Introduction
Chemokines are a large family of chemotractant
cytokines involved in the regulation of cell trafficking which are
believed to play a significant role in influencing tumour
progression (1–3). Chemokine signalling influences the
recruitment and movement of leukocytes and other cell types and can
also provide proliferative, survival and angiogenic stimuli
(4,5). It is well recognised that normal cell
types play an essential role in tumour progression and that
chemokines and other soluble factors derived from tumour cells are
involved in recruitment of these cells and modulation of their
activity. In this manner chemokines produced by tumour cells may
play a role directly or indirectly in tumour neovascularisation and
could also modify anti-tumour immune response. The effect of
malignant transformation upon the regulation of chemokine
expression is of interest because of these implications for
biological therapies which target tumour vasculature and for
immunotherapies.
A major focus of our laboratory has been the
investigation of immunological therapies to induce immunity to
malignant mesothelioma (MM) in syngeneic mouse models of the
disease. Mesothelioma is a particularly aggressive malignancy of
the serosal surfaces which is most frequently associated with
exposure to asbestos (reviewed in ref. 6). The cell lines used in these studies
provide one of the few animal models of cancer in which the
original tumours have been induced by the agent which is believed
to be the causative agent of the human tumour (i.e. asbestos), and
have been extensively characterised both in this laboratory
(7–9) and by others (10,11). A
feature of these murine tumours is a prominent inflammatory
infiltrate composed predominantly of monocytes which may comprise
upward of 50% of the cells (10). A
number of studies have shown that mesothelial cells can regulate
leukocyte trafficking during inflammation through chemokine
production (12–15) and various researchers have
demonstrated in vitro that inflammatory cytokines can
regulate chemokine expression in mesothelial cells (15–17).
However, although IL-8 has been implicated in the progression of MM
in experimental models (18,19)
the function and regulation of chemokine networks has not been
characterised in mesothelial derived malignancy.
The aim of the present study was to determine the
capacity of mesothelioma cells from different mouse strains to
express and regulate chemokine genes in response to inflammatory
mediators. Primary mouse mesothelial cultures from these mouse
strains were used as a basis for comparison and studies using these
also provided useful insights into both mesothelial cell biology
and strain specific chemokine responses.
The CCL chemokines MCP-1/JE (CCL2) and RANTES (CCL5)
have been implicated in the recruitment of tumour associated
macrophages in both mouse tumour models and human disease (2,20). The
murine angiogenic CXC chemokines, GRO-α/KC and MIP-2 are homologues
of the human GRO chemokines and are believed to serve similar
functions in the mouse to IL-8 (CXCL8) in humans (21). Since chemokine production by tumour
cells is likely to be relevant to tumour progression both by
influencing the intratumoural microenvironment and also as a
response to immunotherapy the effects of both Type I (IFN-γ and
TNF-α) and Type II (IL-4) cytokines, each of which may be
elaborated by immune effector cells or has been used in
immunotherapeutic approaches to cancer therapy, were examined. The
availability of both malignant and normal primary cell cultures
derived from two mouse strains, CBA/CaH and Balb/c, allowed
investigation of differences in chemokine gene expression as a
consequence of malignancy and genetic background.
Materials and methods
Mesothelioma cell culture and
reagents
The murine MM cell lines AB1, AB12, AC29, AC34 and
AE17 were used in this study. These cell lines were originally
derived by intraperitoneal inoculation of crocidolite asbestos in
BALB/C mice (AB1 and AB12), CBA/CaH mice (AC29 and AC34) or C57BL/J
mice (AE17) as described previously (7,22). All
cells were cultured and maintained in medium R5, which is RPMI-1640
plus 5% heat-inactivated foetal bovine serum (FBS) (Invitrogen,
Victoria, Australia), 300 mM L-glutamine (Invitrogen), 120 μg/ml
penicillin (Invitrogen) and 100 μg/ml gentamicin (Invitrogen). All
cell cultures were grown at 37°C in a 5% CO2 humidified
atmosphere.
Mesothelial cell culture
Normal mesothelial cells were isolated from the
anterior peritoneal wall of 8–10-week-old female Balb/c or CBA/CaH
mice (Animal Resource Centre, Murdoch, WA, Australia) essentially
as described by Foley-Comer et al(23). Briefly, peritoneal tissue from 3
mice was incubated in 0.25% trypsin and 0.02% EDTA in DMEM medium
(Trace Scientific, Victoria, Australia) at 37°C for 30 min with
gentle agitation. The tissue was removed and cells collected by
centrifugation at 1000 rpm (200 g) for 5 min at room temperature.
Cells were resuspended and transferred to culture flasks in
mesothelial culture medium, consisting of DMEM plus 15% FBS
(Invitrogen), 5 ng/ml epidermal growth factor (Roche Diagnostics,
N.S.W., Australia), 0.4 μg/ml hydrocortisone, 4 mM L-glutamine
(Invitrogen), 120 penicillin 100 U/ml (Invitrogen) and 50 μg/ml
streptomycin (Invitrogen). All cell cultures were grown at 37°C in
a 5% CO2 humidified atmosphere. Peritoneal mesothelial
cells (PMC) demonstrated a characteristic cobblestone morphology in
culture and were used at passages 2–3.
Chemokine expression and release
experiments
To determine the concentration-dependent effect of
cytokines upon MM and PMC, chemokine expression and release,
5×105 cells/well were seeded in 6-well plates and
cultured for 24 h. The cultures were then incubated for 24 h with
various concentrations of one of three cytokines: IFN-γ
(Sigma-Aldrich, N.S.W., Australia, 1–100 ng/ml), TNF-α (Sigma,
1–100 ng/ml) or IL-4 (Sigma, 1–100 ng/ml). Total cellular RNA was
extracted from cells as described below. Culture supernatants were
harvested, clarified by centrifugation and stored at −80°C for
ELISA. All experiments were conducted in triplicate.
Reverse transcription PCR (RT-PCR) and
real-time RT-PCR
Total RNA was prepared from cultures of cell lines
using Ultraspec reagent (Biotecx, TX, USA) according to the
manufacturer’s instructions, resuspended in 1 mM sodium citrate and
stored at −80°C. Prior to RT-PCR, contaminating DNA was removed
from the RNA using RQ1 DNAse (Promega, N.S.W., Australia). First
strand cDNA synthesis was carried out using AMV reverse
transcriptase (Reverse Transcription System, Promega) and 1 μg
total RNA primed with random hexamers in a final volume of 20 μl.
Gene specific PCR primers were designed using the Primer 3 software
(24) and sequences are shown in
Table I. Conventional PCR was
performed in a 25 μl reaction comprising 2 mM MgCl2, DNA
polymerase buffer (Fisher-Biotec, W.A., Australia), 200 μM dNTP,
100 nM each primer, 1U Taq DNA polymerase (Fisher-Biotec), and 2 μl
of cDNA. Amplification reactions were run in PTC-100 (MJ Research,
MA, USA) cyclers and amplified products analysed by agarose gel
electrophoresis, then photographed using a Kodak EDAS 120 digital
camera system.
 | Table IOligonucleotide primers. |
Table I
Oligonucleotide primers.
| Gene | Primer sequence
5′-3′ | GenBank acc. no. |
|---|
| MCP-1/JE |
CAGCACCAGCCAACTCTCACT
AAGGCATCACAGTTCGAGTCA | NM_011333 |
| GRO-α/KC |
CACCATGATCCCAGCCACCCG
TTACTTGGGGACACCTTTTAG | NM_008176 |
| RANTES |
CCCTCACCATCATCCTCACT
CCTTCGAGTGACAAACACGA | NM_013653 |
| MIP-2 |
CACTTCAGCCTAGCGCCAT
GTCAGTTAGCCTTGCCTTTG | NM_009140 |
| MIP-1α |
CCTCTGTCACCTGCTCAACA
GATGAATTGGCGTGGAATCT | NM_011337 |
| CSF1 |
GACCCTCGAGTCAACAGAGC
TGTCAGTCTCTGCCTGGATG | NM_007778 |
| CSF2 |
TGGTCTACAGCCTCTCAGCA
CCGTAGACCCTGCTCGAATA | NM_009969 |
| CCR2 |
GGGTCATGATCCCTATGTGG
TCCATGAGCAGTGGTTTGAA | NM_009915 |
| CXCR2 |
CATCAGCATGGACCGCTAC
GCAGGGCCAGAATTACTGAT | NM_009909 |
Real-time PCR was performed using a RotorGene 2000
real-time amplification instrument (Corbett Research, N.S.W.,
Australia) with Sybr Green I detection chemistry. Reactions were
performed in a 20 μl volume with 2 μl cDNA, 2–4 mM
MgCl2, 200 μM dNTP, 1 U Taq DNA polymerase
(Fisher-Biotec), 0.1–0.5 μM of each primer, 5×10−5 SYBR
Green I (Invitrogen), DNA polymerase buffer (Fisher-Biotec). A
typical protocol comprised 95°C for 5 min followed by 45 cycles of
95°C for 20 sec, 55–60°C for 20 sec, 72°C for 45 sec and 80–87°C
for 15 sec, then an additional 60 sec at 72°C. Fluorescence data
were acquired at 72°C and 80–87°C (optimised for each assay).
Primer and MgCl2 concentration as well as annealing
temperature were optimised for each assay. To confirm amplification
specificity a melt curve analysis was performed at the end of each
run.
Standard curves were generated using serially
diluted cDNA. Real-time PCR assays were conducted in duplicate for
each sample and control. The threshold cycle (CT) was determined
automatically by the RotorGene software (v4.3) using the dynamic
tube normalisation setting. In order to allow for sample-to-sample
variability, gene expression data were normalised to levels of
expression of reference (housekeeping) genes. There were four
reference gene assays for which the primers were: hypoxanthine
phosphoribosyl-transferase 1 (HPRT1): forward
(5′-tgacactggtaaaacaatgca-3′), reverse
(5′-ggtccttttcaccagcaagct-3′); glyceraldehyde-3-phosphate
dehydrogenase (G3PDH): forward (5′-accacagtccatgccatcac-3′),
reverse (5′-tccaccaccctgttgctgta-3′); ubiquitin C (UBC): forward
(5′-aggtcaaacaggaagacagacgta-3′), mouse reverse
(5′-tcacacccaagaacaagcaca-3′); 18S ribosomal RNA (18S): forward
(5′-gtaacc cgttgaaccccatt-3′), reverse (5′-ccatccaatc
gtagtagcg-3′). Primer sequences for the UBC and HPRT1 genes were
obtained from the RTPrimerDB at medgen. ugent.be/rtprimerdb
(25). Real-time assays were
performed on the relevant samples and the most stable reference
genes were determined using the geNorm software (v3.3) and used to
generate a normalisation factor for each sample essentially as
described by Vandesompele et al(26). Relative expression of the target
gene was normalised using this factor and expressed as mean ±
standard deviation relative to a control or calibrator sample.
ELISA
MCP-1 levels in culture supernatants were
quantitated by sandwich ELISA assay. An OptEIA mouse MCP-1 ELISA
set (BD Biosciences, N.S.W., Australia) with a detection limit of
30 pg/ml MCP-1 was used according to the manufacturer’s
instructions.
Results
Expression of CXC and CC chemokine mRNA
in mesothelioma cells
Unstimulated cultures of mouse mesothelioma cell
lines and PMC were surveyed for the expression of specific
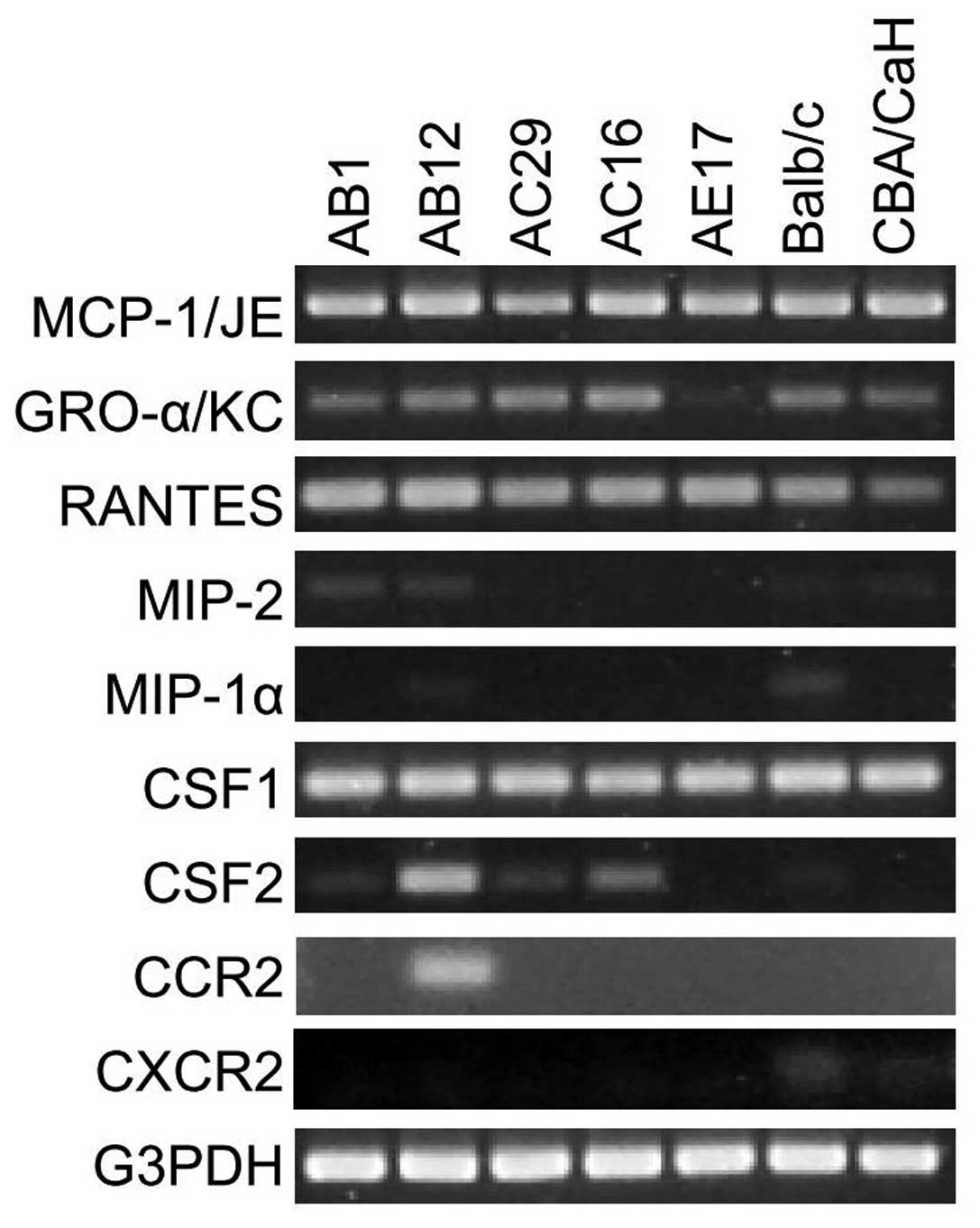
chemokine mRNAs using conventional RT-PCR (Fig. 1). The mRNAs for MCP-1/JE, GRO-α/KC
and RANTES were detectable in all mouse mesothelioma cell lines
(except GRO-α/KC in AE17) and PMC whereas MIP-1α was very faintly
detectable in Balb/c PMC and MIP-2 expression was absent (Fig. 1). We also examined expression of
CSF1 and CSF2, two molecules which can function as monocyte
chemoattractants (27); CSF1 was
detected in all mouse MM and PMC cultures while CSF2 was detected
in AB12 and weakly in AC16.
We also examined the relative basal expression of
MCP-1/JE, GRO-α/KC and CSF1 by performing real-time RT-PCR upon RNA
extracted from cultures of the AB1 and AC29 cell lines as well as
PMC from both Balb/c and CBA/CaH mice. These are the two mouse MM
models used most commonly in our laboratory and elsewhere (10,11).
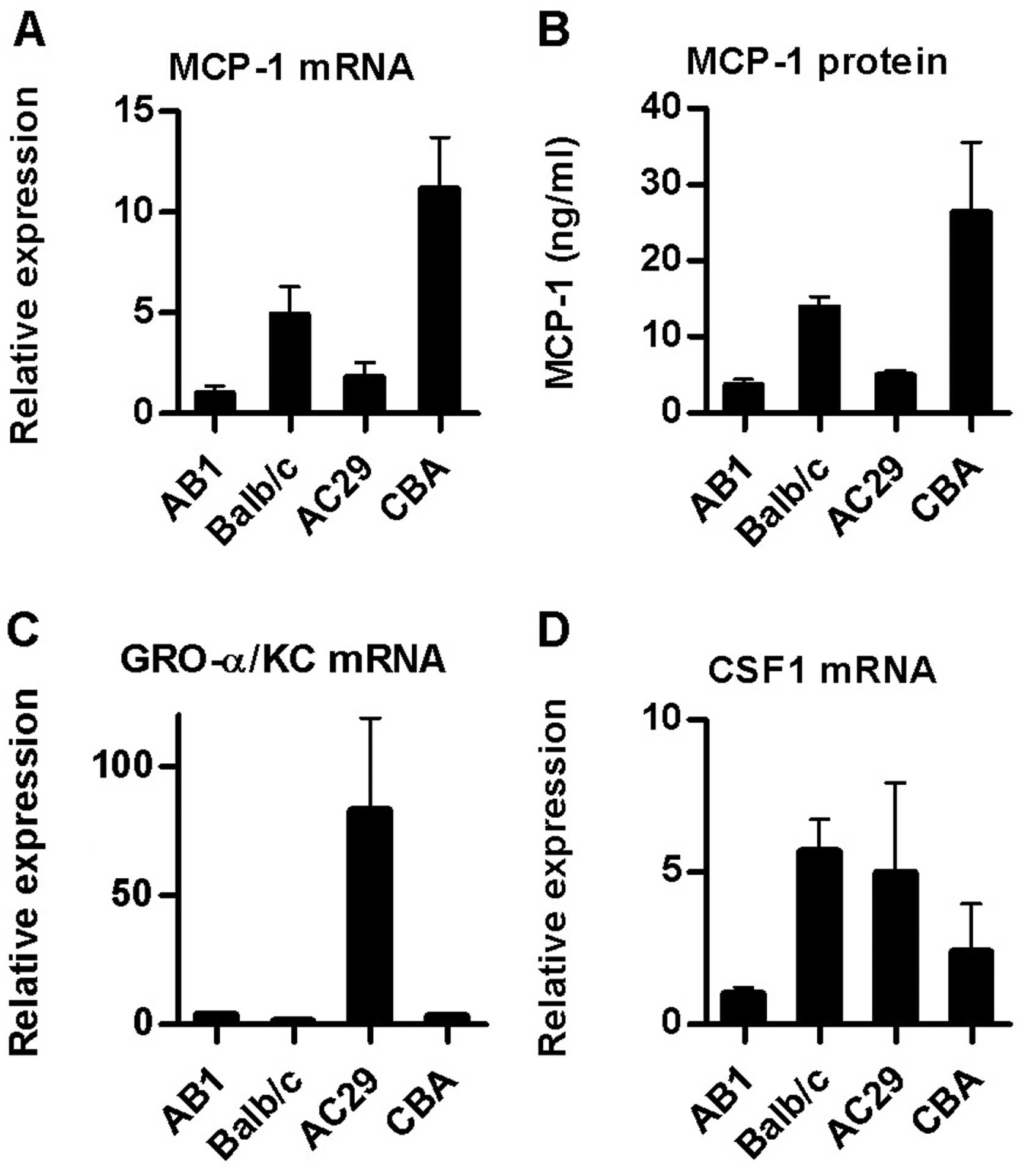
MCP-1/JE mRNA levels were higher in the mouse PMC relative to
mesothelioma cultures (Fig. 2A).
Assay of culture supernatants for MCP-1 protein by ELISA confirmed
these results (Fig. 2B). In
contrast, expression of the CXC chemokine GRO-α/KC was higher in
AC29 cells (26-fold) and to a lesser extent in AB1 (5-fold) than in
normal PMC from the same strain (Fig.
2C).
Expression of the CCR2 and CXCR2
chemokine receptors
Since autocrine chemokine signalling has been
described in some tumour types and mesothelial cell expression of
CCR2 has been reported (28), we
examined expression of CCR2 (receptor for MCP-1) and CXCR2
(receptor for GRO-α) mRNA in murine PMC and MM lines. CCR2 mRNA was
expressed only in the AB12 murine mesothelioma cell line while
CXCR2 mRNA was faintly detectable in PMC cultures but not in any of
the tumour cell lines tested (Fig.
1).
Cytokine regulation of MCP-1 mRNA in
mouse mesothelial and mesothelioma cells
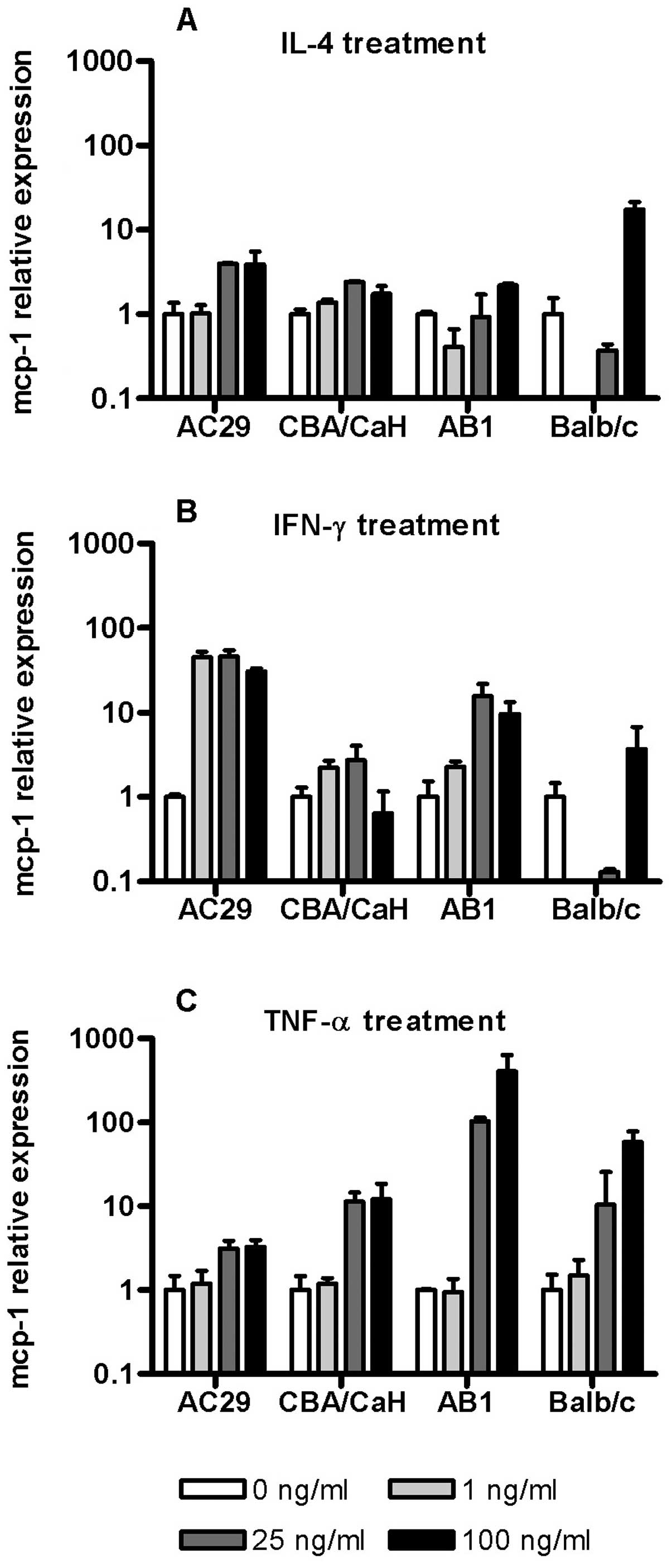
To evaluate the potential for cytokines produced by
tumour cells, stromal cells or inflammatory cells to regulate MCP-1
gene expression we determined the ability of IL-4, IFN-γ and TNF-α
to control MCP-1 expression in mouse AB1 and AC29 MM cell lines as
well as corresponding PMC cultures (Fig. 3). IL-4 induced a modest
dose-dependent increase in MCP-1 mRNA in CBA derived cell but
showed a biphasic response in Balb/c derived cells especially in
PMC with downregulation at low IL-4 concentrations to below the
level of detection while high concentrations resulted in
upregulation (Fig. 3A). A similar
effect was seen when these cells (Balb/c PMC) were exposed to 1–25
ng/ml of IFN-γ (Fig. 3B). Overall
the effects of IFN-γ upon MCP-1 mRNA were more profound in the
tumour cell lines with dose-dependent upregulation especially in
AC29 (45-fold) maximal at 1 ng/ml.
TNF-α consistently upregulated MCP-1 expression at
concentrations ~25 ng/ml in all the cells, although the effects
varied in magnitude (Fig. 3C). In
CBA derived cells PMC were more responsive than tumour cells (AC29)
while in contrast the largest upregulation was seen in AB1 cells
(350-fold). These results indicated that mesothelioma cells retain
and in some cases enhance the ability for cytokine regulated MCP-1
expression. They also reveal strain specific differences in both
normal and tumour cell responses.
MCP-1 release by mouse mesothelial and
mesothelioma cells
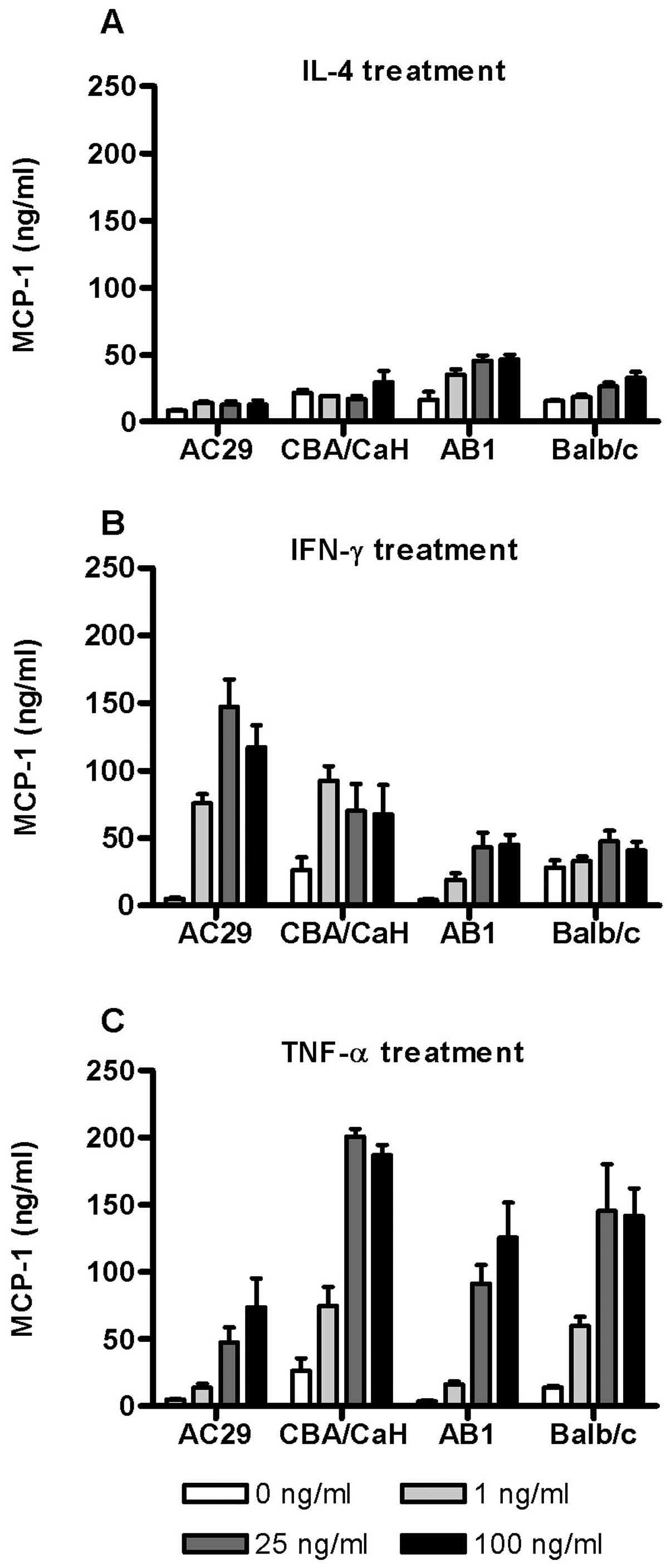
To further examine the dose-dependent effect of
cytokine stimulation on MCP-1 expression, the levels of MCP-1
protein in the corresponding culture supernatants were assayed by
ELISA (Fig. 4). In Balb/c derived
cells (PMC and AB1) IL-4 stimulated a modest dose-dependent release
of MCP-1 but had little effect in those derived from CBA/CaH
(Fig. 4A). In contrast IFN-γ
stimulated substantial upregulation of MCP-1 release by both AC29
and CBA/CaH cells even at 1 ng/ml with lesser effects in Balb/c
cells (Fig. 4B). By comparison
TNF-α induced MCP-1 release in all the cell types assayed (Fig. 4C). The PMC cultures were
consistently more responsive than the corresponding tumour cell
line although substantial upregulation also occurred. In PMC
(Balb/c and CBA/CaH) maximum MCP-1 levels were induced by 25 ng/ml
TNF-α (200 ng/ml and 145 ng/ml, respectively). The results
indicated that on the whole MCP-1 release is consistent with
changes in gene expression.
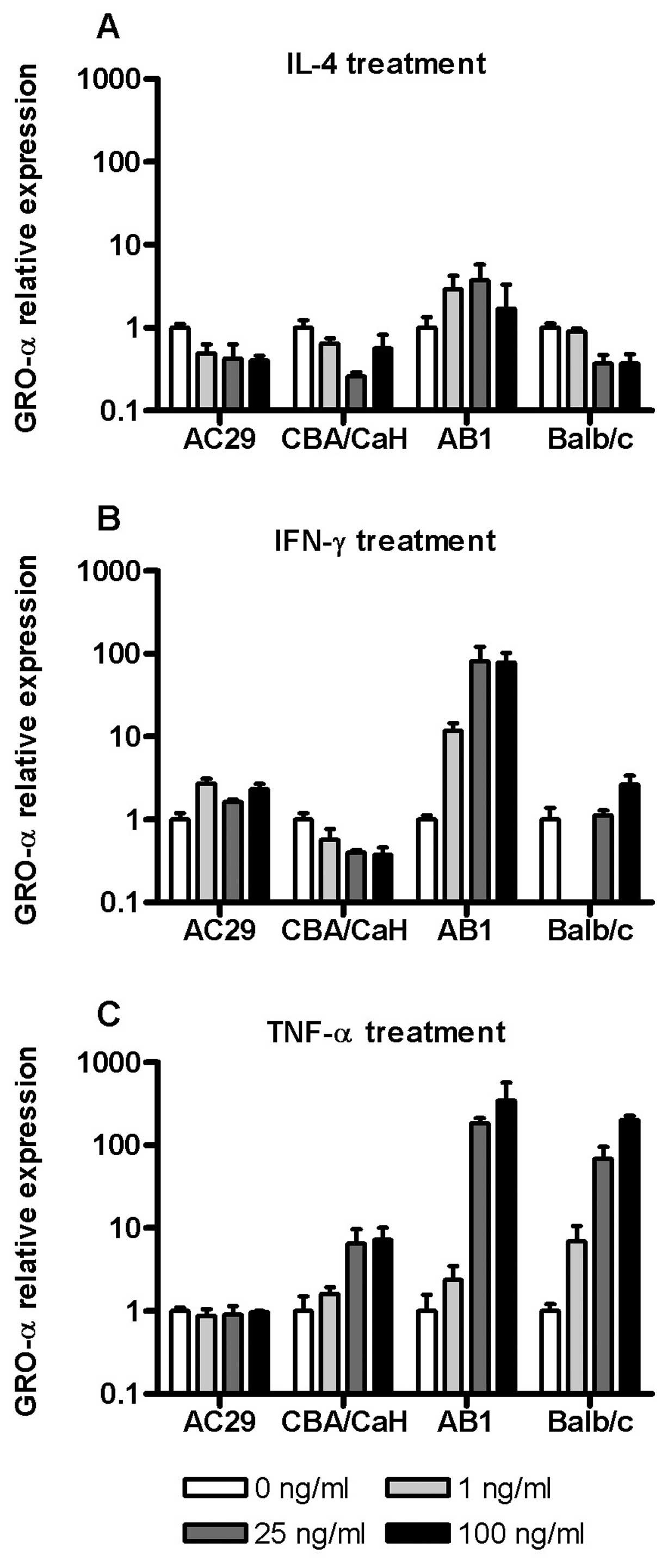
Cytokine regulation of GRO-α/KC and CSF1
mRNA in mouse mesothelial and mesothelioma cells
In addition to the studies of MCP-1 mRNA expression
and protein release described above the mRNA expression of two
other molecules which may impact upon tumourigenesis via chemotaxis
or angiogenesis, GRO-α/KC and CSF1, were also determined in
response to cytokine stimulation. On the whole IL-4 was inhibitory
to GRO-α expression in all cells except AB1 (Fig. 5A). Similarly GRO-α mRNA was
generally insensitive to IFN-γ except in AB1 cells where there was
a large (80-fold) upregulation of this chemokine (Fig. 5B). Of note, in Balb/c PMC we
observed a downregulation of GRO-α mRNA at 1 ng/ml IFN-γ which was
not seen at higher concentrations.
As was found for MCP-1, TNF-α was the most
consistent inducer of GRO-α gene expression with the exception of
AC29 cells which were insensitive (Fig.
5C). GRO-α expression in Balb/c derived PMC and tumour cells
(AB1) were most sensitive to TNF-α with 200- and 400-fold
upregulation, respectively. The expression of CSF-1 mRNA was
relatively insensitive to cytokine exposure in all cells, varying
of the order of 2–3-fold across the dose range (data not
shown).
Discussion
Many cancers express an array of chemokines and
their receptors which may have a role in modulation of the
leukocyte infiltrate. In MM this aspect of tumour biology has
received limited investigation. Macrophages infiltrate many solid
tumours including MM and in animal models of MM this infiltrate is
an early feature of tumour development. This suggests that the
tumour cells may be the source of these recruitment signals.
Previous investigations of mesothelial cells have demonstrated that
cytokines regulate expression and release of chemokines in these
cells and may play an important role in leukocyte recruitment and
trafficking into body cavities (12,14–17).
This prompted us to ask if mesothelioma cells retain this feature
after transformation. To our knowledge the present study is the
first to report on cytokine regulated expression of these molecules
in MM cells. Furthermore, few studies have examined by comparison
the effect of malignant transformation upon regulation of chemokine
gene expression. We found that mesothelioma cells retain the
capacity of their mesothelial counterparts for regulated expression
of these molecules in response to cytokines. In some respects MM
cells displayed a heightened response relative to PMC. As with
other tumour types in which chemokine signalling has been much more
extensively characterised this has implications for the nature of
intercellular signalling within the mesothelioma microenvironment.
In addition to chemokines we examined other factors which might
contribute to monocyte recruitment to mesothelioma tumours and
showed that CSF1 was universally expressed, although expression
levels were not as responsive to cytokines. It was also of
particular interest to be able to compare and contrast regulation
of gene expression between mesothelial and mesothelioma cells
derived from different experimental models.
The CC chemokines MCP-1 and RANTES were detected in
all the mouse MM cell lines examined as well as mouse mesothelial
cultures. Both MCP-1 and RANTES have pro-tumourigenic activities
and have been implicated in monocyte recruitment (3,20). A
prominent macrophage infiltrate is a feature of a number of murine
MM models including AC29 and AB1 (10). Quantitative analysis of MCP-1/JE
basal expression levels in MM cells showed significantly higher
mRNA and protein release in the corresponding mesothelial cultures.
This confounds the notion that constitutive MCP-1 overexpression
contributes to macrophage recruitment in these tumours. However,
cytokine stimulation studies demonstrated that MCP-1 was
significantly upregulated in response particularly to TNF-α.
Previous in vitro studies have demonstrated expression of
functional CCR2 receptors and chemotactic responses to MCP-1 (CCL2)
in human mesothelial cells (28).
More recently, Davidson et al(29) found infrequent chemokine receptor
expression in MM and reactive mesothelium and the absence of CCR2
although CXCR2 was not studied. We did not find expression or
induction of CCR2 mRNA in either murine MM or PMC which was
consistent with this latter study of their reported human
counterparts.
CXC chemokines as a family display a range of
functions including leukocyte chemotaxis and angiogenesis, each of
which has been implicated in tumour progression. IL-8 is the only
chemokine which has been previously implicated in MM progression
through experimental data (18,19)
with evidence of autocrine growth signalling in vitro and of
in vivo tumour growth promotion although this avenue of
research does not appear to have been pursued. The complexity and
interspecies diversity of the chemokine system does not allow for a
direct correlation to be drawn between the roles for human and
murine chemokines. There is no direct mouse homologue for
IL-8/CXCL8, however, it has been demonstrated that the murine
chemokines KC/GRO-α and MIP-2 are functionally analogous (21). Notably, we found almost a 30-fold
overexpression of KC/GRO-α mRNA in the mouse AC29 cell line
relative to mesothelial cultures as well as insensitivity to TNF-α.
Such constitutive overexpression of KC/GRO-α is a feature which has
been implicated in the tumourigenicity of other experimental
tumours (30,31). The absence of expression of the
receptor CXCR2, even after cytokine stimulation, suggests that
autocrine signalling by KC/GRO-α in murine MM cells, at least via
this pathway, may not be prominent in these tumour cells.
In cytokine stimulation studies TNF-α was
consistently the most potent upregulator of chemokine expression
and MCP-1 release with the exception of the mouse AC29 cells. This
has implications for in vivo chemokine production and is
consistent with some current hypotheses regarding the role of
tumour cells in shaping their microenvironment (32,33).
We have shown that in vitro, MM cells are quite resistant to
the cytotoxic effects of TNF-α (34) and in murine tumours TNF-α levels are
quite high (10). Previously in one
murine MM tumour line we observed growth promotion by TNF-α even at
100 ng/ml (34). Both MCP-1 and
RANTES have been shown to induce TNF-α production in macrophages
(20). There is ample evidence
supporting involvement of tumour cell derived chemokines and other
factors in macrophage recruitment and gene expression. The
induction of TNF-α which may in turn participate in local
signalling to enhance tumour cell chemokine production as reported
here and elsewhere (32,33,35),
is one mechanism by which tumour cell-macrophage crosstalk may
enhance tumour growth.
Strain specific differences in the mouse immune
response have been widely reported, however, few studies have
investigated differential chemokine responses in mouse strains
(36–38). Although there were differences in
the basal gene expression levels in PMC cultures from CBA/CaH and
Balb/c mice, differences in the response of PMC from the two
strains to cytokine exposure were more evident (e.g. GRO-α in
response to TNF-α, Fig. 5C). Our
findings support previous suggestions that such strain specific
differences need to be considered when interpreting and designing
studies of mesothelial inflammation in mouse models.
Current evidence regarding the role of inflammatory
mediators and leukocytes in cancer progression indicates that in
many cancers there is a complex interaction with tumour cells which
engenders an environment favourable to tumour growth. The data
presented in this study demonstrate that mesothelioma tumour cells
express a variety of chemokine genes and that these genes are
regulated in response to cytokines known to be present in the
tumour microenvironment. Importantly, this responsiveness may
actually be enhanced in malignant cells. Of particular interest was
TNF-α which upregulated both CC and CXC chemokines in MM cells and
may participate in amplifying paracrine signalling loops. It may
prove possible to target these pathways in MM and our
characterisation here of murine models provides avenues for
investigating these possibilities in vivo.
Acknowledgements
We are grateful to Steven Mutsaers for useful
discussions and assistance with mesothelial cell cultures. We would
like to thank Delia Nelson for critical reading of the manuscript.
Funding assistance from the Cancer Council of Western Australia is
gratefully acknowledged.
References
|
1
|
Robinson SC and Coussens LM: Soluble
mediators of inflammation during tumor development. Adv Cancer Res.
93:159–187. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mantovani A, Allavena P, Sozzani S, Vecchi
A, Locati M and Sica A: Chemokines in the recruitment and shaping
of the leukocyte infiltrate of tumors. Semin Cancer Biol.
14:155–160. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Balkwill F: Chemokine biology in cancer.
Semin Immunol. 15:49–55. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rollins BJ: Inflammatory chemokines in
cancer growth and progression. Eur J Cancer. 42:760–767. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Strieter RM, Burdick MD, Mestas J,
Gomperts B, Keane MP and Belperio JA: Cancer CXC chemokine networks
and tumour angiogenesis. Eur J Cancer. 42:768–778. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Robinson BW, Musk AW and Lake RA:
Malignant mesothelioma. Lancet. 366:397–408. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Davis MR, Manning LS, Whitaker D, Garlepp
MJ and Robinson BW: Establishment of a murine model of malignant
mesothelioma. Int J Cancer. 52:881–886. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fox SA, Loh S, Thean AL and Garlepp MJ:
Identification of differentially expressed genes in murine
mesothelioma cell lines of differing tumorigenicity using
suppression subtractive hybridization. Biochim Biophys Acta.
1688:237–244. 2004. View Article : Google Scholar
|
|
9
|
Leong CC, Marley JV, Loh S, Robinson BW
and Garlepp MJ: The induction of immune responses to murine
malignant mesothelioma by IL-2 gene transfer. Immunol Cell Biol.
75:356–359. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bielefeldt-Ohmann H, Fitzpatrick DR, Marzo
AL, et al: Patho- and immunobiology of malignant mesothelioma:
characterisation of tumour infiltrating leucocytes and cytokine
production in a murine model. Cancer Immunol Immunother.
39:347–359. 1994. View Article : Google Scholar
|
|
11
|
Hegmans JP, Hemmes A, Aerts JG, Hoogsteden
HC and Lambrecht BN: Immunotherapy of murine malignant mesothelioma
using tumor lysate-pulsed dendritic cells. Am J Respir Crit Care
Med. 171:1168–1177. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li FK, Davenport A, Robson RL, et al:
Leukocyte migration across human peritoneal mesothelial cells is
dependent on directed chemokine secretion and ICAM-1 expression.
Kidney Int. 54:2170–2183. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mazar J, Agur T, Rogachev B, et al: CD40
ligand (CD154) takes part in regulation of the transition to
mononuclear cell dominance during peritonitis. Kidney Int.
67:1340–1349. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nasreen N, Mohammed KA, Hardwick J, et al:
Polar production of interleukin-8 by mesothelial cells promotes the
transmesothelial migration of neutrophils: role of intercellular
adhesion molecule-1. J Infect Dis. 183:1638–1645. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Robson RL, McLoughlin RM, Witowski J, et
al: Differential regulation of chemokine production in human
peritoneal mesothelial cells: IFN-gamma controls neutrophil
migration across the mesothelium in vitro and in vivo. J Immunol.
167:1028–1038. 2001. View Article : Google Scholar
|
|
16
|
Mohammed KA, Nasreen N, Ward MJ and Antony
VB: Helper T cell type 1 and 2 cytokines regulate C-C chemokine
expression in mouse pleural mesothelial cells. Am J Respir Crit
Care Med. 159:1653–1659. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Visser CE, Tekstra J, Brouwer-Steenbergen
JJ, et al: Chemokines produced by mesothelial cells: huGRO-alpha,
IP-10, MCP-1 and RANTES. Clin Exp Immunol. 112:270–275. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Galffy G, Mohammed KA, Nasreen N, Ward MJ
and Antony VB: Inhibition of interleukin-8 reduces human malignant
pleural mesothelioma propagation in nude mouse model. Oncol Res.
11:187–194. 1999.PubMed/NCBI
|
|
19
|
Galffy G, Mohammed KA, Dowling PA, Nasreen
N, Ward MJ and Antony VB: Interleukin 8: an autocrine growth factor
for malignant mesothelioma. Cancer Res. 59:367–371. 1999.PubMed/NCBI
|
|
20
|
Robinson SC, Scott KA, Wilson JL, Thompson
RG, Proudfoot AE and Balkwill FR: A chemokine receptor antagonist
inhibits experimental breast tumor growth. Cancer Res.
63:8360–8365. 2003.PubMed/NCBI
|
|
21
|
Bozic CR, Gerard NP, von
Uexkull-Guldenband C, et al: The murine interleukin 8 type B
receptor homologue and its ligands. Expression and biological
characterization. J Biol Chem. 269:29355–29358. 1994.PubMed/NCBI
|
|
22
|
Jackaman C, Bundell CS, Kinnear BF, et al:
IL-2 intratumoral immunotherapy enhances CD8+ T cells
that mediate destruction of tumor cells and tumor-associated
vasculature: a novel mechanism for IL-2. J Immunol. 171:5051–5063.
2003.PubMed/NCBI
|
|
23
|
Foley-Comer AJ, Herrick SE, Al-Mishlab T,
Prele CM, Laurent GJ and Mutsaers SE: Evidence for incorporation of
free-floating mesothelial cells as a mechanism of serosal healing.
J Cell Sci. 115:1383–1389. 2002.PubMed/NCBI
|
|
24
|
Rozen S and Skaletsky H: Primer3 on the
WWW for general users and for biologist programmers. Methods Mol
Biol. 132:365–386. 2000.PubMed/NCBI
|
|
25
|
Pattyn F, Speleman F, De Paepe A and
Vandesompele J: RTPrimerDB: the real-time PCR primer and probe
database. Nucleic Acids Res. 31:122–123. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vandesompele J, De Preter K, Pattyn F, et
al: Accurate normalization of real-time quantitative RT-PCR data by
geometric averaging of multiple internal control genes. Genome
Biol. 3:Research00342002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin EY, Nguyen AV, Russell RG and Pollard
JW: Colony-stimulating factor 1 promotes progression of mammary
tumors to malignancy. J Exp Med. 193:727–740. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nasreen N, Mohammed KA, Galffy G, Ward MJ
and Antony VB: MCP-1 in pleural injury: CCR2 mediates haptotaxis of
pleural mesothelial cells. Am J Physiol Lung Cell Mol Physiol.
278:L591–L598. 2000.PubMed/NCBI
|
|
29
|
Davidson B, Dong HP, Holth A, Berner A and
Risberg B: Chemokine receptors are infrequently expressed in
malignant and benign mesothelial cells. Am J Clin Pathol.
127:752–759. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Loukinova E, Dong G, Enamorado-Ayalya I,
et al: Growth regulated oncogene-alpha expression by murine
squamous cell carcinoma promotes tumor growth, metastasis,
leukocyte infiltration and angiogenesis by a host CXC receptor-2
dependent mechanism. Oncogene. 19:3477–3486. 2000. View Article : Google Scholar
|
|
31
|
Zhou Y, Zhang J, Liu Q, et al: The
chemokine GRO-alpha (CXCL1) confers increased tumorigenicity to
glioma cells. Carcinogenesis. 26:2058–2068. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hagemann T, Wilson J, Burke F, et al:
Ovarian cancer cells polarize macrophages toward a tumor-associated
phenotype. J Immunol. 176:5023–5032. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Szlosarek P, Charles KA and Balkwill FR:
Tumour necrosis factor-alpha as a tumour promoter. Eur J Cancer.
42:745–750. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fox SA, Kusmiaty, Loh SS, Dharmarajan AM
and Garlepp MJ: Cisplatin and TNF-alpha downregulate transcription
of Bcl-xL in murine malignant mesothelioma cells. Biochem Biophys
Res Commun. 337:983–991. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yao PL, Lin YC, Wang CH, et al: Autocrine
and paracrine regulation of interleukin-8 expression in lung cancer
cells. Am J Respir Cell Mol Biol. 32:540–547. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Charles PC, Weber KS, Cipriani B and
Brosnan CF: Cytokine, chemokine and chemokine receptor mRNA
expression in different strains of normal mice: implications for
establishment of a Th1/Th2 bias. J Neuroimmunol. 100:64–73. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Darville T, Andrews CW Jr, Sikes JD,
Fraley PL, Braswell L and Rank RG: Mouse strain-dependent chemokine
regulation of the genital tract T helper cell type 1 immune
response. Infect Immun. 69:7419–7424. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Weinberg JB, Lutzke ML, Alfinito R and
Rochford R: Mouse strain differences in the chemokine response to
acute lung infection with a murine gammaherpesvirus. Viral Immunol.
17:69–77. 2004. View Article : Google Scholar : PubMed/NCBI
|