Introduction
Tumor microenvironment is recognized as the product
of a developing crosstalk between different cells types. In
addition to tumor cells, tumor microenvironment is comprised of
immune cells, fibroblasts, stromal cells and extracellular matrix
(1). Fibroblasts within the tumor
stroma have been termed tumor-associated fibroblasts (TAFs)
(2,3) and play important roles in the
initiation, progression, and metastasis of cancer (4).
Dendritic cells (DCs) were able to recognize tumor
antigens and activate tumor-specific T-cell response (5). However, this response does not usually
occur in most types of human cancer or in animal models, indicating
that the dysfunction or immuno-suppression of host immune system
might be the main mechanisms by which tumors escape from host
immune control (6–8). Previously, we co-incubated DCs with
different tumor cells and found that co-incubation of tumor
cell-DCs down-regulated the abilities of DCs in T cell
proliferation, IL-12 secretion and tumor antigenic-specific CTL
priming (9). Also, Yuan et
al established an in vitro incubation system including
gastric cancer cell line, with sorting CD4+ T cells and
found that gastric cancer cells induced human
CD4+Foxp3+ regulatory T cells (Tregs) through
the production of transforming growth factor β1 (TGF-β1) (10). All these co-incubation systems only
included relative tumor immune cells to observe the direct effect
of tumor cells on immune cells. Although a novel 3-dimensional (3D)
culturing system was employed to recapitulate stromal and
extracellular matrix interactions (11), little is known about the effects of
tumor cells and TAFs on immune cells. Therefore, an in vitro
tumor micro-environmental model established of tumor cells,
fibroblasts and immune cells is urgently needed to explore the
effects of tumor cells and TAFs on DCs.
Foxp3+ Tregs, including thymic-derived
natural Treg and conventional T-derived adaptive Treg cells, are
proposed to play important roles in tumor associated
immuno-suppression. However, the mechanisms of Treg expansion in
tumor microenvironment remain unclear. Programmed cell death 1
ligand 1 (PD-L1) known as B7 homolog 1 (B7H1) and
glucocorticoid-induced TNFR-related protein ligand (GITRL),
expressing DCs, were reported to be candidate immune modulation
molecules (12–15). Yi et al reported that host
APCs augment in vivo expansion of donor natural regulatory T
cells via B7H1/B7.1 in allogeneic recipients (12), which was further confirmed by high
expression of B7H1 molecules on keratinocytes suppressing xeno- and
allo-reactions by inducing Tregs (13). Although B7H1 had the characteristics
of co-inhibitor molecules of modulating inflammatory response, the
role of B7H1 in tumor associated immuno-suppression is still to be
determined. GITRL-dependent expansion of Tregs underlying immune
privilege in corneal allograft was recently described (14), but blockade of GITR-GITRL
interaction was necessary for maintaining Treg function and
prolonging allograft survival was also reported (15). So, the exact functions of GITRL in
inducing tumor associated Treg are still to be clarified.
In the present study, we firstly characterized that
TGF-β were synthesized by macrophage cell line (Raw 264.7),
lymphoma cell line (EL4) and small cell lung cancer cell line
(NCI-H446) by intracellular flow cytometry analysis and Western
blotting respectively, and further confirmed that TGF-β could
efficiently induce Treg generation in vitro; secondly, TGF-β
treatment clearly up-regulated B7H1 and GITRL expressions of DCs;
thirdly and importantly, a tumor microenvironment co-incubation
system was established by co-incubation of seminaphtharhodafluor
(SNARF) labeled Lewis lung cancer (LLC) cells, carboxyfluorescein
succinimidyl ester (CFSE) labeled fibroblasts and
4-chloromethyl-7-hydroxycoumarin (CMHC) labeled DCs; fourthly and
consistent with TGF-β treatment, the co-incubation of LLC cells,
fibroblasts with DCs could augment the expressions of B7H1 and
GITRL molecules of DCs. The data presented here indicate that B7H1
and GITRL molecules might play an important role in TGF-β-induced
Treg expansion of lung cancer microenvironment.
Materials and methods
Reagents
Mouse recombinant GM-CSF, TGF-β and IL-4 were
obtained from R&D (Minneapolis, MN, USA). Fluorescence
conjugated B7H1, GITRL, CD4, CD25, Foxp3 antibodies, CD16/CD32
antibody and isotype control were from eBioscience (San Diego, CA,
USA). Murine TGF-β and secondary HRP-conjugated goat anti-mouse
antibody were bought from Santa Cruz Biotechnology. Murine
anti-CD3, anti-CD28, anti-IL-4, anti-IFN-γ, IL-2, PMA, ionomycin
were obtained from eBioscience. RPMI-1640 medium, Dulbecco’s
modified Eagle’s medium (DMEM) and fetal bovine serum were
purchased from Hyclone (Logan, UT, USA). Murine CD11c and CD4 beads
were from Miltenyi Biotec GmbH (Bergisch Gladbach, Germany).
CellTracker Blue CMHC, Carboxy SNARF-1, CellTrace CFSE were
obtained from Molecular Probes (Eugene, USA).
Animals
Pathogen-free Balb/c mice (female, 6 to 8-week-old,
or newborn) were bought from Shanghai Laboratory Animal Center of
Chinese Academy of Sciences (China) and kept at the Animal Center
of Xiamen University. All animal studies were approved by the
Review Board, Medical College, Xiamen University.
Cell lines
LLC cell line, established from the lung of a mouse
bearing tumor resulting from implantation of primary Lewis lung
carcinoma, was obtained from the Institute of Biochemistry and Cell
Biology, Chinese Academy of Sciences (ATCC CRL-1642). LLC cells,
NCI-H446 were cultured in DMEM medium (Dulbecco’s modified Eagle’s
medium, 4 mM L-glutamine, 1.5 g/l sodium bicarbonate, 4.5 g/l
glucose, 10% FBS). Raw 264.7 and EL4 cells were cultured in
RPMI-1640 medium with 10% FBS.
Fibroblasts separation from murine
skin
Fibroblasts were separated from murine skin as
previously described (16).
Briefly, skin was firstly taken from newborn Balb/c mice and cut to
1 mm2 size with sterile scissors. Then, tissues were
explanted into culture dishes in equil amounts with DMEM (including
10% FBS) medium. Thirdly, the purification of fibroblasts was
completed by digesting cells with trypsin and re-explanting cells
into dishes.
Bone marrow-derived murine DCs
Bone marrow-derived DCs were prepared as previously
described (17). Briefly, bone
marrow mononuclear cells were prepared from bone marrow suspensions
by depletion of red cells and then were cultured at a density of
1×106 cells/ml in RPMI-1640 medium with 10 ng/ml of
GM-CSF and 1 ng/ml of IL-4. Non-adherent cells were gently washed
out on day 4 of culture; the remaining loosely adherent clusters
were used as DCs.
Labeling cells for analysis with
fluorescence
Cells were labeled according to the product
description. Briefly, cells were re-suspended in pre-warmed
PBS/0.1% BSA at a final concentration of 1×106 cells/ml.
Then, CellTracker Blue CMHC, Carboxy SNARF-1 or CellTrace CFSE was
added at the final concentration of 10 μM, respectively, and
incubated at 37°C. For labeling DCs, the incubation time was 30,
15, 30 min for CMHC, CFSE and SNARF, respectively; for labeling
fibroblasts, the incubation time was 15, 30 min for CFSE, SNARF,
respectively; for labeling LLC cells, the incubation time was 30
min for SNARF. After labeling, the dyes were removed from cells
with pre-warmed PBS and the cells were added with fresh, pre-warmed
medium for further culture.
Co-incubation of DCs and fibroblasts
To establish the DC-fibroblast co-incubation system,
1×106 cells/ml DCs were labeled with 10 μM SNARF for 30
min and fibroblasts were labeled with 10 μM CFSE for 15 min. Then,
DCs and fibroblasts were removed from dishes with 0.05% trypsin and
mixed at a ratio of 1:1 for further co-incubation. After 12-h
co-incubation, the cells were observed and images were recorded by
fluorescence microscope at the wavelength of 488 nm.
Co-incubation of LLC, DCs and
fibroblasts
To establish the LLC-DC-fibroblast co-incubation
system, 1×106 cells/ml LLC cells, DCs and fibroblasts
were labeled with SNARF for 30 min, CMHC for 30 min and CFSE for 15
min, respectively, at the final concentration of 10 μM. Then, DCs,
fibroblasts and LLC cells were removed from dishes with 0.05%
trypsin and mixed at a ratio of 2:2:1 for further co-incubation.
After 12 h co-incubation, the cells were observed and images were
recorded by fluorescence microscope at the wavelength of 488
nm.
MACS separation
DCs of CD4+ T cells were separated from
the LLC-fibroblast-DC co-incubation system and mouse splenocytes
according to the methods previously described (18). Briefly, all cells collected from
co-incubation system or splenocytes were counted and re-suspended
in 400 μl buffer per 108 total cells. Then, 100 μl of
CD11c/CD4 beads was mixed with cells and incubated for 15 min at
4–8°C. After incubation, the cells were washed, and re-suspended in
500 μl buffer. The column was prepared by rinsing with appropriate
amount of buffer and the cell suspension was applied onto the
column. After three washes of column, the labeled cells were
collected by removing the column from the separator and pipetting
appropriate buffer and firmly applying the plunger of the column.
The labeled cells were washed and the assay performed.
Flow cytometric measurement
The expressions of surface molecules on DCs were
determined by flow cytometry according to the methods described
previously (17). Briefly, DCs
separated from the LLC-DC-fibroblast co-incubation system or DCs
treated with TGF-β were incubated for 15 min at 4°C with antibody
to CD16/CD32 at a concentration of 1 μg per 1×106 cells
for blockade of Fc receptors. Cell staining was performed on ice
for 30 min with related fluorescence conjugated antibodies and then
cells were washed with ice-cold PBS, containing 0.1%
NaN3 and 0.5% BSA. Flow cytometry was done with
FACSCalibur and data were analyzed with CellQuest software.
TGF-β quantification of tumor cell
lines
TGF-β of tumor cell lines was determined by the
method of staining intracellular antigens for flow cytometry
(18). Briefly, RAW264.7, EL4 and
NCI-H446 cells were collected from the culture medium and washed
with PBS. Then cells were fixed with IC fixation buffer in the dark
at room temperature for 20 min and further washed twice with
permeabilization buffer. TGF-β antibody and secondary antibody were
added to the cells and incubated in the dark at room temperature
for 20 min to detect intracellular TGF-β expression. The cells were
washed with permeabilization buffer and flow cytometry staining
buffer, respectively, and data were acquired by flow cytometry.
Western blot analysis
For analysis of TGF-β expression of RAW264.7, EL4
and NCI-H446 cells, proteins were obtained in lysis buffer as
previously described (18). Protein
lysates (30 μg/ml) were electrophoresed on 12% SDS-PAGE gels,
transferred to PVDF membranes, and blotted with polyclonal TGF-β
antibodies, followed by anti-mouse horseradish peroxidase and
detected by chemiluminescence ECL. As loading controls, antibodies
against β-actin were used.
Treg expansion induced by TGF-β in
vitro
The expansion of TGF-β induced by Tregs was
performed according to previous description (19). Briefly, CD4+ T
lymphocytes were firstly separated from splenocytes by
CD4+ MACS beads. Then, 5×105 CD4+
T cells were cultured in a 24-well plate in RPMI-1640 medium in the
presence of anti-CD3 (5 μg/ml), anti-CD28 (1 μg/ml), anti-IL-4 (1
μg/ml), anti-IFN-γ (1 μg/ml), IL-2 (50 U/ml), with or without TGF-β
(5 ng/ml) for 3 days. PMA (50 ng/ml), ionomycin (1 μM) was used to
stimulate cells for 4–6 h, then the cells were labeled with CD4,
CD25 and Foxp3 antibodies. Flow cytometry was done with FACSCalibur
and data were analyzed with CellQuest software.
Statistical analysis
The data are expressed as average of experimental
data points, and standard error of means (SEM) were determined
using the calculated standard deviation of a data set. Statistical
significance was tested using Student’s t-test and one-way ANOVA
test by Prism software. Statistical differences were considered to
be significant at p<0.05.
Results
TGF-β was expressed by RAW 264.7, EL4 and
NCI-H446 cells
It was reported that TGF-β plays an important role
in mediating tumor-associated immuno-suppression (20). To investigate TGF-β generation of
tumor microenvironment, macrophage cell line (Raw 264.7), lymphoma
cell line (EL4) and small cell lung cancer cell line (NCI-H446)
were stained with TGF-β antibody and TGF-β production was
determined by intracellular staining for flow cytometry and western
blotting, respectively. The results showed that in contrast to
primary lung cells control, Raw 264.7, EL4 and NCI-H446 cells have
high values of geometric mean fluorescence (MFI). The MFI values of
Raw 264.7, EL4 and NCI-H446 cells were 15.50, 17.41 and 16.45,
respectively (Fig. 1A), which
indicated that all these cells could generate TGF-β (Fig. 1B). The results of western blotting
also confirmed that Raw 264.7, EL4 and NCI-H446 cells produced
TGF-β (Fig. 1C).
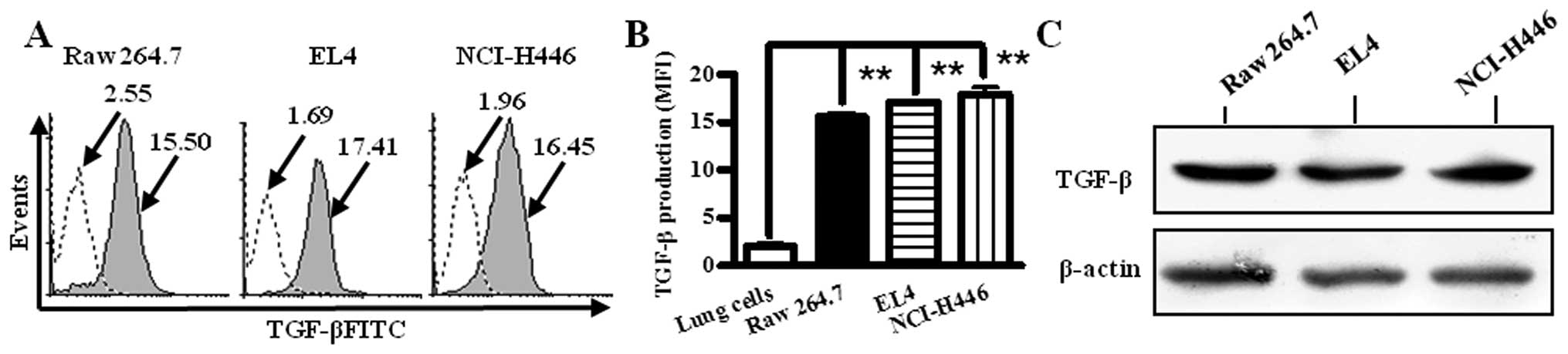 | Figure 1TGF-β is expressed in RAW 264.7, EL4
and NCI-H446 cells. RAW264.7, EL4 and NCI-H446 cells were collected
from the culture medium and fixed with IC fixation buffer. After
washes, the cells were labeled with TGF-β and
fluorescence-conjugated secondary antibodies. Then, the cells were
washed with permeabilization buffer and flow cytometry staining
buffer, respectively, and data were acquired on flow cytometry.
Primary murine lung cells were used as TGF-β negative control.
Numbers in the histogram indicate geometric mean fluorescence of
test samples. (A) Histogram presentation of TGF-β production of
RAW264.7, EL4 and NCI-H446 cells. (B) Histographic presentation of
MFI on expression of TGF-β. Data are given as mean ± SEM, out of 3
experiments. **p<0.001, one-way ANOVA with post
Newman-Keuls test. (C) TGF-β expression of RAW264.7, EL4 and
NCI-H446 cells was assessed by Western blotting. Data are shown of
representative Western blot analyses (n=3). β-actin was used as
loading control. |
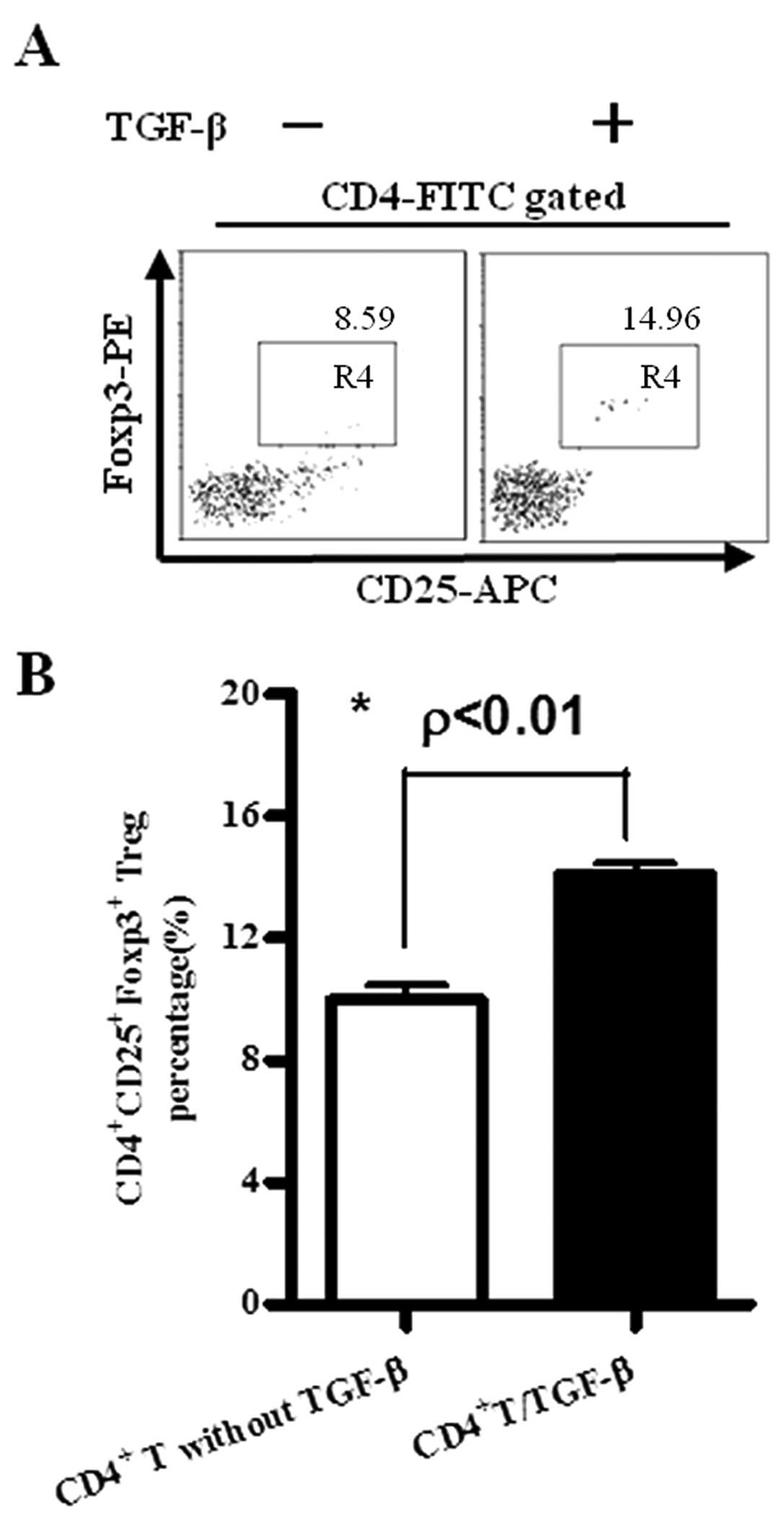
TGF-β is able to efficiently induce Treg
generation in vitro
Tregs play an important role in tumor-associated
immuno-suppression (20–23). As TGF-β could be produced by Raw
264.7, EL4 and NCI-H446 cells (Fig.
1), CD4+ T cells separated from splenocytes by
CD4+ MACS beads were treated with TGF-β and Treg
generation was investigated by flow cytometry. The results showed
that with TGF-β treatment, the percentage of Treg generation was
increased from 8.59 to 14.96% (Fig.
2A), ~74% higher than that of cells without TGF-β treatment,
which indicated that TGF-β could efficiently transform
CD4+CD25−Foxp3− cells into
CD4+CD25+Foxp3+ cells (Fig. 2B).
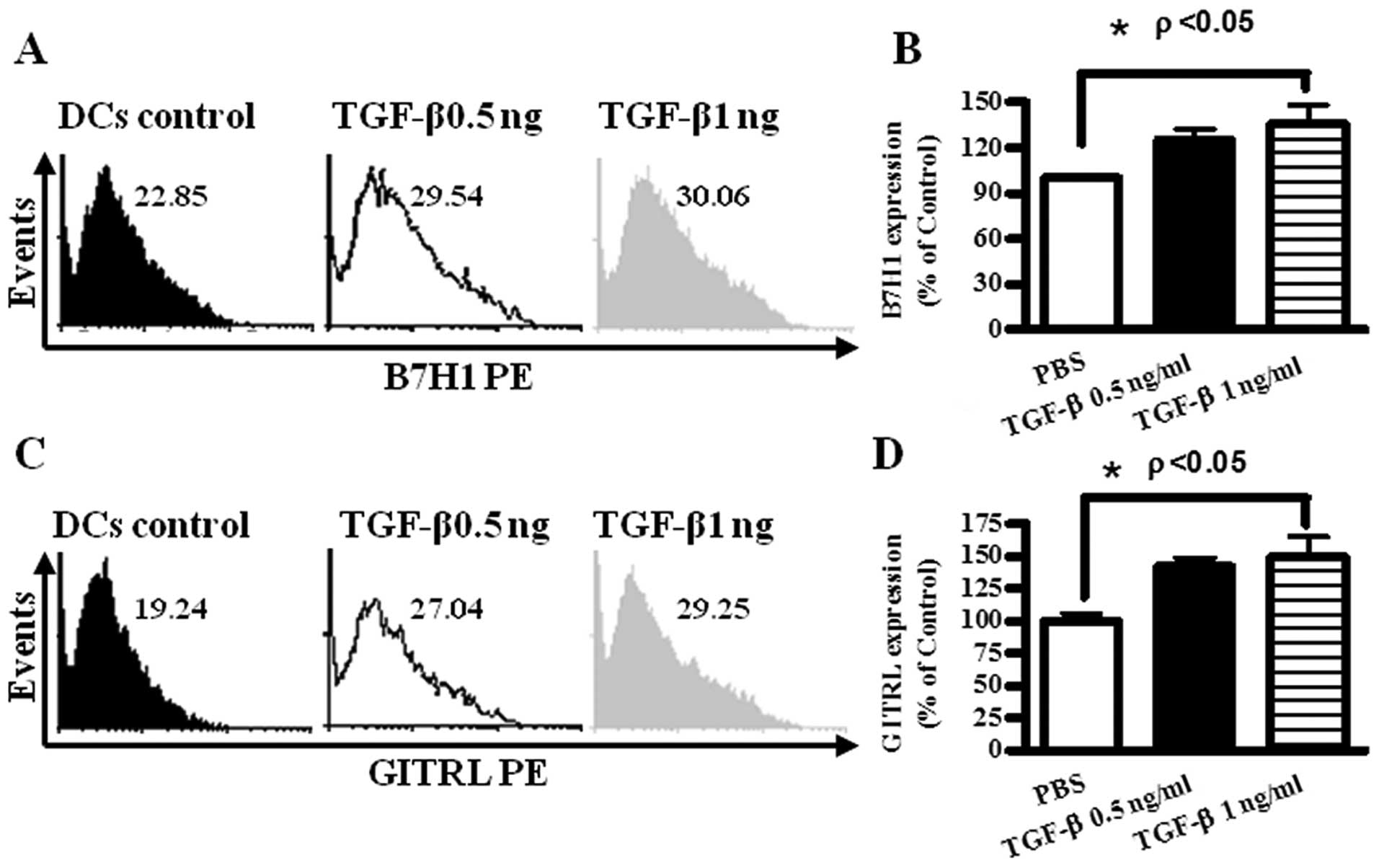
The expressions of B7H1 and GITRL on DCs
were up-regulated by TGF-β treatment
B7H1, GITRL were reported to be candidate immune
modulation molecules (12–15). To investigate the roles of B7H1 and
GITRL in TGF-β induced Treg generation, DCs were treated with
TGF-β, and B7H1 and GITRL expression of DCs were determined by flow
cytometry. The results showed that TGF-β 1 ng/ml treatment could
increase B7H1 and GITRL expression 141.6 and 149.5%, respectively
(Fig. 3).
The establishment of the
tumor-DC-fibroblast co-incubation system in vitro
Tumor cells, fibroblasts and immune cells are the
main components of tumor microenvironment (4). To explore the role of immune cells in
the immune depression induced by tumor cells, fibroblasts separated
from murine skin and DCs induced from bone marrow were labeled with
CFSE and SNARF, respectively, and mixed for further co-incubation.
The results showed that: DCs and fibroblasts could be successfully
labeled with 10 μM SNARF and CFSE, respectively. To establish lung
cancer microenvironment in vitro, LLC, DCs and fibroblasts
were labeled with SNARF, CMHC and CFSE, respectively, and mixed for
further co-incubation. LLC, DCs and fibroblasts were visible as
red, blue and green respectively when co-incubation system was
observed by the fluorescence microscope, indicating that
co-incubation system could be useful for study of DC function in
tumor microenvironment- associated Treg expansion.
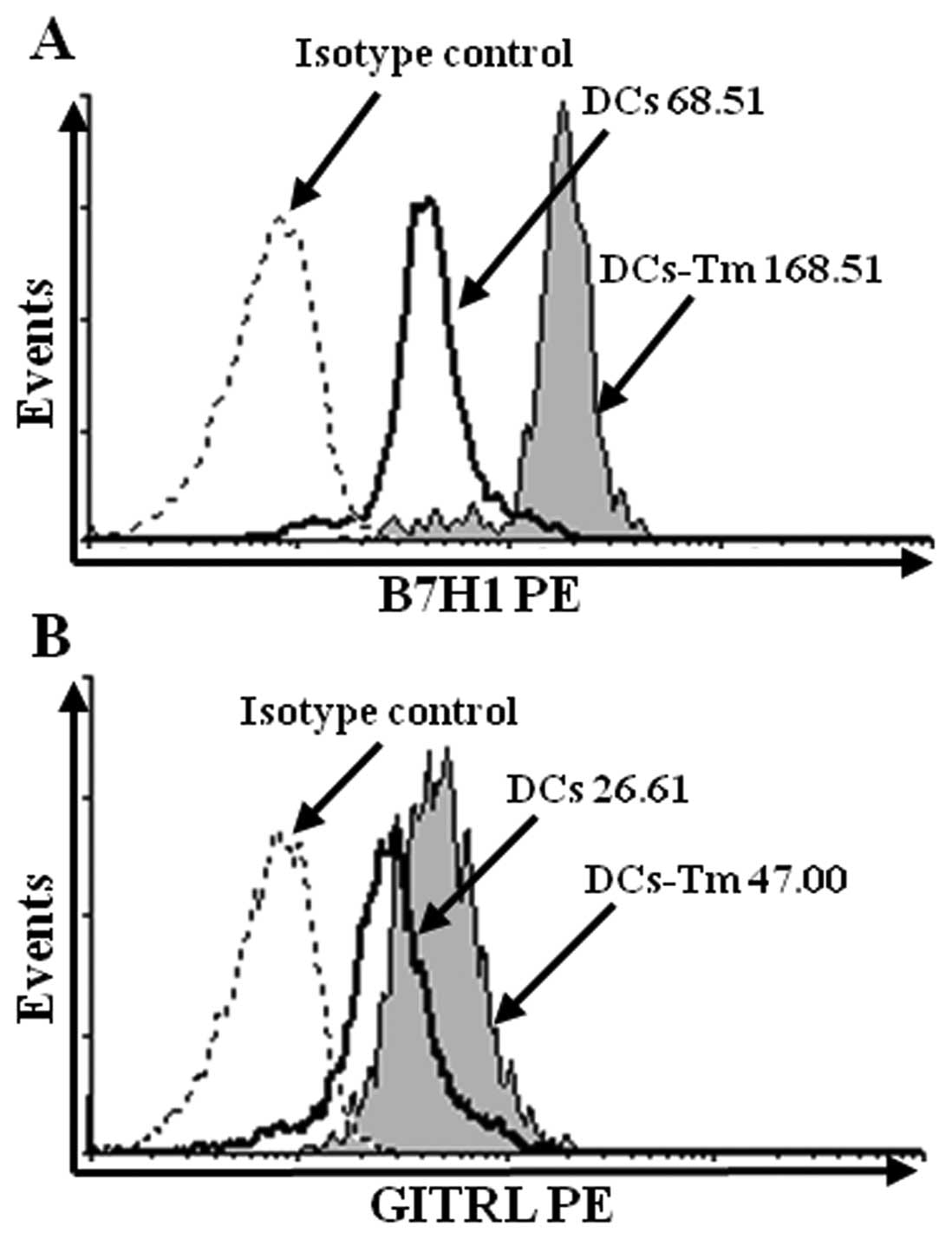
The B7H1 and GITRL expressions on DCs
were up-regulated by the LLC-fibroblast-DC co-incubation
Several reports have described the effects of
co-incubation of tumor cells and DCs on molecules expression of DCs
(7–9), but only DCs and tumor cells were
included in these co-incubation systems. The effects of the
tumor-fibroblast-DC co-incubation are still unknown. To investigate
the effects of tumor microenvironment on B7H1 and GITRL expressions
of DCs, DCs were separated from the LLC-fibroblast-DC co-incubation
system and B7H1 and GITRL expressions were determined by flow
cytometry. The results showed that in contrast to DCs pulsed with
the same amount of LLC lysate, the LLC-fibroblast-DC co-incubation
could increase B7H1 expression from 68.51 to 168.51 and
up-regulated GITRL expression from 26.61 to 46.00, respectively,
which indicated that the LLC-fibroblast-DC co-incubation obviously
augmented B7H1 and GITRL expressions of DCs (Fig. 6A, p<0.001, DCs versus DCs-Tm;
Fig. 6B, p<0.001, DCs versus
DCs-Tm).
Discussion
Cell biology studies indicate that tumor growth is
not just determined by malignant cancer cells themselves, but also
by the tumor microenvironment. In addition to tumor cells, tumor
microenvironment is comprised of immune cells, fibroblasts, stromal
cells and extracellular matrix (1).
Previous studies showed that co-incubation of DCs with different
tumor cells could down-regulate the abilities of DCs mediating
tumor antigenic-specific CTL priming (9) or inducing human Treg expansion
(6). Suppressed DCs functions have
been reported in various tumor microenvironment models, in
tumor-bearing animals, as well as in cancer patients (24–26).
However, these studies explored immune cell dysfunction by tumor
cell and immune cell co-incubation in vitro or direct cell
separation in vivo. Hence, previously little was known about
the effects of tumor microenvironment on DCs by direct tumor
cell-fibroblast-DC contact.
In the present study, we successfully established
lung cancer microenvironment system by LLC, fibroblast and DC
co-incubation in vitro. DCs separated from co-incubation
system were further evaluated by flow cytometry analysis and the
results showed that B7H1, and GITRL expressions were up-regulated
by LLC microenvironment. As TGF-β, an efficient inducer of Treg
expansion, could be generated by Raw 264.7, EL4 and NCI-H446 cells,
the effects of TGF-β on B7H1 and GITRL expressions of DCs were
explored. Our data demonstrated that TGF-β treatment could
up-regulate B7H1, GITRL expressions of DCs, indicating that B7H1
and GITRL might play important roles in TGF-β induced Treg
expansion of lung cancer microenvironment.
Our results showed that Raw 264.7, EL4 and NCI-H446
cells produced TGF-β (Fig. 1),
which is known to shape the tumor microenvironment, exerting both
positive and negative effects on cancer (20–22).
Other sources of TGF-β included inflammatory cells and stromal
fibroblasts (27). At later stages
of tumor progression, cancer cells acquired increasing resistance
to TGF-β growth inhibitory signals (22), which was frequently accompanied by
increased expression of TGF-β by the same cells. Subsequently,
cancer cells started to secrete non-physiological levels of TGF-β
(28). As TGF-β was necessary for
expansion of Treg (10, 29), the increase of Treg percentage from
8.59 to 14.96% with TGF-β treatment (Fig. 2) indicated that the presence of
TGF-β in the tumor microenvironment might impair immune
surveillance by inducing TGF-β expansion.
There is mounting evidence that Foxp3+
Tregs can develop extrathymically under certain conditions
(13,29–31).
Although literature show a potential role for Tregs in the control
of autoimmune or inflammatory diseases (32), the nature of APCs involved in Treg
expansion remains poorly understood. Our previous report documented
that the tumor cell-DC co-incubation decreased CD80, and CD86
expressions of DCs (9). DCs,
especially deficient in CD80 and CD86, appeared to be more
efficient than other APCs in inducing TGF-β expansion (33,34).
Hori et al reported that GITRL-dependent Treg expansion
underlined immune privilege in corneal allografts (14), but that the blockade of GITR-GITRL
interaction was necessary for maintaining Treg function was also
reported (15). In the present
study, DCs were used in lung cancer microenvironment co-incubation
system and its function in Treg expansion was explored. Our data
showed that both LLC microenvironment co-incubation and TGF-β
treatment could augment B7H1 and GITRL expressions on DCs (Figs. 3 and 6), indicating that Treg expansion in tumor
microenvironment might depend on TGF-β inducing B7H1 and GITRL
up-regulation.
The data presented here offered new information on
the immunological alterations associated with tumor
microenvironment, especially the role of TGF-β mediating B7H1 and
GITRL up-regulation in lung cancer microenvironment associated Treg
expansion. This finding is important because it provides a
rationale for further investigation of the mechanisms by which
tumor and tumor associated fibroblasts induce immunosuppression
in vivo.
Acknowledgements
This study was supported by grants from the Natural
Science Foundation of Xiamen (no. 3502Z20104002), the Xiamen
Science and Technology Key program grant (no. 3502Z20100006) and
the grant from the National Laboratory for Oncogenes and Related
Genes of China (90-08-02). We thank Jin Hua Su and Fu Chen for
excellent animal care.
Abbreviations:
|
CMHC
|
4-chloromethyl-7-hydroxycoumarin
|
|
CFSE
|
carboxyfluorescein succinimidyl
ester
|
|
LLC
|
Lewis lung cancer
|
|
SNARF
|
seminaphtharhodafluor
|
|
GITRL
|
glucocorticoid-induced TNFR-related
protein ligand
|
|
B7H1
|
B7 homolog 1
|
|
PD-L1
|
programmed cell death 1 ligand 1
|
|
TGF-β
|
transforming growth factor β
|
References
|
1
|
Witz IP: The tumor microenvironment: the
making of a paradigm. Cancer Microenviron. 2(Suppl 1): 9–17. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mueller MM and Fusenig NE: Friends or foes
- bipolar effects of the tumor stroma in cancer. Nat Rev Cancer.
4:839–849. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Barsky SH, Green WR, Grotendorst GR and
Liotta LA: Desmoplastic breast carcinoma as a source of human
myofibroblasts. Am J Pathol. 115:329–333. 1984.PubMed/NCBI
|
|
4
|
Kalluri R and Zeisberg M: Fibroblasts in
cancer. Nat Rev Cancer. 6:392–401. 2006. View Article : Google Scholar
|
|
5
|
Zou WP: Immunosuppressive networks in the
tumor environment and their therapeutic relevance. Nat Rev Cancer.
5:263–274. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bergmann C, Strauss L, Zeidler R, Lang S
and Whiteside TL: Expansion and characteristics of human T
regulatory type 1 cells in co-cultures simulating tumor
microenvironment. Cancer Immunol Immunother. 56:1429–1442. 2007.
View Article : Google Scholar
|
|
7
|
Zou WP, Machelon V, Coulomb-L’Hermin A, et
al: Stromal-derived factor-1 in human tumors recruits and alters
the function of plasmacytoid precursor dendritic cells. Nat Med.
7:1339–1346. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Iwamoto M, Shinohara H, Miyamoto A, et al:
Prognostic value of tumor-infiltrating dendritic cells expressing
CD83 in human breast carcinomas. Int J Cancer. 104:92–97. 2003.
View Article : Google Scholar
|
|
9
|
Gao F, Hui X, He X, Wan D and Gu J:
Dysfunction of murine dendritic cells induced by incubation with
tumor cells. Cell Mol Immunol. 5:133–140. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yuan XL, Chen L, Zhang TT, et al: Gastric
cancer cells induce human CD4+Foxp3+
regulatory T cells through the production of TGF-β1. World J
Gastroenterol. 17:2019–2027. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sempere LF, Gunn JR and Korc M: A novel
3-dimensional culture system uncovers growth stimulatory actions by
TGFβ in pancreatic cancer cells. Cancer Biol Ther. 12:198–207.
2011.PubMed/NCBI
|
|
12
|
Yi T, Li X, Yao S, et al: Host APCs
augment in vivo expansion of donor natural regulatory T cells via
B7H1/B7.1 in allogeneic recipients. J Immunol. 186:2739–2749. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou X, Zhou Y, Ding Q, et al: High level
expression of B7H1 molecules by keratinocytes suppresses xeno- and
allo-reactions by inducing type I regulatory T cells. Transpl
Immunol. 21:192–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hori J, Taniguchi H, Wang M, Oshima M and
Azuma M: GITR ligand-mediated local expansion of regulatory T cells
and immune privilege of corneal allografts. Invest Ophthalmol Vis
Sci. 51:6556–6565. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim JI, Sonawane SB, Lee MK, et al:
Blockade of GITR-GITRL interaction maintains Treg function to
prolong allograft survival. Eur J Immunol. 40:1369–1374. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Häkkinen L, Koivisto L and Larjava H: An
improved method for culture of epidermal keratinocytes from newborn
mouse skin. Methods Cell Sci. 23:189–196. 2011.PubMed/NCBI
|
|
17
|
Gao FG, Li HT, Li ZJ and Gu JR: Nicotine
stimulated dendritic cells could achieve anti-tumor effects in
mouse lung and liver cancer. J Clin Immunol. 31:80–88. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang MH, Tang H, Guo ZH, et al: Splenic
stroma drives mature dendritic cells to differentiate into
regulatory dendritic cells. Nat Immunol. 5:1124–1133. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ruan Q, Kameswaran V, Tone Y, et al:
Development of Foxp3(+) regulatory T cells is driven by the c-Rel
enhanceosome. Immunity. 31:932–940. 2009.
|
|
20
|
Massague J: TGF-beta and cancer. Cell.
134:215–230. 2008. View Article : Google Scholar
|
|
21
|
Alshaker HA and Matalka KZ: IFN-γ, IL-17
and TGF-β involvement in shaping the tumor microenvironment: the
significance of modulating such cytokines in treating malignant
solid tumors. Cancer Cell Int. 11:332011.
|
|
22
|
Yang L, Pang Y and Moses HL: TGF-β and
immune cells: an important regulatory axis in the tumor
microenvironment and progression. Trends Immunol. 31:220–227.
2010.
|
|
23
|
Siegel PM and Massague J: Cytostatic and
apoptotic actions of TGF-β in homeostasis and cancer. Nat Rev
Cancer. 3:807–821. 2003.
|
|
24
|
Orsini E, Guarini A, Chiaretti S, Mauro FR
and Foa R: The circulating dendritic cell compartment in patients
with chronic lymphocytic leukemia is severely defective and unable
to stimulate an effective T-cell response. Cancer Res.
63:4497–4506. 2003.
|
|
25
|
Sharma S, Stolina M, Yang SC, et al: Tumor
cyclooxygenase 2-dependent suppression of dendritic cell function.
Clin Cancer Res. 9:961–968. 2003.PubMed/NCBI
|
|
26
|
Yang AS and Lattime EC: Tumor-induced
interleukin 10 suppresses the ability of splenic dendritic cell to
stimulate CD4 and CD8 T-cell responses. Cancer Res. 63:2150–2157.
2003.PubMed/NCBI
|
|
27
|
Nam JS, Terabe M, Kang MJ, et al:
Transforming growth factor beta subverts the immune system into
directly promoting tumor growth through interleukin-17. Cancer Res.
68:3915–3923. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gorska AE, Jensen RA, Shyr Y, Aakre ME,
Bhowmick NA and Moses HL: Transgenic mice expressing a
dominant-negative mutant type II transforming growth factor beta
receptor exhibit impaired mammary development and enhanced mammary
tumor formation. Am J Pathol. 163:1539–1549. 2003. View Article : Google Scholar
|
|
29
|
Scholzen A, Mittag D, Rogerson SJ, Cooke
BM and Plebanski M: Plasmodium falciparum-mediated induction of
human CD25hiFoxp3hi CD4 T cells is
independent of direct TCR stimulation and requires IL-2, IL-10 and
TGFβ. PLoS Pathog. 5:e10005432009.PubMed/NCBI
|
|
30
|
Bettelli E, Carrier Y, Gao W, et al:
Reciprocal developmental pathways for the generation of pathogenic
effector TH17 and regulatory T cells. Nature. 441:235–238. 2006.
View Article : Google Scholar
|
|
31
|
Kretschmer K, Apostolou I, Hawiger D,
Khazaie K, Nussenzweig MC and von Boehmer H: Inducing and expanding
regulatory T cell populations by foreign antigen. Nat Immunol.
6:1219–1227. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lafaille C, Kutchukhidze MA, Shen N, Ding
S, Yee YH and Lafaille JJ: Adaptive Foxp3+ regulatory T
cell-dependent and -independent control of allergic inflammation.
Immunity. 29:114–126. 2008.
|
|
33
|
Yamazaki S, Iyoda T, Tarbell K, Olson K,
Velinzon K, Inaba K and Steinman RM: Direct expansion of functional
CD25+ CD4+ regulatory T cells by
antigen-processing dendritic cells. J Exp Med. 198:235–247.
2003.PubMed/NCBI
|
|
34
|
Benson MJ, Pino-Lagos K, Rosemblatt M and
Noelle RJ: All-trans retinoic acid mediates enhanced T reg cell
growth, differentiation, and gut homing in the face of high levels
of co-stimulation. J Exp Med. 204:1765–1774. 2007. View Article : Google Scholar : PubMed/NCBI
|