Introduction
Pancreatic cancer (PC), a highly malignant neoplasm
of the digestive system with a 5-year survival rate of <10%, is
recognized as the ‘king of cancer types’ (1). According to data from the
International Agency for Research on Cancer, ~496,000 new PC cases
and 466,000 related deaths were reported globally in 2020, which
ranks PC as the seventh leading cause of cancer mortality in both
sexes (2). The pathogenesis of PC
is characterized by complex interactions between multiple genetic
and environmental factors. The associated genetic factors include
regulatory molecules, such as tiRNA-Val-CAC-2, which has been shown
to interact with FUBP1 to promote PC metastasis by activating c-MYC
transcription (3); and the
environmental factors encompass smoking and excessive alcohol
consumption. Furthermore, recurrent acute pancreatitis, often
linked to the aforementioned environmental triggers, can augment
long-term PC risk (4). Current
therapeutic modalities remain limited and primarily include
surgical intervention, chemotherapy and radiotherapy (5–7).
However, due to the aggressive biological behaviour of PC, ~80% of
patients present with advanced-stage disease at diagnosis (1), which results in limited opportunities
for radical resection and suboptimal outcomes with conventional
chemo-radiotherapeutic approaches.
Gemcitabine (GEM) remains one of the most widely
used chemotherapeutic agents in PC management (8). Its mechanism of action involves
incorporation into DNA during synthesis, thereby inducing strand
fragmentation and cell cycle arrest (9). However, in the hypoxic tumour
microenvironment (TME), multiple resistance mechanisms have emerged
that undermine GEM efficacy. Specifically, these mechanisms include
reduced drug uptake (for example, loss of the human equilibrative
nucleoside transporter 1), enhanced DNA repair capacity and
metabolic reprogramming (10).
These resistance mechanisms contribute to the limited clinical
efficacy of GEM, which is characterized by an overall response rate
of<20% and frequent development of rapid tumour resistance
(8,11). Therefore, elucidating the molecular
mechanisms underlying chemoresistance and identifying effective
therapeutic targets to overcome drug resistance have become key
challenges in contemporary PC treatment research.
Although recent advancements have been made in
targeted therapies and immunotherapies for PC, its complex TME
remains a pivotal factor contributing to therapeutic failure
(12). The highly fibrotic stroma
and aberrant vascular proliferation in PC result in persistent
intratumoural hypoxia, which fosters a hypoxic microenvironment.
This hypoxic niche, a recognized hallmark of PC, drives tumour
progression, metastasis and chemoresistance through
hypoxia-inducible factor (HIF)-mediated activation of multiple
oncogenic signalling pathways (including the VEGF pathway, the
PI3K/Akt/mTOR pathway and the Notch/Wnt/β-catenin pathways)
(13–16). Previous studies have revealed that
hypoxia markedly enhances the exosomal secretion capacity of tumour
cells (17–19).
Exosomes (Exos), extracellular vesicles measuring
40–160 nm in diameter, serve as molecular carriers within the TME
by transporting bioactive cargo (for example, proteins, lipids and
nucleic acids) to mediate intercellular communication. Under
hypoxic conditions, tumour-derived Exos can modulate drug
sensitivity through the horizontal transfer of non-coding RNAs,
particularly microRNAs (miRNAs). For example, hypoxia-induced Exos
carrying miR-210-3p promote proliferation, migration, invasion,
epithelial-mesenchymal transition and apoptosis resistance in
triple-negative breast cancer (TNBC) cells through the activation
of the nuclear factor I X-Wnt/β-catenin signalling axis (20). Nevertheless, the mechanistic role of
hypoxia-derived exosomal miRNAs in GEM resistance remains to be
elucidated, which represents a key frontier in contemporary PC
research.
miR-301a, a key member of the miRNA family, is
markedly upregulated across multiple malignancies, such as breast
cancer and gastric cancer, and participates in oncogenic processes
including proliferation, invasion and immune evasion through
targeted gene regulation (21,22).
Its tumour-promoting effects may be mediated through the
suppression of tumour suppressor genes such as PTEN and SMAD4
(23,24). Our preliminary investigations
demonstrated that miR-301a acts as a key regulator involved in
hypoxia-induced gemcitabine resistance in PC. Furthermore, hypoxia
induces the upregulation of miR-301a within tumour-derived Exos
(25,26). These Exos can be internalized by
neighbouring malignant cells, subsequently modulating their
biological behaviours, although the precise molecular mechanisms
remain to be fully elucidated.
Acyl-CoA synthetase long chain family member 4
(ACSL4), a key enzyme in fatty acid metabolism, catalyses the
conjugation of long-chain fatty acids with coenzyme A to generate
acyl-CoA derivatives, thereby regulating membrane phospholipid
remodelling, signal transduction and energy metabolism (27). Dysregulated ACSL4 expression has
been implicated in the progression of multiple cancer types. For
example, in TNBC, aberrant ACSL4 expression mediates cell membrane
phospholipid remodelling, inducing lipid raft localization and
integrin β1 activation in a CD47-dependent manner, which promotes
tumour metastasis via focal adhesion kinase phosphorylation
(28). In hepatocellular carcinoma,
ACSL4 enhances tumour cell proliferation and metastasis by
regulating de novo lipid synthesis (29). Notably, ACSL4 has emerged as a key
regulator of ferroptosis, an iron-dependent form of cell death
driven by lipid peroxidation (30).
MiRNAs have also been revealed to modulate ACSL4-mediated
processes: MiR-23a-3p promotes sorafenib resistance in
hepatocellular carcinoma by inhibiting ACSL4 expression and
blocking ferroptosis (31), whereas
miR-20a-5p alleviates ferroptosis in renal ischaemia-reperfusion
injury by directly targeting the 3′ untranslated region (3′-UTR) of
ACSL4 mRNA (32). Notably, emerging
evidence suggests that ACSL4 also contribute to GEM resistance in
PC, although the underlying mechanisms remain largely undefined
(33).
The present study aimed to investigate the
functional impact of hypoxic tumour-derived exosomal miR-301a on
GEM resistance in PC and elucidate its underlying molecular
mechanisms. The present study provides novel insights into the
chemoresistance landscape of PC and establishes a theoretical
foundation for the potential development of exosomal miRNA-based
combinatorial therapeutic strategies in the future.
Materials and methods
Cell culture and transfection
Cell line information
Descriptions of the original cell line derivation
are shown in Table I (34).
 | Table I.Cell line characteristics. |
Table I.
Cell line characteristics.
| Cell line | Derivation | Molecular
subtype | Aggressiveness
level | Key genetic
features |
|---|
| BxPC-3 | Primary tumour | Classical |
Low-to-moderate | Wild-type KRAS,
SMAD4 intact |
| CFPAC-1 | Liver
metastasis | Basal-like | Moderate | KRAS G12V, TP53
mutant |
| AsPC-1 | Ascites | Basal-like | High | KRAS G12V, TP53
mutant |
| MIA PaCa-2 | Primary tumour | Classical | High | KRAS G12C, TP53
mutant |
| PANC-1 | Primary tumour | Basal-like | Highly
aggressive | KRAS G12D, SMAD4
deleted |
Cell culture
All the cell lines used in the present study
[ascites pancreatic cancer 1 (AsPC-1), BxPC-3, cystic fibrosis
pancreatic adenocarcinoma (CFPAC-1), malignant inflammatory
adenocarcinoma pancreatic carcinoma-2 (MIA PaCa-2) and PANC-1)]
were purchased from the Shanghai Cell Bank at the Chinese Academy
of Sciences. MIA PaCa-2 and PANC-1 cells were cultured in DMEM
(cat. no. C11995500BT; Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (cat. no. FSP500; Shanghai ExCell
Biology, Inc.) and 1% penicillin/streptomycin (cat. no. C0222;
Beyotime Institute of Biotechnology). AsPC-1 and BxPC-3 cells were
cultured in RPMI-1640 (cat no. C11875500BT; Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS and 1%
penicillin/streptomycin. CFPAC-1 cells were cultured in Iscove's
Modified Dulbecco's Medium (cat. no. BL312A; Biosharp Life
Sciences) supplemented with 10% FBS and 1% penicillin/streptomycin.
All the cell lines were cultured in an incubator at 37°C in an
atmosphere containing 5% CO2. During hypoxic treatment,
the cells were cultured in a hypoxic incubator (37°C; 5%
CO2; 1% O2).
Cell transfection
The miR-301a inhibitor, miR-301a mimics and their
respective negative control (NC) oligonucleotides were synthesized
by Anhui General Bioscience. The sequences are presented in
Table II. Transfection was
performed using Lipofectamine® 2000 transfection reagent
(cat. no. 11668019; Invitrogen; Thermo Fisher Scientific, Inc.), a
widely used cationic lipid transfection reagent, for which the
manufacturer's protocol recommends a 4–6 h incubation period in a
37°C incubator for optimal nucleic acid delivery while minimizing
cytotoxicity. However, based on our long-term research experience
on the effects and molecular mechanisms of non-coding RNAs in the
malignant biology of pancreatic tumours, the present study extended
the transfection time to 6–8 h to balance transfection efficiency
and cell viability. The specific steps were as follows: Cells under
good growth conditions, characterized by 50–80% confluence (in the
logarithmic growth phase), ≥90% cell viability, normal cell
morphology and no microbial contamination, were evenly plated in a
6-well plate, 1 day before transfection. Transfection was initiated
when the cell confluence reached 60–70%. Preparation of
transfection mixtures: For each well, 5 µl of
Lipofectamine® 2000 was added to 250 µl of Opti-MEM
Reduced Serum Medium (cat. no. L530JV; Shanghai Basal Media
Technologies Co., Ltd.) in a sterile centrifuge tube and gently
mixed. This mixture was incubated at room temperature for 5 min to
allow the Lipofectamine® 2000 to activate. Meanwhile, in
another sterile centrifuge tube, 20 nM of the miR-301a inhibitor,
mimics or NC oligonucleotide was diluted in 250 µl of Opti-MEM
reduced serum medium. The diluted oligonucleotide mixture was then
slowly added to the activated Lipofectamine®
2000-Opti-MEM mixture and mixed gently by pipetting up and down
several times. The resulting transfection mixture was incubated at
room temperature for 20 min to form stable
Lipofectamine®-oligonucleotide complexes. The culture
medium in each well was carefully aspirated and replaced with 1.5
ml of serum-free medium. The prepared transfection mixture was then
added dropwise to each well and the plates were gently swirled to
ensure the even distribution of the complexes. After 6–8 h, the
medium containing serum and antibiotics was replaced, and the cells
were further cultured for 24 or 48 h for subsequent
experiments.
 | Table II.Inhibitor and mimics sequences. |
Table II.
Inhibitor and mimics sequences.
| RNA | Direction | Primer sequences
(5′-3′) |
|---|
| miR-301a
inhibitor | - |
GCUUUGACAAUACUAUUGCACUG |
| Inhibitor NC | - |
UCUACUCUUUCUAGGAGGUUGUGA |
| miR-301a
mimics | S |
CAGUGCAAUAGUAUUGUCAAAGC |
|
| AS |
GCUUUGACAAUACUAUUGCACUG |
| Mimics NC | S |
UCACAACCUCCUAGAAAGAGUAGA |
|
| AS |
UCUACUCUUUCUAGGAGGUUGUGA |
Observation of cell morphology
After 24 and 48 h of culture under normoxic or
hypoxic conditions, the morphological changes of PC cells (AsPC-1,
BxPC-3, CFPAC-1, MIA PaCa-2 and PANC-1) were examined using an
inverted light microscope (brightfield mode). Images of cells were
captured at ×20 magnification.
Western blotting
AsPC-1, BxPC-3, CFPAC-1, MIA PaCa-2 and PANC-1 cells
were used to assess HIF-1α protein expression under normoxic and
hypoxic conditions. MIA PaCa-2, PANC-1 and BxPC-3 cells were used
to evaluate the expression levels of ACSL4 protein after
overexpression of miR-301a. MIA PaCa-2 and PANC-1 cells were
utilized to assess the expression levels of ACSL4 protein following
the knockdown of miR-301a. Protein extraction and quantification:
Collected cells were lysed in RIPA buffer (cat. no. BL1321A;
Biosharp Life Sciences) supplemented with 1% PMSF (cat. no.
BL1426A; Biosharp Life Sciences) on ice for 15 min. The lysates
were centrifuged at 12,000 × g for 15 min at 4°C and the
supernatants were collected. The protein concentrations were
determined using a BCA assay kit (cat. no. SW201-02; Seven
Biotech). For western blot analysis, 20 µg of protein was resolved
by 7.5% SDS-PAGE using the Colour PAGE Gel Rapid Preparation Kit
(cat. no. PG111; Shanghai Epizyme Biopharmaceutical Technology Co.,
Ltd.; Ipsen Pharma) and transferred to PVDF membranes (cat. no.
IPVH00010; MilliporeSigma). Each membrane was then blocked in 10%
skim milk at room temperature for 2 h. Subsequently, anti-HIF-1α
(1:500; cat. no. 610958; BD Biosciences), anti-ACSL4 (1:2,000; cat.
no. ab155282; Abcam) and anti-β-actin antibodies (1:4,000; cat. no.
60008; Proteintech Group, Inc.) were added and incubated on a
shaker at 4°C overnight. The membranes were incubated with
horseradish peroxidase-labelled mouse (1:5,000; cat. no. PR30012;
Proteintech Group, Inc.)/rabbit (1:5,000; cat. no. PR30011;
Proteintech Group, Inc.) secondary antibodies at room temperature
for 1 h. Lastly, a high-sensitivity ECL detection reagent
(Ultrasensitive ECL Detection Kit; cat. no. PK10003; Proteintech
Group, Inc.) was used for visualization of the protein signals and
imaged with the Chemiluminescence Imaging System (ChemiDoc MP;
Bio-Rad Laboratories, Inc.; Tanon 5200 Multi; Tanon Science and
Technology Co., Ltd.).
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
FreeZol Reagent (cat. no. R711-01; Vazyme Biotech
Co., Ltd.) was used to extract the total RNA from human pancreatic
cancer cell lines (AsPC-1, BxPC-3, CFPAC-1, MIA PaCa-2 and PANC-1)
according to the instructions, and the RNA concentration was
determined and reverse transcribed following the manufacturer's
protocol (Evo M-MLV RT Kit for qPCR; cat. no. AG11707; Hunan
Accurate Biotechnology Co., Ltd.) into complementary DNA (cDNA),
which was then amplified using cDNA as a template. Specific RT-qPCR
experiments were performed to detect miR-301a expression using a
universal high-specificity miRNA quantitative PCR kit (MonAmp™
miRNA Universal Super Specificity qPCR Mix; cat. no. MQ00901; Monad
Biotech Co., Ltd.) according to the manufacturer's protocol. The
following thermocycling conditions were performed: Initial
denaturation at 95°C for 10 min; followed by 40 cycles of
denaturation at 95°C for 10 sec and combined annealing/extension at
60°C for 30 sec (two-step PCR). U6 was used as an internal
reference and the relative expression was calculated using the
2−ΔΔCq method (35). All
RT-qPCR primers were designed against Homo sapiens (human)
genes and synthesized by Anhui General Bioscience. The primer
sequences are listed in Table
III.
 | Table III.RT-qPCR primer sequences. |
Table III.
RT-qPCR primer sequences.
| Gene | Primer
use/direction | Primer sequences
(5′-3′) |
|---|
| miR-301a | RT primer |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAC[GCTTTG] |
| miR-301a | Forward |
GCCAGTGCAATAGTATTGTCAAAG |
| miR-301a | Reverse |
GTGCAGGGTCCGAGGT |
| U6 | Forward |
CGCTTCGGCAGCACATATAC |
| U6 | Reverse |
CAGGGGCCATGCTAATCTT |
Cytotoxicity assay
AsPC-1, BxPC-3, CFPAC-1, MIA PaCa-2 and PANC-1 cells
were used to validate the hypoxic effect on gemcitabine resistance.
The MIA PaCa-2, PANC-1 and BxPC-3 and cells were used to validate
the effect of miR-301a on gemcitabine resistance. After the cells
were transfected with the miR-301a inhibitor for 24 h, they were
digested with trypsin and resuspended in a 96-well plate
(5×103 cells/well). After 6 h of adherence, the GEM drug
(Eli Lilly and Company) at various concentrations (0–50 µΜ; 0 µΜ as
the control group) was added. At 48 h after drug addition, the
original culture medium was discarded and fresh medium containing
10% Cell Counting Kit-8 (CCK-8; cat. no. GK10001; GLPBIO Technology
LLC) was added to each well, followed by incubation at 37°C for 1
h. The absorbance at 450 nm was measured using a microplate reader
to calculate the cell viability. Cell viability=[(As-Ab)/(Ac-Ab)]
×100% [As, experimental wells (containing cells, medium, CCK-8
solution and drug solution); Ac, control wells (containing cells,
medium and CCK-8 solution, but not containing drugs); Ab, blank
wells (containing medium and CCK-8 solution, but not containing
cells and drugs)].
Isolation and identification of
exosomes
The PANC-1 cells were used for the isolation and
identification of exosomes.
Extraction and identification of
exosomes using ultracentrifugation
Supernatants from cells treated under hypoxic and
normoxic conditions were collected, filtered through a 0.22 µm
filter and transferred into 50 ml centrifuge tubes. The tubes were
centrifuged at 300 × g for 10 min at 4°C to remove the cells. The
resulting supernatants were centrifuged at 4°C and 3,000 × g for 30
min to eliminate cellular debris. The clarified supernatants were
transferred into ultracentrifuge tubes and centrifuged at, 10,000 ×
g for 40 min at 4°C. The supernatants from this step were
subsequently transferred into new ultracentrifuge tubes and
centrifuged at 100,000 × g at 4°C for 2 h. After
ultracentrifugation, the supernatants were discarded and the
precipitate was resuspended in 100 ul PBS (cat. no. BL302A;
Biosharp Life Sciences) to obtain exosomes. The isolated exosomes
were stored at −80°C for subsequent experiments.
Transmission electron microscopy (TEM) was used to
observe the morphology and size of the exosomes, with the sample
preparation steps as follows: Exosome pellets were first fixed with
2.5% glutaraldehyde (in 0.1 M PBS, Ph 7.4) at 4°C for 1 h, then
post-fixed with 1% osmium tetroxide at 4°C for 2 h. After
dehydration using gradient concentrations of ethanol (50, 70, 90
and 100%), the samples were embedded in Epon 812 epoxy resin and
incubated at room temperature overnight, before being cut into
70–100 nm ultrathin sections. The sections were then stained with
2% uranyl acetate at room temperature for 10 min, followed by 2%
lead citrate at room temperature for 10 min. Nanoparticle tracking
analysis (NTA) was used to detect the distribution of exosome
sizes.
Exosome uptake experiment
The hypoxia-derived exosome pellet was resuspended
in PBS and labelled with the membrane phospholipid dye PKH67
(green; cat. no. PKH67GL-1KT; MilliporeSigma) according to the
manufacturer's protocol. Exosomes were incubated with 4 µl of PKH67
at room temperature for 5 min and the reaction was terminated by
adding 10% FBS. Unbound dye was removed by ultracentrifugation at
100,000 × g at 4°C for 2 h and labelled exosomes were resuspended
in serum-free DMEM. Normoxic PC cells were treated with
PKH67-labelled hypoxic exosomes for 12 h at 37°C. Following
co-culture, the cells were fixed at room temperature and stained
with DAPI (cat. no. C1002; Beyotime Institute of Biotechnology) at
room temperature for 5 min to visualize the cell nuclei. Images of
the stained samples were captured under a laser confocal
microscope.
Bioinformatics analysis
TargetScan (targetscan.org/vert_80/) and starBase
(rnasysu.com/encori/) databases were used to predict the binding
sites between miRNAs and the target genes. TargetScan relies on
conserved seed matches in the 3′-UTR of genes annotated in RefSeq
and University of California, Santa Cruz genome alignments.
StarBase integrates 2,725 cross-linking and immunoprecipitation
(CLIP)-seq datasets [e.g., photoactivatable
ribonucleoside-enhanced-CLIP, a modified CLIP technique that uses
photoactivatable nucleosides to improve the accuracy of mapping
RNA-protein interaction sites; and high-throughput sequencing-CLIP,
an early CLIP derivative that combines cross-linking
immunoprecipitation with high-throughput sequencing to identify
genome-wide RNA targets of RNA-binding proteins] and 100
degradome-seq datasets to prioritize experimentally supported
miRNA-target interactions.
Statistical analysis
GraphPad Prism (version 9.5.0; Dotmatics) was used
for statistical analysis and plotting. All the experiments were
repeated three times. The quantitative data that conformed to a
normal distribution are expressed as the means ± SE. An unpaired
t-test was used for comparisons between two groups. For >2
groups one-way ANOVA was conducted first, followed by two post-hoc
tests based on comparison goals: Tukey's honestly significant
difference test for pairwise comparisons among all groups (to
detect differences between every pair of groups), and Dunnett's
test for pairwise comparisons between each experimental group and
the control group (to focus on control-centred differences and
reduce type I errors). P<0.05 was considered to indicate a
statistically significant difference.
Results
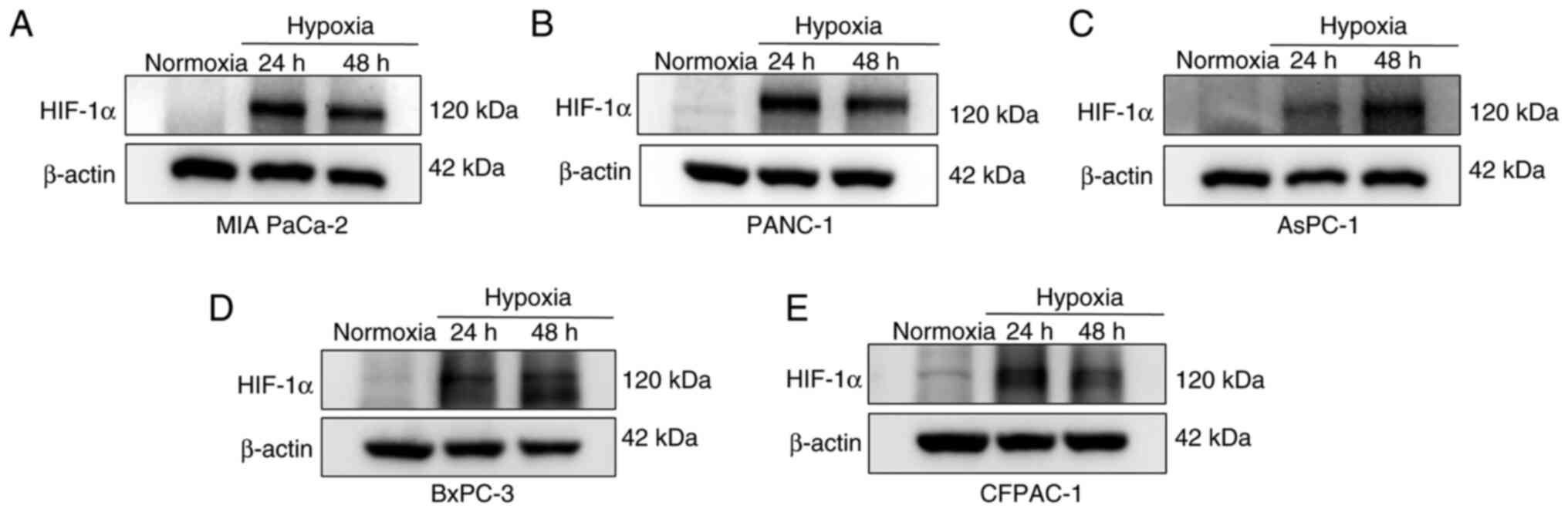
Hypoxia induces a substantial
accumulation of HIF-1α in PC cells
A physically hypoxic environment was used to
establish a hypoxic model. PC cells (AsPC-1, BxPC-3, CFPAC-1, MIA
PaCa-2 and PANC-1) were cultured under hypoxic conditions for 24
and 48 h, with normoxic conditions serving as the baseline.
Detection of basal HIF-1α under normoxia serves two key purposes:
i) Baseline control to establish the natural expression level of
HIF-1α in oxygen-rich conditions, as even low, non-inducible HIF-1α
levels can occur in certain cell types (such as HCT116, human
colorectal cancer cells and CFPAC-1 cells) under normoxia (36); and ii) hypoxia model validation, by
comparing normoxic and hypoxic samples, the present study confirmed
that observed HIF-1α upregulation was hypoxia-induced and not an
experimental artifact. Western blotting was used to detect the
expression levels of HIF-1α protein in PC cells under normoxic and
hypoxic conditions. Compared with the results under normoxia, there
was a notable accumulation of HIF-1α protein in the PC cell lines
after 24 and 48 h under hypoxic conditions (Fig. 1). Notably, HIF-1α accumulation
exhibited notable cell line-specific dynamics: BxPC-3, CFPAC-1, MIA
PaCa-2 and PANC-1 cells displayed marked HIF-1α induction at 24 h,
with levels plateauing or decreasing marginally by 48 h. AsPC-1
cells represented an outlier, with HIF-1α expression increasing
from 24 to 48 h.
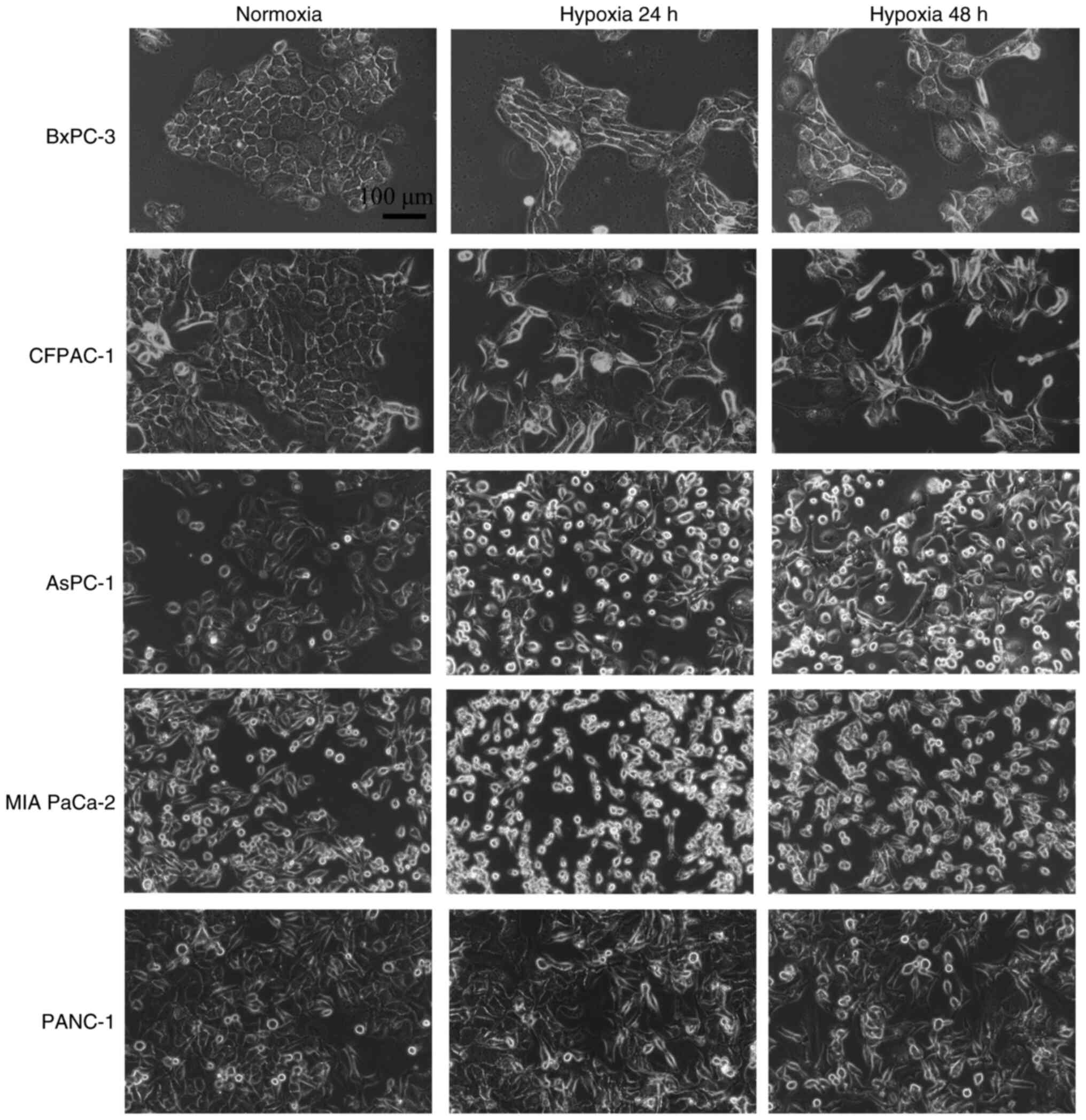
Morphological changes in PC cells
under hypoxic conditions
PC cells (AsPC-1, BxPC-3, CFPAC-1, MIA PaCa-2 and
PANC-1) were cultured under normoxic conditions or hypoxic
conditions for 24 and 48 h and their morphologies were examined
under an inverted microscope. Compared with the normoxic state
observations, the BxPC-3 and CFPAC-1 cells under hypoxic conditions
displayed notable morphological changes characterized by a shift
from an epithelial morphology (oval shape with indistinct cell
boundaries) to a mesenchymal morphology (a spindle-shaped
morphology with well-defined cell margins). By contrast, AsPC-1,
MIA PaCa-2 and PANC-1 cells presented minimal morphological
alterations under hypoxia. While AsPC-1 and MIA PaCa-2 cells
occasionally demonstrated subtle cellular rounding or cytoplasmic
vacuolization at 48 h, potentially reflecting hypoxic stress or
early apoptotic changes, these changes did not represent a fully
mesenchymal phenotype and lacked the elongated, spindle-shaped
morphology observed in BxPC-3 and CFPAC-1 cells (Fig. 2).
Hypoxia upregulates the expression
levels of miR-301a and enhances GEM resistance in PC cells
The expression level of miR-301 was detected through
RT-qPCR experiments, which revealed that miR-301a in PC cells under
hypoxic conditions was significantly upregulated compared with that
in the normoxic group (Fig. 3A-E).
Notably, MIA PaCa-2 and BxPC-3 cells presented a unique temporal
pattern of miR-301a expression. Under hypoxia, the miR-301a levels
significantly increased at 24 h [1.4-fold in MIA PaCa-2 cells
(P<0.01) and 1.8-fold in BxPC-3 cells (P<0.001; Fig. 3A and D). However, by 48 h, the
expression had declined to baseline levels.
![Hypoxia upregulates the expression
levels of miR-301a and enhances GEM resistance in PC cells. (A-E)
Relative expression levels of miR-301a in PC cell lines [(A) MIA
PaCa-2, (B) PANC-1, (C) AsPC-1, (D) BxPC-3 and (E) CFPAC-1] were
detected using RT-qPCR following culture under normoxia conditions
and hypoxic conditions for 24 and 48 h. (F-J) Cell viability of PC
cells [(F) MIA PaCa-2, (G) PANC-1, (H) BxPC-3, (I) AsPC-1 and (J)
CFPAC-1] in both hypoxic and normoxic treatment groups was measured
using CCK-8 cytotoxicity assays upon exposure to various
concentrations of GEM. *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001. NO, normoxic; HO, hypoxic; PC, pancreatic cancer;
miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR;
CCK-8, Cell Counting Kit-8; GEM, gemcitabine; CFPAC-1, cystic
fibrosis pancreatic adenocarcinoma; AsPC-1, ascites pancreatic
cancer 1; MIA PaCa-2, malignant inflammatory adenocarcinoma
pancreatic carcinoma-2.](/article_images/ol/30/6/ol-30-06-15352-g02.jpg) | Figure 3.Hypoxia upregulates the expression
levels of miR-301a and enhances GEM resistance in PC cells. (A-E)
Relative expression levels of miR-301a in PC cell lines [(A) MIA
PaCa-2, (B) PANC-1, (C) AsPC-1, (D) BxPC-3 and (E) CFPAC-1] were
detected using RT-qPCR following culture under normoxia conditions
and hypoxic conditions for 24 and 48 h. (F-J) Cell viability of PC
cells [(F) MIA PaCa-2, (G) PANC-1, (H) BxPC-3, (I) AsPC-1 and (J)
CFPAC-1] in both hypoxic and normoxic treatment groups was measured
using CCK-8 cytotoxicity assays upon exposure to various
concentrations of GEM. *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001. NO, normoxic; HO, hypoxic; PC, pancreatic cancer;
miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR;
CCK-8, Cell Counting Kit-8; GEM, gemcitabine; CFPAC-1, cystic
fibrosis pancreatic adenocarcinoma; AsPC-1, ascites pancreatic
cancer 1; MIA PaCa-2, malignant inflammatory adenocarcinoma
pancreatic carcinoma-2. |
CCK-8 cytotoxicity assays were employed to measure
the cell viability in both the hypoxic and normoxic treatment
groups following their exposure to various concentrations of GEM.
The results (Fig. 3F-J) revealed
that the cell viability of the hypoxic treatment group was
significantly greater compared with that of the normoxic group at
different concentrations (P<0.05).
Hypoxia increases the number of
exosomes and facilitates their transfer to normoxic cells
After the hypoxia model was established, PANC-1
cells were selected for exosome experiments due to their robust
miR-301a upregulation under hypoxia. As shown in Fig. 3B, PANC-1 cells exhibited a 2.18-fold
increase in miR-301a expression at 48 h compared with normoxia
(P<0.0001), which makes PANC-1 cells ideal for modelling
exosome-mediated miR-301a transfer. Equal numbers of PANC-1 cells
were cultured under normoxic and hypoxic conditions for 48 h.
Exosomes from the supernatants of normoxic and hypoxic PANC-1 cells
were collected and identified. TEM was used to observe the
morphology of the isolated exosomes, which revealed that both
normoxic and hypoxic PANC-1-derived exosomes exhibited a
characteristic cup-shaped double-layered membrane structure
(Fig. 4A). NTA was used to analyse
the size distribution of the exosomes and the findings revealed
that the concentration of exosome particles in the ‘Hypoxia-Exos’
group was significantly greater compared with that in the
‘Normoxia-Exos’ group (P<0.0001) and that this secretion could
be inhibited (P<0.0001) by the exosome inhibitor GW4869
(Fig. 4B and C).
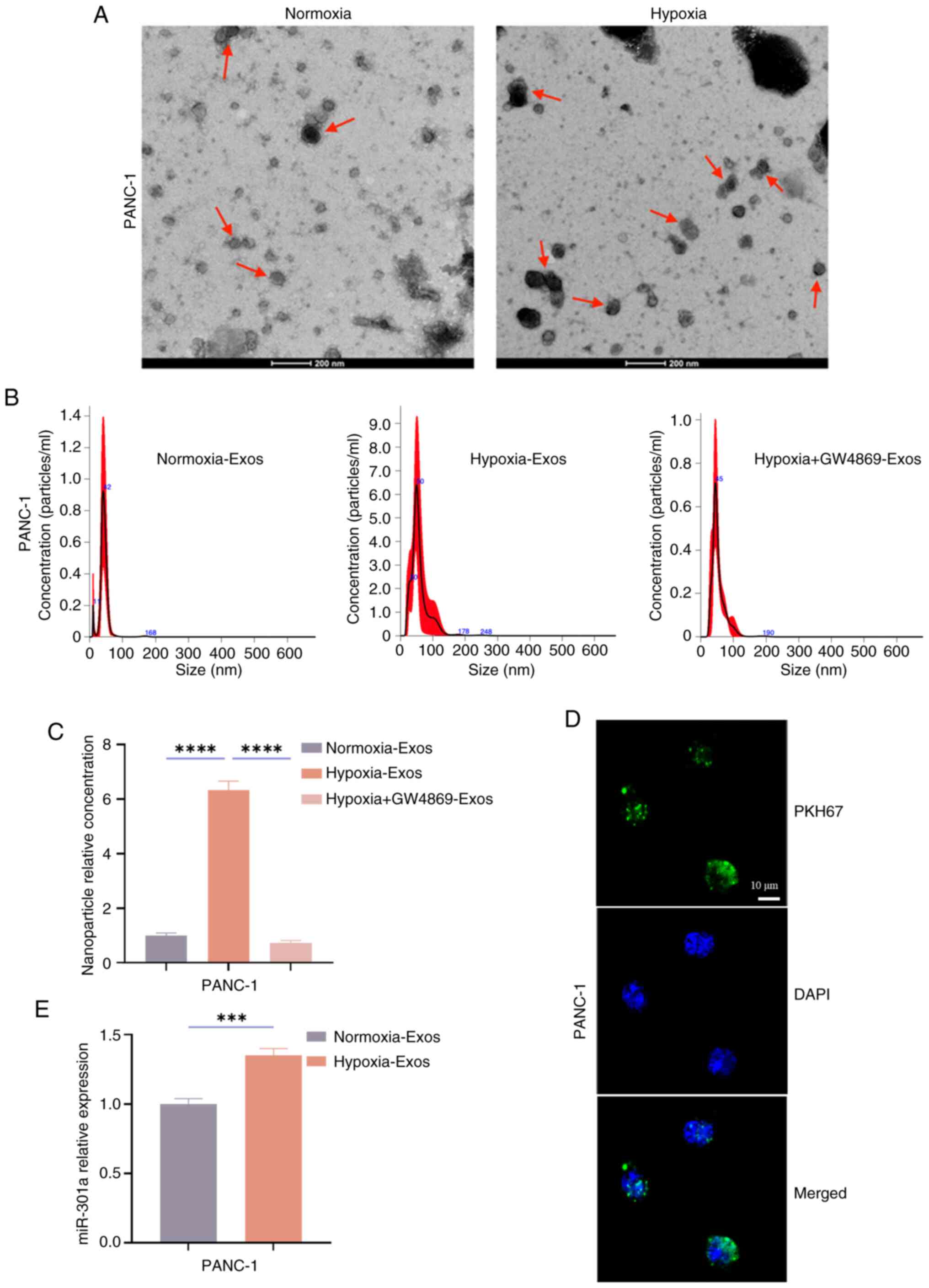 | Figure 4.Hypoxia induces an increase in
exosomes and facilitates their transfer to normoxic cells. (A) The
exosomes from PANC-1 cells cultured under normoxic and hypoxia
condition for 48 h were collected and their morphology was observed
by TEM; red arrows indicate the exosomes (scale bar, 200 nm). (B
and C) PANC-1 cells were cultured under normoxic and hypoxia
condition for 48 h and exosome inhibitor GW4869 was added to
hypoxia culture, exosomes were collected and NTA was used to
analyse the relative particle concentration in the Normoxia-Exos,
Hypoxia-Exos and Hypoxia + GW4869-Exos group. (D) Hypoxic
PANC-1-derived exosomes were collected and labelled with the
membrane phospholipid dye PKH67. After co-culture with normoxic
PANC-1 cells for 12 h, laser confocal microscopy was employed to
observe the endocytosis of labelled exosomes by normoxic PANC-1
cells (scale bar, 10 µm). (E) Following co-culturing the collected
normoxic and hypoxic-derived exosomes with normoxic PANC-1 cells
for 24 h, RT-qPCR was performed to detect the relative expression
level of the miR-301a. ***P<0.001 and ****P<0.0001. TEM,
transmission electron microscopy; NTA, nanoparticle tracking
analysis; RT-qPCR, reverse transcription-quantitative PCR; miR,
microRNA. |
To investigate intercellular exosome transfer,
hypoxic PANC-1-derived exosomes were labelled with the membrane dye
PKH67 and co-cultured with normoxic PANC-1 recipient cells for 12
h. Laser confocal microscopy demonstrated that hypoxic-derived
exosomes could be endocytosed by normoxic PANC-1 cells (Fig. 4D). After the collected normoxic and
hypoxic-derived exosomes were co-cultured with normoxic PANC-1
cells for 24 h, RT-qPCR was performed to detect the relative
expression level of the miR-301a gene. The results revealed that
the expression levels of miR-301a in the hypoxic Exos-treated group
was significantly greater compared with that in the Normoxic
Exos-treated group (P<0.001; Fig.
4E). These results indicated that hypoxia induces an increase
in the number of exosomes and facilitates the transfer of a large
amount of miR-301a to normoxic cells.
Effect of miR-301a on GEM resistance
in PC cells under normoxia
To explore the effect of hypoxia-derived exosomal
miR-301a on normoxic PC cells, a miR-301a inhibitor was transfected
into PC cells. Due to their high basal expression of miR-301a, MIA
PaCa-2, PANC-1 and BxPC-3 cells were selected for these experiments
and RT-qPCR was used to verify the knockdown efficiency of
miR-301a. The results demonstrated that the knockdown efficiency of
miR-301a in the three cell lines was >70% (P<0.05; Fig. 5A). Subsequently, CCK-8 cytotoxicity
assays were performed to detect the resistance of PC cells to GEM
in both the inhibitor NC group and the miR-301a inhibitor group.
Notably, the GEM concentration gradients in Fig. 5B-D were tailored to the drug
sensitivity of each cell line, as determined by preliminary
dose-response experiments. The results indicated that, when the
cells were treated with different drug concentrations, the
viability of the cells in the miR-301a inhibitor group was lower
compared with that of the cells in the inhibitor NC group
(P<0.05; Fig. 5B-D).
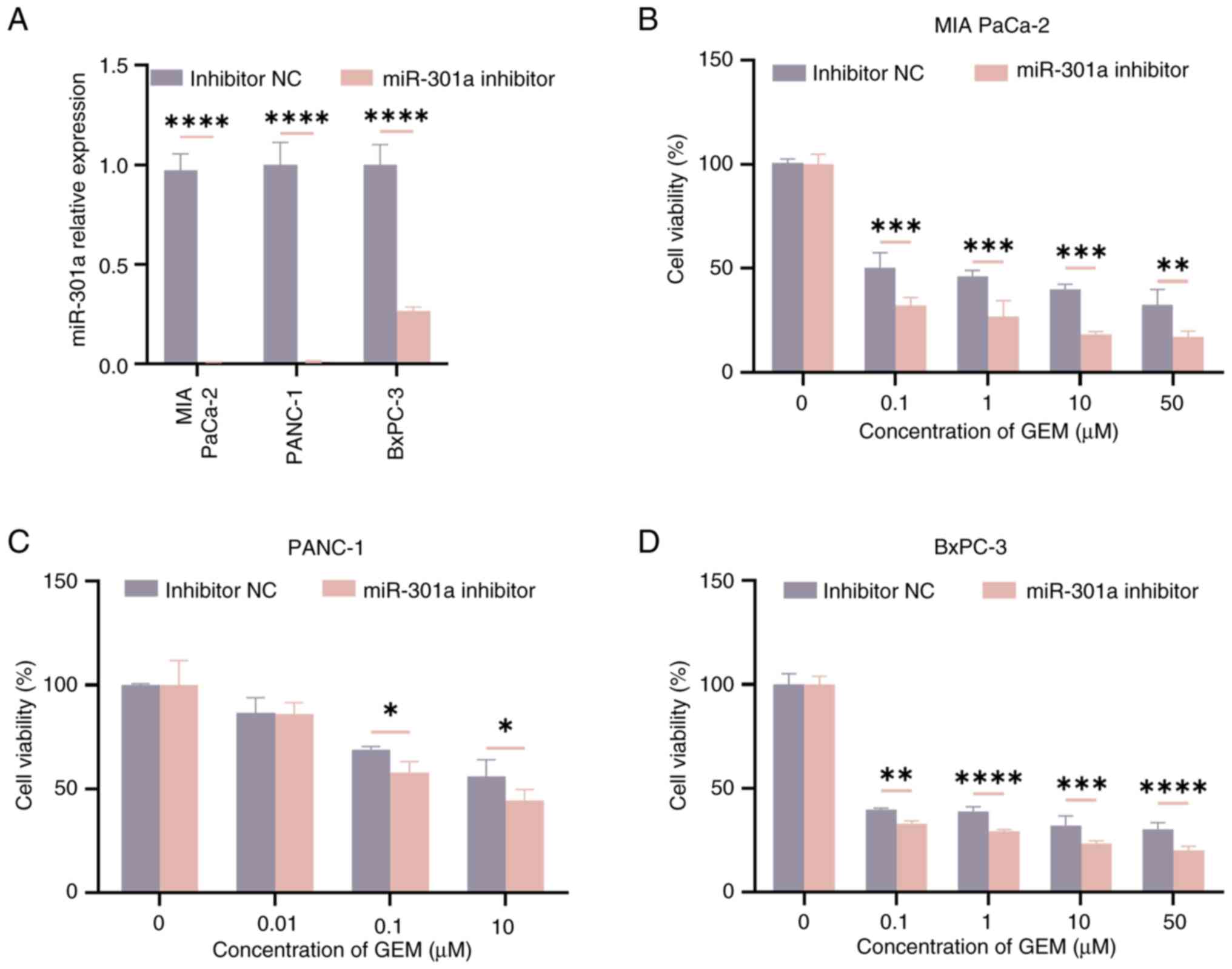 | Figure 5.Regulatory mechanism of
hypoxia-derived exosome miR-301a on GEM resistance in normoxic PC
cells. (A) PC cells were transfected with inhibitor NC and miR-301a
inhibitor for 24 h and then the relative expression level of
miR-301a was detected by RT-qPCR. (B) MIA PaCa-2, (C) PANC-1, and
(D) BxPC-3 cells were treated with a gradient concentration of GEM
for 48 h. CCK-8 cytotoxicity assays were conducted to detect cell
viability in both the inhibitor NC group and the miR-301a inhibitor
group. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001.
GEM, gemcitabine; miR, microRNA; PC, pancreatic cancer; RT-qPCR,
reverse transcription-quantitative PCR; NC, negative control;
CCK-8, Cell Counting Kit-8. |
Regulatory mechanism of
hypoxia-derived exosomal miR-301a on GEM resistance in normoxic PC
cells
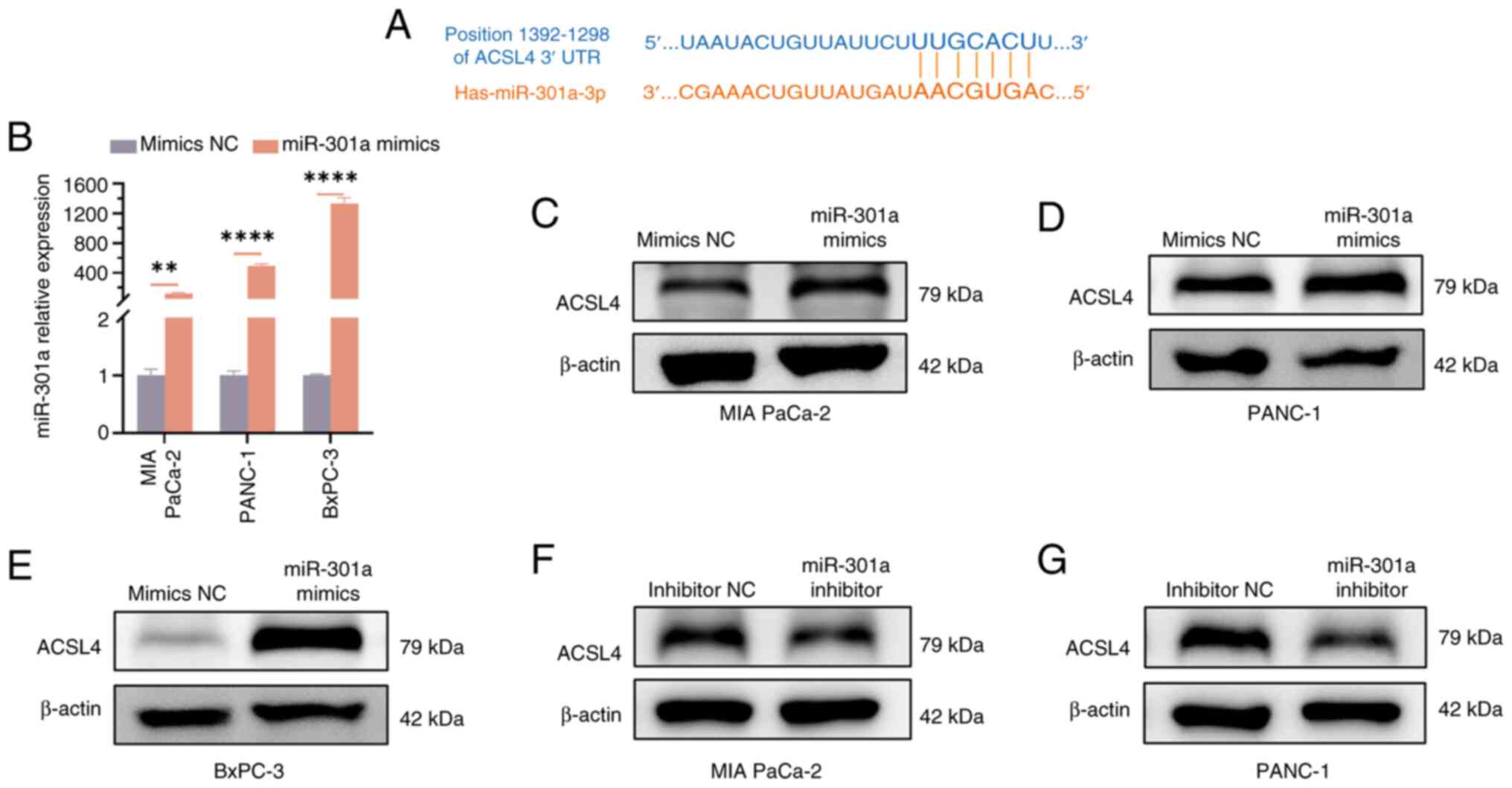
It has been reported that ACSL4 is associated with
GEM resistance (33). Further
bioinformatics analysis revealed a potential binding site between
miR-301a and ACSL4 (Fig. 6A). To
determine whether GEM resistance in PC cells is enhanced by
miR-301a through the regulation of ACSL4 expression, miR-301a
mimics were first transfected into PC cells and the effect of
miR-301a overexpression was verified using RT-qPCR. The results
revealed that the expression levels of miR-301a were significantly
increased in all three studied PC cell lines (P<0.05; Fig. 6B). Furthermore, western blotting
results indicated that when the expression level of miR-301a was
upregulated, the protein expression level of ACSL4 also increased
significantly (Fig. 6C-E).
Conversely, when miR-301a expression was inhibited, the ACSL4
protein level decreased markedly (Fig.
6F and G). These results suggested that hypoxia-derived
exosomal miR-301a further influences the GEM resistance of PC cells
by regulating the expression level of ACSL4 under normoxic
conditions.
Discussion
PC is a highly malignant tumour of the digestive
system, with persistently high incidence and mortality rates. As a
first-line chemotherapy drug for PC, GEM has been widely used in
clinical practice, yet the emergence of drug resistance has notably
limited its efficacy (8,37). In recent years, the role of the TME,
especially the hypoxic microenvironment, in tumour chemotherapy
resistance has attracted increasing attention (38). Previous studies have reported that
the hypoxic microenvironment increases glucose metabolism and
pyrimidine synthesis in tumour cells by inducing the upregulation
of HIF-1α, thereby leading to the resistance of PC cells to GEM
(39). Yoo et al (40) reported that hypoxia can also
regulate metabolic reprogramming through HIF-2α-mediated generation
of SLC1A5 variants, further enhancing the resistance of PC cells to
GEM. In the present study, a hypoxic model for PC cells was
established using physical hypoxia and the effectiveness of the
model was validated by detecting the level of HIF-1α protein.
Compared with that under normoxia, the protein level of HIF-1α was
markedly elevated after hypoxia treatment for 24 and 48 h.
Additionally, morphological changes in PC cells were observed under
hypoxic conditions. Further evaluation of cytotoxicity through
CCK-8 assays revealed that hypoxia treatment significantly reduced
the sensitivity of PC cells to GEM, which was consistent with
previously reported findings that hypoxia promoted chemotherapy
resistance in tumours (41).
Exosomes can carry various bioactive molecules, such
as proteins, lipids and nucleic acids (for example, mRNAs, miRNAs),
and participate in intercellular communication and material
exchange. In various disease models, hypoxic stimulation not only
leads to an increase in the secretion of exosomes but also causes
alterations in their contents, which further affects the biological
effects on recipient cells. For example, in pancreatic
neuroendocrine tumours (17) and
chondrosarcomas (18,42), the hypoxic microenvironment induces
the production of several exosomes (such as carcinoembryonic
antigen-related cell adhesion molecule 5, long non-coding RNA RAMP2
antisense RNA 1) by cells, resulting in M2 polarization of
tumour-associated macrophages, which in turn promotes tumour
invasion and metastasis. Furthermore, Liu et al (19) reported that the number of exosomes
derived from hypoxic bone marrow mesenchymal stem cells increased
and could enhance fracture healing.
Exosomes released by tumour cells serve key roles in
tumour progression and drug resistance, particularly through their
small RNA molecules, such as miRNAs, which can regulate the
expression levels of target genes and influence the biological
characteristics of cells. Victor Ambros and Gary Ruvkun were
awarded the Nobel Prize in 2024 for their discovery of miRNAs and
their role in post-transcriptional gene regulation (43), which underscores the importance of
miRNAs in intercellular communication and gene expression
regulation and provides a theoretical foundation for the present
study experiments. Zhao et al (44) demonstrated that exosomal miR-934 can
induce M2 polarization of macrophages, thereby promoting liver
metastasis of colorectal cancer. Furthermore, exosomal miR-522
secreted by cancer-associated fibroblasts in the tumour
microenvironment is involved in chemotherapy resistance in gastric
cancer (45). Through a literature
review, it was found that miR-301a is differentially expressed in a
variety of malignant tumours and affects tumorigenesis and
development through different regulatory mechanisms. A previous
study has demonstrated that miR-301a is abnormally upregulated in
breast cancer and further promotes breast cancer cell
proliferation, metastasis, and cell cycle progression through the
cytoplasmic polyadenylation element binding protein 1/sirtuin
1/Sox2 axis, accelerating the malignant progression of breast
cancer (21). Furthermore, miR-301a
can promote the proliferation of prostate cancer cells by
inhibiting the expression of p21 and SMAD4 (46). Qi et al (47) demonstrated that miR-301a is highly
expressed in PC. Rather than verifying or further exploring the
differential expression and regulatory mechanisms of miR-301a
itself, the present study focused on the impact and underlying
mechanisms of exosomal miR-301a on GEM drug resistance in PC under
hypoxic conditions, which aimed to provide a novel strategy for PC
treatment. These results indicated that the hypoxic
microenvironment markedly enhanced the resistance of PC cells to
GEM, accompanied by a marked increase in exosome secretion and
notable upregulation of miR-301a expression. Notably, MIA PaCa-2
and BxPC-3 cells presented a unique temporal pattern of miR-301a
expression. Under hypoxia, miR-301a levels increased significantly
at 24 h. However, by 48 h, expression had declined to baseline
levels. A literature review revealed that HIF-2α can regulate the
expression levels of miR-301a in PC cells under hypoxic conditions
(48). Furthermore, compared with
that in the 24 h hypoxic treatment, the protein level of HIF-2α
decreases at 48 h of hypoxic treatment. Therefore, it can be
hypothesized that the upregulation of miR-301a expression in MIA
PaCa-2 and BxPC-3 cell lines at 24 h and its downregulation at 48 h
are due to the regulation of HIF-2α. Our research group previously
reported that miR-301a expression was markedly increased in hypoxic
exosomes (25). Using exosome
co-culture experiments, it was verified that the number of
hypoxia-induced exosomes increased and that these exosomes could be
delivered to normoxic PC cells. To explore the role of exosomal
miR-301a, miR-301a knockdown experiments were conducted and the key
role of miR-301a in the resistance of PC to GEM was confirmed.
Compared with the NC group, miR-301a knockdown markedly reduced the
resistance of normoxic PC cells to GEM. These findings suggested
that the upregulation of miR-301a in hypoxic-derived exosomes is
one of the key mechanisms underlying GEM resistance in normoxic PC.
According to previous studies, miRNAs primarily act by binding to
the 3′-UTR of mRNAs, inducing mRNA degradation or inhibiting their
translation, thereby downregulating the expression of target genes.
In certain cases, they bind to the 5′-UTRs of mRNAs to positively
regulate target genes.
Previous studies have reported that miRNAs can
participate in disease progression by targeting and regulating
ACSL4 expression. For example, miR-23a-3p inhibits ferroptosis in
hepatocellular carcinoma by targeting ACSL4, thereby participating
in the resistance of hepatocellular carcinoma cells to sorafenib
(31). Shi et al (32) reported that miR-20a-5p alleviates
renal ischaemia/reperfusion injury by inhibiting ACSL4 expression.
Other studies have demonstrated that ACSL4 is also involved in PC
resistance to GEM, but the specific mechanism has not been fully
elucidated (33,49). In the present study, using
bioinformatics analysis, potential binding sites between miR-301a
and ACSL4 were identified, which led to the hypothesis that
miR-301a acts by binding to the 3′-UTR of ACSL4 mRNA. However,
validation experiments revealed that miR-301a overexpression
increased ACSL4 protein levels, whereas miR-301a knockdown
decreased ACSL4 expression, findings that contradict the
traditional negative regulatory pattern of miRNAs. It was
speculated that miR-301a might bind to the 5′-UTR of ACSL4 mRNA.,
but after consulting the bioinformatics databases, it was revealed
that there was no binding site for miR-301a on the 5′-UTR of ACSL4
mRNA. Based on these results, the present study proposes that
miR-301a may not directly target ACSL4 but instead it regulates
ACSL4 through an intermediate molecule, which provides a novel
direction for future research. Subsequent studies could focus on
identifying this potential intermediate molecule, with the aim of
uncovering novel therapeutic strategies and targets for PC and
other tumours.
In summary, a key mechanism was revealed in the
present study, whereby hypoxia-induced upregulation of miR-301a is
transferred to normoxic PC cells via exosomes and contributes to
GEM resistance by regulating ACSL4. This finding not only provides
novel insights into the molecular mechanisms underlying GEM
resistance in PC but also offers potential targets for the
development of novel therapeutic strategies against PC in the
future. The specific interaction mechanism between miR-301a and
ACSL4, as well as potential treatment approaches targeting this
axis, remains to be explored in future research.
Notwithstanding the aforementioned findings, the
present study has certain limitations to note. First, the
intermediate molecule mediating miR-301a-induced regulation of
ACSL4 was not identified, leaving the exact regulatory cascade
unresolved. Second, in vivo validation using animal models
(such as orthotopic PC xenografts) is lacking, which limits the
translational relevance of the in vitro results. Finally, no
clinical specimens were analysed to determine the association of
miR-301a/ACSL4 expression with GEM response or patient outcomes,
thus hindering evaluation of their clinical potential as biomarkers
or targets. Addressing these limitations in future work will help
strengthen the clinical applicability of the current findings.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Research Fund of Anhui
Institute of Translational Medicine (grant no. 2022zhyx-C71) and
the Graduate Scientific Research and Practice Innovation Project of
Anhui Medical University (grant no. YJS20230037).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
WQ performed all cell culture, exosome isolation and
molecular biology experiments, including RT-qPCR, western blot and
CCK-8 assays. WQ conducted data analysis, prepared the initial data
summaries and drafted the manuscript. WaT conducted bioinformatics
prediction and analysis on the potential target genes of miR-301a,
and performed subsequent experimental validation using western
blotting, while MY provided key reagents, including PC cell lines
and exosome inhibitors and offered expertise in hypoxia modelling.
WeT and MY participated in data analysis and figure preparation.
WeT made substantial contributions to the analysis and
interpretation of experimental data (specifically verifying the
reliability of exosome isolation efficiency and miR-301a detection
data), revised the key intellectual content of the manuscript, and
reviewed the consistency of experimental descriptions and data
presentation. LZ designed and conducted supplementary experiments
(including CCK analysis for BxPC-3, AsPC-1 and CFPAC-1 cells
treated with GEM under normoxia/hypoxia) to address reviewer
feedback, obtained funding and helped respond to reviewers,
including clarifying methodologies and data interpretation. GL
conceived and designed the present study, supervised all
experimental work and interpreted the results, oversaw the overall
validation of data integrity and the accuracy of research outcomes.
GL revised the manuscript for intellectual content and ensured the
integrity of the data and the accuracy of the findings. WQ and GL
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Kratzer TB, Giaquinto AN, Sung
H and Jemal A: Cancer statistics, 2025. CA Cancer J Clin. 75:10–45.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xiong Q, Zhang Y, Xu Y, Yang Y, Zhang Z,
Zhou Y, Zhang S, Zhou L, Wan X, Yang X, et al: tiRNA-Val-CAC-2
interacts with FUBP1 to promote pancreatic cancer metastasis by
activating c-MYC transcription. Oncogene. 43:1274–1287. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Munigala S, Almaskeen S, Subramaniam DS,
Bandi S, Bowe B, Xian H, Sheth SG, Burroughs TE and Agarwal B:
Acute pancreatitis recurrences augment long-term pancreatic cancer
risk. Am J Gastroenterol. 118:727–737. 2023. View Article : Google Scholar
|
|
5
|
Topal H, Aerts R, Laenen A, Collignon A,
Jaekers J, Geers J and Topal B: Survival after minimally invasive
vs open surgery for pancreatic adenocarcinoma. JAMA Netw Open.
5:e22481472022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Padrón LJ, Maurer DM, O'Hara MH, O'Reilly
EM, Wolff RA, Wainberg ZA, Ko AH, Fisher G, Rahma O, Lyman JP, et
al: Sotigalimab and/or nivolumab with chemotherapy in first-line
metastatic pancreatic cancer: Clinical and immunologic analyses
from the randomized phase 2 PRINCE trial. Nat Med. 28:1167–1177.
2022. View Article : Google Scholar
|
|
7
|
Li K, Tandurella JA, Gai J, Zhu Q, Lim SJ,
Thomas DL II, Xia T, Mo G, Mitchell JT, Montagne J, et al:
Multi-omic analyses of changes in the tumor microenvironment of
pancreatic adenocarcinoma following neoadjuvant treatment with
anti-PD-1 therapy. Cancer Cell. 40:1374–1391. 2022. View Article : Google Scholar
|
|
8
|
Liu Y, Guo X, Xu P, Song Y, Huang J, Chen
X, Zhu W, Hao J and Gao S: Clinical outcomes of second-line
chemotherapy in patients with advanced pancreatic adenocarcinoma: A
real-world study. Cancer Biol Med. 21:799–812. 2024.PubMed/NCBI
|
|
9
|
Beutel AK and Halbrook CJ: Barriers and
opportunities for gemcitabine in pancreatic cancer therapy. Am J
Physiol Cell Physiol. 324:C540–C552. 2023. View Article : Google Scholar
|
|
10
|
Patzak MS, Kari V, Patil S, Hamdan FH,
Goetze RG, Brunner M, Gaedcke J, Kitz J, Jodrell DI, Richards FM,
et al: Cytosolic 5′-nucleotidase 1A is overexpressed in pancreatic
cancer and mediates gemcitabine resistance by reducing
intracellular gemcitabine metabolites. EBioMedicine. 40:394–405.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Neoptolemos JP, Palmer DH, Ghaneh P,
Psarelli EE, Valle JW, Halloran CM, Faluyi O, O'Reilly DA,
Cunningham D, Wadsley J, et al: Comparison of adjuvant gemcitabine
and capecitabine with gemcitabine monotherapy in patients with
resected pancreatic cancer (ESPAC-4): A multicentre, open-label,
randomised, phase 3 trial. Lancet. 389:1011–1024. 2017. View Article : Google Scholar
|
|
12
|
Hosein AN, Brekken RA and Maitra A:
Pancreatic cancer stroma: An update on therapeutic targeting
strategies. Nat Rev Gastroenterol Hepatol. 17:487–505. 2020.
View Article : Google Scholar
|
|
13
|
Wicks EE and Semenza GL: Hypoxia-inducible
factors: Cancer progression and clinical translation. J Clin
Invest. 132:e1598392022. View Article : Google Scholar
|
|
14
|
Mao Y, Wang J, Wang Y, Fu Z, Dong L and
Liu J: Hypoxia induced exosomal Circ-ZNF609 promotes pre-metastatic
niche formation and cancer progression via miR-150-5p/VEGFA and
HuR/ZO-1 axes in esophageal squamous cell carcinoma. Cell Death
Discov. 10:1332024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song H, Qiu Z, Wang Y, Xi C, Zhang G, Sun
Z, Luo Q and Shen C: HIF-1α/YAP signaling rewrites glucose/iodine
metabolism program to promote papillary thyroid cancer progression.
Int J Biol Sci. 19:225–241. 2023. View Article : Google Scholar
|
|
16
|
Zhang Q, Xiong L, Wei T, Liu Q, Yan L,
Chen J, Dai L, Shi L, Zhang W, Yang J, et al: Hypoxia-responsive
PPARGC1A/BAMBI/ACSL5 axis promotes progression and resistance to
lenvatinib in hepatocellular carcinoma. Oncogene. 42:1509–1523.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ye M, Lu F, Gu D, Xue B, Xu L, Hu C, Chen
J, Yu P, Zheng H, Gao Y, et al: Hypoxia exosome derived CEACAM5
promotes tumor-associated macrophages M2 polarization to accelerate
pancreatic neuroendocrine tumors metastasis via MMP9. FASEB J.
38:e237622024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hou SM, Lin CY, Fong YC and Tang CH:
Hypoxia-regulated exosomes mediate M2 macrophage polarization and
promote metastasis in chondrosarcoma. Aging (Albany NY).
15:13163–13175. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu W, Li L, Rong Y, Qian D, Chen J, Zhou
Z, Luo Y, Jiang D, Cheng L, Zhao S, et al: Hypoxic mesenchymal stem
cell-derived exosomes promote bone fracture healing by the transfer
of miR-126. Acta Biomater. 103:196–212. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang M, Zheng Y, Hao Q, Mao G, Dai Z, Zhai
Z, Lin S, Liang B, Kang H and Ma X: Hypoxic BMSC-derived exosomal
miR-210-3p promotes progression of triple-negative breast cancer
cells via NFIX-Wnt/β-catenin signaling axis. J Transl Med.
23:392025. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jia Y, Zhao J, Yang J, Shao J and Cai Z:
miR-301 regulates the SIRT1/SOX2 pathway via CPEB1 in the breast
cancer progression. Mol Ther Oncolytics. 22:13–26. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xia X, Wang S, Ni B, Xing S, Cao H, Zhang
Z, Yu F, Zhao E and Zhao G: Hypoxic gastric cancer-derived exosomes
promote progression and metastasis via MiR-301a-3p/PHD3/HIF-1α
positive feedback loop. Oncogene. 39:6231–6244. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alves Â, Ferreira M, Eiras M, Lima L,
Medeiros R, Teixeira AL and Dias F: Exosome-derived
hsa-miR-200c-3p, hsa-miR-25-3p and hsa-miR-301a-3p as potential
biomarkers and therapeutic targets for restoration of PTEN
expression in clear cell renal cell carcinoma. Int J Biol Macromol.
302:1406072025. View Article : Google Scholar
|
|
24
|
Zhong M, Huang Z, Wang L, Lin Z, Cao Z, Li
X, Zhang F, Wang H, Li Y and Ma X: Malignant transformation of
human bronchial epithelial cells induced by arsenic through
STAT3/miR-301a/SMAD4 loop. Sci Rep. 8:132912018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang X, Luo G, Zhang K, Cao J, Huang C,
Jiang T, Liu B, Su L and Qiu Z: Hypoxic tumor-derived exosomal
miR-301a mediates M2 macrophage polarization via PTEN/PI3K gamma to
promote pancreatic cancer metastasis. Cancer Res. 78:4586–4598.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Luo G, Xia X, Wang X, Zhang K, Cao J,
Jiang T, Zhao Q and Qiu Z: miR-301a plays a pivotal role in
hypoxia-induced gemcitabine resistance in pancreatic cancer. Exp
Cell Res. 369:120–128. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ding K, Liu C, Li L, Yang M, Jiang N, Luo
S and Sun L: Acyl-CoA synthase ACSL4: An essential target in
ferroptosis and fatty acid metabolism. Chin Med J (Engl).
136:2521–2537. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qiu Y, Wang X, Sun Y, Jin T, Tang R, Zhou
X, Xu M, Gan Y, Wang R, Luo H, et al: ACSL4-mediated membrane
phospholipid remodeling induces integrin β1 activation to
facilitate triple-negative breast cancer metastasis. Cancer Res.
84:1856–1871. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen J, Ding C, Chen Y, Hu W, Yu C, Peng
C, Feng X, Cheng Q, Wu W, Lu Y, et al: ACSL4 reprograms fatty acid
metabolism in hepatocellular carcinoma via c-Myc/SREBP1 pathway.
Cancer Lett. 502:154–165. 2021. View Article : Google Scholar
|
|
30
|
Grube J, Woitok MM, Mohs A, Erschfeld S,
Lynen C, Trautwein C and Otto T: ACSL4-dependent ferroptosis does
not represent a tumor-suppressive mechanism but ACSL4 rather
promotes liver cancer progression. Cell Death Dis. 13:7042022.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu Y, Chan YT, Tan HY, Zhang C, Guo W, Xu
Y, Sharma R, Chen ZS, Zheng YC, Wang N and Feng Y: Epigenetic
regulation of ferroptosis via ETS1/miR-23a-3p/ACSL4 axis mediates
sorafenib resistance in human hepatocellular carcinoma. J Exp Clin
Cancer Res. 41:32022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shi L, Song Z, Li Y, Huang J, Zhao F, Luo
Y, Wang J, Deng F, Shadekejiang H, Zhang M, et al: MiR-20a-5p
alleviates kidney ischemia/reperfusion injury by targeting
ACSL4-dependent ferroptosis. Am J Transplant. 23:11–25. 2023.
View Article : Google Scholar
|
|
33
|
Qi R, Bai Y, Li K, Liu N, Xu Y, Dal E,
Wang Y, Lin R, Wang H, Liu Z, et al: Cancer-associated fibroblasts
suppress ferroptosis and induce gemcitabine resistance in
pancreatic cancer cells by secreting exosome-derived
ACSL4-targeting miRNAs. Drug Resist Updat. 68:1009602023.
View Article : Google Scholar
|
|
34
|
Deer EL, González-Hernández J, Coursen JD,
Shea JE, Ngatia J, Scaife CL, Firpo MA and Mulvihill SJ: Phenotype
and genotype of pancreatic cancer cell lines. Pancreas. 39:425–435.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li W, Zhou C, Yu L, Hou Z, Liu H, Kong L,
Xu Y, He J, Lan J, Ou Q, et al: Tumor-derived lactate promotes
resistance to bevacizumab treatment by facilitating autophagy
enhancer protein RUBCNL expression through histone H3 lysine 18
lactylation (H3K18la) in colorectal cancer. Autophagy. 20:114–130.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Siegel RL, Giaquinto AN and Jemal A:
Cancer statistics, 2024. CA Cancer J Clin. 74:12–49. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang L, Zhao S, Liu X, Zhang Y, Zhao S,
Fang X and Zhang J: Hypoxic cancer-associated fibroblast exosomal
circSTAT3 drives triple negative breast cancer stemness via
miR-671-5p/NOTCH1 signaling. J Transl Med. 23:8142025. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shukla SK, Purohit V, Mehla K, Gunda V,
Chaika NV, Vernucci E, King RJ, Abrego J, Goode GD, Dasgupta A, et
al: MUC1 and HIF-1alpha signaling crosstalk induces anabolic
glucose metabolism to impart gemcitabine resistance to pancreatic
cancer. Cancer Cell. 32:71–87. 2017. View Article : Google Scholar
|
|
40
|
Yoo HC, Park SJ, Nam M, Kang J, Kim K, Yeo
JH, Kim JK, Heo Y, Lee HS, Lee MY, et al: A variant of SLC1A5 is a
mitochondrial glutamine transporter for metabolic reprogramming in
cancer cells. Cell Metab. 31:267–283. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ding J, Xie Y, Liu Z, Zhang Z, Ni B, Yan
J, Zhou T and Hao J: Hypoxic and acidic tumor
microenvironment-driven AVL9 promotes chemoresistance of pancreatic
ductal adenocarcinoma via the AVL9-IκBα-SKP1 complex.
Gastroenterology. 168:539–555. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cheng C, Zhang Z, Cheng F and Shao Z:
Exosomal lncRNA RAMP2-AS1 derived from chondrosarcoma cells
promotes angiogenesis through miR-2355-5p/VEGFR2 axis. Onco Targets
Ther. 13:3291–3301. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Organization NP, . Nobel Prize in
Physiology or Medicine. 2024.https://www.nobelprize.org/prizes/medicine/2024/press–release/January
5–2025
|
|
44
|
Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu
Y, Zhang Z, Cai S, Xu Y, Li X, et al: Tumor-derived exosomal
miR-934 induces macrophage M2 polarization to promote liver
metastasis of colorectal cancer. J Hematol Oncol. 13:1562020.
View Article : Google Scholar
|
|
45
|
Zhang H, Deng T, Liu R, Ning T, Yang H,
Liu D, Zhang Q, Lin D, Ge S, Bai M, et al: CAF secreted miR-522
suppresses ferroptosis and promotes acquired chemo-resistance in
gastric cancer. Mol Cancer. 19:432020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li X, Li J, Cai Y, Peng S, Wang J, Xiao Z,
Wang Y, Tao Y, Li J, Leng Q, et al: Hyperglycaemia-induced miR-301a
promotes cell proliferation by repressing p21 and Smad4 in prostate
cancer. Cancer Lett. 418:211–220. 2018. View Article : Google Scholar
|
|
47
|
Qi B, Wang Y, Zhu X, Gong Y, Jin J, Wu H,
Man X, Liu F, Yao W and Gao J: miR-301a-mediated crosstalk between
the Hedgehog and HIPPO/YAP signaling pathways promotes pancreatic
cancer. Cancer Biol Ther. 26:24577612025. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang KD, Hu B, Cen G, Yang YH, Chen WW,
Guo ZY, Wang XF, Zhao Q and Qiu ZJ: MiR-301a transcriptionally
activated by HIF-2α promotes hypoxia-induced epithelial-mesenchymal
transition by targeting TP63 in pancreatic cancer. World J
Gastroenterol. 26:2349–2373. 2020. View Article : Google Scholar
|
|
49
|
Li J, Ma C, Cao P, Guo W, Wang P, Yang Y,
Ding B, Yin F, Li Z, Wang Y, et al: A CD147-targeted small-molecule
inhibitor potentiates gemcitabine efficacy by triggering
ferroptosis in pancreatic ductal adenocarcinoma. Cell Rep Med.
6:1022922025. View Article : Google Scholar : PubMed/NCBI
|















![Hypoxia upregulates the expression
levels of miR-301a and enhances GEM resistance in PC cells. (A-E)
Relative expression levels of miR-301a in PC cell lines [(A) MIA
PaCa-2, (B) PANC-1, (C) AsPC-1, (D) BxPC-3 and (E) CFPAC-1] were
detected using RT-qPCR following culture under normoxia conditions
and hypoxic conditions for 24 and 48 h. (F-J) Cell viability of PC
cells [(F) MIA PaCa-2, (G) PANC-1, (H) BxPC-3, (I) AsPC-1 and (J)
CFPAC-1] in both hypoxic and normoxic treatment groups was measured
using CCK-8 cytotoxicity assays upon exposure to various
concentrations of GEM. *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001. NO, normoxic; HO, hypoxic; PC, pancreatic cancer;
miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR;
CCK-8, Cell Counting Kit-8; GEM, gemcitabine; CFPAC-1, cystic
fibrosis pancreatic adenocarcinoma; AsPC-1, ascites pancreatic
cancer 1; MIA PaCa-2, malignant inflammatory adenocarcinoma
pancreatic carcinoma-2.](/article_images/ol/30/6/ol-30-06-15352-g02.jpg)