Metabolic reprogramming is a fundamental
characteristic of cancer development and progression (1), and it is primarily marked by a notable
increase in aerobic glycolysis (2).
This atypical activation of aerobic glycolysis not only meets the
energy and biosynthetic demands of rapidly proliferating malignant
tumor cells but also contributes to the formation of a hypoxic,
acidic and nutrient-deprived tumor microenvironment (TME). Such an
environment facilitates tumor immune evasion and presents a notable
challenge to the antitumor immune response (3,4).
Hexokinase 2 (HK2), the initial and critical rate-limiting enzyme
in glycolysis, is markedly upregulated in various solid tumors
[such as colorectal cancer (5),
triple-negative breast cancer (6),
hepatocellular carcinoma (7),
pancreatic cancer (8), esophageal
cancer (9), etc.], where it serves
a pivotal role in catalyzing glycolytic processes and influencing
the malignant behavior of tumors; it is also closely related to
poor patient prognosis (10–18).
The regulatory function of HK2 in immune evasion has increasingly
attracted attention for research (3,19–21).
Previous research indicates that the functions of HK2 encompass
multiple immune escape mechanisms (3,20,22).
The expression and post-translational modification of HK2 are
modulated by a variety of signal transduction pathways and
oncogenic factors, which further facilitate immune evasion through
both glycolysis-dependent and independent mechanisms. These
mechanisms impact immune cell distribution (23), modulate the expression of immune
checkpoint molecules (20) and
contribute to the establishment of an immunosuppressive
microenvironment (3). Furthermore,
HK2-mediated hyperactive glycolysis in tumor cells competes with
immune cells for glucose, induces metabolic reprogramming in immune
cells, suppresses the activation and function of immune cells, and
releases large quantities of metabolites to establish an
immunosuppressive niche, thereby impairing immune molecule
expression and further weakening the antitumor immune response
(24,25). Although small-molecule inhibitors
[such as 3-bromopyruvic acid (26)
and lonidamine (27)] and
gene-editing tools that target HK2 can reverse metabolism-dependent
immunosuppression, their clinical application is constrained by
off-target toxicity, compensatory metabolism and delivery
efficiency (21,28–30).
Emerging strategies focusing on subcellular localization and
non-catalytic intervention may offer novel directions to overcome
these clinical translation bottlenecks (3).
At present, an expanding body of research has
substantiated the multifaceted role of HK2 in bridging tumor
metabolic reprogramming and immune evasion; however, comprehensive
analyses remain limited. In light of this, the present review aims
to summarize the research advancements regarding the regulation of
immune escape in malignant tumors by HK2 and the role of HK2 in
metabolic-immune interactions, with the objective of providing
references and theoretical underpinnings for the development of
novel immunotherapeutic strategies.
HK serves as the rate-limiting enzyme that initiates
the glycolytic pathway by catalyzing the phosphorylation of glucose
to glucose-6-phosphate (G-6-P), thereby facilitating energy
production and biosynthesis (31).
In mammals, five isoforms of HK have been identified, HK1-4 and HK
domain-containing 1, with HK2 being markedly upregulated in
malignancies, and strongly associated with tumor aerobic glycolysis
and malignant progression (14,32).
Unlike other isomers, HK2 has unique structural features: Although
its secondary structure is very similar to that of HK1 and HK3,
both composed of two symmetrical α-helical regions linked to
glucokinase-catalyzed equivalent domains, HK2 is the only isomer
that maintains catalytic activity in both domains (2), which allows for more efficient
glycolysis (3). The N-terminal
hydrophobic domain of HK2 directly anchors to the voltage-dependent
anion channel (VDAC) located on the outer mitochondrial membrane.
This crucial interaction results in the formation of a
transmembrane-coupled complex with the adenine nucleotide
translocator on the inner membrane. Such a configuration
effectively prevents inhibition by G-6-P and establishes a
‘metabolic compartmentalization’ effect, thereby facilitating
efficient coordination between glycolysis and mitochondrial energy
metabolism within localized cellular regions (33). As a result, ATP produced by the
mitochondria is directly utilized by HK2 to initiate glycolysis,
creating a metabolic relay. This mechanism rapidly supplies the
energy, metabolic intermediates and precursors necessary for tumor
cell proliferation within a short period (34), thereby serving a pivotal role in
cancer cell proliferation, drug resistance and immune evasion
(35). Concurrently, HK2 localized
to the mitochondrial outer membrane serves a critical role in
inhibiting the opening of the mitochondrial permeability transition
pore, stabilizing the mitochondrial membrane potential, suppressing
the release of cytochrome c and blocking apoptotic signaling
pathways, thereby facilitating tumor cell survival (36,37).
Furthermore, HK2 is subject to O-GlcNAcylation, a process that
enhances cell proliferation, migration and invasion in non-small
cell lung cancer, and mitigates mitochondrial damage, and thus,
facilitates tumor progression (38).
Activation of the PI3K/AKT signaling pathway
facilitates cellular metabolic reprogramming (4) and is frequently associated with
immunosuppressive effects (21).
Research indicates that the AKT-induced nuclear translocation of
HK2 enhances the transcription of programmed death-ligand 1 (PD-L1)
through hypoxia-inducible factor 1-α (HIF-1α), which promotes
immune evasion in gastric cancer cells (39). AKT further activates the downstream
mechanistic target of rapamycin complex 1 (mTORC1), leading to the
stabilization of HIF-1α, which subsequently binds to
hypoxia-response elements (HREs) within the HK2 promoter. This
interaction establishes a positive feedback loop that upregulates
HK2 transcription, induces M2 macrophage polarization and
suppresses the functions of T cells and natural killer (NK) cells
(40). The inhibition of PI3K
activity or the application of 2-deoxy-D-glucose (2-DG) can reverse
these effects (41). A clinical
study has corroborated a positive association between the
expression of PI3K/AKT and HK2 in hepatocellular carcinoma (HCC),
with a marked increase in PD-L1 positivity observed in groups
exhibiting high co-expression levels (42). The programmed cell death protein 1
(PD-1)/PD-L1 pathway inhibits T cell glycolysis and promotes
autophagy through negative regulation of HK2 activity (43). Inhibitors of
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit β,
such as AZD8186, disrupt the AKT-HK2 signaling pathway, leading to
decreased lactate production and the restoration of T cell
functionality (44). In glioma
models, the combination of HK2 inhibitors with PD-1 antibodies has
been demonstrated to enhance the infiltration of CD8+ T
cells (3,5). Future research should focus on
developing precise stratification strategies based on the
subcellular localization of HK2 and investigate the potential of
PI3K isoform-specific inhibitors.
The MAPK signaling pathway facilitates signal
transduction through a hierarchical kinase cascade, predominantly
involving the extracellular signal-regulated kinases 1 and 2
(ERK1/2), c-Jun amino-terminal kinases 1 to 3 (JNK1 to −3), p38 (α,
β, γ and δ), and ERK5 families (45). HK2 activates ERK1/2 via the Raf/MEK
pathway, leading to the induction of cyclin A1 expression, the
downregulation of p27, the initiation of DNA replication stress,
the release of damage-associated molecular patterns, the activation
of chronic inflammatory responses and the recruitment of
immunosuppressive cells (46,47).
Furthermore, HK2 enhances the expression of pyruvate dehydrogenase
kinase 1 through the JNK-c-Jun pathway, thereby inhibiting
CD8+ T cell infiltration, or modulates the
nucleocytoplasmic shuttling of heterogeneous nuclear
ribonucleoprotein A1 (hnRNPA1) via p38 MAPKβ (48). hnRNPA1 interacts with the 3′
untranslated region of PD-L1 mRNA to maintain its stability,
consequently suppressing T cell activity (49) and fostering the development of an
immunosuppressive microenvironment. In the context of colorectal
cancer, fructose attenuates MAPK/signal transducer and activator of
transcription (STAT)1 signaling through the HK2- inositol
1,4,5-trisphosphate receptor type 3-Ca2+ axis, thereby
inhibiting the polarization of M1-type tumor-associated macrophages
(TAMs) and further diminishing antitumor immune responses (50). In melanoma, the splicing of hnRNPA1
regulated by HK2 leads to the production of immunosuppressive human
leukocyte antigen G mRNA isoforms, which subsequently diminish the
cytotoxic efficacy of NK cells (51). Additionally, the pro-inflammatory
mediator, macrophage migration inhibitory factor, enhances HK2
expression through the MAPK-ERK signaling pathway, establishing an
immunosuppressive positive feedback loop that synergistically
promotes immune evasion by the tumor (52).
The NF-κB signaling pathway is activated by chronic
inflammation, oncogenic mutations or microenvironmental stressors
(53). HK2 interacts with the NF-κB
pathway through mechanisms that are both dependent and independent
of its metabolic functions. A seminal study conducted by Guo et
al (3) demonstrated that HK2
possessed non-metabolic roles in the cytoplasm, functioning as a
protein kinase to directly phosphorylate the threonine 291 residue
of the NF-κB inhibitor nuclear factor of κ light polypeptide gene
enhancer in B-cells inhibitor α (IκBα). This phosphorylation event
facilitated the degradation of IκBα via the µ-calpain-proteasome
pathway, thereby releasing the NF-κB dimer (RelA/p50) to
translocate into the nucleus and initiate the transcription of
immune checkpoint proteins, including PD-L1 (3). Concurrently, G-6-P, produced through
HK2 catalysis, activates the mTORC1 signaling pathway, which
promotes the phosphorylation of the IκB kinase complex and
indirectly increases NF-κB activity (54). Simultaneously, NF-κB directly
interacts with the HK2 promoter region, thereby augmenting its
transcriptional activity and activating downstream
glycolysis-related genes, which collectively constitute a
self-reinforcing regulatory network known as the ‘NF-κB-HK2-Warburg
effect’ (42).
In pancreatic ductal adenocarcinoma models, the
HK2-IκBα-NF-κB axis markedly upregulates PD-L1 expression,
diminishes CD8+ T cell infiltration and stabilizes
β-catenin, thereby enhancing the self-renewal capacity of
CD133+ tumor stem cells and further mediating
immunosuppression (55). In
sarcoma, the canonical NF-κB pathway upregulates HK2 expression,
promoting lactate production and suppressing mitochondrial
oxidative phosphorylation, which results in the acidification of
the TME (56). Lactate contributes
to the establishment of an immunosuppressive microenvironment by
activating regulatory T cells (Tregs) and M2 macrophages, while
inhibiting dendritic cell (DC) maturation (57,58).
Preclinical studies have demonstrated that a combined therapeutic
approach utilizing the HK2-specific inhibitor 2-DG and the NF-κB
pathway inhibitor BAY11-7082 enhanced antitumor immune responses
through dual mechanisms: The suppression of PD-L1 protein
expression and the reversal of T cell exhaustion phenotypes
(59,60). This synergistic therapeutic effect
lays the theoretical groundwork for the clinical advancement of
innovative immunometabolic combination therapies.
The TGF-β pathway facilitates the differentiation of
immunosuppressive cells and diminishes immune cell cytotoxicity
through mechanisms that are both SMAD-dependent and independent,
thereby promoting an immune-tolerant phenotype (61). In models characterized by energy
deficiency induced by oligomycin A-mediated inhibition of oxidative
phosphorylation, highly metastatic HCT116 cells stimulated by TGF-β
contribute to the formation of an immunosuppressive
microenvironment through the upregulation of HK2 expression
(62). Furthermore, TGF-β modulates
HK2 expression by activating the SMAD3 pathway, while HK2 directly
interacts with TGF-β receptor I (TβRI) via its structural domain,
stabilizing the TβRI-TβRII complex and enhancing the
phosphorylation efficiency of SMAD2/3 (31,63).
This receptor-kinase interaction creates a positive feedback loop
that intensifies TGF-β-induced epithelial-mesenchymal transition
and stromal fibrosis, thereby physically obstructing T cell
infiltration into tumor beds (64–66).
Furthermore, the TGF-β/SMAD signaling pathway directly inhibits the
transcription of major histocompatibility complex class I (MHC-I)
heavy chains and antigen processing-associated transporters
(67). Concurrently, HK2 activates
histone deacetylases through metabolic byproducts, leading to
further silencing of the epigenetic regulatory regions of MHC-I
genes and impairment of the recognition and binding of
tumor-specific antigens by immune cells (68).
The JAK/STAT pathway, a pivotal axis for cytokine
signal transduction, facilitates tumor immunosuppression through
aberrant activation of STAT3/STAT5 and alters the TME via metabolic
reprogramming (69). In colorectal
cancer, upregulation of HK2 enhances STAT3 phosphorylation, reduces
the infiltration of immune cells such as CD8+ T cells
and NK cells, and markedly increases PD-L1 levels, and these
effects are reversed by JAK/STAT3 inhibitors (5). Mechanistically, phosphorylated STAT3
directly binds to the PD-L1 promoter, and HK2 indirectly promotes
PD-L1 transcription by augmenting STAT3 activity (70).
SUMOylation has been identified as a crucial factor
in the cellular response to mitochondrial stress, acting as a
sensor and mediator of stress signals to maintain mitochondrial
homeostasis (65). This regulatory
mechanism is particularly relevant in the context of HK2,
suggesting that deSUMOylation likely enhances the interaction
between HK2 and VDAC1, thereby activating JAK2 kinase activity and
establishing an HK2-JAK2/STAT3-immunosuppressive positive feedback
loop (71,72). Epigenetic modifications serve a
notable role in this process. Specifically, methyltransferase-like
3 stabilizes HK2 mRNA through N6-methyladenosine
modification, and its upregulation enhances HK2 translation via
insulin-like growth factor 2 mRNA-binding protein 2, while
concurrently activating STAT3-dependent PD-L1 transcription
(73). Furthermore, the
accumulation of lactate mediated by HK2 inhibits glycolysis in T
cells, diminishes interferon γ (IFN-γ) secretion and activates the
JAK-STAT pathway through paracrine signaling, thereby further
impairing antitumor immunity (74)
(Table I).
The phenomenon of tumor immune escape is intricately
linked to the immune subtypes present within the TME (75). However, the specific role of HK2
within these subtypes remains poorly understood. A comprehensive
analysis utilizing data from The Cancer Genome Atlas and the
International Union of Immunological Societies Immunology Databases
revealed that HK2 expression is markedly elevated in
immunosuppressive tumors, including pancreatic and liver cancers.
This elevated expression is associated with the accumulation of
Tregs and M2-type macrophages (76). Such an immunosuppressive milieu may
further facilitate tumor cell immune escape through metabolic
modulation mediated by HK2 (38).
Conversely, in immune-active tumors, such as melanoma, a negative
association between HK2 expression and T helper type 1 (Th1)
cytokines (such as IFN-γ and C-X-C motif chemokine ligand 9)
suggests that HK2 may alter the composition and functionality of
immune cells by modulating the expression of chemokines and
antigen-presenting molecules (77).
Furthermore, research has demonstrated that knockout of HK2 can
markedly increase the expression of MHC-II and its
antigen-presenting capabilities in DCs, while also modulating the
Th17/Treg balance in CD4+ T cells. This modulation
subsequently influences the specificity and intensity of the immune
response (78,79). These observations underscore the
pivotal role of HK2 in determining immune cell subtypes.
Consequently, these findings imply that HK2 is not only critical
within tumor cells but also potentially modulates immune responses
by affecting the functionality of immune cells within the tumor
immune microenvironment.
Lactate, a central metabolic product of HK2-driven
glycolysis, has transitioned from being considered merely a
‘metabolic waste product’ (80) to
a multifunctional mediator, acting as a signaling molecule, energy
substrate and immunoregulatory agent that governs tumor immune
evasion (81,82). Tumor cells continuously produce
lactate through HK2 hyperactivation, which increases extracellular
acidification rates, contributes to the formation of an acidic TME
and establishes an immunosuppressive niche through multiple
pathways (such as inhibiting the function of effector immune cells,
promoting the expansion of inhibitory immune cells, disrupting
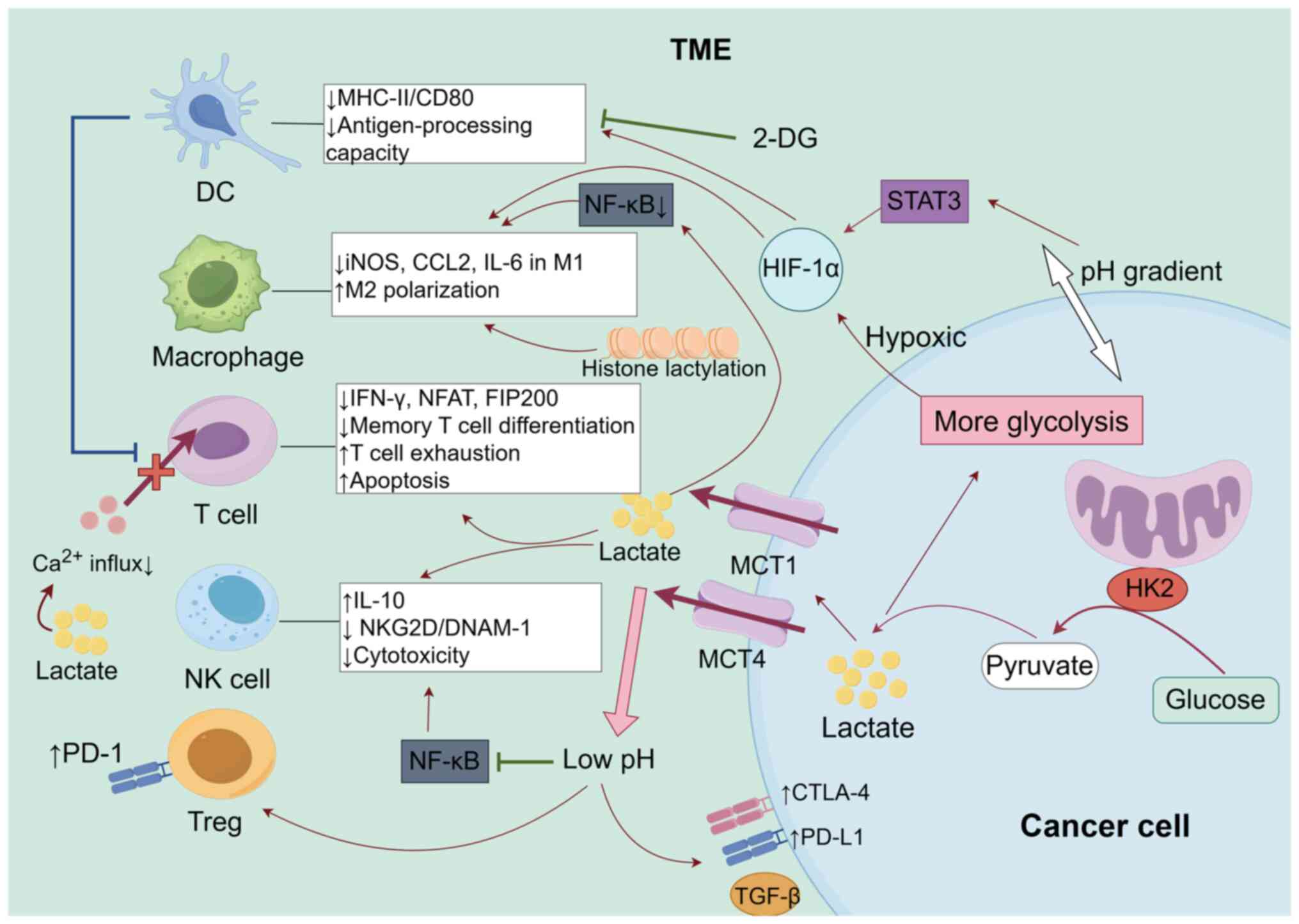
antigen presentation and upregulating checkpoints) (30). As shown in Fig. 1, the elevated lactate levels
mediated by HK2 result in a reduction in the calcium influx in T
cells, inhibition of IFN-γ secretion and prevention of the nuclear
translocation of activated T cell nuclear factors, as well as the
expression of the autophagy factor focal adhesion kinase family
interacting protein of 200 kDa (57,83,84).
These processes contribute to T cell exhaustion, apoptosis and a
diminished potential for memory T cell differentiation (85,86).
Application of the HK2 inhibitor 2-DG counteracts these effects,
thereby restoring the antitumor immune function of T cells
(87). HK2-driven glycolysis
creates intracellular and extracellular pH gradients, which
directly induce the expansion of Tregs and upregulate PD-1
expression. Furthermore, pH gradients downregulate major
MHC-II/CD80 expression through HIF-1α, thereby impairing the
antigen-processing capacity of DCs and the polarization of helper T
cells (88–91). Lactate also upregulates interleukin
(IL)-10 and downregulates acidification-dependent natural killer
group 2D (NKG2D) and DNAX accessory molecule-1 (DNAM-1) by
suppressing the activation of NF-κB (92), leading to suppressed cytotoxicity in
NK cells (93,94). Additionally, lactate reduces the
expression of inducible nitric oxide synthase, C-C motif chemokine
ligand (CCL)2 and IL-6 in M1 macrophages by inhibiting the activity
of the NF-κB signaling pathway, while the STAT3/HIF-1α axis
promotes macrophage M2 polarization, facilitating immune escape
(95,96). Furthermore, the elevation of lactate
mediated by HK2 upregulates inhibitory molecules such as PD-L1,
cytotoxic T-lymphocyte-associated protein 4 and TGF-β (97–99),
induces histone lysine lactylation, and establishes a
‘glycolysis-lactylation-immunosuppression’ positive feedback loop
(100,101).
The interaction of HK2 with mitochondrial VDAC1
leads to localized microenvironmental hypoxia, which accelerates
lactate accumulation and abnormal angiogenesis, thereby further
exacerbating hypoxia (100,102).
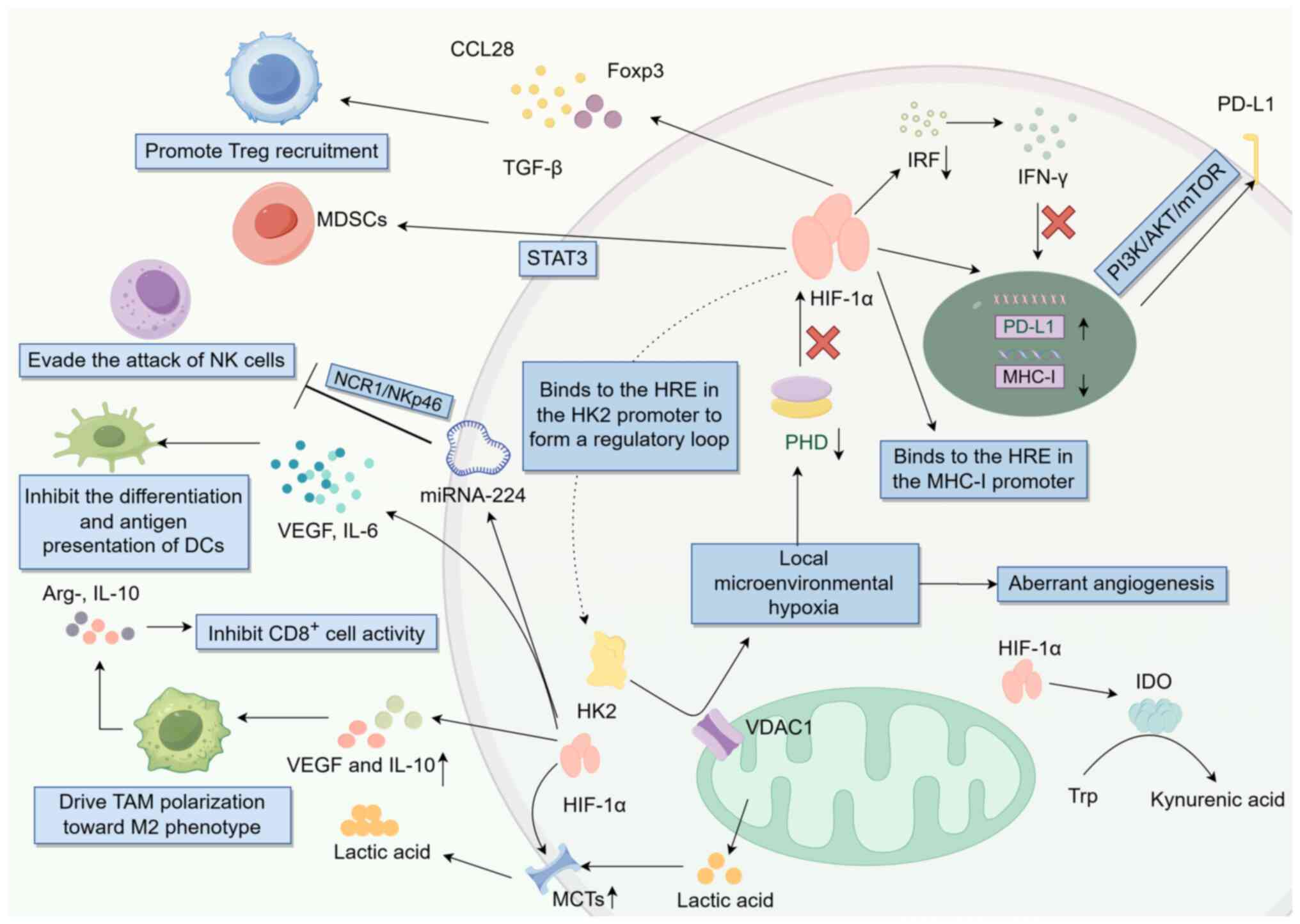
As shown in Fig. 2, intratumoral
hypoxia predominantly inhibits the prolyl hydroxylase-mediated
stabilization of HIF-1α (103).
Activation of HIF-1α also enhances HK2 expression (104). The hypoxia-HIF-1α axis directly
interacts with HREs in the HK2 promoter, amplifying the hypoxia
process in the TME and forming an immunosuppressive feedforward
loop (105,106). Clinical investigations have
demonstrated that the HK2-mediated upregulation of HIF-1α in breast
and lung cancer is positively associated with PD-L1 expression, and
patients with high HK2 expression exhibit reduced response rates to
immune checkpoint inhibitors (107,108). Mechanistically, the activation of
HIF-1α by HK2 facilitates its binding to HREs within the PD-L1
promoter, leading to upregulation of PD-L1 transcription (109). Additionally, this activation
promotes the post-translational modifications of PD-L1 through the
activation of the PI3K/AKT/mTOR signaling pathway (110). Furthermore, HK2-mediated
activation of HIF-1α results in the upregulation of microRNA-224,
which suppresses the natural cytotoxicity triggering receptor
1/NKp46 pathway and thereby protects tumors from NK cell-mediated
attack (111). HIF-1α also induces
the secretion of CCL28 and TGF-β by tumor cells, facilitating the
recruitment of Tregs to tumor sites (112,113), and sustains immunosuppressive
function of CCL28 and TGF-β by enhancing the expression of forkhead
box P3 (Foxp3) (114).
Concurrently, HIF-1α promotes the polarization of TAMs towards the
M2 phenotype through the upregulation of vascular endothelial
growth factor (VEGF) and IL-10. These M2 TAMs subsequently secrete
arginase 1, IL-10 and other factors (such as TGF-β, PD-L1) that
inhibit the activity of CD8+ T cells (115). Furthermore, HIF-1α activates the
STAT3 pathway, leading to the expansion of myeloid-derived
suppressor cells, which further suppress the antitumor effects of
NK and T cells (116).
HK2-activated HIF-1α directly interacts with HREs in the MHC-I gene
promoter or recruits histone deacetylases to induce chromatin
compaction, thereby inhibiting MHC-I transcription (30). HIF-1α inhibits the IFN-γ-mediated
upregulation of MHC-I by suppressing the activity of IFN regulatory
factors (117). Additionally,
HIF-1α impairs DC differentiation and antigen-presenting
capabilities through the induction of VEGF and IL-6, resulting in
inadequate T cell priming (118).
HK2 further enhances the expression of HIF-1α and monocarboxylate
transporters, facilitating HK2-mediated lactate efflux and
exacerbating the immunosuppressive TME (119). HIF-1α also upregulates CD73
expression, which catalyzes the conversion of ATP to adenosine,
subsequently inhibiting T cell activation and NK cell cytotoxicity
via adenosine A2A receptor interaction (120–122). Furthermore, HIF-1α induces the
expression of indoleamine 2,3-dioxygenase, leading to tryptophan
depletion and kynurenine acid production in the microenvironment,
thereby suppressing T cell proliferation and promoting Treg
differentiation (123,124).
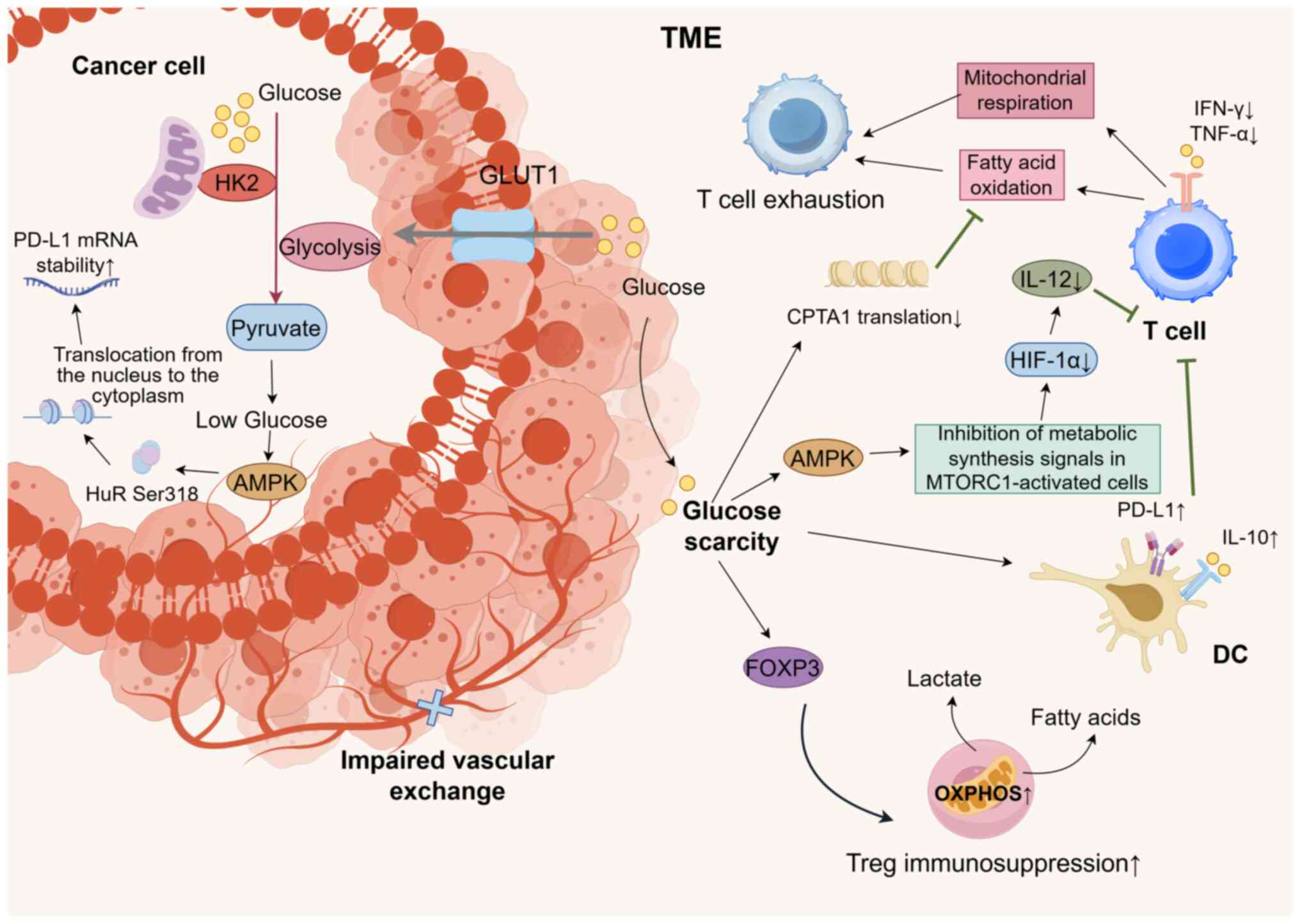
The synergistic effects of the HK2-mediated
glycolytic enhancement and impaired vascular exchange result in
limited glucose availability within the TME, causing intratumoral
hypoglycemia (125). Under these
glucose-deprivation conditions, the TME selectively impairs
glucose-dependent immune cells (126). As shown in Fig. 3, the hypoglycemic TME driven by HK2
activates AMP-activated protein kinase (AMPK), inhibits the signal
of MTORC1-activated cell metabolic synthesis, downregulates HIF-1α
expression, reduces IL-12 secretion and fails to effectively
activate naïve T cells (127–129). The secretion of effector cytokines
such as IFN-γ and tumor necrosis factor-α is also diminished
(130). Under conditions of
HK2-induced glucose deprivation, T cells are compelled to shift
towards fatty acid oxidation and mitochondrial respiration.
However, this metabolic shift is inefficient due to the suppression
of carnitine palmitoyltransferase 1A translation mediated by
glucose deprivation, ultimately leading to T cell exhaustion
(131). Hypoglycemia induces DCs
to adopt a tolerogenic phenotype characterized by elevated PD-L1
and IL-10 expression, further inhibiting T cell responses (128). By contrast, Tregs within the
hypoglycemic TME have enhanced oxidative phosphorylation through
Foxp3-mediated metabolic reprogramming, relying on fatty acid and
lactate metabolism to maintain their immunosuppressive functions.
This metabolic adaptation provides Tregs with a competitive
advantage in nutrient competition, thereby exacerbating antitumor
immune suppression (132).
Furthermore, glucose deprivation triggers the activation of the
AMPK pathway in tumor cells. AMPK phosphorylates Hu antigen R (HuR)
at Ser318, facilitating its translocation from the nucleus to the
cytoplasm (133). In melanoma
cells, the cytoplasmic localization of HuR increases from 20 to 60%
under hypoglycemic conditions, which enhances its binding affinity
to PD-L1 mRNA, and thus, stabilizes PD-L1 mRNA (134).
Advancements in the development of novel HK2
inhibitors have reached the clinical research phase, in which they
have shown notable antitumor activity (Table II). Small molecule inhibitors, such
as benserazide and benitrobenrazide (BNBZ), have been shown to
decrease glucose uptake, lactic acid production and intracellular
ATP levels by specifically binding to HK2 and inhibiting its
enzymatic function (135,136). This mechanism induces apoptosis in
tumor cells and suppresses tumor cell proliferation. BNBZ has
exhibited notable antitumor effects in xenograft mouse models using
SW1990 and SW480 cells, highlighting its potential as a promising
HK2 inhibitor (136). Similarly,
benserazide has been shown to effectively reduce tumor growth in a
SW480 cell xenograft mouse model without significant toxic effects
(135). The nanoparticle
formulation of benserazide further enhances its antitumor efficacy
and specificity in targeting tumors (135).
VDA-1102 (Almavid™), the first small molecule
targeting the HK2-VDAC1 interaction, disrupts HK2 mitochondrial
localization to restore cancer cell apoptosis. In a phase II trial
for cutaneous T-cell lymphoma and actinic keratosis, VDA-1102
achieved an objective response rate (ORR) >60% with manageable
toxicity (137). The subcutaneous
formulation of VDA-1102 exhibited a stable plasma concentration
(>24 h) and dose-linear pharmacokinetics in pediatric patients
with brain tumors, indicating broad-spectrum potential in solid
tumors (138).
Ketoconazole, a United States Food and Drug
Administration-approved antifungal agent and HK2 inhibitor,
disrupts the interaction between HK2 and aminoacyl transfer RNA
synthetase complex-interacting multifunctional protein 2 (AIMP2).
This reduces autophagic lysosome-dependent degradation of AIMP2,
restoring AIMP2-mediated apoptosis and synergizing with
radiotherapy to suppress HCC growth. This drug-repurposing strategy
overcomes radioresistance with translational advantages in terms of
safety and cost (139).
A novel series of 2,6-disubstituted glucosamines has
been identified as potent and selective HK2 inhibitors. These
compounds demonstrate significant selectivity over HK1 and have
been shown to inhibit glycolysis in cancer cell lines, underscoring
their potential as therapeutic agents (140). Ikarugamycin, a polycyclic
tetramide macrolide compound isolated from Actinomycetes associated
with mangroves, has been shown to inhibit the glycolytic flux in
pancreatic cancer cells by targeting HK2 and exhibits antitumor
activity in murine models without exhibiting significant
cytotoxicity (141). Another
promising development in the field is the identification of natural
HK2 inhibitors from Ganoderma sinense. For example, a
specific steroid compound,
(22E,24R)-6β-methoxyergosta-7,9(11),22-triene-3β,5α-diol, has been
identified as having a high binding affinity for HK2, suggesting
its potential as a drug candidate for cancer therapy (142). This discovery is complemented by a
broader study that systematically reviews the characteristics of
HK2 inhibitors, emphasizing the need for compounds with improved
efficacy and selectivity due to the poor performance of existing
inhibitors when used alone (143).
RNA interference-based therapy, which silences the
expression of HK2, has been shown to inhibit tumor cell
proliferation and metastasis, thereby demonstrating notable
therapeutic potential (144–148). The clinical potential of HK2
inhibitors is further supported by studies on their role in
inducing apoptosis and endoplasmic reticulum stress in cancer
cells. For instance, the HK2 inhibitor 3-bromopyruvate has been
shown to inhibit the survival and proliferation of colon cancer
cells, suggesting its utility in combination therapies (149). Additionally, the use of azole
antifungals, such as ketoconazole and posaconazole, has
demonstrated efficacy in targeting HK2-expressing glioblastoma
cells, providing a basis for repurposing these compounds in
clinical trials (150). These
findings collectively underscore the therapeutic promise of HK2
inhibitors and their potential to advance to clinical applications
(Table II).
As the role of HK2 in tumor immune evasion becomes
clearer, its utility as a predictive biomarker for immunotherapy is
gaining attention. In bladder cancer research, the immune
expression of glycolysis-related proteins, including HK2, has been
assessed. The findings indicated that high HK2 expression was
closely associated with independent prognostic factors for
progression-free survival and overall survival, suggesting its
utility as a prognostic biomarker for patients with this disease
(151). In colorectal cancer, HK2
expression is associated with tumor size, depth of invasion, liver
metastasis and TNM stage, effectively predicting the recurrence
risk and overall mortality. Thus, HK2 serves as an important
prognostic biomarker (12).
Additionally, in patients with lung cancer, HK2 expression serves
as a biomarker for identifying a novel subset of circulating tumor
cells associated with poor prognosis. This finding is instrumental
for prognostic evaluation prior to treatment (152). Positron emission tomography
utilizing 18F-fluorodeoxyglucose (18F-FDG) is
a widely employed technique for assessing tumor glucose metabolism.
HK2 expression is associated with the uptake of 18F-FDG
by tumors (153). Consequently,
HK2 detection can facilitate the early diagnosis, staging and
evaluation of the treatment response in tumors (10). Furthermore, the development of a
ternary scoring system incorporating HK2, PD-L1 and HIF-1α for
predicting the response of patients with liver cancer is a
promising approach that leverages the interplay between metabolic
pathways, immune checkpoints and hypoxic conditions within the TME
(154). This model is
substantiated by several studies that highlight the critical roles
of these biomarkers in HCC prognosis and treatment response
(155–157).
The integration of single-cell transcriptomics and
spatial transcriptomics has revolutionized the current
understanding of cell dynamics and tissue structure, providing
unprecedented insights into the study of gene expression patterns
and cell interactions in natural spatial environments (158,159). This two-pronged research strategy
is particularly important when monitoring the changes in HK2; it
can achieve single-cell resolution analysis of HK2 expression
profiles while maintaining spatial information, thereby deeply
analyzing the expression distribution of HK2 in complex tissue
environments and its interaction with other factors. This strategy
can also provide in-depth insights into the metabolic heterogeneity
within tumors (160,161). Based on single-cell transcriptome
and spatial transcriptome techniques, the HK2-Glycolytic Activity
Index shows good stability in various tumor types and is associated
with the characteristics of immune infiltration in the tumor
microenvironment, having the potential to guide the selection of
CAR-T cell therapy (20).
Furthermore, advanced computational tools, such as the Significant
Process Inference Algorithm, enhance the detection of significant
expression patterns and biological processes, even the subtle
changes in HK2 expression across different cell populations
(162). Furthermore, the
application of deep learning models, such as KanCell, enhances the
analysis of cellular heterogeneity by integrating single-cell
RNA-sequencing and spatial transcriptomics data (163). These models can effectively
capture non-linear relationships and optimize computational
efficiency, providing a more accurate depiction of HK2 expression
patterns and their implications in disease microenvironments
(164). By leveraging these
advanced technologies, researchers can gain a deeper understanding
of the molecular mechanisms underlying HK2 regulation, and the
impact on cellular metabolism and disease progression.
Current research on tumor immune escape is focused
on several advanced fields. A comprehensive investigation into the
interactions among diverse cellular and molecular mechanisms within
the TME, particularly the metabolic interactions between immune
cells and tumor cells, will provide a theoretical foundation for
the development of more effective therapeutic strategies. Studies
have identified that TAMs facilitate tumor immune escape through
multiple signaling pathways, such as IL-4/STAT6, MAPK/ERK and
TLR-NF-κB (165–167). Therapeutic strategies aimed at
targeting TAMs, including colony stimulating factor 1 receptor
inhibitors and CD40 agonists, have progressed to clinical trials
(nos. NCT04169672 and NCT04059588), illustrating their potential to
mitigate immunosuppression and augment the effectiveness of
immunotherapy (78,168,169). Conversely, the utilization of
advanced technologies, such as single-cell sequencing and
high-dimensional flow cytometry, offers a deeper understanding of
the heterogeneity inherent in tumor immune evasion, thereby
providing a more precise foundation for personalized treatment
approaches (170).
The multifaceted role of HK2 in linking tumor
metabolic reprogramming and immune escape is becoming increasingly
evident. HK2, with its distinctive dual catalytic active domain and
N-terminal mitochondrial anchoring capability, facilitates the
‘metabolic compartmentalization’ effect and ‘metabolic relay’,
thereby mediating efficient glycolysis. HK2 has a bidirectional
regulatory network with critical oncogenic pathways, including the
PI3K/AKT, MAPK, NF-κB, TGF-β and JAK/STAT pathways, thereby
enhancing the glycolysis-dependent immune evasion of tumors. The
metabolic competition induced by the Warburg effect facilitates the
influence of HK2 on the function of immune cells in the TME,
leading to the formation of hypoxic and hypoglycemic niches. This
process is characterized by abundant lactic acid excretion, which
collectively influences immune cell distribution, alters immune
cell function and modulates the expression of immune checkpoints,
thereby further facilitating tumor immune escape. The protein
kinase function, subcellular localization regulation of HK2 and its
involvement in different immune subtypes have gained notable
attention, offering a novel perspective for a comprehensive
analysis of its non-glycolytic functions. Emerging HK2 inhibitors,
along with multi-dimensional predictive models and their
integration with immunotherapy, have demonstrated promising
potential for clinical translation. Future research should focus on
elucidating immune subtype-specific mechanisms, developing highly
selective targeted therapeutics and conducting comprehensive
analyses of HK2-related molecular characteristics. These efforts
should aim to advance the clinical implementation of HK2-targeted
intervention strategies in precision immunotherapy.
Not applicable.
The present study was supported by the National Natural Science
Foundation of China (grant no. 82505450), the Top Talent Support
Program for Young and Middle-Aged People of Wuxi Health Committee
(grant no. HB2023067), the Natural Science Foundation of Jiangsu
Province (grant no. BK20240309), and the Natural Science Foundation
of Nanjing University of Chinese Medicine (grant no.
XZR2023095).
Not applicable.
YQ participated in conceptualization, data curation
and writing of the original draft. XZ participated in
conceptualization, acquisition of funding, and reviewing and
editing of the manuscript. DN and QT participated in reviewing and
editing the manuscript. CJ participated in reviewing and editing
the manuscript, and acquiring resources. Data authentication is not
applicable. All authors have read and approved the final version of
the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
During the preparation of this work, artificial
intelligence tools (ChatGPT) were used to improve the readability
and language of the manuscript, and subsequently, the authors
revised and edited the content produced by the artificial
intelligence tools as necessary, taking full responsibility for the
ultimate content of the present manuscript.
|
1
|
Galassi C, Chan TA, Vitale I and Galluzzi
L: The hallmarks of cancer immune evasion. Cancer Cell.
42:1825–1863. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Paul S, Ghosh S and Kumar S: Tumor
glycolysis, an essential sweet tooth of tumor cells. Semin Cancer
Biol. 86:1216–1230. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guo D, Tong Y, Jiang X, Meng Y, Jiang H,
Du L, Wu Q, Li S, Luo S, Li M, et al: Aerobic glycolysis promotes
tumor immune evasion by hexokinase2-mediated phosphorylation of
IκBα. Cell Metab. 34:1312–1324. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu L, Jin Y, Zhao X, Tang K, Zhao Y, Tong
L, Yu X, Xiong K, Luo C, Zhu J, et al: Tumor aerobic glycolysis
confers immune evasion through modulating sensitivity to T
cell-mediated bystander killing via TNF-α. Cell Metab.
35:1580–1596. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qing S and Shen Z: High expression of
hexokinase 2 promotes proliferation, migration and invasion of
colorectal cancer cells by activating the JAK/STAT pathway and
regulating tumor immune microenvironment. Nan Fang Yi Ke Da Xue Xue
Bao. 45:542–553. 2025.(In Chinese). PubMed/NCBI
|
|
6
|
Li C, Tang Y, Zhang R, Shi L, Chen J,
Zhang P, Zhang N and Li W: Inhibiting glycolysis facilitated
checkpoint blockade therapy for triple-negative breast cancer.
Discov Oncol. 16:5502025. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cheng M, Wang B, Duan L, Jin Y, Zhang W
and Li N: HOTAIR knockdown increases the sensitivity of
hepatocellular carcinoma cells to sorafenib by disrupting
miR-145-5p/HK2 axis-mediated mitochondrial function and glycolysis.
Front Biosci (Landmark Ed). 30:373682025. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cui R, Wang G, Liu F, Wang Y, Zhao Z,
Mutailipu M, Mu H, Jiang X, Le W, Yang L and Chen B:
Neurturin-induced activation of GFRA2-RET axis potentiates
pancreatic cancer glycolysis via phosphorylated hexokinase 2.
Cancer Lett. 621:2175832025. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lyu SI, Simon AG, Jung JO, Fretter C,
SchrÖder W, Bruns CJ, Schmidt T, Quaas A and Knipper K: Hexokinase
2 as an independent risk factor for worse patient survival in
esophageal adenocarcinoma and as a potential therapeutic target
protein: A retrospective, single-center cohort study. Oncol Lett.
28:4952024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo W, Kuang Y, Wu J, Wen D, Zhou A, Liao
Y, Song H, Xu D, Wang T, Jing B, et al: Hexokinase 2 depletion
confers sensitization to metformin and inhibits glycolysis in lung
squamous cell carcinoma. Front Oncol. 10:522020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jin Z, Gu J, Xin X, Li Y and Wang H:
Expression of hexokinase 2 in epithelial ovarian tumors and its
clinical significance in serous ovarian cancer. Eur J Gynaecol
Oncol. 35:519–524. 2014.PubMed/NCBI
|
|
12
|
Katagiri M, Karasawa H, Takagi K, Nakayama
S, Yabuuchi S, Fujishima F, Naitoh T, Watanabe M, Suzuki T, Unno M
and Sasano H: Hexokinase 2 in colorectal cancer: A potent
prognostic factor associated with glycolysis, proliferation and
migration. Histol Histopathol. 32:351–360. 2017.PubMed/NCBI
|
|
13
|
Yoshino H, Enokida H, Itesako T, Kojima S,
Kinoshita T, Tatarano S, Chiyomaru T, Nakagawa M and Seki N:
Tumor-suppressive microRNA-143/145 cluster targets hexokinase-2 in
renal cell carcinoma. Cancer Sci. 104:1567–1574. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Botzer LE, Maman S, Sagi-Assif O, Meshel
T, Nevo I, Yron I and Witz IP: Hexokinase 2 is a determinant of
neuroblastoma metastasis. Br J Cancer. 114:759–766. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao H, Zhou Y and Chen X: Tregs and
platelets play synergistic roles in tumor immune escape and
inflammatory diseases. Crit Rev Immunol. 42:59–69. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu Y, Wu K, Shi L, Xiang F, Tao K and
Wang G: Prognostic significance of the metabolic marker
hexokinase-2 in various solid tumors: A meta-analysis. PLoS One.
11:e01662302016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kwee SA, Hernandez B, Chan O and Wong L:
Choline kinase alpha and hexokinase-2 protein expression in
hepatocellular carcinoma: Association with survival. PLoS One.
7:e465912012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Suh DH, Kim MA, Kim H, Kim MK, Kim HS,
Chung HH, Kim YB and Song YS: Association of overexpression of
hexokinase II with chemoresistance in epithelial ovarian cancer.
Clin Exp Med. 14:345–353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang XT, Xie L, Hu YT, Zhao YY, Wang RY,
Yan Y, Zhu XZ and Liu LL: T. pallidum achieves immune evasion by
blocking autophagic flux in microglia through hexokinase 2. Microb
Pathog. 199:1072162025. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin J, Fang W, Xiang Z, Wang Q, Cheng H,
Chen S, Fang J, Liu J, Wang Q, Lu Z and Ma L: Glycolytic enzyme HK2
promotes PD-L1 expression and breast cancer cell immune evasion.
Front Immunol. 14:11899532023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang L, Jiang C, Zhong Y, Sun K, Jing H,
Song J, Xie J, Zhou Y, Tian M, Zhang C, et al: STING is a
cell-intrinsic metabolic checkpoint restricting aerobic glycolysis
by targeting HK2. Nat Cell Biol. 25:1208–1222. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li J, Xu S, Zhan Y, Lv X, Sun Z, Man L,
Yang D, Sun Y and Ding S: CircRUNX1 enhances the Warburg effect and
immune evasion in non-small cell lung cancer through the
miR-145/HK2 pathway. Cancer Lett. 28:2176392025. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li R, Mei S, Ding Q, Wang Q, Yu L and Zi
F: A pan-cancer analysis of the role of hexokinase II (HK2) in
human tumors. Sci Rep. 12:188072022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ho PC, Bihuniak JD, Macintyre AN, Staron
M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, et
al: Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T
cell responses. Cell. 162:1217–1228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chang CH, Qiu J, O'Sullivan D, Buck MD,
Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ,
et al: Metabolic competition in the tumor microenvironment is a
driver of cancer progression. Cell. 162:1229–1241. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shi ZY, Yang C, Lu LY, Lin CX, Liang S, Li
G, Zhou HM and Zheng JM: Inhibition of hexokinase 2 with 3-BrPA
promotes MDSCs differentiation and immunosuppressive function. Cell
Immunol. 385:1046882023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
İpek ӦS, Sucu BO, Selvi S, Alkan FK,
Tiryaki B, Alkan HK, Sayyah E, Tolu İ, Güzel M, Durdağı S, et al:
Anti-cancer efficacy of novel lonidamine derivatives: Design,
synthesis, in vitro, in vivo, and computational studies targeting
hexokinase-2. Eur J Med Chem. 296:1178902025. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gu QL, Zhang Y, Fu XM, Lu ZL, Yu Y, Chen
G, Ma R, Kou W and Lan YM: Toxicity and metabolism of
3-bromopyruvate in Caenorhabditis elegans. J Zhejiang Univ Sci B.
21:77–86. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pajak B, Siwiak E, Sołtyka M, Priebe A,
Zieliński R, Fokt I, Ziemniak M, Jaśkiewicz A, Borowski R,
Domoradzki T and Priebe W: 2-Deoxy-d-glucose and its analogs: From
diagnostic to therapeutic agents. Int J Mol Sci. 21:2342019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rho H, Terry AR, Chronis C and Hay N:
Hexokinase 2-mediated gene expression via histone lactylation is
required for hepatic stellate cell activation and liver fibrosis.
Cell Metab. 35:1406–1423. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Patra KC, Wang Q, Bhaskar PT, Miller L,
Wang Z, Wheaton W, Chandel N, Laakso M, Muller WJ, Allen EL, et al:
Hexokinase 2 is required for tumor initiation and maintenance and
its systemic deletion is therapeutic in mouse models of cancer.
Cancer Cell. 24:213–228. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu Y, Li M, Zhang Y, Wu C, Yang K, Gao S,
Zheng M, Li X, Li H and Chen L: Structure based discovery of novel
hexokinase 2 inhibitors. Bioorg Chem. 96:1036092020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bustamante E and Pedersen PL:
Mitochondrial hexokinase of rat hepatoma cells in culture:
Solubilization and kinetic properties. Biochemistry. 19:4972–4977.
1980. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Deberardinis RJ, Sayed N, Ditsworth D and
Thompson CB: Brick by brick: Metabolism and tumor cell growth. Curr
Opin Genet Dev. 18:54–61. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ganapathy-Kanniappan S and Geschwind JF:
Tumor glycolysis as a target for cancer therapy: Progress and
prospects. Mol Cancer. 12:1522013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nawaz MH, Ferreira JC, Nedyalkova L, Zhu
H, Carrasco-López C, Kirmizialtin S and Rabeh WM: The catalytic
inactivation of the N-half of human hexokinase 2 and structural and
biochemical characterization of its mitochondrial conformation.
Biosci Rep. 38:BSR201716662018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee HJ, Li CF, Ruan D, He J, Montal ED,
Lorenz S, Girnun GD and Chan CH: Non-proteolytic ubiquitination of
Hexokinase 2 by HectH9 controls tumor metabolism and cancer stem
cell expansion. Nat Commun. 10:26252019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Panpan SI, Wei GE, Kaiming WU and Zhang R:
O-GlcNAcylation of hexokinase 2 modulates mitochondrial dynamics
and enhances the progression of lung cancer. Mol Cell Biochem.
480:2633–2643. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huang C, Chen B, Wang X, Xu J, Sun L, Wang
D, Zhao Y, Zhou C, Gao Q, Wang Q, et al: Gastric cancer mesenchymal
stem cells via the CXCR2/HK2/PD-L1 pathway mediate
immunosuppression. Gastric Cancer. 26:691–707. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ranjbar A, Soltanshahi M, Taghiloo S and
Asgarian-Omran H: Glucose metabolism in acute myeloid leukemia cell
line is regulated via combinational PI3K/AKT/mTOR pathway
inhibitors. Iran J Pharm Res. 22:e1405072023.PubMed/NCBI
|
|
41
|
Collins NB, Al Abosy R, Milsler BC, Bi K,
Zhao Q, Quigley M, Ishizuka JJ, Yates KB, Pope HW, Manguso R, et
al: PI3K activation allows immune evasion by promoting an
inhibitory myeloid tumor microenvironment. J Immunother Cancer.
10:e0034022022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen L, Lin X, Lei Y, Xu X, Zhou Q, Chen
Y, Liu H, Jiang J, Yang Y, Zheng F and Wu B: Aerobic glycolysis
enhances HBx-initiated hepatocellular carcinogenesis via
NF-κBp65/HK2 signalling. J Exp Clin Cancer Res. 41:3292022.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kawasaki Y, Sato K, Mashima K, Nakano H,
Ikeda T, Umino K, Morita K, Izawa J, Takayama N, Hayakawa H, et al:
Mesenchymal stromal cells inhibit aerobic glycolysis in activated t
cells by negatively regulating hexokinase II activity through
PD-1/PD-L1 interaction. Transplant Cell Ther. 27:231.e231–231.e238.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lynch JT, Polanska UM, Delpuech O, Hancox
U, Trinidad AG, Michopoulos F, Lenaghan C, McEwen R, Bradford J,
Polanski R, et al: Inhibiting PI3Kβ with AZD8186 regulates key
metabolic pathways in PTEN-null tumors. Clin Cancer Res.
23:7584–7595. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cargnello M and Roux PP: Activation and
function of the MAPKs and their substrates, the MAPK-activated
protein kinases. Microbiol Mol Biol Rev. 75:50–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ji P, Bäumer N, Yin T, Diederichs S, Zhang
F, Beger C, Welte K, Fulda S, Berdel WE, Serve H, et al: DNA damage
response involves modulation of Ku70 and Rb functions by cyclin A1
in leukemia cells. Int J Cancer. 121:706–713. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cui N, Li L, Feng Q, Ma HM, Lei D and
Zheng PS: Hexokinase 2 promotes cell growth and tumor formation
through the raf/mek/erk signaling pathway in cervical cancer. Front
Oncol. 10:5812082020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Courteau L, Crasto J, Hassanzadeh G, Baird
SD, Hodgins J, Liwak-Muir U, Fung G, Luo H, Stojdl DF, Screaton RA
and Holcik M: Hexokinase 2 controls cellular stress response
through localization of an RNA-binding protein. Cell Death Dis.
6:e18372015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lian C, Zhang C, Tian P, Tan Q, Wei Y,
Wang Z, Zhang Q, Zhang Q, Zhong M, Zhou L, et al: Epigenetic reader
ZMYND11 noncanonical function restricts HNRNPA1-mediated stress
granule formation and oncogenic activity. Signal Transduct Target
Ther. 9:2582024. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yan H, Wang Z, Teng D, Chen X, Zhu Z, Chen
H, Wang W, Wei Z, Wu Z, Chai Q, et al: Hexokinase 2 senses fructose
in tumor-associated macrophages to promote colorectal cancer
growth. Cell Metab. 36:2449–2467. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dunnett L, Das S, Venditti V and Prischi
F: Enhanced identification of small molecules binding to hnRNPA1
via cryptic pockets mapping coupled with X-ray fragment screening.
J Biol Chem. 301:1083352025. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yang S, Tang W, Azizian A, Gaedcke J,
Ohara Y, Cawley H, Hanna N, Ghadimi M, Lal T, Sen S, et al:
MIF/NR3C2 axis regulates glucose metabolism reprogramming in
pancreatic cancer through MAPK-ERK and AP-1 pathways.
Carcinogenesis. 45:582–594. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang T, Ma C, Zhang Z, Zhang H and Hu H:
NF-κB signaling in inflammation and cancer. MedComm (2020).
2:618–653. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jiang Q, Xu J and Wang H:
Intestine-derived exosomes regulates inflammatory response of HK-2
cells through miR-146a-regulated NF-κB pathway. Drug Eval Res.
47:2326–2333. 2024.
|
|
55
|
Tong Y, Liu X, Wu L, Xiang Y, Wang J,
Cheng Y, Zhang C, Han B, Wang L and Yan D: Hexokinase 2
nonmetabolic function-mediated phosphorylation of IκBα enhances
pancreatic ductal adenocarcinoma progression. Cancer Sci.
115:2673–2685. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Londhe P, Yu PY, Ijiri Y, Ladner KJ,
Fenger JM, London C, Houghton PJ and Guttridge DC: Classical NF-κB
metabolically reprograms sarcoma cells through regulation of
hexokinase 2. Front Oncol. 8:1042018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Brand A, Singer K, Koehl GE, Kolitzus M,
Schoenhammer G, Thiel A, Matos C, Bruss C, Klobuch S, Peter K, et
al: LDHA-associated lactic acid production blunts tumor
immunosurveillance by T and NK cells. Cell Metab. 24:657–671. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Colegio OR, Chu NQ, Szabo AL, Chu T,
Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC,
Phillips GM, et al: Functional polarization of tumour-associated
macrophages by tumour-derived lactic acid. Nature. 513:559–563.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sharan L, Pal A, Babu SS, Kumar A and
Banerjee S: Bay 11-7082 mitigates oxidative stress and
mitochondrial dysfunction via NLRP3 inhibition in experimental
diabetic neuropathy. Life Sci. 359:1232032024. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Dent AT and Wilks A: Contributions of the
heme coordinating ligands of the Pseudomonas aeruginosa outer
membrane receptor HasR to extracellular heme sensing and transport.
J Biol Chem. 295:10456–10467. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Derynck R, Turley SJ and Akhurst RJ: TGFβ
biology in cancer progression and immunotherapy. Nat Rev Clin
Oncol. 18:9–34. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhong C, Wang W, Yao Y, Lian S, Xie X, Xu
J, He S, Luo L, Ye Z, Zhang J, et al: TGF-β secreted by cancer
cells-platelets interaction activates cancer metastasis potential
by inducing metabolic reprogramming and bioenergetic adaptation. J
Cancer. 16:1310–1323. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Roh JI, Kim Y, Oh J, Kim Y, Lee J, Lee J,
Chun KH and Lee HW: Hexokinase 2 is a molecular bridge linking
telomerase and autophagy. PLoS One. 13:e01931822018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lu J, Liu Q, Wang L, Tu W, Chu H, Ding W,
Jiang S, Ma Y, Shi X, Pu W, et al: Increased expression of latent
TGF-β-binding protein 4 affects the fibrotic process in scleroderma
by TGF-β/SMAD signaling. Lab Invest. 97:591–601. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Jiang YL, Li X, Tan YW, Fang YJ, Liu KY,
Wang YF, Ma T, Ou QJ and Zhang CX: Docosahexaenoic acid inhibits
the invasion and migration of colorectal cancer by reversing EMT
through the TGF-β1/Smad signaling pathway. Food Funct.
15:9420–9433. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mariathasan S, Turley SJ, Nickles D,
Castiglioni A, Yuen K, Wang Y, Kadel EE III, Koeppen H, Astarita
JL, Cubas R, et al: TGFβ attenuates tumour response to PD-L1
blockade by contributing to exclusion of T cells. Nature.
554:544–548. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Jeon HS and Jen J: TGF-beta signaling and
the role of inhibitory Smads in non-small cell lung cancer. J
Thorac Oncol. 5:417–419. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Santarpia M, González-Cao M, Viteri S,
Karachaliou N, Altavilla G and Rosell R: Programmed cell death
protein-1/programmed cell death ligand-1 pathway inhibition and
predictive biomarkers: Understanding transforming growth
factor-beta role. Transl Lung Cancer Res. 4:728–742.
2015.PubMed/NCBI
|
|
69
|
Ayele TM, Muche ZT, Teklemariam AB, Kassie
AB and Abebe EC: Role of JAK2/STAT3 signaling pathway in the
tumorigenesis, chemotherapy resistance, and treatment of solid
tumors: A systemic review. J Inflamm Res. 15:1349–1364. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ren D, Hua Y, Yu B, Ye X, He Z, Li C, Wang
J, Mo Y, Wei X, Chen Y, et al: Predictive biomarkers and mechanisms
underlying resistance to PD1/PD-L1 blockade cancer immunotherapy.
Mol Cancer. 19:192020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Shangguan X, He J, Ma Z, Zhang W, Ji Y,
Shen K, Yue Z, Li W, Xin Z, Zheng Q, et al: SUMOylation controls
the binding of hexokinase 2 to mitochondria and protects against
prostate cancer tumorigenesis. Nat Commun. 12:18122021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Xue YN, Yu BB, Li JL, Guo R, Zhang LC, Sun
LK, Liu YN and Li Y: Zinc and p53 disrupt mitochondrial binding of
HK2 by phosphorylating VDAC1. Exp Cell Res. 374:249–258. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wang Q, Guo X, Li L, Gao Z, Su X, Ji M and
Liu J: N6-methyladenosine METTL3 promotes cervical
cancer tumorigenesis and Warburg effect through YTHDF1/HK2
modification. Cell Death Dis. 11:9112020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Klinke DJ II, Cheng N and Chambers E:
Quantifying crosstalk among interferon-γ, interleukin-12, and tumor
necrosis factor signaling pathways within a TH1 cell model. Sci
Signal. 5:ra322012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhang H, Li S, Wang D, Liu S, Xiao T, Gu
W, Yang H, Wang H, Yang M and Chen P: Metabolic reprogramming and
immune evasion: The interplay in the tumor microenvironment.
Biomark Res. 12:962024. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ferreira JC, Khrbtli AR, Shetler CL,
Mansoor S, Ali L, Sensoy O and Rabeh WM: Linker residues regulate
the activity and stability of hexokinase 2, a promising anticancer
target. J Biol Chem. 296:1000712021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Peláez R, Fernández-García P, Herrero P
and Moreno F: Nuclear import of the yeast hexokinase 2 protein
requires α/β-importin-dependent pathway. J Biol Chem.
287:3518–3529. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Li S, Zhang M, Gao Y, Zhao C, Liao S, Zhao
X, Ning Q and Tang S: Mechanisms of tumor-associated macrophages
promoting tumor immune escape. Carcinogenesis. 46:bgaf0232025.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Gou Q, Che S, Chen M, Chen H, Shi J and
Hou Y: PPARγ inhibited tumor immune escape by inducing PD-L1
autophagic degradation. Cancer Sci. 114:2871–2881. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Rabinowitz JD and Enerbäck S: Lactate: The
ugly duckling of energy metabolism. Nat Metab. 2:566–571. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Brooks GA: The science and translation of
lactate shuttle theory. Cell Metab. 27:757–785. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhang W, Wang G, Xu ZG, Tu H, Hu F, Dai J,
Chang Y, Chen Y, Lu Y, Zeng H, et al: Lactate is a natural
suppressor of rlr signaling by targeting MAVS. Cell. 178:176–189.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Chang CH, Curtis JD, Maggi LB Jr, Faubert
B, Villarino AV, O'Sullivan D, Huang SC, van der Windt GJ, Blagih
J, Qiu J, et al: Posttranscriptional control of T cell effector
function by aerobic glycolysis. Cell. 153:1239–1251. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Xia H, Wang W, Crespo J, Kryczek I, Li W,
Wei S, Bian Z, Maj T, He M, Liu RJ, et al: Suppression of FIP200
and autophagy by tumor-derived lactate promotes naïve T cell
apoptosis and affects tumor immunity. Sci Immunol. 2:eaan46312017.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Arruga F, Gyau BB, Iannello A, Vitale N,
Vaisitti T and Deaglio S: Immune response dysfunction in chronic
lymphocytic leukemia: Dissecting molecular mechanisms and
microenvironmental conditions. Int J Mol Sci. 21:18252020.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Corbet C and Feron O: Tumour acidosis:
From the passenger to the driver's seat. Nat Rev Cancer.
17:577–593. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Sukumar M, Liu J, Ji Y, Subramanian M,
Crompton JG, Yu Z, Roychoudhuri R, Palmer DC, Muranski P, Karoly
ED, et al: Inhibiting glycolytic metabolism enhances CD8+ T cell
memory and antitumor function. J Clin Invest. 123:4479–4488. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Nasi A, Fekete T, Krishnamurthy A, Snowden
S, Rajnavölgyi E, Catrina AI, Wheelock CE, Vivar N and Rethi B:
Dendritic cell reprogramming by endogenously produced lactic acid.
J Immunol. 191:3090–3099. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Fischer K, Hoffmann P, Voelkl S,
Meidenbauer N, Ammer J, Edinger M, Gottfried E, Schwarz S, Rothe G,
Hoves S, et al: Inhibitory effect of tumor cell-derived lactic acid
on human T cells. Blood. 109:3812–3819. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Lin S, Sun L, Lyu X, Ai X, Du D, Su N, Li
H, Zhang L, Yu J and Yuan S: Lactate-activated macrophages induced
aerobic glycolysis and epithelial-mesenchymal transition in breast
cancer by regulation of CCL5-CCR5 axis: A positive metabolic
feedback loop. Oncotarget. 8:110426–110443. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zappasodi R, Serganova I, Cohen IJ, Maeda
M, Shindo M, Senbabaoglu Y, Watson MJ, Leftin A, Maniyar R, Verma
S, et al: CTLA-4 blockade drives loss of T(reg) stability in
glycolysis-low tumours. Nature. 591:652–658. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Liu N, Luo J, Kuang D, Xu S, Duan Y, Xia
Y, Wei Z, Xie X, Yin B, Chen F, et al: Lactate inhibits ATP6V0d2
expression in tumor-associated macrophages to promote
HIF-2α-mediated tumor progression. J Clin Invest. 129:631–646.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Gao Y, Zhou H, Liu G, Wu J, Yuan Y and
Shang A: Tumor microenvironment: Lactic acid promotes tumor
development. J Immunol Res. 2022:31193752022. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Langin D: Adipose tissue lipolysis
revisited (again!): Lactate involvement in insulin antilipolytic
action. Cell Metab. 11:242–243. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Pan Y, Yu Y, Wang X and Zhang T:
Corrigendum: Tumor-associated macrophages in tumor immunity. Front
Immunol. 12:7757582021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
El-Kenawi A, Gatenbee C, Robertson-Tessi
M, Bravo R, Dhillon J, Balagurunathan Y, Berglund A, Vishvakarma N,
Ibrahim-Hashim A, Choi J, et al: Acidity promotes tumour
progression by altering macrophage phenotype in prostate cancer. Br
J Cancer. 121:556–566. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Liu Q, Du F, Huang W, Ding X, Wang Z, Yan
F and Wu Z: Epigenetic control of Foxp3 in intratumoral T-cells
regulates growth of hepatocellular carcinoma. Aging (Albany NY).
11:2343–2351. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Yu J, Chai P, Xie M, Ge S, Ruan J, Fan X
and Jia R: Histone lactylation drives oncogenesis by facilitating
m6A reader protein YTHDF2 expression in ocular melanoma.
Genome Biol. 22:852021. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Jiang J, Huang D, Jiang Y, Hou J, Tian M,
Li J, Sun L, Zhang Y, Zhang T, Li Z, et al: Lactate modulates
cellular metabolism through histone lactylation-mediated gene
expression in non-small cell lung cancer. Front Oncol.
11:6475592021. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Certo M, Tsai CH, Pucino V, Ho PC and
Mauro C: Lactate modulation of immune responses in inflammatory
versus tumour microenvironments. Nat Rev Immunol. 21:151–161. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Wang N, Wang W, Wang X, Mang G, Chen J,
Yan X, Tong Z, Yang Q, Wang M, Chen L, et al: Histone lactylation
boosts reparative gene activation post-myocardial infarction. Circ
Res. 131:893–908. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Mathupala SP, Ko YH and Pedersen PL:
Hexokinase II: Cancer's double-edged sword acting as both
facilitator and gatekeeper of malignancy when bound to
mitochondria. Oncogene. 25:4777–4786. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Majmundar AJ, Wong WJ and Simon MC:
Hypoxia-inducible factors and the response to hypoxic stress. Mol
Cell. 40:294–309. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zhang LF, Lou JT, Lu MH, Gao C, Zhao S, Li
B, Liang S, Li Y, Li D and Liu MF: Suppression of miR-199a
maturation by HuR is crucial for hypoxia-induced glycolytic switch
in hepatocellular carcinoma. EMBO J. 34:2671–2685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Bao MH and Wong CC: Hypoxia, metabolic
reprogramming, and drug resistance in liver cancer. Cells.
10:17152021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Reyes A, Duarte LF, Farías MA, Tognarelli
E, Kalergis AM, Bueno SM and González PA: Impact of hypoxia over
human viral infections and key cellular processes. Int J Mol Sci.
22:79542021. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Zheng H, Ning Y, Zhan Y, Liu S, Yang Y,
Wen Q and Fan S: Co-expression of PD-L1 and HIF-1α predicts poor
prognosis in patients with non-small cell lung cancer after
surgery. J Cancer. 12:2065–2072. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Bandopadhyay S and Patranabis S:
Mechanisms of HIF-driven immunosuppression in tumour
microenvironment. J Egypt Natl Canc Inst. 35:272023. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
You L, Wang X, Wu W, Nepovimova E, Wu Q
and Kuca K: HIF-1α inhibits T-2 toxin-mediated ‘immune evasion’
process by negatively regulating PD-1/PD-L1. Toxicology.
480:1533242022. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Yang Z, Su W, Wei X, Pan Y, Xing M, Niu L,
Feng B, Kong W, Ren X, Huang F, et al: Hypoxia inducible factor-1α
drives cancer resistance to cuproptosis. Cancer Cell. 43:937–954.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Chen CH, Li SX, Xiang LX, Mu HQ, Wang SB
and Yu KY: HIF-1α induces immune escape of prostate cancer by
regulating NCR1/NKp46 signaling through miR-224. Biochem Biophys
Res Commun. 503:228–234. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Facciabene A, Peng X, Hagemann IS, Balint
K, Barchetti A, Wang LP, Gimotty PA, Gilks CB, Lal P, Zhang L and
Coukos G: Tumour hypoxia promotes tolerance and angiogenesis via
CCL28 and T(reg) cells. Nature. 475:226–230. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Peng J, Wang X, Ran L, Song J, Luo R and
Wang Y: Hypoxia-inducible factor 1α regulates the transforming
growth factor β1/SMAD family member 3 pathway to promote breast
cancer progression. J Breast Cancer. 21:259–266. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Clambey ET, McNamee EN, Westrich JA,
Glover LE, Campbell EL, Jedlicka P, de Zoeten EF, Cambier JC,
Stenmark KR, Colgan SP and Eltzschig HK: Hypoxia-inducible factor-1
alpha-dependent induction of FoxP3 drives regulatory T-cell
abundance and function during inflammatory hypoxia of the mucosa.
Proc Natl Acad Sci USA. 109:E2784–E2793. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Sormendi S and Wielockx B: Hypoxia pathway
proteins as central mediators of metabolism in the tumor cells and
their microenvironment. Front Immunol. 9:402018. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Balamurugan K: HIF-1 at the crossroads of
hypoxia, inflammation, and cancer. Int J Cancer. 138:1058–1066.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Murthy A, Gerber SA, Koch CJ and Lord EM:
Intratumoral hypoxia reduces IFN-γ-mediated immunity and mhc class
I induction in a preclinical tumor model. Immunohorizons.
3:149–160. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Bhandari T, Olson J, Johnson RS and Nizet
V: HIF-1α influences myeloid cell antigen presentation and response
to subcutaneous OVA vaccination. J Mol Med (Berl). 91:1199–1205.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Ruiz-Iglesias A and Mañes S: The
importance of mitochondrial pyruvate carrier in cancer cell
metabolism and tumorigenesis. Cancers (Basel). 13:14882021.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Zhang T, Yu-Jing L and Ma T: The
immunomodulatory function of adenosine in sepsis. Front Immunol.
13:9365472022. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Haskó G, Linden J, Cronstein B and Pacher
P: Adenosine receptors: Therapeutic aspects for inflammatory and
immune diseases. Nat Rev Drug Discov. 7:759–770. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Chambers AM and Matosevic S:
Immunometabolic dysfunction of natural killer cells mediated by the
hypoxia-CD73 axis in solid tumors. Front Mol Biosci. 6:602019.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Zhao F, Xiao C, Evans KS, Theivanthiran T,
DeVito N, Holtzhausen A, Liu J, Liu X, Boczkowski D, Nair S, et al:
Paracrine Wnt5a-β-catenin signaling triggers a metabolic program
that drives dendritic cell tolerization. Immunity. 48:147–160.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Godin-Ethier J, Hanafi LA, Piccirillo CA
and Lapointe R: Indoleamine 2,3-dioxygenase expression in human
cancers: Clinical and immunologic perspectives. Clin Cancer Res.
17:6985–6991. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Irshad Z, Xue M, Ashour A, Larkin JR,
Thornalley PJ and Rabbani N: Activation of the unfolded protein
response in high glucose treated endothelial cells is mediated by
methylglyoxal. Sci Rep. 9:78892019. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Siska PJ, Beckermann KE, Mason FM,
Andrejeva G, Greenplate AR, Sendor AB, Chiang YJ, Corona AL, Gemta
LF, Vincent BG, et al: Mitochondrial dysregulation and glycolytic
insufficiency functionally impair CD8 T cells infiltrating human
renal cell carcinoma. JCI Insight. 2:e934112017. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Blagih J, Coulombe F, Vincent EE, Dupuy F,
Galicia-Vázquez G, Yurchenko E, Raissi TC, van der Windt GJ,
Viollet B, Pearce EL, et al: The energy sensor AMPK regulates T
cell metabolic adaptation and effector responses in vivo. Immunity.
42:41–54. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Liu X, Zhao Y, Wu X, Liu Z and Liu X: A
novel strategy to fuel cancer immunotherapy: Targeting glucose
metabolism to remodel the tumor microenvironment. Front Oncol.
12:9311042022. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Liu X, Mo W, Ye J, Li L, Zhang Y, Hsueh
EC, Hoft DF and Peng G: Regulatory T cells trigger effector T cell
DNA damage and senescence caused by metabolic competition. Nat
Commun. 9:2492018. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Scharping NE and Delgoffe GM: Tumor
microenvironment metabolism: A new checkpoint for anti-tumor
immunity. Vaccines (Basel). 4:462016. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Jiang Y, Li Y and Zhu B: T-cell exhaustion
in the tumor microenvironment. Cell Death Dis. 6:e17922015.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Bader JE, Voss K and Rathmell JC:
Targeting metabolism to improve the tumor microenvironment for
cancer immunotherapy. Mol Cell. 78:1019–1033. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Geraghty KM, Chen S, Harthill JE, Ibrahim
AF, Toth R, Morrice NA, Vandermoere F, Moorhead GB, Hardie DG and
MacKintosh C: Regulation of multisite phosphorylation and 14-3-3
binding of AS160 in response to IGF-1, EGF, PMA and AICAR. Biochem
J. 407:231–241. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Zhang Q, Yang Z, Hao X, Dandreo LJ, He L,
Zhang Y, Wang F, Wu X and Xu L: Niclosamide improves cancer
immunotherapy by modulating RNA-binding protein HuR-mediated PD-L1
signaling. Cell Biosci. 13:1922023. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Li W, Zheng M, Wu S, Gao S, Yang M, Li Z,
Min Q, Sun W, Chen L, Xiang G and Li H: Benserazide, a
dopadecarboxylase inhibitor, suppresses tumor growth by targeting
hexokinase 2. J Exp Clin Cancer Res. 36:582017. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zheng M, Wu C, Yang K, Yang Y, Liu Y, Gao
S, Wang Q, Li C, Chen L and Li H: Novel selective hexokinase 2
inhibitor Benitrobenrazide blocks cancer cells growth by targeting
glycolysis. Pharmacol Res. 164:1053672021. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
VDA-1102, a first-in-class VDAC/HK
modulator entering phase 1/2 drug development for treatment of
actinic keratosis, cutaneous squamous cell carcinoma. J AM ACAD
DERMATOL. 74:2016.
|
|
138
|
Lozzi F, Lanna C, Mazzeo M, Garofalo V,
Palumbo V, Mazzilli S, Diluvio L, Terrinoni A, Bianchi L and
Campione E: Investigational drugs currently in phase II clinical
trials for actinic keratosis. Expert Opin Investig Drugs.
28:629–642. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Zheng Y, Zhan Y, Zhang Y, Zhang Y, Liu Y,
Xie Y, Sun Y, Qian J, Ding Y, Ding Y and Fang Y: Hexokinase 2
confers radio-resistance in hepatocellular carcinoma by promoting
autophagy-dependent degradation of AIMP2. Cell Death Dis.
14:4882023. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Lin H, Zeng J, Xie R, Schulz MJ, Tedesco
R, Qu J, Erhard KF, Mack JF, Raha K, Rendina AR, et al: Discovery
of a novel 2,6-disubstituted glucosamine series of potent and
selective hexokinase 2 inhibitors. ACS Med Chem Lett. 7:217–222.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Jiang SH, Dong FY, Da LT, Yang XM, Wang
XX, Weng JY, Feng L, Zhu LL, Zhang YL, Zhang ZG, et al:
Ikarugamycin inhibits pancreatic cancer cell glycolysis by
targeting hexokinase 2. FASEB J. 34:3943–3955. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Bao F, Yang K, Wu C, Gao S, Wang P, Chen L
and Li H: New natural inhibitors of hexokinase 2 (HK2): Steroids
from Ganoderma sinense. Fitoterapia. 125:123–129. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Wang S, Zhuang Y, Xu J, Tong Y, Li X and
Dong C: Advances in the study of hexokinase 2 (HK2) inhibitors.
Anticancer Agents Med Chem. 23:736–746. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Xu Z, Zhang D, Zhang Z, Luo W, Shi R, Yao
J, Li D, Wang L and Liao B: MicroRNA-505, suppressed by oncogenic
long non-coding RNA LINC01448, acts as a novel suppressor of
glycolysis and tumor progression through inhibiting HK2 expression
in pancreatic cancer. Front Cell Dev Biol. 8:6250562020. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Ye J, Xiao X, Han Y, Fan D, Zhu Y and Yang
L: MiR-3662 suppresses cell growth, invasion and glucose metabolism
by targeting HK2 in hepatocellular carcinoma cells. Neoplasma.
67:773–781. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Zheng C, Li R, Zheng S, Fang H, Xu M and
Zhong L: The knockdown of lncRNA DLGAP1-AS2 suppresses osteosarcoma
progression by inhibiting aerobic glycolysis via the miR-451a/HK2
axis. Cancer Sci. 114:4747–4762. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Zhu W, Huang Y, Pan Q, Xiang P, Xie N and
Yu H: MicroRNA-98 suppress warburg effect by targeting HK2 in colon
cancer cells. Dig Dis Sci. 62:660–668. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Guo Y, Liang F, Zhao F and Zhao J:
Resibufogenin suppresses tumor growth and Warburg effect through
regulating miR-143-3p/HK2 axis in breast cancer. Mol Cell Biochem.
466:103–115. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Zhong J, Lu S, Jia X, Li Q, Liu L, Xie P,
Wang G, Lu M, Gao W, Zhao T, et al: Role of endoplasmic reticulum
stress in apoptosis induced by HK2 inhibitor and its potential as a
new drug combination strategy. Cell Stress Chaperones. 27:273–283.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Agnihotri S, Mansouri S, Burrell K, Li M,
Mamatjan Y, Liu J, Nejad R, Kumar S, Jalali S, Singh SK, et al:
Ketoconazole and posaconazole selectively target HK2-expressing
glioblastoma cells. Clin Cancer Res. 25:844–855. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Afonso J, Gonçalves C, Costa M, Ferreira
D, Santos L, Longatto-Filho A and Baltazar F: Glucose metabolism
reprogramming in bladder cancer: Hexokinase 2 (HK2) as prognostic
biomarker and target for bladder cancer therapy. Cancers (Basel).
15:9822023. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Yang L, Yan X, Chen J, Zhan Q, Hua Y, Xu
S, Li Z, Wang Z, Dong Y, Zuo D, et al: Hexokinase 2 discerns a
novel circulating tumor cell population associated with poor
prognosis in lung cancer patients. Proc Natl Acad Sci USA.
118:e20122281182021. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Xu S, Catapang A, Doh HM, Bayley NA, Lee
JT, Braas D, Graeber TG and Herschman HR: Hexokinase 2 is
targetable for HK1 negative, HK2 positive tumors from a wide
variety of tissues of origin. J Nucl Med. 60:212–217. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Shurin MR and Umansky V: Cross-talk
between HIF and PD-1/PD-L1 pathways in carcinogenesis and therapy.
J Clin Invest. 132:e1594732022. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Qiu J, Zhong F, Zhang Z, Pan B, Ye D,
Zhang X, Yao Y, Luo Y, Wang X and Tang N: Hypoxia-responsive lncRNA
MIR155HG promotes PD-L1 expression in hepatocellular carcinoma
cells by enhancing HIF-1α mRNA stability. Int Immunopharmacol.
136:1124152024. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Xu H, Chen Y, Li Z, Zhang H, Liu J and Han
J: The hypoxia-inducible factor 1 inhibitor LW6 mediates the
HIF-1α/PD-L1 axis and suppresses tumor growth of hepatocellular
carcinoma in vitro and in vivo. Eur J Pharmacol. 930:1751542022.
View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Dai X, Pi G, Yang SL, Chen GG, Liu LP and
Dong HH: Association of PD-L1 and HIF-1α coexpression with poor
prognosis in hepatocellular carcinoma. Transl Oncol. 11:559–566.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Desta GM and Birhanu AG: Advancements in
single-cell RNA sequencing and spatial transcriptomics:
Transforming biomedical research. Acta Biochim Pol. 72:139222025.
View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Shen X, Zhao Y, Wang Z and Shi Q: Recent
advances in high-throughput single-cell transcriptomics and spatial
transcriptomics. Lab Chip. 22:4774–4791. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Elosua-Bayes M, Nieto P, Mereu E, Gut I
and Heyn H: SPOTlight: Seeded NMF regression to deconvolute spatial
transcriptomics spots with single-cell transcriptomes. Nucleic
Acids Res. 49:e502021. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Kleshchevnikov V, Shmatko A, Dann E,
Aivazidis A, King HW, Li T, Elmentaite R, Lomakin A, Kedlian V,
Gayoso A, et al: Cell2location maps fine-grained cell types in
spatial transcriptomics. Nat Biotechnol. 40:661–671. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Biran H, Hashimshony T, Lahav T, Efrat O,
Mandel-Gutfreund Y and Yakhini Z: Detecting significant expression
patterns in single-cell and spatial transcriptomics with a flexible
computational approach. Sci Rep. 14:261212024. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Wang Z, Dai R, Wang M, Lei L, Zhang Z, Han
K, Wang Z and Guo Q: KanCell: Dissecting cellular heterogeneity in
biological tissues through integrated single-cell and spatial
transcriptomics. J Genet Genomics. 52:689–705. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Ravindran U and Gunavathi C: Deep learning
assisted cancer disease prediction from gene expression data using
WT-GAN. BMC Med Inform Decis Mak. 24:3112024. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Hu B, Wang Z, Zeng H, Qi Y, Chen Y, Wang
T, Wang J, Chang Y, Bai Q, Xia Y, et al: Blockade of DC-SIGN(+)
tumor-associated macrophages reactivates antitumor immunity and
improves immunotherapy in muscle-invasive bladder cancer. Cancer
Res. 80:1707–1719. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Yang H, Zhang Q, Xu M, Wang L, Chen X,
Feng Y, Li Y, Zhang X, Cui W and Jia X: CCL2-CCR2 axis recruits
tumor associated macrophages to induce immune evasion through PD-1
signaling in esophageal carcinogenesis. Mol Cancer. 19:412020.
View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Wolf-Dennen K, Gordon N and Kleinerman ES:
Exosomal communication by metastatic osteosarcoma cells modulates
alveolar macrophages to an M2 tumor-promoting phenotype and
inhibits tumoricidal functions. Oncoimmunology. 9:17476772020.
View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Rodriguez-Perdigon M, Haeni L,
Rothen-Rutishauser B and Rüegg C: Dual CSF1R inhibition and CD40
activation demonstrates anti-tumor activity in a 3D macrophage-
HER2(+) breast cancer spheroid model. Front Bioeng Biotechnol.
11:11598192023. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Kinouchi M, Miura K, Mizoi T, Ishida K,
Fujibuchi W, Sasaki H, Ohnuma S, Saito K, Katayose Y, Naitoh T, et
al: Infiltration of CD40-positive tumor-associated macrophages
indicates a favorable prognosis in colorectal cancer patients.
Hepatogastroenterology. 60:83–88. 2013.PubMed/NCBI
|
|
170
|
Shvefel SC, Pai JA, Cao Y, Pal LR, Bartok
O, Levy R, Zemanek MJ, Weller C, Herzog E, Yao W, et al: Temporal
genomic analysis of homogeneous tumor models reveals key regulators
of immune evasion in melanoma. Cancer Discov. 28:OF1–OF25.
2024.
|