Introduction
Breast cancer is one of the most prevalent malignant
tumors among women and poses a significant threat to both health
and survival (1). In recent
decades, the burden of breast cancer has intensified in developing
countries due to rising incidence rates and persistently high
mortality (2). By 2040, it is
projected that population growth and aging alone will contribute to
>3 million new breast cancer cases and 1 million related deaths
annually (3). Current treatment
modalities include surgery, targeted therapies, immunotherapy,
radiotherapy and chemotherapy (4).
Among the chemotherapeutic agents, paclitaxel (PTX) plays a crucial
role in breast cancer management (5). However, the development of PTX
resistance during treatment remains a major clinical challenge,
often leading to therapeutic failure (6). Understanding the mechanisms underlying
PTX resistance and identifying effective reversal strategies is
therefore of great clinical importance.
Small breast epithelial mucin (SBEM) is a secreted
protein, the gene expression of which has been associated with
breast cancer prognosis (7).
Elevated SBEM expression has been linked to poorer clinical
outcomes, suggesting its potential as a prognostic biomarker
(8). Research indicates that SBEM
contributes to the invasion and metastasis of breast cancer cells
(9), potentially by regulating
extracellular matrix degradation, modulating cell adhesion and
altering the tumor microenvironment (10). However, the exact biological role of
SBEM in breast cancer remains unclear. SBEM has been identified as
a potential predictive biomarker for adjuvant chemotherapy in
breast cancer (11). PTX, a drug
frequently employed in combination chemotherapy regimens, still
faces limitations in its clinical application. This is due to
notable individual variations in drug sensitivities (5). To the best of our knowledge, no
studies have clearly revealed the association between SBEM and the
response to PTX, especially the role of SBEM in predicting PTX
sensitivity.
The mitogen-activated protein kinase (MAPK)
signaling pathway plays a central role in the initiation,
progression and metastasis of breast cancer (12). Dysregulated MAPK activation promotes
tumor cancer cell proliferation, invasion, metastasis and
resistance to multiple chemotherapeutic agents (13), including PTX. Numerous studies have
confirmed that abnormal MAPK signaling is a key mechanism driving
PTX resistance (14–16). MAPK activity is primarily regulated
by a family of proteins known as dual-specificity phosphatases
(DUSPs) (17), also referred to as
MAPK phosphatases (MKPs), which are involved in signal
transduction, cell growth regulation and metabolism (18). Among this protein family, MKP7, also
known as dual specificity phosphatase 16 (DUSP16), is primarily
cytoplasmic and inhibits the activity of both p38 and JNK MAPKs
(19,20). DUSP16 also regulates ERK and p38MAP
activation and has been shown to influence AMPK signaling (21–23).
Additionally, abnormal DUSP16 expression is associated with
tumorigenesis, progression and metastasis (24). Despite this, the regulatory
relationship between SBEM and DUSP16 as well as their combined
influence on the AMPK signaling pathway remains largely
unexplored.
The primary objective of the present study was to
explore whether SBEM, by interacting with DUSP16 and activating the
AMPK signaling pathway, contributes to PTX resistance in patients
with breast cancer. Through a series of experiments and analyses,
the study aimed to determine if such a mechanism exists and leads
to the observed PTX resistance. This research also provides novel
insights into future treatment strategies and the improvement of
patient prognosis.
Materials and methods
Molecular docking
The 3D structure of the SBEM protein was predicted
using AlphaFold software 2 (https://alphafold.ebi.ac.uk). PrankWeb (https://prankweb.cz/), a machine learning-based tool
employing a Random Forest algorithm trained on known binding sites,
was used to predict potential active sites (25). Through multi-sequence alignment
analysis using BioXM 2.7.1, MUCL1 (UniProt ID: Q96DR8) and MUCL3
(UniProt ID: Q3MIW9) were examined to identify conserved regions.
Subsequently, the 2D structure of the SBEM protein was analyzed
using PyMOL 2.5 (https://pymol.sourceforge.net). Briefly, based on the
protein structure confidence, 2D structure stability, active site
prediction and sequence alignment, Met1 was selected as the lattice
center and a docking box was established. The specific box
information was as follows: center_x=6.439; center_y=−0.223;
center_z=−12.452; size_x=73.15; size_y=73.15; and size_z=73.15. The
SDF format of PTX was obtained from the PubChem database
(https://pubchem.ncbi.nlm.nih.gov) and
converted to the PDB format using Open Babel (http://openbabel.org/index.html). The protein
structure was dehydrated, hydrogenated and its charge calculated
before being converted to the PDBQT format using AutoDockTools
1.5.7 (https://autodocksuite.scripps.edu/adt/). The ligand
was also hydrogenated, its torsional degrees of freedom were
determined and it was converted to the PDBQT format. The docking
box coordinates were defined and molecular docking was performed
using AutoDock Vina (https://vina.scripps.edu/). Visualization and 3D
analysis diagrams were generated using PyMOL 2.1.0.
Cell culture
Human breast cancer cell lines (SUM190PT, BT474,
AU565, SKBR3 and MCF7) and the normal mammary epithelial cell line
MCF10A were purchased from Qingqi (Shanghai) Biotechnology
Development Co., Ltd. All cell lines were authenticated by short
tandem repeat profiling to prevent cross-contamination. Cells were
thawed from liquid nitrogen, resuspended and cultured in Dulbecco's
Modified Eagle Medium (DMEM) (Beyotime Biotechnology; cat. no.
C0891) supplemented with 10% fetal bovine serum (Beyotime
Biotechnology; cat. no. C0235) and 1% penicillin-streptomycin. The
cells were placed in a cell incubator (Beyotime Biotechnology; cat.
no. E2392) at 37°C with 5% CO2.
Cell transfection
Exponentially growing SUM190PT and SKBR3 cells were
treated with 2 ml trypsin digestion solution (Wuhan Servicebio
Technology Co., Ltd.; cat. no. G4011) and incubated at 37°C in a 5%
CO2 atmosphere (Suzhou Jiemei Electronics Co., Ltd.;
cat. no. CI-191C) for 3 min. Following incubation, 3 ml
high-glucose DMEM (Wuhan Servicebio Technology Co., Ltd.; cat. no.
G4524) was added and the cells were centrifuged at 1,200 × g for 3
min at 25°C. The supernatant was discarded and the pellet was
resuspended in 3 ml DMEM. The cells were then seeded into 6-well
plates at a density of 1×105 cells/well and incubated
overnight. The following day, small interfering RNA (siRNA) and
overexpression vectors were mixed with Lipofectamine™ 3000
transfection reagent (Thermo Fisher Scientific, Inc.; cat. no.
L3000001) and incubated at room temperature for 30 min. The
transfection mixture was then added to the wells according to the
experimental requirements and incubated at 37°C for 5 h. siSBEM was
synthesized by GenScript Biotech Corporation, with a synthesis
amount of 2.5 nmol diluted to 100 µmol/l. The overexpression vector
backbone employed in this study was pcDNA3.1(+) (Thermo Fisher
Scientific, Inc.; cat. no. V79020). For cell transfection, 4 µg of
the pcDNA3.1(+) vector, at a concentration of 1 µg/µl, was
introduced into the cells. The siRNA interference and
overexpression primer sequences are shown in Tables SI and SII, respectively.
Construction of PTX-resistant SUM190PT
and SKBR3 cell lines
PTX-resistant breast cancer cells were developed
based on previously described methods (26). Details of the drug-induction
protocol are shown in Table SIII.
To confirm the stability of the resistance phenotype, the PTX
(MedChemExpress; cat. no. HY-B0015) concentration was gradually
increased until a final concentration of 500 nmol/l was reached.
The cells were then maintained at this concentration and
subcultured for at least 10 generations. IC50 values
were measured at the 5th, 10th and 20th generations post-withdrawal
of PTX. When the IC50 values showed minimal variation
and were significantly higher than those of the parental cell
lines, the resistance model was deemed successfully established.
The parental lines are denoted as SUM190PT and SKBR3, while their
drug resistant counterparts are referred to as SUM190PT/PTX and
SKBR3/PTX. Resistance success was confirmed by comparing cell
viability and resistance coefficients before and after PTX
exposure. Resistant cell lines were continuously cultured in 500
nmol/l PTX to maintain drug resistance.
Cell experimental grouping
To assess whether SBEM knockdown could reverse drug
resistance in SUM190PT/PTX cells, the following groups were
established: SUM190PT (control), SUM190PT + PTX, SUM190PT/PTX + PTX
+ si-negative control (NC) and SUM190PT/PTX + PTX + siSBEM. To
evaluate whether SBEM overexpression induces PTX resistance, SKBR3
cells, which have relatively low SBEM expression, were divided
into: SKBR3 (control), SKBR3/PTX + PTX, SKBR3 + PTX + Vector and
SKBR3 + PTX + oeSBEM. AMPK activator 13 (MedChemExpress; cat. no.
HY-155363), an AMPK signaling pathway activator (27), was used to investigate the role of
SBEM in regulating AMPK signaling. For this, SUM190PT/PTX cells
were divided into four groups: Control, AMPK activator 13, siSBEM
and AMPK activator 13 + siSBEM. Both SUM190PT and SKBR3 cells were
treated with PTX at a concentration of 500 nmol/l for 48 h.
Drug-resistant cell lines SUM190PT/PTX and SKBR3/PTX were
maintained in medium containing 500 nmol/l PTX. Cells transfected
with siRNA or overexpression plasmids were incubated at 37°C for 48
h. In all groups involving AMPK activator 13, the concentration was
set at 5 µmol/l with an incubation time at 37°C for 24 h (27).
Cell counting kit-8 (CCK-8) assay
Following transfection, breast cancer cells and
drug-resistant cell lines in the exponential growth phase were
harvested, digested and resuspended. Then, 3,000 cells per well
were seeded into 96-well plates and incubated overnight. PTX was
applied at varying concentrations (125, 250, 500, 1,000, 2,000,
4,000 and 8,000 nmol/l) and the cells were incubated for 48 h.
Subsequently, 10 µl CCK-8 solution (Wuhan Servicebio Technology
Co., Ltd.; cat. no. G4103) was added to each well and incubated for
an additional 3 h. Optical density at 450 nm was measured using a
microplate reader (Thermo Fisher Scientific, Inc.; cat. no.
A51119500C). IC50 values and PTX resistance coefficients
were calculated using GraphPad Prism version 9.5.0 (Dotmatics).
Total RNA extraction
Cell suspensions were collected and centrifuged at
1,200 × g for 3 min at 25°C. The supernatant was discarded and 1 ml
RNA extraction reagent (Wuhan Servicebio Technology Co., Ltd.; cat.
no. G3013) was added, followed by a 5-min incubation at room
temperature. Next, 100 µl chloroform (Wuhan Servicebio Technology
Co., Ltd.; cat. no. G3014) was added and the mixture was shaken
vigorously for 15 sec, then allowed to stand for 5 min at room
temperature. The sample was centrifuged at 12,000 × g for 15 min at
4°C, and the aqueous phase was transferred to a new centrifuge
tube. Then, 500 µl isopropanol (Shanghai Macklin Biochemical Co.,
Ltd.; cat. no. 67-63-0) was added and incubated for 10 min at room
temperature. After centrifugation at 12,000 × g for 15 min at 4°C,
the supernatant was discarded. The RNA pellet was washed with 1 ml
75% ethanol (Shanghai Macklin Biochemical Co., Ltd.; cat. no.
E809061), air-dried and resuspended in 50 µl sterile water (Wuhan
Servicebio Technology Co., Ltd.; cat. no. G4700). The RNA
concentration was quantified for use in subsequent experiments.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RT-qPCR was conducted according to the instructions
of the SweScript One-Step RT-PCR Kit (Wuhan Servicebio Technology
Co., Ltd.; cat. no. G3335). The PCR cycling program was as follows:
50°C for 30 min, 98°C for 2 min, followed by 39 cycles of 98°C for
15 sec, 55°C for 20 sec and 72°C for 20 sec. A final extension step
was carried out at 72°C for 5 min. The primer sequence (5′-3′)
information is as follows: SBEM forward, TCTGCCCAGAATCCGACAAC, and
reverse GGGCTTCATCATCAGCAGGA; GAPDH forward, CTGGGCTACACTGAGCACC,
and reverse AAGTGGTCGTTGAGGGCAATG. GAPDH was used as the internal
reference gene and the 2−∆∆Cq was used as the relative
expression level of the target gene mRNA (28).
Western blotting
Breast cancer cell suspensions were collected and
lysed using RIPA lysis buffer (Wuhan Servicebio Technology Co.,
Ltd.; cat. no. G2002). Lysates were incubated on ice for 30 min and
then centrifuged at 12,000 × g for 15 min at 25°C. The supernatant
was collected and proteins were denatured in a 95°C water bath for
15 min. Protein concentrations were determined using a BCA protein
quantification kit (Wuhan Servicebio Technology Co., Ltd.; cat. no.
G2026). Cell lysis and protein-loading buffers (Beyotime Institute
of Biotechnology; cat. no. P0015) were added to normalize the
protein concentrations across samples. Then, 10 µl (30 µg) of each
protein solution was loaded into the wells of the gel (10%).
Electrophoresis was performed at 80 V for 30 min, followed by 120 V
for 60 min. After electrophoresis, the proteins were transferred to
PVDF membranes at 260 mA for 2 h. PVDF membranes were blocked with
5% skim milk (Wuhan Servicebio Technology Co., Ltd.; cat. no.
GC310001) at room temperature for 2 h, then washed three times with
TBST (0.1% Tween 20) (Wuhan Servicebio Technology Co., Ltd.; cat.
no. G0004) for 5 min each. Primary antibody solutions were added
and incubated overnight at 4°C. After incubation, secondary
antibody solutions were applied and incubated at room temperature
for 1 h. Details of the antibodies, including manufacturer cat. no.
and dilution ratios, are provided in Table SIV. After washing with TBST,
chemiluminescent detection was performed using a luminescent
substrate (Wuhan Servicebio Technology Co., Ltd.; cat. no. G2161)
and images were captured using a gel imaging system (Mona
Biotechnology Co., Ltd.; cat. no. GD50202). Images were analyzed
with ImageJ 1.5.2a software (National Institutes of Health) for
subsequent statistical analyses.
Cell apoptosis
Exponentially growing breast cancer cells were
treated as aforementioned in the Cell experimental grouping
subsection, then centrifuged and resuspended as previously
described. According to the instructions of the flow cytometry
apoptosis detection kit (Wuhan Servicebio Technology Co., Ltd.;
cat. no. G1511), 5 µl FITC solution was added to each cell
suspension, followed by incubation in the dark at room temperature
for 30 min. Subsequently, PI solution was added and incubated in
the dark for an additional 5 min. Cells were analyzed using a
Attune™ NxT flow cytometer (Thermo Fisher Scientific, Inc.; cat.
no. A24858) and Attune™ NxT software (version 6; Thermo Fisher
Scientific, Inc.) within 2 h of staining.
Co-immunoprecipitation (Co-IP)
Exponentially growing breast cancer cells were
treated as aforementioned in the Cell experimental grouping
subsection, then centrifuged and resuspended as previously
described. Cell lysis buffer was added according to the Co-IP kit
protocol (Beyotime Institute of Biotechnology; cat. no. P2179M) and
the samples were incubated at room temperature for 30 min. The
lysates were then centrifuged at 1,200 × g for 3 min at 25°C and
the supernatant was collected. Protein A/G magnetic beads
pre-conjugated with the primary antibody were added (20 µl of
magnetic bead suspension to every 500 µl of protein sample) and the
mixtures were incubated overnight at room temperature. After
incubation, the beads were washed with washing buffer and the
immunocomplexes bound to the beads were eluted using the elution
buffer. The eluted target proteins were analyzed via western
blotting. Antibody details are provided in Table SIV.
Statistical analysis
Data were analyzed using SPSS version 22.0 (IBM
Corp.). All experiments were independently repeated at least three
times. Results are expressed as mean ± standard deviation.
Comparisons between two groups were performed using unpaired
t-tests. Differences among multiple groups were assessed using
one-way analysis of variance followed by Tukey's post-hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
SBEM induces PTX drug resistance
The 2D structural formula of PTX is shown in
Fig. 1A. The SBEM mRNA expression
levels in various breast cancer cell and normal breast epithelial
cell lines were assessed using RT-qPCR. Compared with MCF10A cells,
SBEM mRNA expression was highest in SUM190PT cells and lowest in
SKBR3 cells (Fig. 1B). Since SBEM
was highly expressed in the SUM190PT cell line, a knockdown cell
line using this cell line was initially planned. However, as the
present study focuses on drug-resistant strains, a SBEM-knockdown
cell line using the SUM190PT/PTX drug-resistant strain was
ultimately constructed. Additionally, SKBR3 cells, which have low
SBEM expression, were selected to create a SBEM-overexpressing cell
line. Knockdown and overexpression experiments in cell lines
representing the two extremes of SBEM expression were conducted to
more clearly observe the impact of SBEM modulation on cellular
behaviors, such as viability and PTX resistance. This maximized the
differences in experimental outcomes and enhanced the
persuasiveness of the findings.
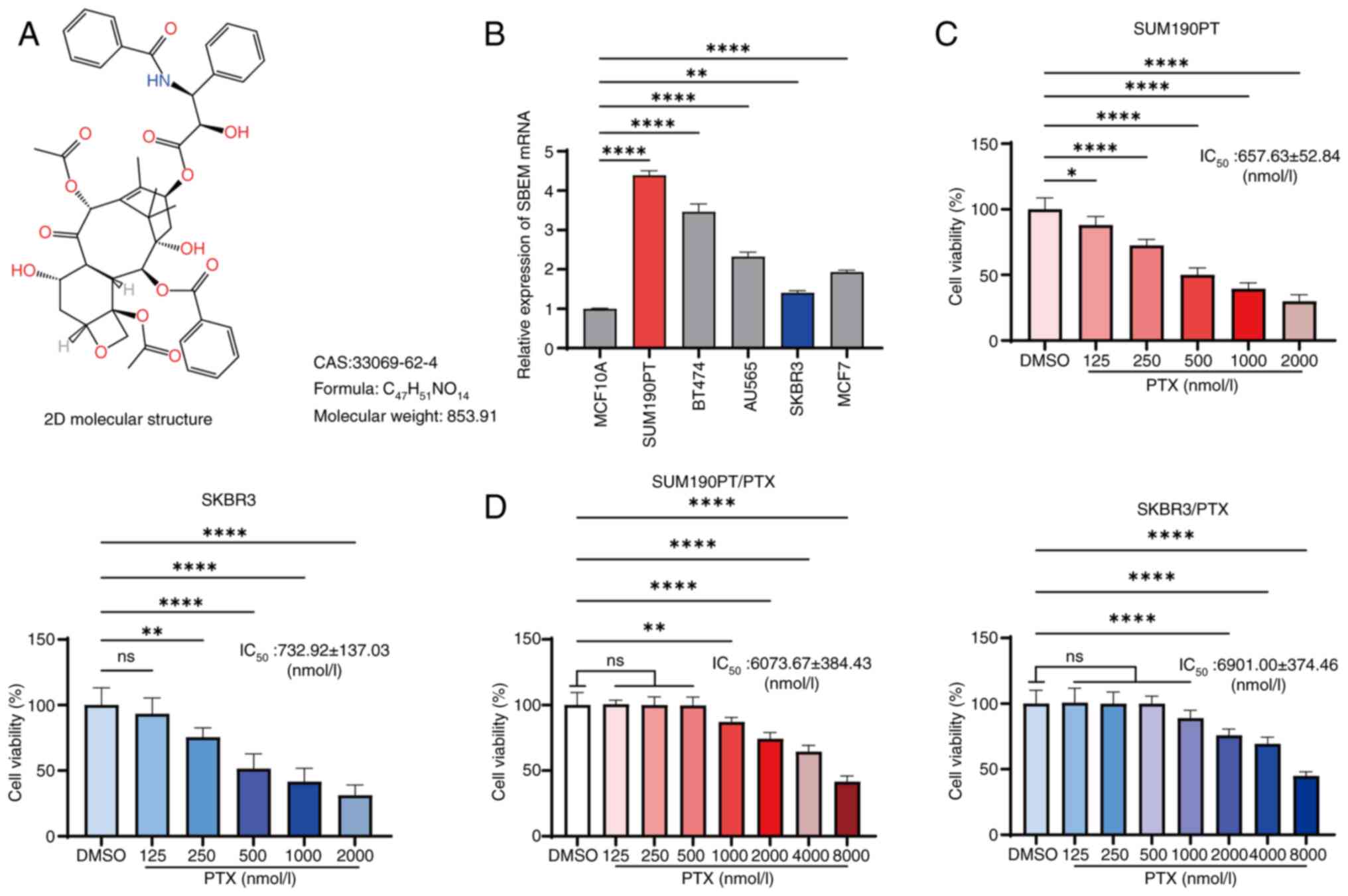 | Figure 1.Inhibition of breast cancer cell
viability by PTX and the construction of PTX-resistant strains. (A)
2D molecular structure of PTX. (B) Relative expression levels of
SBEM mRNA in MCF10A, SUM190PT, BT474, AU565, SKBR3 and MCF7 cells
were measured using reverse transcription-quantitative polymerase
chain reaction. (C) CCK-8 assay showing changes in the viability of
SUM190PT and SKBR3 cells treated with varying PTX concentrations.
(D) CCK-8 assay showing changes in the viability of SUM190PT/PTX
and SKBR3/PTX cells treated with varying PTX concentrations.
*P<0.05, **P<0.01, ****P<0.0001. ns, not significant; PTX,
paclitaxel; SBEM, small breast epithelial mucin; CCK-8, Cell
Counting Kit-8. |
The establishment of PTX-resistant cell lines was
validated using the CCK-8 assay. After 48 h of PTX treatment, the
SUM190PT and SKBR3 cell viabilities were significantly inhibited,
with IC50 values of 657.63±52.84 nmol/l and
732.92±137.03 nmol/l, respectively (Fig. 1C). By contrast, the PTX-resistant
SUM190PT/PTX and SKBR3/PTX cells exhibited respective
IC50 values of 6073.67±384.43 nmol/l and 6901.00±374.46
nmol/l, with corresponding resistance coefficients of 8.95±1.33 and
9.04±1.46 (Fig. 1D). These data
confirmed the successful generation of the PTX-resistant cell
lines.
To further explore the association and potential
mechanism of SBEM in PTX resistance, cell lines with upregulated or
downregulated SBEM expression were established (Fig. 2A and B). Among the siRNAs tested,
siSBEM-1 most effectively inhibited both SBEM mRNA and protein
expression and was therefore selected for subsequent experiments.
The effects of SBEM knockdown and overexpression on the
proliferation of SUM190PT/PTX and SKBR3 cells were assessed using
the CCK-8 assay. The results demonstrated that SBEM knockdown
restored PTX sensitivity in SUM190PT/PTX cells, whereas SBEM
overexpression reduced PTX sensitivity (Fig. 2C and D). These findings suggest that
SBEM expression may be associated with PTX sensitivity in breast
cancer cells.
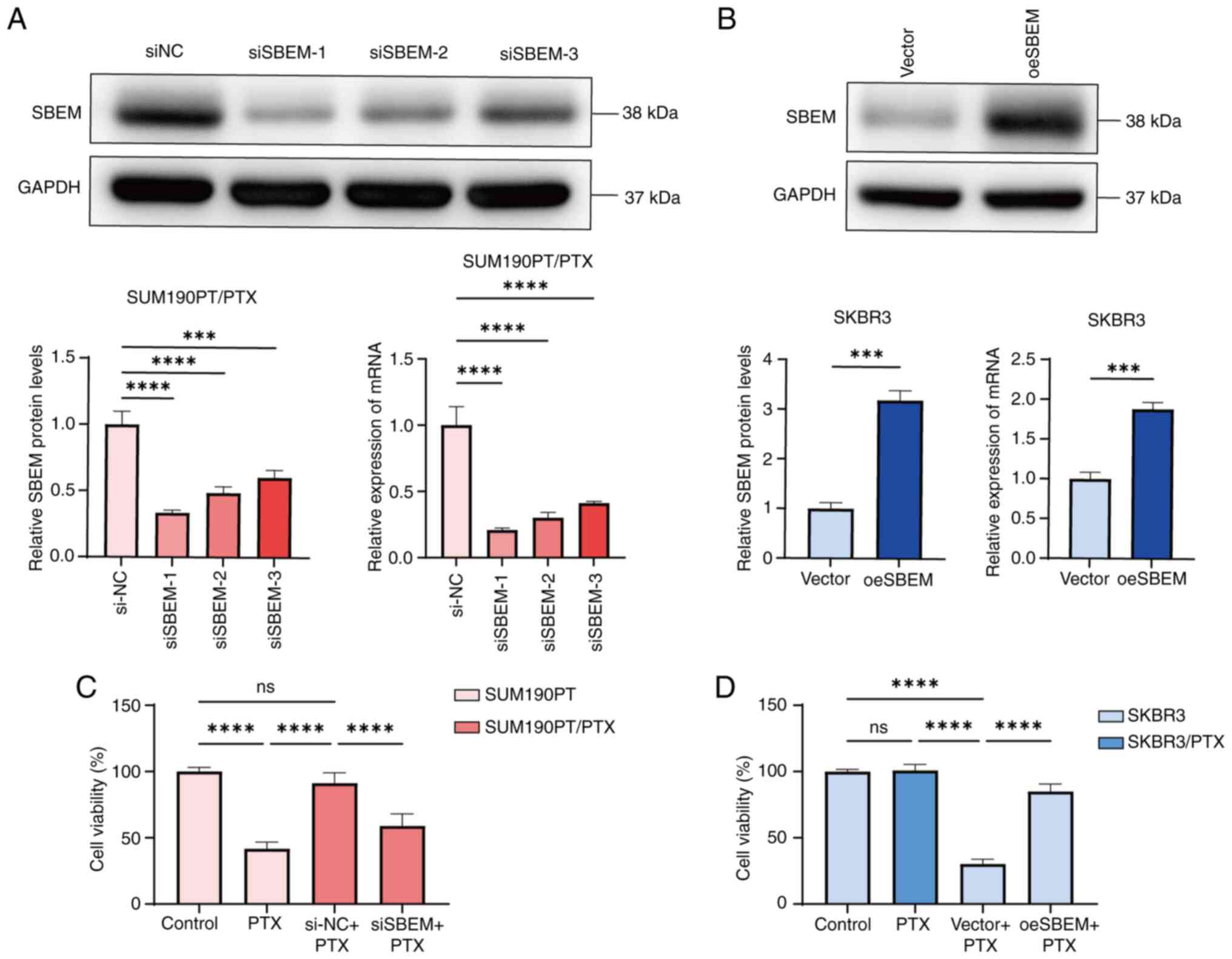 | Figure 2.Changes in SBEM expression and the
effect on PTX resistance. (A) Western blotting and RT-qPCR analyses
of SBEM protein and mRNA levels in SUM190PT/PTX cells 48 h
post-transfection with siSBEM. (B) Western blotting and RT-qPCR
analyses of SBEM protein and mRNA levels in SKBR3 cells 48 h
post-transfection with the SBEM overexpression plasmid. (C)
SUM190PT cells were treated with PTX (500 nmol/l) for 48 h. In the
case of SUM190PT/PTX cells, the PTX concentration in the
drug-resistant medium was maintained at 500 nmol/l. Subsequently,
siRNA (SBEM) was transfected for 48 h. The CCK-8 assay was used to
detect the changes in cell viability of SUM190PT and SUM190PT/PTX
cells after treatment. (D) SKBR3 cells were treated with PTX (500
nmol/l) for 48 h. Subsequently, the cells were transfected with the
SBEM-overexpressing plasmid and incubated for another 48 h. For
SKBR3/PTX cells, the PTX concentration in the drug-resistant medium
was maintained at 500 nmol/l. The CCK-8 assay was used to detect
the changes in cell viability of SKBR3 and SKBR3/PTX cells after
treatment. ***P<0.001, ****P<0.0001. ns, not significant;
PTX, paclitaxel; SBEM, small breast epithelial mucin; RT-qPCR,
reverse transcription-quantitative polymerase chain reaction;
CCK-8, Cell Counting Kit-8; si, small interfering (RNA); NC,
negative control; oe, overexpression. |
SBEM has binding sites for PTX and
interacts with DUSP16
While it was demonstrated that SBEM may be
associated with PTX sensitivity, its precise regulatory mechanism
remains unclear. The following analyses were conducted to screen
the rational domains of the SBEM protein. The prediction results
showed that the protein ipTM=-pTM=0.23, with a relatively low
overall confidence level (Fig.
S1A). In addition, amino acids 1–20 were predicted to form the
signal peptide region (Fig. S1B).
Next, PrankWeb was used, which is based on machine learning; it
learns the characteristics of a large number of known binding sites
through a Random Forest algorithm to predict the protein structure
(28). The results are shown in
Fig. S1C and the red area
represents the active site. The score of this active site (residue
site: 1) is >1 (score: 1.45) and the average AlphaFold score is
>70 (score: 83.21), both of which indicate a high level of
confidence in this site (27).
Additionally, it was noted that SBEM is also known as MUCL1. MUCL
has two subtypes: MUCL1 and MUCL3. To detect the conserved
sequences, a sequence alignment between MUCL1 (ID: Q96DR8) and
MUCL3 (ID: Q3MIW9) was performed (Fig.
S1D). The alignment results were consistent with the prediction
of the active site, both of which are located in the N-terminal
region. The molecular docking results showed that PTX can bind
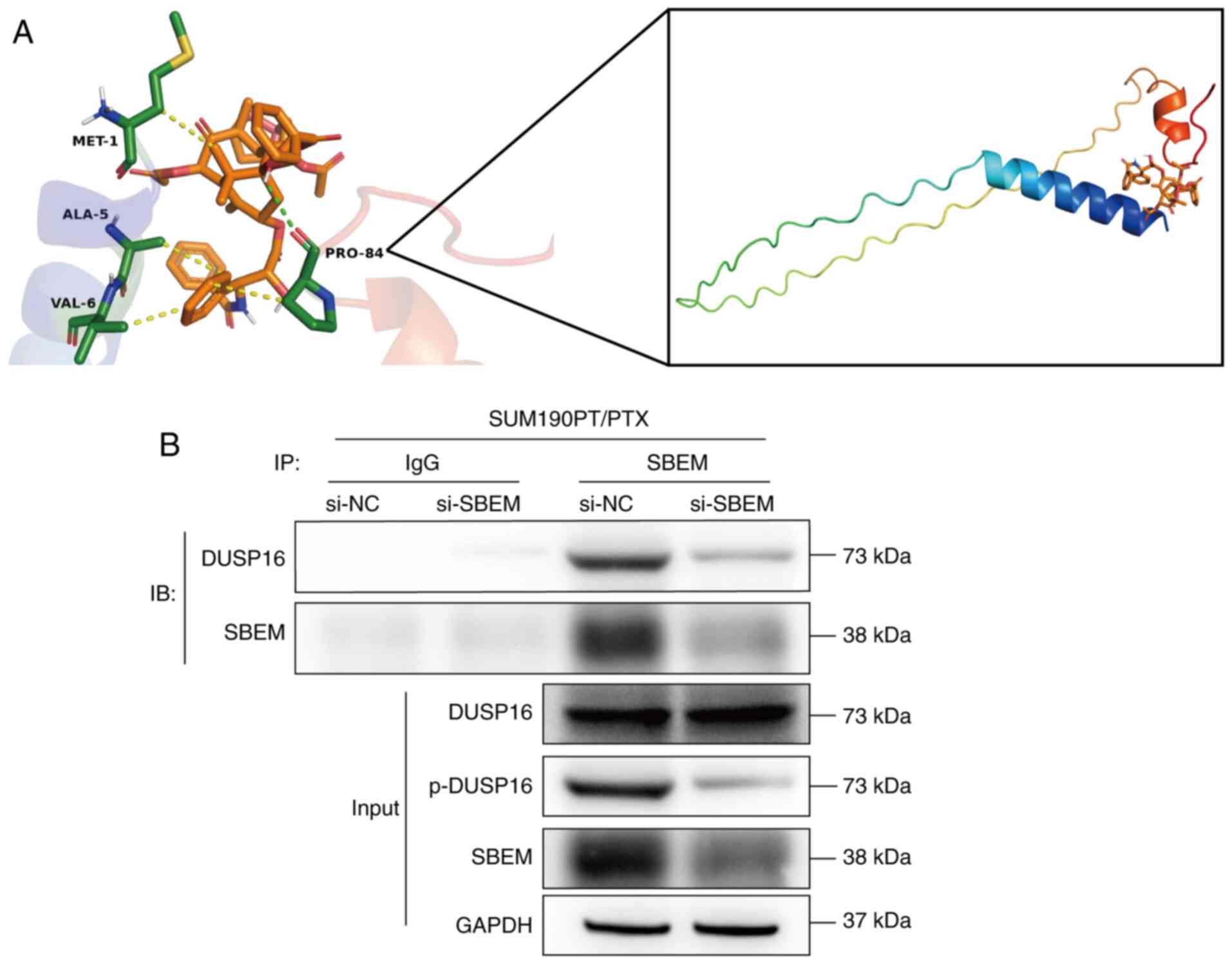
tightly to SBEM (Fig. 3A). The
strongest calculated binding free energy was-6.4 kcal/mol. In
addition, each docking free energy was <-5.0 kcal/mol,
demonstrating a good binding between the two. PTX was predicted to
not only bind to the Pro84 amino acid of SBEM through a hydrogen
bond but also form hydrophobic interactions with Met1, Ala5, Val6
and Pro84. Most notably, the distance of each interaction was <4
Å, indicating a relatively strong bond energy (Table SV).
Additionally, to verify the interaction between SBEM
and DUSP16, a Co-IP assay was performed. DUSP16 was detected in the
SBEM IP, supporting a physical interaction between the two
proteins. Furthermore, SBEM knockdown led to a reduction in the
phosphorylated (p-)DUSP16 levels in the input group. However, no
significant change in total DUSP16 protein expression was observed
compared with the siNC group, suggesting that SBEM may regulate
DUSP16 phosphorylation rather than its expression (Fig. 3B).
Downregulation of SBEM restores PTX
sensitivity in breast cancer cells
Following SBEM knockdown, the apoptosis rate of
SUM190PT/PTX cells (in culture medium containing 500 nmol/l PTX)
significantly increased. The apoptosis rate of the SBEM-knockdown
group increased approximately three-fold, rising from 16.53% in the
control group (siNC + PTX) to 44.25% in the knockdown group (siSBEM
+ PTX) (Fig. 4A). By contrast, SBEM
overexpression in SKBR3 cells (in culture medium containing 500
nmol/l PTX) significantly decreased apoptosis (Fig. 4B). To further investigate this
observation, the expression of apoptosis-related proteins was
analyzed.
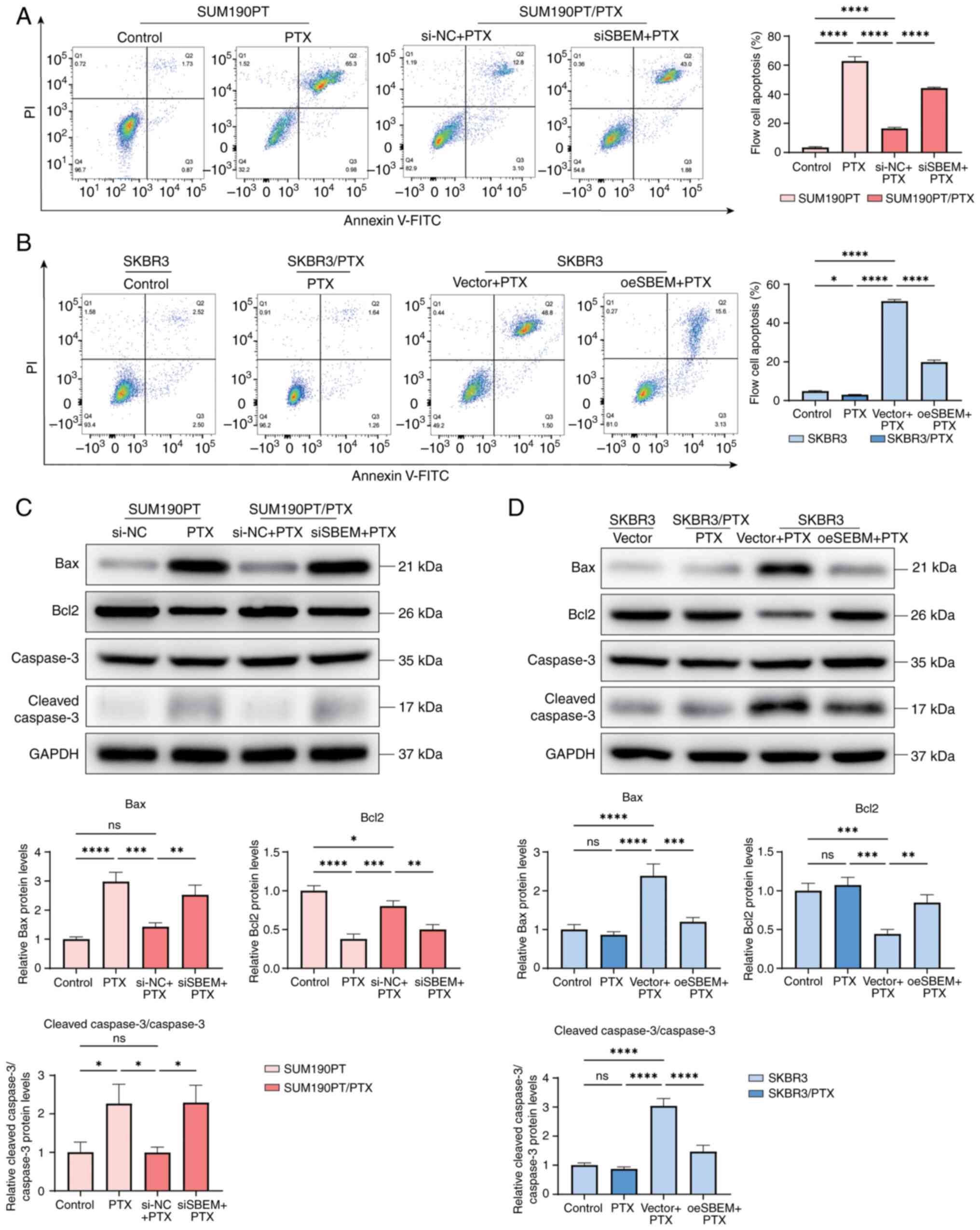 | Figure 4.SBEM modulates PTX-induced apoptosis
in breast cancer cells. (A) Flow cytometry analysis of apoptosis in
SUM190PT/PTX and SUM190PT cells following various treatments. (B)
Flow cytometry analysis of apoptosis in SKBR3/PTX and SKBR3 cells
following various treatments. (C) Western blot analysis of Bax,
Bcl2, Caspase-3 and cleaved-Caspase-3 in SUM190PT/PTX and SUM190PT
cells. (D) Western blot analysis of Bax, Bcl2, Caspase-3 and
cleaved-Caspase-3 in SKBR3/PTX and SKBR3 cells. *P<0.05,
**P<0.01, ***P<0.001, ****P<0.0001. ns, not significant;
PTX, paclitaxel; SBEM, small breast epithelial mucin; si, small
interfering (RNA); NC, negative control; oe, overexpression. |
In SUM190PT/PTX cells treated with PTX, compared
with the siNC + PTX group, the siSBEM + PTX group had increased
expression levels of Bax and cleaved-Caspase-3/Caspase-3, while the
Bcl2 expression was decreased (Fig.
4C). Conversely, in SKBR3 cells treated with PTX, compared with
the Vector + PTX group, overexpression of SBEM in the oeSBEM + PTX
group decreased the expression of Bax and
cleaved-Caspase-3/Caspase-3 and increased the expression of Bcl2
(Fig. 4D).
Downregulation of SBEM inhibits
p-DUSP16 expression and the MAPK pathway
To elucidate the mechanism by which SBEM affects PTX
sensitivity, the expression levels of DUSP16 and MAPK-related
proteins were analyzed. In SUM190PT/PTX cells, compared with the
siNC + PTX group, knockdown of SBEM in the siSBEM + PTX group
decreased the ratios of p-DUSP16/DUSP16 and p-MAPK/MAPK (Fig. 5A). By contrast, in SKBR3 cells
treated with PTX, compared with the Vector + PTX group,
upregulation of SBEM in the oeSBEM + PTX group increased these
ratios (Fig. 5B). These findings
suggest that SBEM enhances DUSP16 phosphorylation and activates the
MAPK signaling pathway, thereby potentially contributing to PTX
resistance.
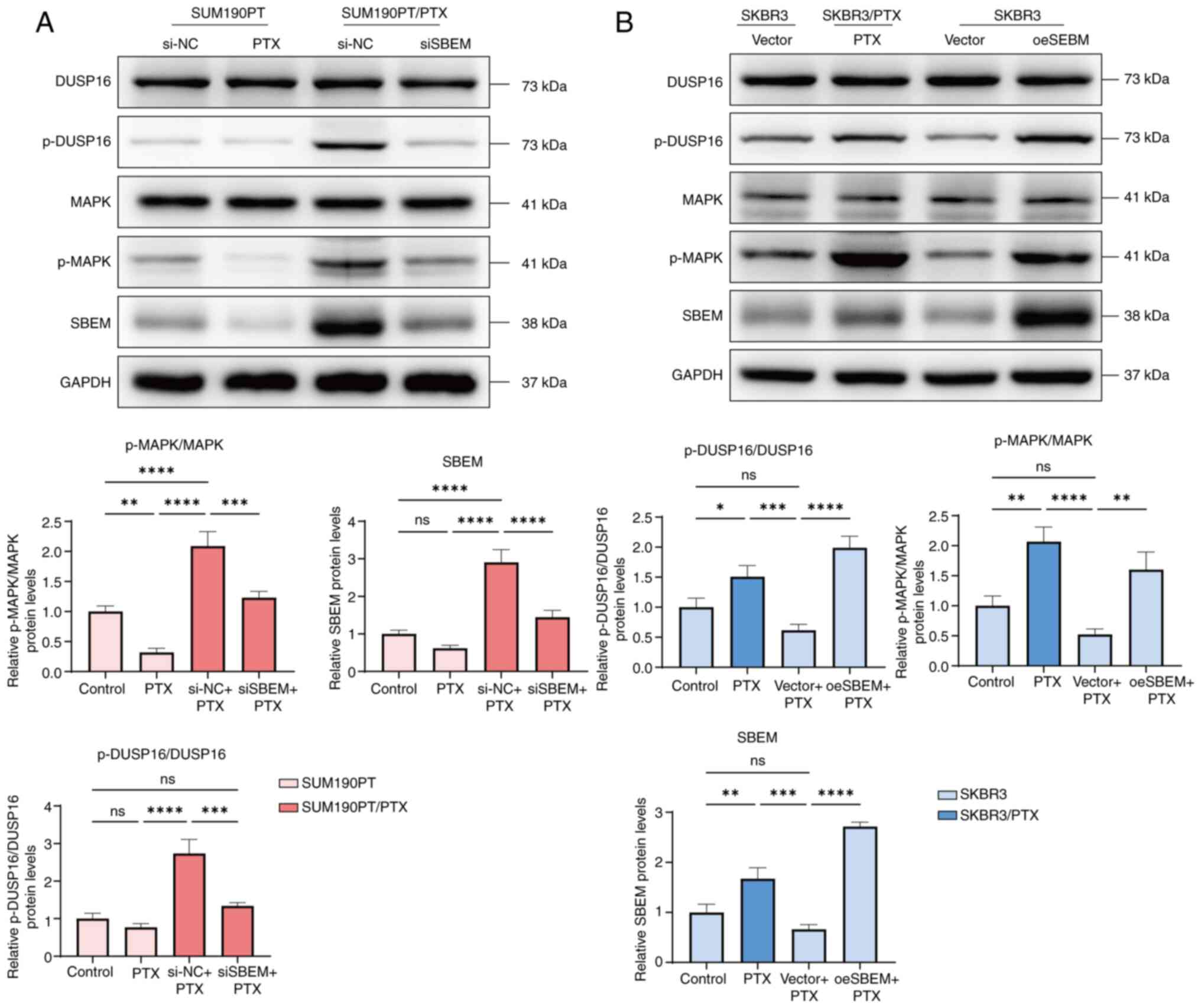 | Figure 5.SBEM regulates p-DUSP16 and the MAPK
signaling pathway. (A) Western blotting of DUSP16, p-DUSP16, MAPK,
p-MAPK and SBEM in SUM190PT and SUM190PT/PTX cells. (B) Western
blotting of DUSP16, p-DUSP16, MAPK, p-MAPK and SBEM in SKBR3 and
SKBR3/PTX cells. *P<0.05, **P<0.01, ***P<0.001,
****P<0.0001. ns, not significant. PTX, paclitaxel; SBEM, small
breast epithelial mucin; DUSP16, dual specificity phosphatase 16;
p-, phosphorylated; si, small interfering (RNA); NC, negative
control; oe, overexpression, |
SBEM induces PTX resistance in breast
cancer cells by activating the MAPK pathway
To further investigate the regulatory effect of SBEM
on the MAPK pathway, SUM190PT/PTX cells were treated with the MAPK
pathway activator, AMPK activator 13. The results showed that in
SUM190PT/PTX cells treated with PTX, compared with the siSBEM
group, the level of apoptosis in the siSBEM + AMPK activator 13
group decreased significantly while the cell viability increased
significantly. The results indicated that AMPK activator 13
counteracted the inhibitory effects of SBEM on the viability and
induction of apoptosis in SUM190PT/PTX cells (Fig. 6A and B). Furthermore, compared with
the siSBEM group, the addition of AMPK activator 13 (siSBEM + AMPK
activator 13 group) significantly reduced the levels of Bax and
cleaved-Caspase-3 and increased the expression of Bcl2 (Fig. 6C). The effect of AMPK activator 13
on apoptosis-related proteins reversed the impact of siSBEM.
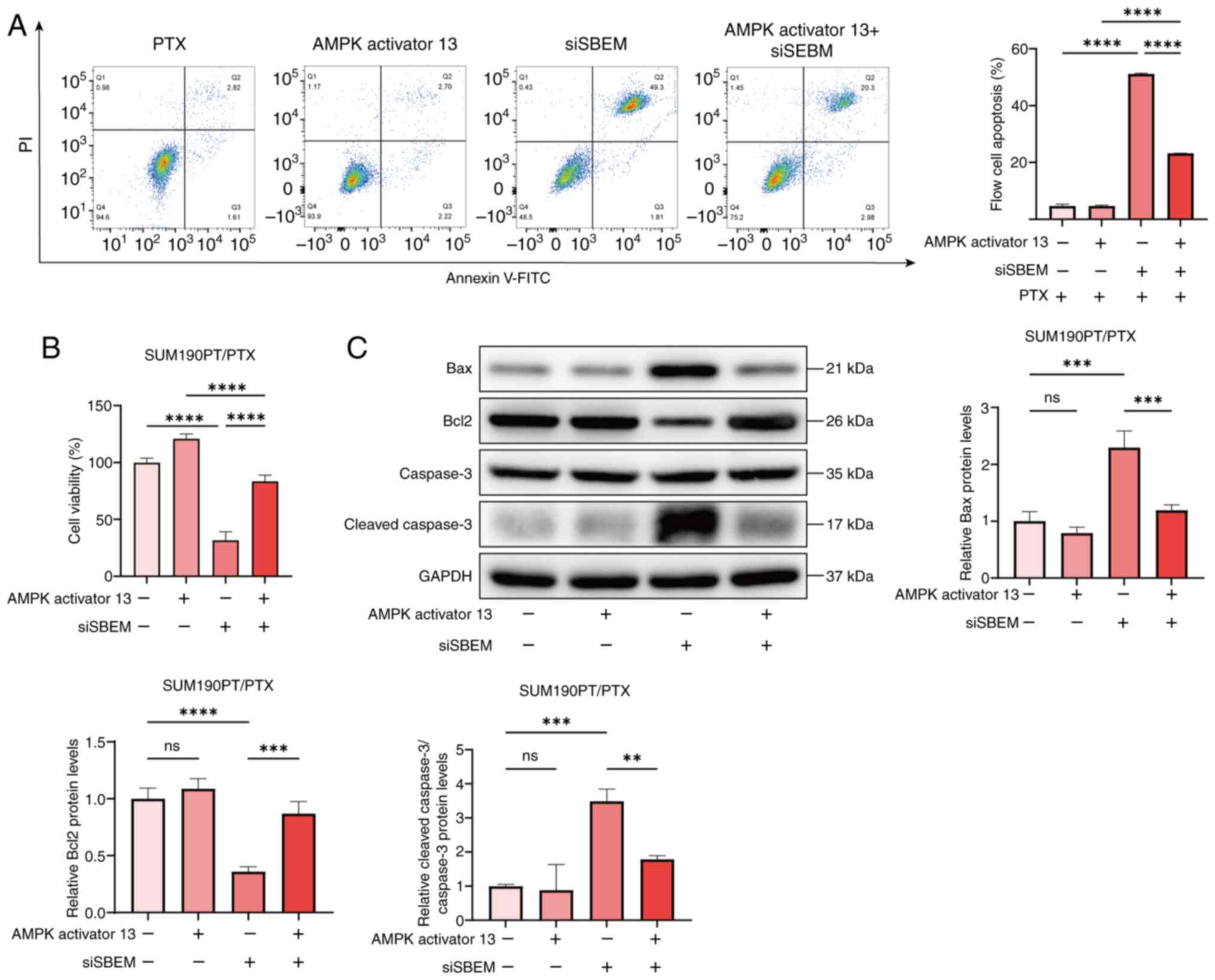 | Figure 6.SBEM induces PTX resistance in breast
cancer cells by activating the AMPK pathway. (A) Flow cytometry
analysis of apoptosis in SUM190PT/PTX cells transfected with siRNA
for 48 h and treated with AMPK activator 13 (5 µmol/l) for 24 h.
(B) The treatment of SUM190PT/PTX cells was the same as
aforementioned. After the treatment, the change in cell viability
was determined by the CCK-8 assay. (C) The treatment of
SUM190PT/PTX cells was the same as aforementioned. After the
treatment, the relative expression levels of Bax, Bcl2, Caspase-3
and cleaved-Caspase-3 proteins were detected by western blotting.
**P<0.01, ***P<0.001, ****P<0.0001. ns, not significant;
PTX, paclitaxel; SBEM, small breast epithelial mucin; CCK-8, Cell
Counting Kit-8; si, small interfering (RNA). |
In addition, in SUM190PT/PTX cells treated with PTX,
compared with the siSBEM group, the p-DUSP16/DUSP16 level decreased
and the p-MAPK/MAPK level increased in the siSBEM + AMPK activator
13 group (Fig. 7). These findings
suggest that SBEM may contribute to drug resistance in breast
cancer cells by activating the MAPK pathway.
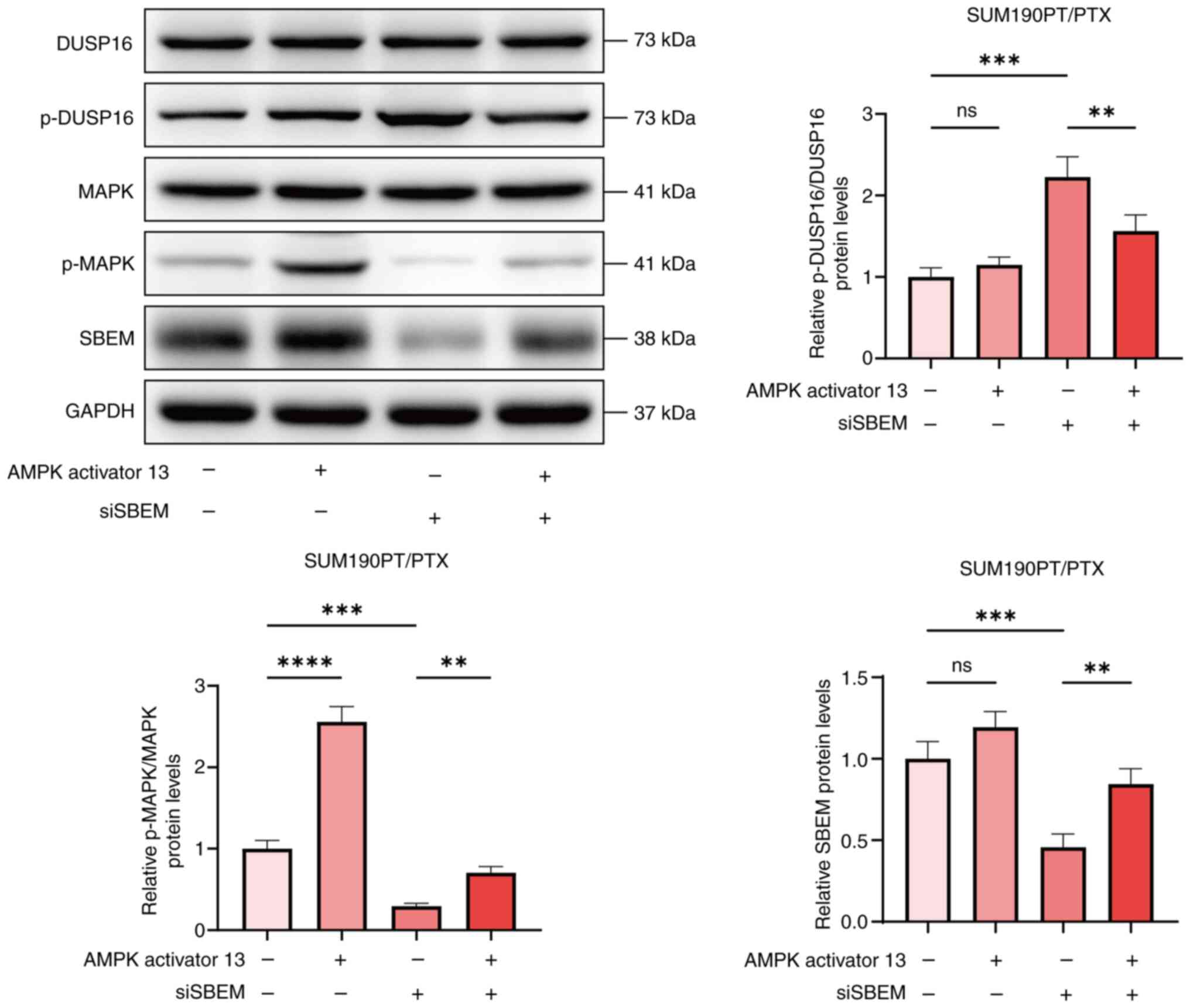 | Figure 7.AMPK activator 13 restores drug
sensitivity to PTX. Western blotting of DUSP16, p-DUSP16, MAPK,
p-MAPK and SBEM protein levels in SUM190PT/PTX cells transfected
with siRNA for 48 h and treated with AMPK activator 13 for 24 h.
**P<0.01, ***P<0.001, ****P<0.0001. ns, not significant;
PTX, paclitaxel; SBEM, small breast epithelial mucin; DUSP16, dual
specificity phosphatase 16; p-, phosphorylated; si, small
interfering (RNA). |
Discussion
PTX, a key chemotherapeutic agent for breast cancer,
is frequently associated with the development of drug resistance,
making it crucial to investigate the underlying mechanisms of this
resistance (29). In the present
study, a predicted binding interaction between SBEM and PTX was
identified and it was demonstrated that downregulation of SBEM can
restore PTX sensitivity. However, the specific binding sites
(single or multiple) between PTX and SBEM remain unclear. In
future, we plan to utilize protein structure-based approaches such
as site-directed mutagenesis and crystallographic analysis to
systematically elucidate the interaction between PTX and SBEM. This
will further validate and refine the findings of the present study.
Apoptosis, a form of programmed cell death (30), is one of the primary mechanisms by
which PTX exerts its anticancer effects. However, tumor cells often
develop resistance to PTX, diminishing its therapeutic efficacy
(31,32). The present study demonstrated that
overexpression of SBEM in SKBR3 cells conferred resistance to PTX
and reduced its cytotoxic effect. Conversely, SBEM knockdown in
SUM190PT/PTX cells restored apoptosis upon PTX treatment,
indicating that SBEM inhibits PTX-induced apoptosis. Bax, a member
of the Bcl2 protein family, plays a crucial role in PTX-induced
apoptosis by promoting mitochondrial membrane permeabilization
(33). During apoptosis, Bax
translocates from the cytoplasm to the mitochondrial membrane and
forms homodimers, increasing mitochondrial outer membrane
permeability and leading to the release of cytochrome c and other
pro-apoptotic factors, which activate the Caspase cascade and
ultimately induce apoptosis (34,35).
In line with this mechanism, the findings of the present study
revealed that SBEM downregulation significantly increased the
expression of Bax and cleaved-Caspase-3 proteins while reducing
Bcl2 expression, thereby promoting apoptosis in SUM190PT/PTX
cells.
PTX has been shown to activate the AMPK signaling
pathway, which plays an important role in its anticancer effects
(36,37). To further investigate the mechanism
of SBEM-induced PTX resistance, the present study focused on the
involvement of DUSP16 and the AMPK signaling pathways. AMPK, a
cellular energy sensor, is activated under conditions of energy
deficiency; it promotes catabolic pathways and inhibits anabolic
processes. The MAPK family, which includes ERK, JNK and p38,
responds to growth factors, stress and inflammatory signals to
regulate cell proliferation, apoptosis, and differentiation
(38). DUSP16 is mainly located
downstream of the MAPK signaling pathway, and its function is to
inhibit the activities of p38 and JNK MAPKs (19,20).
The results of the present study revealed an association between
SBEM and DUSP16 expression. Overexpression of SBEM significantly
increased the levels of p-DUSP16 and p-AMPK. Moreover, treatment
with an AMPK activator reversed the effects of siSBEM on breast
cancer apoptosis and associated protein expression. Therefore, we
hypothesize that SBEM may regulate PTX resistance by activating the
AMPK signaling pathway and inducing DUSP16 phosphorylation. The
results of the Co-IP experiment further confirmed that SBEM and
DUSP16 may interact with each other. However, based on the current
results, it cannot be determined whether SBEM and DUSP16 affect
protein expression through direct binding or indirect binding.
Therefore, further experimental verification is needed.
The major contribution of the present study is the
identification of SBEM as a regulator of DUSP16 phosphorylation and
the MAPK signaling pathway, ultimately promoting PTX resistance in
breast cancer cells. However, several limitations should be
acknowledged. In vivo validation using xenograft tumor mouse
models is necessary. Furthermore, SBEM expression should be
compared with established drug resistance markers, such as
P-glycoprotein and βIII-tubulin (39,40).
Although a positive association was observed between SBEM and
phosphorylated DUSP16 and MAPK proteins, the direct binding
relationship between SBEM and phosphorylated DUSP16 remains to be
elucidated and will be a key focus of future research.
Additionally, PTX resistance involves complex signaling networks,
necessitating further extensive investigation. At present, to the
best of our knowledge, the relationship between SBEM expression and
PTX resistance has not been examined in clinical breast cancer
samples. Future work will focus on clinical collaboration to
collect and analyze samples from patients with PTX-resistant breast
cancer, to further validate the clinical relevance and therapeutic
potential of targeting SBEM.
In conclusion, the results of the present study
demonstrated that downregulation of SBEM enhanced the sensitivity
of breast cancer cells to PTX by targeting DUSP16 and inhibiting
the MAPK signaling pathway. These findings suggest that SBEM could
serve as a therapeutic target to overcome PTX resistance. Thus, the
present study provides a theoretical basis for the use of SBEM
inhibitors in combination with PTX to improve treatment efficacy in
breast cancer.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by Wu Jieping Medical Foundation for
Special Fund of Clinical Scientific Research (grant no. 320.6750)
and Beijing Health League Foundation for Clinical and Medical
Research of Medical Research and Development Foundation
Project.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
LL conceived, designed and supervised the study. NL
and XL performed the data analysis and drafted the manuscript. QY,
SL and JW collected the data. HS and YD were responsible for
conceptualization and data curation. XL revised the manuscript. NL
and XL confirm the authenticity of all the raw data. All the
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wilkinson L and Gathani T: Understanding
breast cancer as a global health concern. Br J Radiol.
95:202110332022. View Article : Google Scholar
|
|
2
|
DeSantis CE, Ma J, Gaudet MM, Newman LA,
Miller KD, Goding Sauer A, Jemal A and Siegel RL: Breast cancer
statistics, 2019. CA Cancer J Clin. 69:438–451. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arnold M, Morgan E, Rumgay H, Mafra A,
Singh D, Laversanne M, Vignat J, Gralow JR, Cardoso F, Siesling S
and Soerjomataram I: Current and future burden of breast cancer:
Global statistics for 2020 and 2040. Breast. 66:15–23. 2022.
View Article : Google Scholar
|
|
4
|
Burstein HJ, Curigliano G, Thürlimann B,
Weber WP, Poortmans P, Regan MM, Senn HJ, Winer EP and Gnant M;
Panelists of the St Gallen Consensus Conference, : Customizing
local and systemic therapies for women with early breast cancer:
The St. Gallen International Consensus Guidelines for treatment of
early breast cancer 2021. Ann Oncol. 32:1216–1235. 2021. View Article : Google Scholar
|
|
5
|
Abu Samaan TM, Samec M, Liskova A, Kubatka
P and Büsselberg D: Paclitaxel's mechanistic and clinical effects
on breast cancer. Biomolecules. 9:7892019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dan VM, Raveendran RS and Baby S:
Resistance to intervention: Paclitaxel in breast cancer. Mini Rev
Med Chem. 21:1237–1268. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y, Lun X and Guo W: Expression of
TRPC1 and SBEM protein in breast cancer tissue and its relationship
with clinicopathological features and prognosis of patients. Oncol
Lett. 20:3922020.
|
|
8
|
Hao H, Yang L, Wang B, Sang Y and Liu X:
Small breast epithelial mucin as a useful prognostic marker for
breast cancer patients. Open Life Sci. 18:202207842023. View Article : Google Scholar
|
|
9
|
Hao H, Wang B, Yang L, Sang Y, Xu W, Liu
W, Zhang L and Jiang D: miRNA-186-5p inhibits migration, invasion
and proliferation of breast cancer cells by targeting SBEM. Aging
(Albany NY). 15:6993–7007. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li QH, Liu ZZ, Ge Y, Liu X, Xie XD, Zheng
ZD, Ma YH and Liu B: Small breast epithelial mucin promotes the
invasion and metastasis of breast cancer cells via promoting
epithelial-to-mesenchymal transition. Oncol Rep. 44:509–518. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu ZZ, Xie XD, Qu SX, Zheng ZD and Wang
YK: Small breast epithelial mucin (SBEM) has the potential to be a
marker for predicting hematogenous micrometastasis and response to
neoadjuvant chemotherapy in breast cancer. Clin Exp Metastasis.
27:251–259. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wen S, Hou Y, Fu L, Xi L, Yang D, Zhao M,
Qin Y, Sun K, Teng Y and Liu M: Cancer-associated fibroblast
(CAF)-derived IL32 promotes breast cancer cell invasion and
metastasis via integrin β3-p38 MAPK signalling. Cancer Lett.
442:320–332. 2019. View Article : Google Scholar
|
|
13
|
Butti R, Das S, Gunasekaran VP, Yadav AS,
Kumar D and Kundu GC: Receptor tyrosine kinases (RTKs) in breast
cancer: Signaling, therapeutic implications and challenges. Mol
Cancer. 17:342018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ren C, Han X, Lu C, Yang T, Qiao P, Sun Y
and Yu Z: Ubiquitination of NF-κB p65 by FBXW2 suppresses breast
cancer stemness, tumorigenesis, and paclitaxel resistance. Cell
Death Differ. 29:381–392. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou Y, Pang J, Liu H, Cui W, Cao J and
Shi G: Fibronectin type III domain-containing protein 5 promotes
autophagy via the AMPK/mTOR signaling pathway in hepatocellular
carcinoma cells, contributing to nab-paclitaxel chemoresistance.
Med Oncol. 40:532022. View Article : Google Scholar
|
|
16
|
Zhao PW, Cui JX and Wang XM: Upregulation
of p300 in paclitaxel-resistant TNBC: Implications for cell
proliferation via the PCK1/AMPK axis. Pharmacogenomics J. 24:52024.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Abedini MR, Muller EJ, Bergeron R, Gray DA
and Tsang BK: Akt promotes chemoresistance in human ovarian cancer
cells by modulating cisplatin-induced, p53-dependent ubiquitination
of FLICE-like inhibitory protein. Oncogene. 29:11–25. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Caunt CJ and Keyse SM: Dual-specificity
MAP kinase phosphatases (MKPs): Shaping the outcome of MAP kinase
signalling. FEBS J. 280:489–504. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cargnello M and Roux PP: Activation and
function of the MAPKs and their substrates, the MAPK-activated
protein kinases. Microbiol Mol Biol Rev. 75:50–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Roux PP and Blenis J: ERK and p38
MAPK-activated protein kinases: A family of protein kinases with
diverse biological functions. Microbiol Mol Biol Rev. 68:320–344.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hoornaert I, Marynen P, Goris J, Sciot R
and Baens M: MAPK phosphatase DUSP16/MKP-7, a candidate tumor
suppressor for chromosome region 12p12-13, reduces BCR-ABL-induced
transformation. Oncogene. 22:7728–7736. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Low HB and Zhang Y: Regulatory roles of
MAPK phosphatases in cancer. Immune Netw. 16:85–98. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lu H, Tran L, Park Y, Chen I, Lan J, Xie Y
and Semenza GL: Reciprocal regulation of DUSP9 and DUSP16
expression by HIF1 controls ERK and p38 MAP kinase activity and
mediates chemotherapy-induced breast cancer stem cell enrichment.
Cancer Res. 78:4191–4202. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Keyse SM: Dual-specificity MAP kinase
phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 27:253–261.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Polák L, Škoda P, Riedlová K, Krivák R,
Novotný M and Hoksza D: PrankWeb 4: A modular web server for
Protein-ligand binding site prediction and downstream analysis.
Nucleic Acids Res. 53:W466–W471. 2025. View Article : Google Scholar
|
|
26
|
Shi Y, Wang J, Tao S, Zhang S, Mao L, Shi
X, Wang W, Cheng C, Shi Y and Yang Q: miR-142-3p improves
paclitaxel sensitivity in resistant breast cancer by inhibiting
autophagy through the GNB2-AKT-mTOR pathway. Cell Signal.
103:1105662023. View Article : Google Scholar
|
|
27
|
Mishra T, Gupta S, Rai P, Khandelwal N,
Chourasiya M, Kushwaha V, Singh A, Varshney S, Gaikwad AN and
Narender T: Anti-adipogenic action of a novel oxazole derivative
through activation of AMPK pathway. Eur J Med Chem. 262:1158952023.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu Y, Wang A, Zhang S, Kim J, Xia J,
Zhang F, Wang D, Wang Q and Wang J: Paclitaxel-loaded ginsenoside
Rg3 liposomes for drug-resistant cancer therapy by dual targeting
of the tumor microenvironment and cancer cells. J Adv Res.
49:159–173. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fleisher TA: Apoptosis. Ann Allergy Asthma
Immunol. 78:245–950. 1997. View Article : Google Scholar
|
|
31
|
Wu M, Xue L, Chen Y, Tang W, Guo Y, Xiong
J, Chen D, Zhu Q, Fu F and Wang S: Inhibition of checkpoint kinase
prevents human oocyte apoptosis induced by chemotherapy and allows
enhanced tumour chemotherapeutic efficacy. Hum Reprod.
38:1769–1783. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang BR, Han JB, Jiang Y, Xu S, Yang R,
Kong YG, Tao ZZ, Hua QQ, Zou Y and Chen SM: CENPN suppresses
autophagy and increases paclitaxel resistance in nasopharyngeal
carcinoma cells by inhibiting the CREB-VAMP8 signaling axis.
Autophagy. 20:329–348. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Habib TN, Altonsy MO, Ghanem SA, Salama MS
and Hosny MA: Optimizing combination therapy in prostate cancer:
Mechanistic insights into the synergistic effects of Paclitaxel and
Sulforaphane-induced apoptosis. BMC Mol Cell Biol. 25:52024.
View Article : Google Scholar
|
|
34
|
Lin YW, Lin TT, Chen CH, Wang RH, Lin YH,
Tseng TY, Zhuang YJ, Tang SY, Lin YC, Pang JY, et al: Enhancing
efficacy of Albumin-bound paclitaxel for human lung and colorectal
cancers through autophagy receptor sequestosome 1
(SQSTM1)/p62-mediated nanodrug delivery and cancer therapy. ACS
Nano. 17:19033–19051. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Albuquerque T, Neves AR, Paul M, Biswas S,
Vuelta E, García-Tuñón I, Sánchez-Martin M, Quintela T and Costa D:
A Potential effect of circadian rhythm in the Delivery/therapeutic
performance of Paclitaxel-dendrimer nanosystems. J Funct Biomater.
14:3622023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim JH, Lee JO, Kim N, Lee HJ, Lee YW, Kim
HI, Kim SJ, Park SH and Kim HS: Paclitaxel suppresses the viability
of breast tumor MCF7 cells through the regulation of EF1α and
FOXO3a by AMPK signaling. Int J Oncol. 47:1874–1880. 2015.
View Article : Google Scholar
|
|
37
|
Tang Z, Zhang Y, Yu Z and Luo Z: Metformin
suppresses stemness of Non-small-cell lung cancer induced by
paclitaxel through FOXO3a. Int J Mol Sci. 24:166112023. View Article : Google Scholar
|
|
38
|
Yuan J, Dong X, Yap J and Hu J: The MAPK
and AMPK signalings: Interplay and implication in targeted cancer
therapy. J Hematol Oncol. 13:1132020. View Article : Google Scholar
|
|
39
|
Chu J, Panfen E, Wang L, Marino A, Chen
XQ, Fancher RM, Landage R, Patil O, Desai SD, Shah D, et al:
Evaluation of encequidar as an intestinal P-gp and BCRP specific
inhibitor to assess the role of intestinal P-gp and BCRP in
Drug-drug interactions. Pharm Res. 40:2567–2584. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rieske P, Krynska B and Azizi SA: Human
Fibroblast-derived cell lines have characteristics of embryonic
stem cells and cells of Neuro-ectodermal origin. Differentiation.
73:474–483. 2005. View Article : Google Scholar : PubMed/NCBI
|