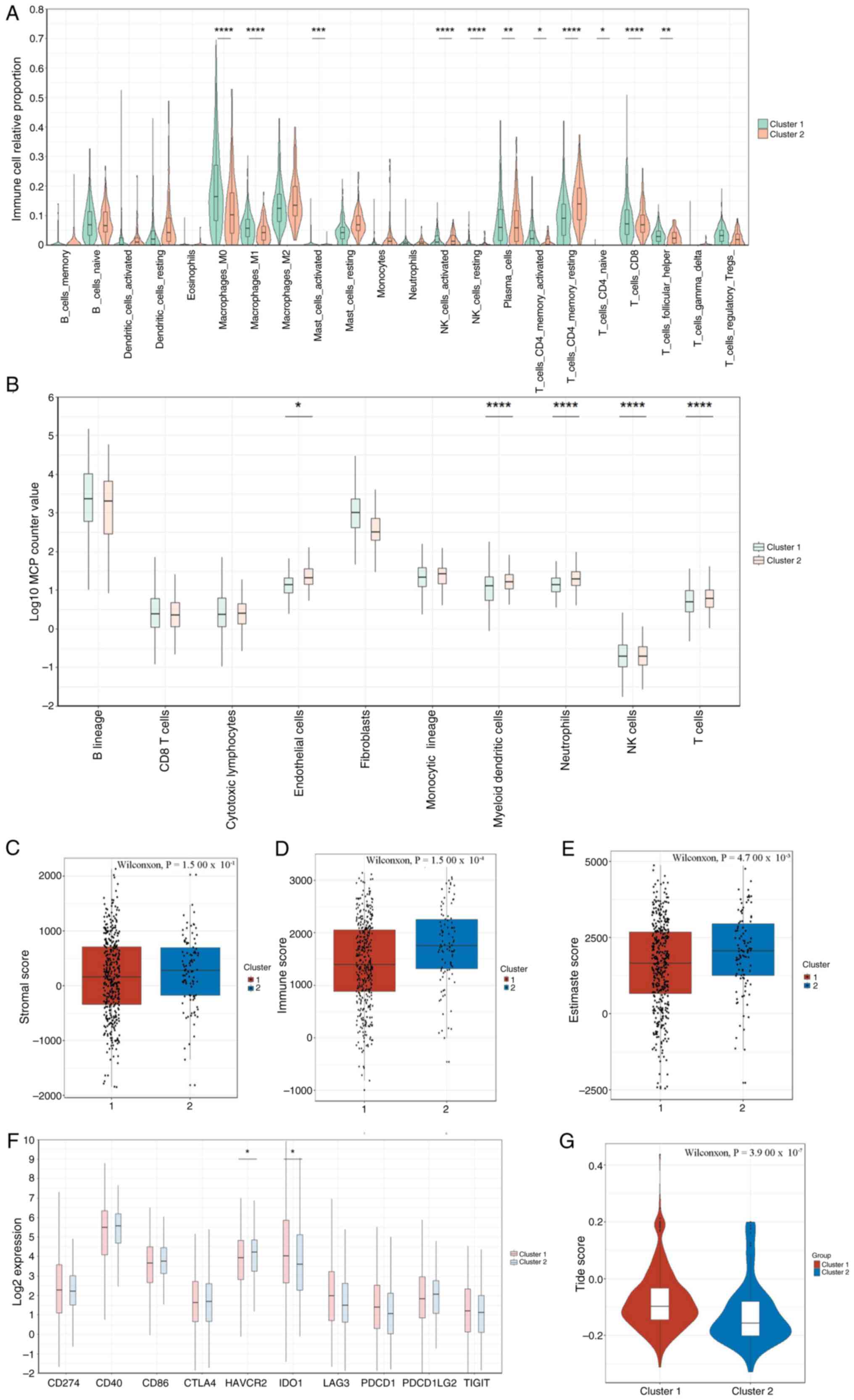
ESTIMATE. CIBERSORT analysis indicated that Subtype 1 showed higher
levels of macrophage M1 and M2, B cells, CD4+ T cells
and T-cell infiltration, while memory B cells and mast cell levels
were relatively lower (P<0.05; Fig.
7A and G). MCP-Counter results also revealed higher levels of
myeloid dendritic cells, neutrophils, NK cells and T cells in
Subtype 1 (P<0.05; Fig. 7B),
indicating a more active immune status in this subtype.
Furthermore, the ESTIMATE algorithm demonstrated markedly higher
stromal, immune and ESTIMATE scores in Subtype 2 (P<0.001;
Fig. 7C-E), which may be associated
with its more complex TME. Considering the critical therapeutic
significance of ICIs, comparative profiling of 10 immune-modulatory
targets was conducted across molecular subtypes. Differential
expression analysis revealed Cluster A displayed elevated
expression of immunoregulatory markers including HAVCR2 (log2FC,
1.8) and IDO1 (log2FC, 2.3) relative to Cluster B
(Benjamini-P<0.001; Fig. 7F).
Notably, Cluster A demonstrated markedly higher TIDE prediction
indices (Welch's t-test, P<0.001; Fig. 7E), implying enhanced ICI
responsiveness in this molecular subgroup.
Construction and validation of
ERS-related prognostic feature models
To comprehensively elucidate the potential impact of
ERSGs on the prognosis of patients with lung cancer, the current
study meticulously developed and constructed a prognostic signature
model based on ERSGs. Initially, using univariate Cox regression
analysis, genes with significant prognostic effects were
preliminarily identified from an initial pool of 67 candidate
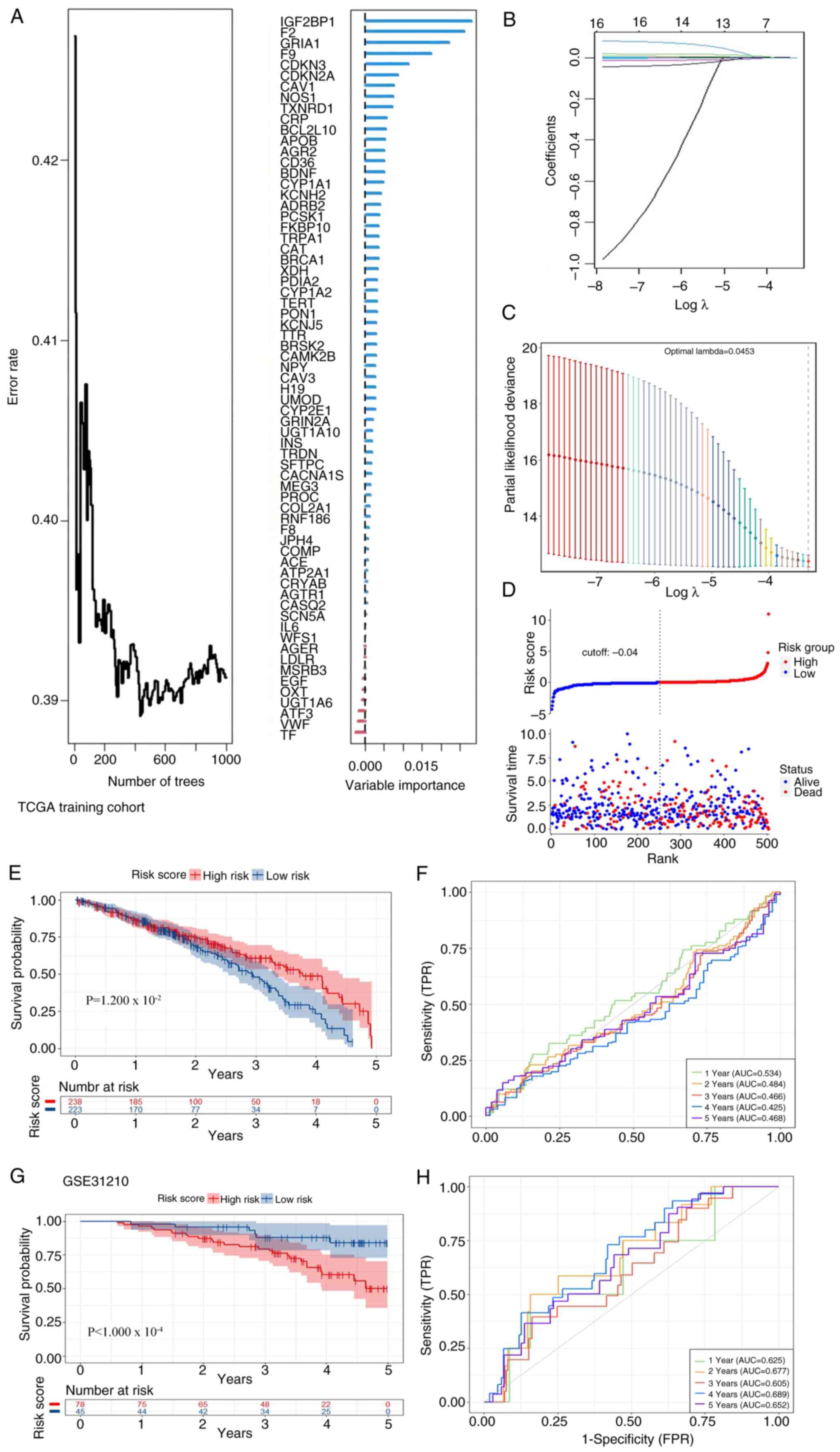
genes. Subsequently, the RSF algorithm was employed to further
optimize gene selection, eliminating genes with low or negative
contributions to the model (Fig. 8A and
B), thereby enhancing the efficiency and accuracy of the model.
Building on this, the LASSO regression method was used to construct
a prognostic signature model comprising 16 pivotal ERSGs (Fig. 8C and D). By integrating the
expression data of these genes, the model calculates an
individualized risk score for each patient using the following
formula: Risk Score=F9* 5.61 + F2* 0.00052 + IGF2BP1* 0.050 +
GRTA1* (−0.496) + BCL2L10 * 0.0188 + CDKN2A* (−0.0006) + CDKN3*
0.0005 + TXNRD1* 0.00006 + CAV1* 0.0001 + CAT* (−0.0002) + TRPA1*
0.020 + NOS1* 0.009 + BDNF* 0.066 + KCNH2* (−0.002) + TERT* 0.062 +
CRP* 0.008. This risk stratification algorithm provides a
quantitative metric for evaluating clinical prognosis. To verify
the discriminatory capacity of the predictive model, external
validation was performed using TCGA-LUAD (n=498) and GSE31210
(n=226) cohorts. In TCGA-LUAD, cases were dichotomized into
elevated- and reduced-risk subgroups via median risk score
thresholding. Significant survival status disparities
(χ2; 12.7) and temporal survival advantages were
observed between subgroups (Fig.
8E), corroborated by Kaplan-Meier estimator analysis (log-rank,
P=0.032; Fig. 8F). Temporal
predictive performance was quantified through ROC analysis,
yielding AUCs of 0.534 (95% CI; 0.48-0.59), 0.484 (0.43-0.54),
0.466 (0.41-0.52), 0.425 (0.37-0.48) and 0.468 (0.41-0.53) for
1–5-year intervals, respectively. Cross-cohort validation in
GSE31210 confirmed robust prognostic differentiation (log-rank,
P=1.4×10−4; Fig. 8G and
H), substantiating the pan-cohort applicability and predictive
consistency of this ERS-based prognostic classifier.
SMR and colocalization analysis
reveals the key role of BDNF in LUAD and its potential drug
target
To further investigate the ERSGs closely associated
with LUAD expression, the current study uniquely integrated SMR and
colocalization analyses. By integrating eQTL data from blood
samples with LUAD GWAS, 15 cis-eQTL probes were identified
associated with ERSGs. Notably, only the BDNF gene passed the
stringent SMR test, revealing a significant association between
BDNF and LUAD (p-SMR=0.03; P_HEIDI >0.05) (28), strongly suggesting that BDNF may
contribute to the onset and progression of LUAD through pleiotropy
or direct causality (Fig. 9A and
B). The SMR effect plot is particularly striking, as it
visually demonstrates a positive association between BDNF
expression levels and LUAD risk. To further substantiate this
finding, a colocalization analysis was conducted to assess the
colocalization of BDNF, as detected by both SMR and HEIDI tests,
between blood eQTL data and LUAD phenotypes. The analysis revealed
significant colocalization evidence between BDNF and LUAD traits
(PPH4, 0.134), strongly supporting the hypothesis that BDNF is a
critical ERSG in LUAD and underscoring the need for further
in-depth exploration of BDNF in subsequent studies (Fig. 9C).
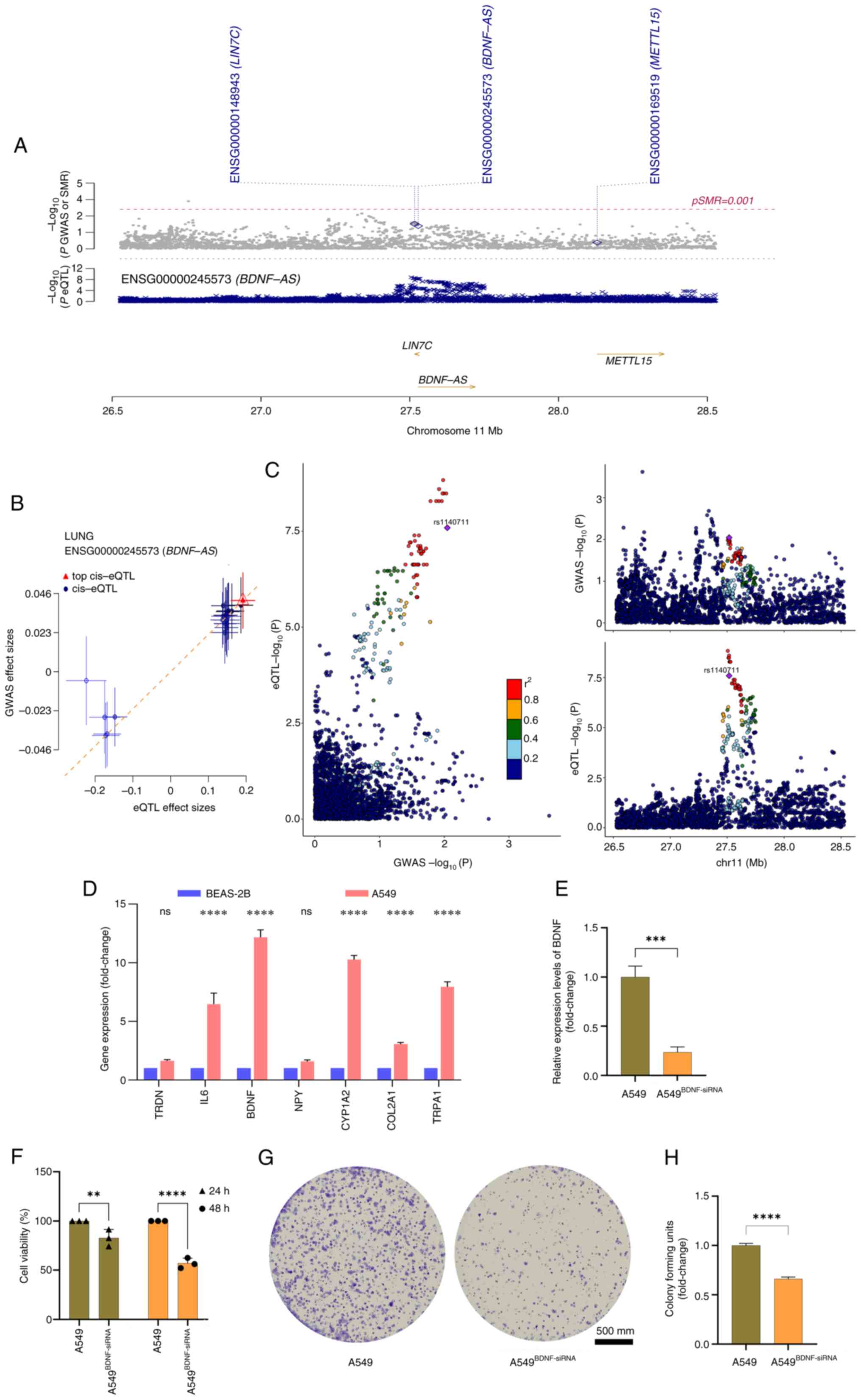 | Figure 9.Integrative genomics prioritization
of causal ERSGs. (A) SMR visualization at BDNF locus
(hg38:11p14.1). Top: Probe passing SMR-HEIDI joint test (HEIDI;
P>0.01). Bottom: GWAS variants (gray circles) vs. lung eQTLs
(red crosses, GTEx v8). (B) Causal effect estimates (Bayes
factor>10) between gene expression and LUAD risk. (C)
Colocalization probability analysis (PP.H4>0.8) showing shared
causal variants (LD, r2 color gradient; lead
variant=purple square). (D) Quantitative PCR validation of ERSGs in
A549 (malignant) vs. BEAS-2B (normal) cells (GAPDH-normalized,
triplicates), ****P<0.001. (E) BDNF silencing efficiency (siRNA
vs. scramble, ΔΔCq method), ***P<0.005. (F) Proliferation
kinetics (CCK-8 assay) at 24/48 h post-transfection, **P<0.01,
****P<0.001. (G and H) Clonogenic capacity assessment (crystal
violet staining) with quantitative histograms (triplicate
experiments, Mann-Whitney ****P<0.001). ERSGs, endoplasmic
reticulum stress related genes; SMR, summary-Mendelian
randomization; BDNF, brain-derived neurotrophic factor; GWAS,
genome-wide association studies; eQTLs, cis-expression quantitative
trait loci; LUAD, lung adenocarcinoma; siRNA, small interfering
RNA. |
In light of the potential significance of BDNF in
cancer therapy, the present study extended its scope to include
molecular docking analyses, with the objective of identifying
potential drugs targeting BDNF, building on prior studies that have
highlighted the potential of BDNF as a therapeutic target in cancer
(32). Through RT-qPCR analysis,
BDNF expression levels were compared between human NSCLC A549 cells
and normal human lung epithelial BEAS-2B cells, revealing a
significant overexpression of BDNF in A549 cells (P<0.001;
Fig. 9D). To investigate the
specific mechanisms of BDNF in LUAD, a BDNF knockdown model was
successfully established (A549BDNF-siRNA) in A549 cells using
loss-of-function experiments utilizing siRNA (33). Results of RT-qPCR confirmed the
knockdown efficiency, thereby ensuring the reliability of the
experimental model (Fig. 9E and F).
CCK-8 cell viability assays demonstrated that BDNF knockdown
markedly suppressed the proliferation of A549 cells (P<0.01;
Fig. 9G). This finding was further
corroborated by colony formation assays, which revealed a marked
decrease in the colony-forming ability of A549BDNF-siRNA cells
(P<0.05; Fig. 9H). These
molecular docking results not only offered valuable insights for
the development of targeted therapies against BDNF but also
establish a robust foundation for subsequent experimental
validation.
Expression pattern and functional
validation of BDNF in LUAD
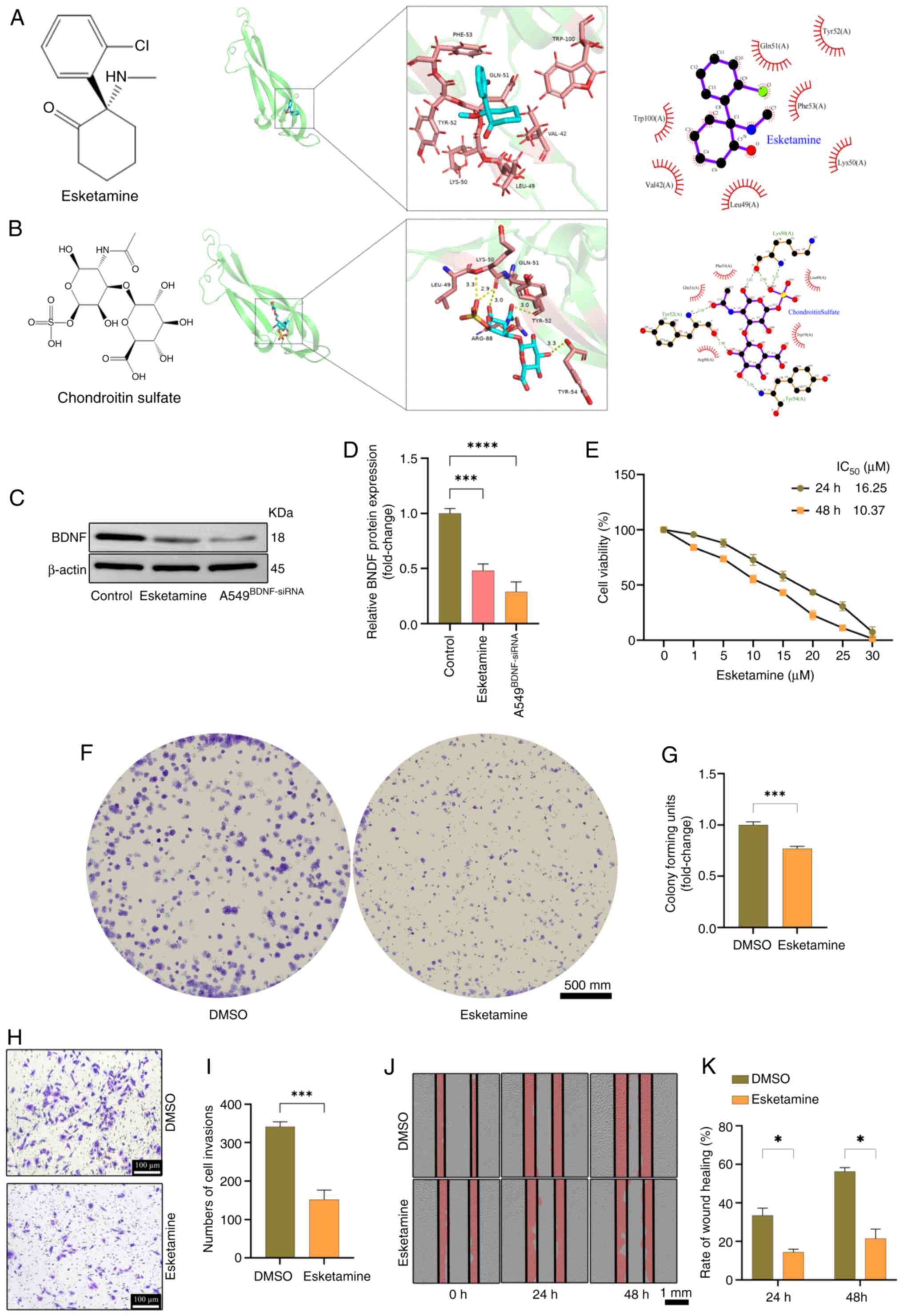
The present study sought to elucidate the expression
patterns and functional implications of BDNF in LUAD. Molecular
docking analyses of BDNF were performed with approved drugs in
Drugbank (https://go.drugbank.com/). Of the two
drugs tested, Esketamine (34)
demonstrated the most favorable binding energy (−5.3 kcal/mol),
closely followed by chondroitin sulfate (35) with a binding energy of-4.9 kcal/mol
(Fig. 10A and B). Esketamine was
selected as a potential therapeutic to investigate its effects on
A549 cells. Esketamine can downregulate the expression of BDNF
(Fig. 10C and D). The impact of
Esketamine on A549 cell proliferation was validated using CCK-8 and
colony formation assays (Fig.
10E-G). The effects of Esketamine on invasion and migration
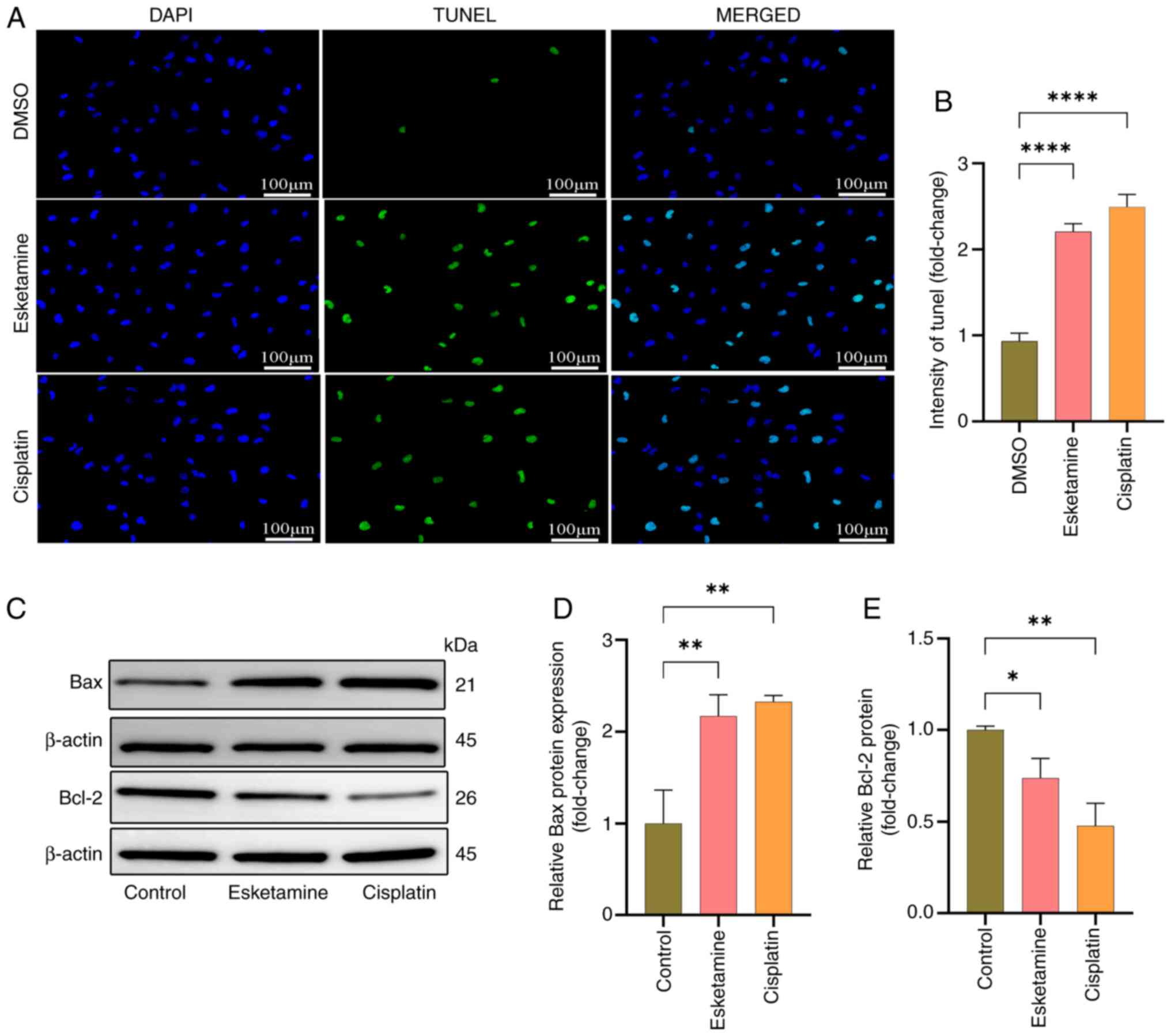
were corroborated by Transwell and wound healing assays (Fig. 10H-K). The TUNEL assay was employed
to detect the apoptotic rate of A549 cells, while western blotting
was used to assess the expression of the apoptotic marker proteins
Bax and Bcl-2, confirming that Esketamine could induce apoptosis in
A549 cells (Fig. 11). These
findings not only validate the oncogenic role of BDNF in LUAD but
also provide compelling experimental evidence for the development
of targeted therapies based on BDNF.
Discussion
In light of the critically low 5-year survival rate
of <20% among patients with LUAD (36), there is an urgent need to explore
innovative and efficacious therapeutic strategies to enhance
patient prognosis. Emerging research highlights that the TME,
serving as a critical nexus for malignant tumor progression and
immune cell infiltration, instigates substantial ERS. This process
not only orchestrates tumor growth and metastasis but also exerts a
profound influence on the efficacy of antitumor immune responses
(37). Consequently, the precise
modulation of ERS to induce direct tumoricidal effects and
potentiate antitumor immune responses presents novel therapeutic
avenues for LUAD treatment (38).
Building on existing research and the comprehensive analysis
undertaken in the present study, it is strongly contended that
ERSGs occupy a central role in delineating the characteristics of
the TME and in predicting prognosis for patients with LUAD.
The current study adeptly integrated a spectrum of
biological analysis strategies to systematically elucidate the
specific expression patterns of ERSGs in LUAD samples. By
harnessing authoritative database resources such as GEO, TCGA and
Gene Cards, 67 ERSGs were screened and identified. Through an
exhaustive analysis of single-cell sequencing and bulk RNA
transcriptome data, it was not only revealed the differential
expression profiles of ERSGs in LUAD samples, but also their robust
associations with LUAD using SMR and colocalization analyses were
substantiated. This multi-layered, multi-dimensional approach
effectively mitigated data noise and bias, thereby providing robust
data support for a deeper understanding of the mechanisms
underlying the role of ERSGs in LUAD progression.
Notably, the heterogeneity of the TME exerts a
profound influence on the efficacy of immunotherapy (39), with varying ERSG expression levels
emerging as pivotal determinants shaping immune responses within
this intricate milieu (40). The
present study revealed that elevated ERSG expression is frequently
associated with diminished immune activity, characterized by
disrupted immune cell interactions and aggravated T-cell
exhaustion. Genomic instability in LUAD promotes sustained ERS,
activating the UPR and selectively upregulating pro-survival ERSGs
such as BDNF. This creates a permissive microenvironment for tumor
progression. Specifically, cell populations with elevated
ERS-scores exhibited heightened cytotoxicity and attenuated T-cell
exhaustion, further corroborating the critical role of ERSGs in
orchestrating the TME. Furthermore, molecular subtype analysis
based on RNA data uncovered a positive association between
subgroups with low ERSG expression and increased levels of immune
cell infiltration, implying that ERSGs could serve as potential
targets for modulating tumor immune evasion.
To accurately identify pivotal molecules among
ERSGs, the current study used SMR and colocalization analyses,
concentrating on eQTL data of ERSGs in blood, ultimately
pinpointing BDNF as the central gene within this group. The
combined validation through SMR and colocalization analyses not
only confirmed the significant expression of BDNF in LUAD samples,
but also demonstrated its strong colocalization with LUAD traits,
further underscoring the prominence of BDNF as a research priority
among ERSGs. BDNF, as a multifunctional protein, is widely
acknowledged for its central role in the regulation of apoptosis
(41). Previous studies have
indicated that BDNF facilitates tumor cell proliferation and
invasion by activating the TrkB/PLCγ1 signaling pathway (42). While BDNF acts as a tumor suppressor
in neuroblastoma through TrkB-induced differentiation, its role in
LUAD is distinctly oncogenic. This divergence likely stems from
LUAD-specific ERS hyperactivation, where chronic UPR reprograms
BDNF into a pro-survival factor, corroborated by the observation of
BDNF/ERSG co-amplification exclusively in LUAD. The present study
validated BDNF overexpression in LUAD through cell experiments and
demonstrated that BDNF knockdown markedly inhibited various
malignant biological behaviors of LUAD cells, including
proliferation, migration and clonogenicity, thereby corroborating
the SMR and colocalization findings and reinforcing the potential
of BDNF as a therapeutic target. Moreover, molecular docking
analysis identified clinically approved drugs such as Esketamine
and cisplatin as promising candidates for inhibiting BDNF
expression and promoting apoptosis in LUAD cells, thereby offering
new avenues for targeted therapeutic strategies.
Although the current study has made significant
progress, certain limitations must be acknowledged. First, analyses
based on limited database samples may introduce certain biases.
Second, SMR and colocalization analyses did not encompass the
entirety of ERSG probe data, potentially leading to the omission of
key genes. While Esketamine demonstrates potent anti-BDNF effects,
its non-selective NMDA receptor antagonism may cause dose-dependent
neurotoxicity or dissociative effects in clinical settings. Future
work should explore tumor-targeted delivery systems to mitigate
systemic exposure risks. Clinical pharmacokinetic data confirm the
rapid distribution and hepatic clearance of Esketamine, aligning
with Q3W dosing regimens in ongoing oncology trials. While NMDA
receptor binding may cause dissociation, tumor-targeted liposomal
delivery reduced neurotoxicity 4-fold in PDAC models without
compromising BDNF inhibition. Future research should endeavor to
expand sample sizes, refine analytical methods and delve deeper
into the comprehensive mechanisms of ERSGs in LUAD. Additionally,
conducting in vivo experiments to validate in vitro
findings will be an essential step in advancing ERSG research
towards clinical application.
The present study investigated the role of ERSGs in
LUAD through a multi-omics integrated analysis and, in conjunction
with experimental validation, identified the potential therapeutic
value of the BDNF gene and the BDNF-inhibiting drug Esketamine in
hindering the progression of LUAD.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by The Second Batch of
Traditional Chinese medicine Backbone Talent Training Object
Project in Hunan Province during the 14th Five-Year Plan period
(grant no. 202403), The Natural Science Foundation of Hunan
province (grant no. 2022JJ4414) and Key Project of Hunan Province
Traditional Chinese Medicine Research plan (grant no. 2021206).
Availability of data and materials
The data generated in the present study are included
in the figures and/or tables of this article.
Authors' contributions
XD, YT, GT and HN conceived and designed the study
and contributed to the acquisition of research materials. XD and YT
performed the experiments and conducted the statistical analysis.
XD, YT, GT and HN participated in data interpretation and were
involved in drafting the manuscript, critically revising it for
important intellectual content. The study was supported by funding
obtained by XD, GT and YT. XD and YT confirm the authenticity of
all the raw data. All authors agree to be accountable for all
aspects of the work. All authors have read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Thai AA, Solomon BJ, Sequist LV, Gainor JF
and Heist RS: Lung cancer. Lancet. 398:535–554. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang Y, Vaccarella S, Morgan E, Li M,
Etxeberria J, Chokunonga E, Manraj SS, Kamate B, Omonisi A and Bray
F: Global variations in lung cancer incidence by histological
subtype in 2020: A population-based study. Lancet Oncol.
24:1206–1218. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bi KW, Wei XG, Qin XX and Li B: BTK Has
potential to be a prognostic factor for lung adenocarcinoma and an
indicator for tumor microenvironment remodeling: A study based on
TCGA data mining. Front Oncol. 10:4242020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Succony L, Rassl DM, Barker AP, McCaughan
FM and Rintoul RC: Adenocarcinoma spectrum lesions of the lung:
Detection, pathology and treatment strategies. Cancer Treat Rev.
99:1022372021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu J, Zhang Y, Li M, Shao Z, Dong Y, Li Q,
Bai H, Duan J, Zhong J, Wan R, et al: A single-cell characterised
signature integrating heterogeneity and microenvironment of lung
adenocarcinoma for prognostic stratification. EBioMedicine.
102:1050922024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun R, Hou Z, Zhang Y and Jiang B: Drug
resistance mechanisms and progress in the treatment of EGFR-mutated
lung adenocarcinoma. Oncol Lett. 24:4082022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Liu B, Min Q, Yang X, Yan S, Ma Y,
Li S, Fan J, Wang Y, Dong B, et al: Spatial transcriptomics
delineates molecular features and cellular plasticity in lung
adenocarcinoma progression. Cell Discov. 9:962023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
McLaughlin M and Vandenbroeck K: The
endoplasmic reticulum protein folding factory and its chaperones:
New targets for drug discovery? Br J Pharmacol. 162:328–345. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saaoud F, Lu Y, Xu K, Shao Y, Praticò D,
Vazquez-Padron RI, Wang H and Yang X: Protein-rich foods, sea
foods, and gut microbiota amplify immune responses in chronic
diseases and cancers-targeting PERK as a novel therapeutic strategy
for chronic inflammatory diseases, neurodegenerative disorders, and
cancer. Pharmacol Ther. 255:1086042024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Oakes SA: Endoplasmic reticulum stress
signaling in cancer cells. Am J Pathol. 190:934–946. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin L, Lin G, Lin H, Chen L, Chen X, Lin
Q, Xu Y and Zeng Y: Integrated profiling of endoplasmic reticulum
stress-related DERL3 in the prognostic and immune features of lung
adenocarcinoma. Front Immunol. 13:9064202022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen X and Cubillos-Ruiz JR: Endoplasmic
reticulum stress signals in the tumour and its microenvironment.
Nat Rev Cancer. 21:71–88. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cubillos-Ruiz JR, Bettigole SE and
Glimcher LH: Tumorigenic and immunosuppressive effects of
endoplasmic reticulum stress in cancer. Cell. 168:692–706. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cao T, Zhang W, Wang Q, Wang C, Ma W,
Zhang C, Ge M, Tian M, Yu J, Jiao A, et al: Cancer SLC6A6-mediated
taurine uptake transactivates immune checkpoint genes and induces
exhaustion in CD8+ T cells. Cell. 187:2288–2304.e27.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fan C, Yang Y, Liu Y, Jiang S, Di S, Hu W,
Ma Z, Li T, Zhu Y, Xin Z, et al: Icariin displays anticancer
activity against human esophageal cancer cells via regulating
endoplasmic reticulum stress-mediated apoptotic signaling. Sci Rep.
6:211452016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang L, Xiong Y, Zhang J, Feng Y and Xu
A: Systematic proteome-wide Mendelian randomization using the human
plasma proteome to identify therapeutic targets for lung
adenocarcinoma. J Transl Med. 22:3302024. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Deng B, Liao F, Liu Y, He P, Wei S, Liu C
and Dong W: Comprehensive analysis of endoplasmic reticulum
stress-associated genes signature of ulcerative colitis. Front
Immunol. 14:11586482023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qiu WR, Qi BB, Lin WZ, Zhang SH, Yu WK and
Huang SF: Predicting the lung adenocarcinoma and its biomarkers by
integrating gene expression and DNA methylation data. Front Genet.
13:9269272022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang S, Xiong Y, Zhao L, Gu K, Li Y, Zhao
F, Li J, Wang M, Wang H, Tao Z, et al: UCSCXenaShiny: an R/CRAN
package for interactive analysis of UCSC Xena data. Bioinformatics.
38:527–529. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Z, Wang Y, Chang M, Wang Y, Liu P, Wu
J, Wang G, Tang X, Hui X, Liu P, et al: Single-cell transcriptomic
analyses provide insights into the cellular origins and drivers of
brain metastasis from lung adenocarcinoma. Neuro Oncol.
25:1262–1274. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang J, Zhang J, Zhang F, Lu S, Guo S,
Shi R, Zhai Y, Gao Y, Tao X, Jin Z, et al: Identification of a
disulfidptosis-related genes signature for prognostic implication
in lung adenocarcinoma. Comput Biol Med. 165:1074022023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu F, Cai J, Wen C and Tan H: Co-sparse
non-negative matrix factorization. Front Neurosci. 15:8045542022.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu S, Zheng Z, Hu W and Lei C:
Conditional cancer-specific survival for inflammatory breast
cancer: Analysis of SEER, 2010 to 2016. Clin Breast Cancer.
23:628–639.e2. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qin Y, Liu Y, Xiang X, Long X, Chen Z,
Huang X, Yang J and Li W: Cuproptosis correlates with
immunosuppressive tumor microenvironment based on pan-cancer
multiomics and single-cell sequencing analysis. Mol Cancer.
22:592023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Q, Qiao W, Zhang H, Liu B, Li J, Zang
C, Mei T, Zheng J and Zhang Y: Nomogram established on account of
Lasso-Cox regression for predicting recurrence in patients with
early-stage hepatocellular carcinoma. Front Immunol.
13:10196382022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Song M, Zhang Q, Song C, Liu T, Zhang X,
Ruan G, Tang M, Xie H, Zhang H, Ge Y, et al: The advanced lung
cancer inflammation index is the optimal inflammatory biomarker of
overall survival in patients with lung cancer. J Cachexia
Sarcopenia Muscle. 13:2504–2514. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhai T: Druggable genome-wide Mendelian
randomization for identifying the role of integrated stress
response in therapeutic targets of bipolar disorder. J Affect
Disord. 362:843–852. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shao Y, Wang Z, Wu J, Lu Y, Chen Y, Zhang
H, Huang C, Shen H, Xu L and Fu Z: Unveiling immunogenic cell
death-related genes in colorectal cancer: an integrated study
incorporating transcriptome and Mendelian randomization analyses.
Funct Integr Genomics. 23:3162023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bisht A, Tewari D, Kumar S and Chandra S:
Network pharmacology, molecular docking, and molecular dynamics
simulation to elucidate the mechanism of anti-aging action of
Tinospora cordifolia. Mol Divers. 28:1743–1763. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jaganathan R and Kumaradhas P: Binding
mechanism of anacardic acid, carnosol and garcinol with PCAF: A
comprehensive study using molecular docking and molecular dynamics
simulations and binding free energy analysis. J Cell Biochem.
124:731–742. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu Y, Zhang C and Zhao D, Li W, Zhao Z,
Yao S and Zhao D: BDNF Acts as a prognostic factor associated with
tumor-infiltrating Th2 cells in pancreatic adenocarcinoma. Dis
Markers. 2021:78420352021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu L, Zhou S, Hong W, Lin N, Wang Q and
Liang P: Characterization of an endoplasmic reticulum
stress-associated lncRNA prognostic signature and the
tumor-suppressive role of RP11-295G20.2 knockdown in lung
adenocarcinoma. Sci Rep. 14:122832024. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang T, Weng H, Zhou H, Yang Z, Tian Z, Xi
B and Li Y: Esketamine alleviates postoperative depression-like
behavior through anti-inflammatory actions in mouse prefrontal
cortex. J Affect Disord. 307:97–107. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Siebert JR and Osterhout DJ: Select
neurotrophins promote oligodendrocyte progenitor cell process
outgrowth in the presence of chondroitin sulfate proteoglycans. J
Neurosci Res. 99:1009–1023. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nakagawa K, Garon EB, Seto T, Nishio M,
Ponce Aix S, Paz-Ares L, Chiu CH, Park K, Novello S, Nadal E, et
al: Ramucirumab plus erlotinib in patients with untreated,
EGFR-mutated, advanced non-small-cell lung cancer (RELAY): A
randomised, double-blind, placebo-controlled, phase 3 trial. Lancet
Oncol. 20:1655–1669. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Salvagno C, Mandula JK, Rodriguez PC and
Cubillos-Ruiz JR: Decoding endoplasmic reticulum stress signals in
cancer cells and antitumor immunity. Trends Cancer. 8:930–943.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qiao L, Shao X, Gao S, Ming Z, Fu X and
Wei Q: Research on endoplasmic reticulum-targeting fluorescent
probes and endoplasmic reticulum stress-mediated nanoanticancer
strategies: A review. Colloids Surf B Biointerfaces.
208:1120462021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cao LL and Kagan JC: Targeting innate
immune pathways for cancer immunotherapy. Immunity. 56:2206–2217.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang H, Li Z, Tao Y, Ou S, Ye J, Ran S,
Luo K, Guan Z, Xiang J, Yan G, et al: Characterization of
endoplasmic reticulum stress unveils ZNF703 as a promising target
for colorectal cancer immunotherapy. J Transl Med. 21:7132023.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang YH, Huo BL, Li C, Ma G and Cao W:
Knockdown of long noncoding RNA SNHG7 inhibits the proliferation
and promotes apoptosis of thyroid cancer cells by downregulating
BDNF. Eur Rev Med Pharmacol Sci. 23:4815–4821. 2019.PubMed/NCBI
|
|
42
|
Xu Y, Jiang WG, Wang HC, Martin T, Zeng
YX, Zhang J and Qi YS: BDNF activates TrkB/PLCγ1 signaling pathway
to promote proliferation and invasion of ovarian cancer cells
through inhibition of apoptosis. Eur Rev Med Pharmacol Sci.
23:5093–5100. 2019.PubMed/NCBI
|