Introduction
Breast cancer is the most common cancer and the
leading cause of cancer death among women all over the world
(1). Development and progression of
breast cancer depend on the balance of oncogenes and tumor
suppressor genes (2).
Identification and characterization of these genes will lead to
discovery of new markers and potential therapeutic targets for
prevention and treatment of breast cancer (3).
Wilms’ tumor 1 (WT1) gene was originally identified
as a tumor suppressor gene, which is responsible for Wilms tumor
(4,5). However, accumulating evidence
indicated that WT1 may play an oncogenic role in leukemogenesis and
tumorigenesis (6–11). It was shown that the growth of
leukemic cells and a variety of solid cancer cells was inhibited by
knockdown of WT1 expression, while forced expression of WT1
promoted cell growth and motility, suppressed apoptosis and induced
leukemia in WT1-transgenic mice (9).
WT1 protein and mRNA were firstly found to be
expressed in the normal breast tissue, but absent in >90% of the
breast cancer (12), and WT1
inhibited proliferation and tumorige- nesis of breast cancer cells,
indicating WT1 served as a tumor suppressor gene in breast cancer
(13–15). On the contrary, a number of studies
demonstrated that wild-type WT1 gene plays an important role in the
development of breast cancer (16).
Loeb et al reported that WT1 could be detected in 87% of
primary breast carcinomas, but not in normal breast epithelium
(17). Miyoshi et al found
that WT1 mRNA significantly correlated with the poor prognosis of
breast cancer (18). In addition,
WT1 could promote the proliferation and restrain the apoptosis of
breast cancer cells (19–21), all of which indicate that WT1 might
serve as an oncogene in breast cancer. Therefore, it is necessary
to clarify the oncogenic or tumor suppressive role of WT1 in breast
cancer. Moreover, relationship between WT1 expression and
clinicopathological parameters and prognosis of breast cancer was
inconsistent so far (12,18,22,23),
and the possible correlation between WT1 expression and molecular
subtypes has not been reported.
In this study, we analyzed the WT1 mRNA expression
in a microarray dataset of 266 early breast cancer patients and
investigated its possible relationship with the clinicopathological
parameters, molecular subtypes and prognosis so as to explore the
role of WT1 gene in breast cancer.
Materials and methods
Tumor samples
The publicly available microarray dataset, GSE21653,
was collected from National Center for Biotechnology Information
(NCBI) Gene Expression Omnibus (GEO). GSE21653 involved 266 early
breast cancer patients, who underwent initial surgery in Institut
Paoli-Calmettes (IPC) institution between 1992 and 2004 (24). They included 227 cases previously
reported and 39 additional cases, all of which were similarly
profiled using Affymetrix U133 Plus 2.0 human oligonucleotide
microarrays (25).
Clinicopathological characteristics of the patients have been
described previously (24,25). Among them, a total of 252 patients
had the DFS information.
Microarray analysis
Regarding the Affymetrix-based datasets, GSE21653,
we used Robust Multichip Average (RMA) with the nonparametric
quantile algorithm as normalization parameter (26). RMA was applied to the raw data from
the IPC series. Quantile normalization or RMA was done in R using
bioconductor (27) and associated
packages. Then the log2 transformed data for the
following WT1 probes were evaluated. Patients were stratified based
on the main available prognostic factors for breast cancer, such as
pathological tumor size (pT), histological grades (Grade 1, G1;
Grade 2, G2; Grade 3, G3) according to Searff-Bloom-Richardson
(SBR) staging system, Ki67, estrogen receptor (ER), molecular
subtypes (basal-like, ERBB2, luminal A, luminal B and normal-like;
luminal: luminal A and luminal B) and disease-free survival
(DFS).
Statistical analysis
T-test was used to investigate whether the WT1 gene
expression values were significantly different between any two
compared groups of different disease characteristics with P<0.05
(28,29). DFS analyses were performed during
the period from the date of diagnosis to the first observation of
any metastasis. DFS was estimated according to the Kaplan-Meier
method and analyzed by log-rank (Mantel-Cox) test. Hazard ratio
(HR) and 95% confidence interval (CI) were estimated by use of a
stratified Cox regression analysis. Multivariate analysis was done
by incorporating all variables with a P-value <0.05 in
univariate analysis.
All statistical tests were two-sided at the 5% level
of significance. Statistical analysis was done using the survival
package (version 2.36) in the R software (version 2.12.1).
Results
Relationship between WT1 mRNA expression
and clinicopathological factors and molecular subtypes of breast
cancer
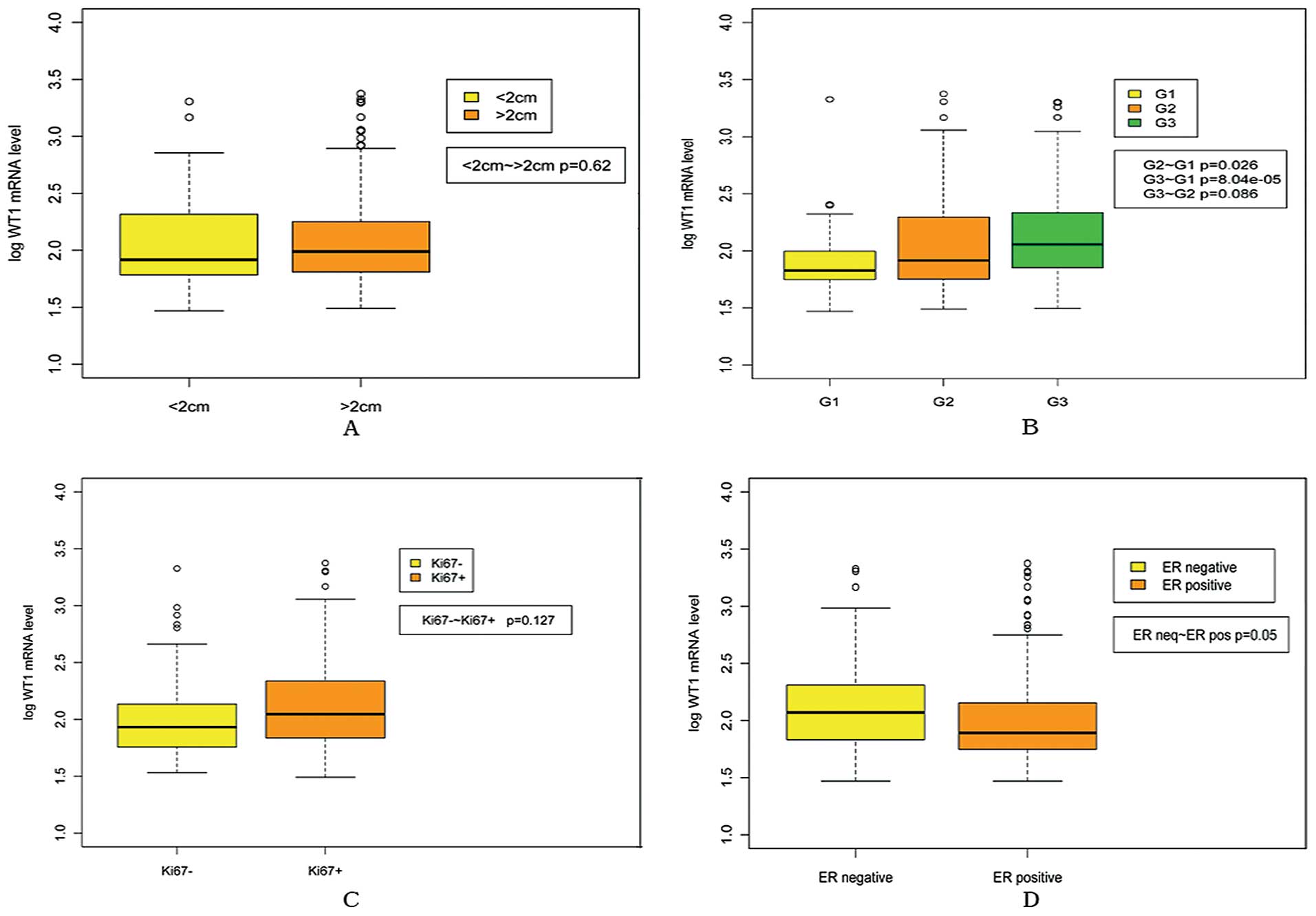
As shown in Fig. 1,
WT1 mRNA expression increased in relation to histological grades
(G2 vs. G1, P=0.026; G3 vs. G1, P=8.04e-5). In addition, WT1 mRNA
expression was significantly higher in ER negative group than in ER
positive group (P=0.050). However, no correlation was found between
WT1 mRNA expression and pT (<2 cm vs. >2 cm, P=0.620) and
Ki67 expression (Ki67 positive vs. Ki67 negative, P=0.127). As
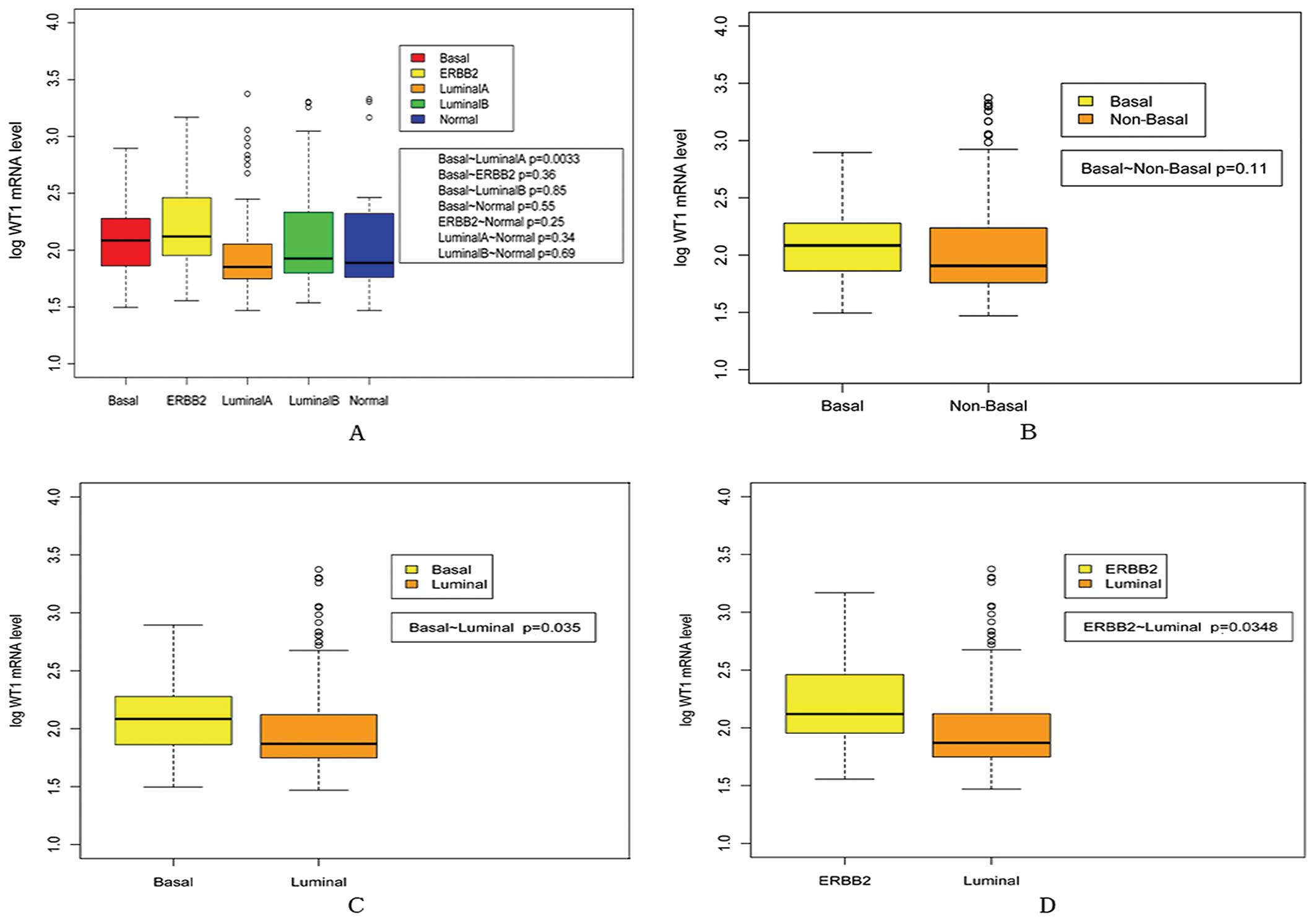
shown in Fig. 2, WT1 mRNA
expression was significantly higher in basal-like and ERBB2
subtypes than in luminal subtype (P=0.003 for basal-like vs.
luminal A, P=0.035 for basal-like vs. luminal, P=0.035 for ERBB2
vs. luminal).
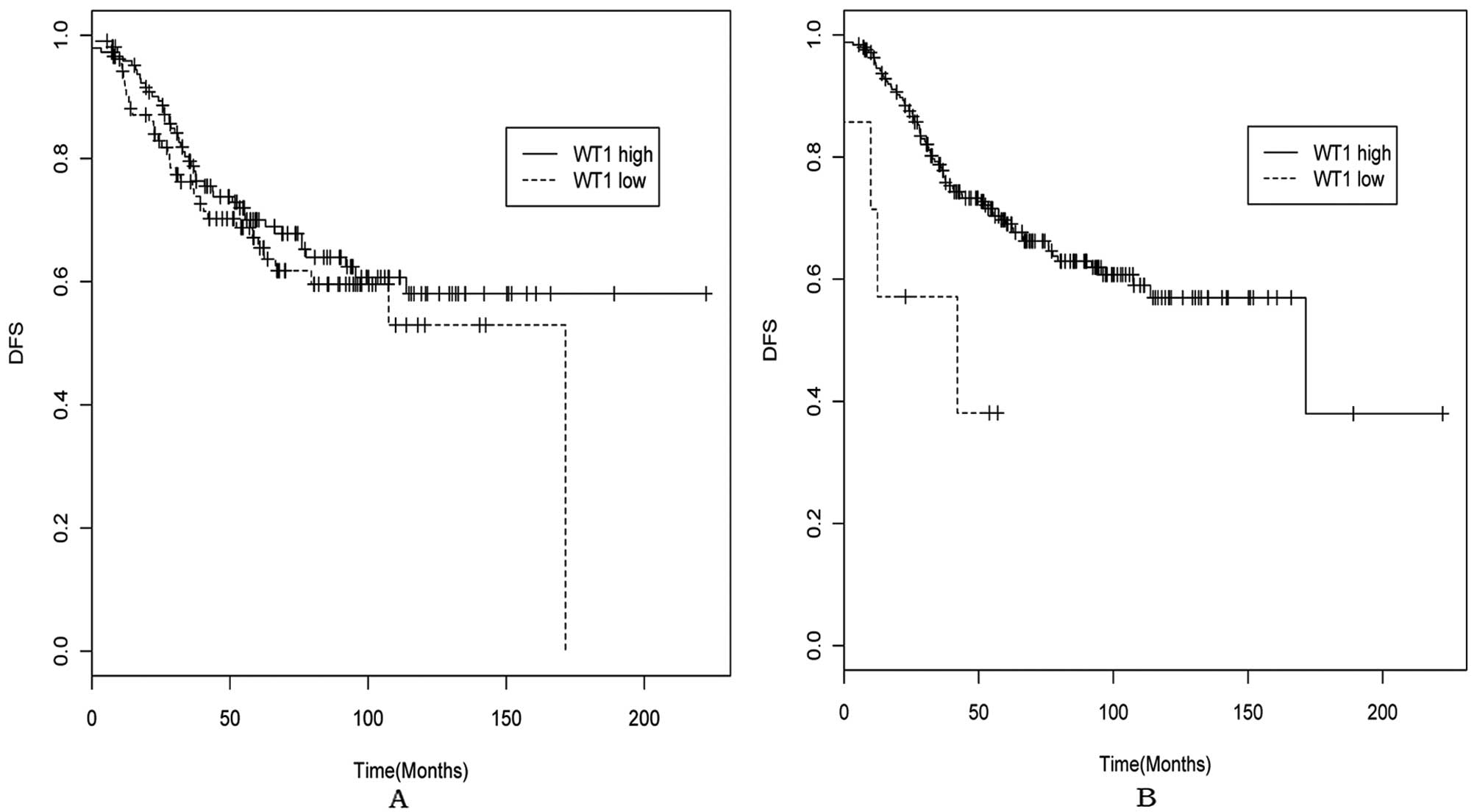
Univariate analysis of WT1 mRNA
expression and DFS in GSE21653
Patients were divided into the WT1 high and low mRNA
expression groups according to the cutoff value of 2.1, which
corresponded to the mean of the WT1 mRNA levels in total breast
cancer tissues and the cutoff value of 3.1, which corresponded to
the mean +2 standard deviation of the WT1 mRNA levels of the
normal-like breast subtype, considering its intrinsic gene
expression patterns (30,31) and WT1 expression level in this study
(Fig. 2A). Kaplan-Meier analyses of
the probability that patients would remain free of distant
metastases were carried out between WT1 high and low expression
groups. As shown in Fig. 3 and
Table I, there was no significant
difference of DFS between the WT1 high and low expression group
(HR=1.226, 95% CI=0.794–1.895, P=0.357) with the 2.1 cutoff value,
however, WT1 high expression group showed a significantly lower DFS
than the low expression group (HR=3.294, 95% CI=1.198–9.055,
P=0.014) with the 3.1 cutoff value.
 | Table IUnivariate analysis of relationship
between WT1 mRNA expression and DFS. |
Table I
Univariate analysis of relationship
between WT1 mRNA expression and DFS.
| Cutoff value | Survival | HR | 95% CI | Log-rank
(P-value) | High level (N) | Low level (N) | N |
|---|
| 2.1 | DFS | 1.226 | (0.794, 1.895) | 0.357 | 106 | 146 | 252 |
| 3.1 | DFS | 3.294 | (1.198, 9.055) | 0.014 | 7 | 245 | 252 |
Multivariate analysis of WT1 mRNA
expression and DFS in GSE21653
Using the Cox proportional hazards model,
multivariate analysis demonstrated that grades are the strongest
predictor of the likelihood of DFS (HR=2.611, 95% CI=1.133–6.019,
P=0.024 for G2 vs. G1; HR=2.993, 95% CI=1.249–7.171, P=0.014 for G3
vs. G1), when using the 2.1 cutoff value (Table II). However, with the 3.1 cutoff
value, multivariate analysis demonstrated that both grades
(HR=3.121, 95% CI=1.236–7.883, P=0.016 for G3 vs. G1) and WT1 mRNA
expression (HR=3.454, 95% CI=1.203–9.918, P=0.021 for high
expression group vs. low expression group) are independent
prognostic indicators for breast cancer (Table III).
 | Table IIMultivariable proportional-hazards
analysis of DFS based on 2.1 cutoff value. |
Table II
Multivariable proportional-hazards
analysis of DFS based on 2.1 cutoff value.
| Variable | HR (95% CI) | P-value |
|---|
| WT1 values (high vs.
low) | 1.093
(0.687–1.740) | 0.708 |
| Grade (G2 vs.
G1) | 2.611
(1.133–6.019) | 0.024 |
| Grade (G3 vs.
G1) | 2.993
(1.249–7.171) | 0.014 |
| pT (<2 cm vs.
>2 cm) | 1.162
(0.662–2.040) | 0.601 |
| ER (ER positive vs.
ER negative) | 0.675
(0.380–1.197) | 0.179 |
| Subtype (basal vs.
non-basal) | 1.357
(0.703–2.619) | 0.363 |
 | Table IIIMultivariable proportional-hazards
analysis of DFS based on 3.1 cutoff value. |
Table III
Multivariable proportional-hazards
analysis of DFS based on 3.1 cutoff value.
| Variable | Hazard ratio (95%
CI) | P-value |
|---|
| WT1 values (high
vs. low) | 3.454
(1.203–9.918) | 0.021 |
| Grade (G2 vs.
G1) | 2.385
(0.961–5.924) | 0.061 |
| Grade (G3 vs.
G1) | 3.121
(1.236–7.883) | 0.016 |
| pT (<2 cm vs.
>2 cm) | 0.967
(0.523–1.789) | 0.916 |
| ER (ER positive vs.
ER negative) | 0.628
(0.330–1.196) | 0.157 |
| Subtype (basal vs.
non-basal) | 1.203
(0.572–2.534) | 0.626 |
Discussion
The WT1 gene, located at chromosome 11p13, encodes a
DNA-binding protein, which contains an NH2-terminal glutamine and
proline-rich domain involved in transcriptional repression and
activation and a C-terminal domain composed of four
Cys-Cys-His-His-type zinc finger domains (ZF) involved in DNA and
RNA binding and protein-protein interactions (32). WT1 regulates a diverse array of
genes through ZF domain at GC-rich sites (32), playing an important role in cell
growth and development (6–11,33).
Though originally isolated as a tumor suppressor
gene responsible for Wilms’ tumor, WT1 is found to be overexpressed
in primary human leukemia and a variety of solid tumors, including
lung, colon, liver, thyroid and pancreatic ductal cancer (9,34,35).
In addition, knockdown of WT1 by shRNA induced mitochondrial damage
and the resultant apoptosis in several WT1-expressing solid tumor
cells, indicating that WT1 might play an oncogenic role in these
tumors (36).
Up to now, the role and function of WT1 in breast
cancer have been not clarified. As for expression of WT1 in breast
tissues, the groups of Silberstein and Loeb obtained contrary
results in the clinical reports (12,17).
As for function of WT1 in breast cancer cells, a promoting or
repressing effect of WT1 on breast cancer cells was observed in the
experimental studies, making it hard to conclude on role of WT1 in
breast cancer. In addition, some studies have focused on WT1
expression in breast cancer, but results are not clear enough to
demonstrate its possible relationship with tumor biology.
In this study, we found that WT1 expression was
higher in ER-negative patients than in ER-positive patients, which
may be explained by the fact that overexpression of WT1 directly
resulted in the down-regulation of ER expression (22). In addition, our results also
demonstrated that WT1 tended to be overexpressed in tumors with
high histological grades. It has been well established that breast
cancer patients with ERBB2-overexpression and basal-like molecular
subtypes had worse prognosis than luminal subtypes (31). Of note, our present data showed that
WT1 mRNA expression was much higher in patients with ERBB2 and
basal tumors. To the best of our knowledge, this is the first
report exploring the possible relationship between WT1 and
molecular subtypes, and the preliminary results suggest a potential
role of WT1 in progression of breast cancer. Though Miyoshi et
al (18) found that the
prognosis of patients with high WT1 expression was significantly
worse than that in patients with low WT1 expression, Camcı et
al failed to confirm this finding in a later study (23). In the present study, we concluded
that WT1 mRNA high expression predicted poor prognosis of breast
cancer patients when using 3.1 as cutoff value, which might be due
to the facts that WT1 could promote proliferation, invasion,
migration, response to hypoxia and drug resistance of cancer cells
(19–22,35,37,38).
Conclusively, our study demonstrates association
between WT1 mRNA levels and histological grade, ER status,
molecular subtype and clinical outcome of breast cancer, consistent
with the hypothesis that WT1 plays an oncogenic role in breast
cancer. However, further research is required to confirm the
current findings and clarify the functions and relevant mechanisms
of WT1 in human breast cancer.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (No. 81102030). We thank Miss
Jun-Lan Liu, from Breast Disease Center, Southwest Hospital, Third
Military Medical University, Chongqing, China, for language editing
of the manuscript. We also would like to thank for the kind help of
bioinformatics analysis from Fenghe (Shanghai) Information
Technology Co., Ltd.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global Cancer Statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gumireddy K, Li A, Gimotty PA,
Klein-Szanto AJ, Showe LC, Katsaros D, et al: KLF17 is a negative
regulator of epithelial-mesenchymal transition and metastasis in
breast cancer. Nat Cell Biol. 11:1297–1304. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Haber DA, Buckler AJ, Glaser T, Call KM,
Pelletier J, Sohn RL, et al: An internal deletion within an 11p13
zinc finger gene contributes to the development of Wilms’ tumor.
Cell. 61:1257–1269. 1990.PubMed/NCBI
|
|
5
|
Little M and Wells C: A clinical overview
of WT1 gene mutations. Hum Mutat. 9:209–225. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Loeb DM and Sukumar S: The role of WT1 in
oncogenesis: tumor suppressor or oncogene? Int J Hematol.
76:117–126. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hohenstein P and Hastie ND: The many
facets of the Wilms’ tumour gene, WT1. Hum Mol Genet. 15:R196–R201.
2006.PubMed/NCBI
|
|
8
|
Yang L, Han Y, Suarez Saiz F and Minden
MD: A tumor suppressor and oncogene: the WT1 story. Leukemia.
21:868–876. 2007.PubMed/NCBI
|
|
9
|
Sugiyama H: WT1 (Wilms’ tumor gene 1):
biology and cancer immunotherapy. Jpn J Clin Oncol. 40:377–387.
2010.
|
|
10
|
Huff V: Wilms’ tumours: about tumour
suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer.
11:111–121. 2011.
|
|
11
|
Oji Y, Ogawa H, Tamaki H, Oka Y, Tsuboi A,
Kim EH, et al: Expression of the Wilms’ tumor gene WT1 in solid
tumors and its involvement in tumor cell growth. Jpn J Cancer Res.
90:194–204. 1999.
|
|
12
|
Silberstein GB, Van Horn K, Strickland P,
Roberts CT Jr and Daniel CW: Altered expression of the WT1 wilms
tumor suppressor gene in human breast cancer. Proc Natl Acad Sci
USA. 94:8132–8137. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang TF, Yu SQ, Guan LS and Wang ZY:
Inhibition of breast cancer cell growth by the Wilms’ tumor
suppressor WT1 is associated with a destabilization of
beta-catenin. Anticancer Res. 23:3575–3584. 2003.
|
|
14
|
Reizner N, Maor S, Sarfstein R,
Abramovitch S, Welshons WV, Curran EM, et al: The WT1 Wilms’ tumor
suppressor gene product interacts with estrogen receptor-alpha and
regulates IGF-I receptor gene transcription in breast cancer cells.
J Mol Endocrinol. 35:135–144. 2005.
|
|
15
|
Wang L and Wang ZY: The Wilms’ tumor
suppressor WT1 inhibits malignant progression of neoplastigenic
mammary epithelial cells. Anticancer Res. 28:2155–2160. 2008.
|
|
16
|
Oji Y, Miyoshi Y, Kiyotoh E, Koga S,
Nakano Y, Ando A, et al: Absence of mutations in the Wilms’ tumor
gene WT1 in primary breast cancer. Jpn J Clin Oncol. 34:74–77.
2004.
|
|
17
|
Loeb DM, Evron E, Patel CB, Sharma PM,
Niranjan B, Buluwela L, et al: Wilms’ tumor suppressor gene (WT1)
is expressed in primary breast tumors despite tumor-specific
promoter methylation. Cancer Res. 61:921–925. 2001.
|
|
18
|
Miyoshi Y, Ando A, Egawa C, Taguchi T,
Tamaki Y, Tamaki H, et al: High expression of Wilms’ tumor
suppressor gene predicts poor prognosis in breast cancer patients.
Clin Cancer Res. 8:1167–1171. 2002.
|
|
19
|
Zapata-Benavides P, Tuna M,
Lopez-Berestein G and Tari AM: Downregulation of Wilms’ tumor 1
protein inhibits breast cancer proliferation. Biochem Biophys Res
Commun. 295:784–790. 2002.
|
|
20
|
Tuna M, Chavez-Reyes A and Tari AM:
HER2/neu increases the expression of Wilms’ Tumor 1 (WT1) protein
to stimulate S-phase proliferation and inhibit apoptosis in breast
cancer cells. Oncogene. 24:1648–1652. 2005.
|
|
21
|
Caldon CE, Lee CS, Sutherland RL and
Musgrove EA: Wilms’ tumor protein 1: an early target of progestin
regulation in T-47D breast cancer cells that modulates
proliferation and differentiation. Oncogene. 27:126–138. 2008.
|
|
22
|
Han Y, Yang L, Suarez-Saiz F, San-Marina
S, Cui J and Minden MD: Wilms’ tumor 1 suppressor gene mediates
antiestrogen resistance via down-regulation of estrogen
receptor-alpha expression in breast cancer cells. Mol Cancer Res.
6:1347–1355. 2008.
|
|
23
|
Camcı C, Kalender ME, Paydaş S, Sevinç A,
Zorludemir S and Suner A: Prognostic significance of Wilms Tumor 1
(WT1) protein expression in breast cancer. Gaziantep Med J.
17:67–72. 2011.
|
|
24
|
Sabatier R, Finetti P, Cervera N,
Lambaudie E, Esterni B, Mamessier E, et al: A gene expression
signature identifies two prognostic subgroups of basal breast
cancer. Breast Cancer Res Treat. 126:407–420. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Finetti P, Cervera N, Charafe-Jauffret E,
Chabannon C, Charpin C, Chaffanet M, et al: Sixteen-kinase gene
expression identifies luminal breast cancers with poor prognosis.
Cancer Res. 68:767–776. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U, et al: Exploration,
normalization, and summaries of high density oligonucleotide array
probe level data. Biostatistics. 4:249–264. 2003. View Article : Google Scholar
|
|
27
|
Nie H, Neerincx PB, van der Poel J,
Ferrari F, Bicciato S, Leunissen JA, et al: Microarray data mining
using Bioconductor packages. BMC Proc. 3:S92009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Crawford JR and Garthwaite PH: Single-case
research in neuropsychology: A comparison of five forms of t-test
for comparing a case to controls. Cortex. Jul 23–2011.(Epub ahead
of print).
|
|
29
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, et al: Molecular portraits of human breast
tumours. Nature. 406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, et al: Gene expression patterns of breast
carcinomas distinguish tumor subclasses with clinical implications.
Proc Natl Acad Sci USA. 98:10869–10874. 2001.PubMed/NCBI
|
|
32
|
Stoll R, Lee BM, Debler EW, Laity JH,
Wilson IA, Dyson HJ, et al: Structure of the Wilms tumor suppressor
protein zinc finger domain bound to DNA. J Mol Biol. 372:1227–1245.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Martínez-Estrada OM, Lettice LA, Essafi A,
Guadix JA, Slight J, Velecela V, et al: Wt1 is required for
cardiovascular progenitor cell formation through transcriptional
control of Snail and E-cadherin. Nat Genet. 42:89–93.
2010.PubMed/NCBI
|
|
34
|
Sera T, Hiasa Y, Mashiba T, Tokumoto Y,
Hirooka M, Konishi I, et al: Wilms’ tumour 1 gene expression is
increased in hepatocellular carcinoma and associated with poor
prognosis. Eur J Cancer. 44:600–608. 2008.
|
|
35
|
Perugorria MJ, Castillo J, Latasa MU, Goni
S, Segura V, Sangro B, et al: Wilms’ tumor 1 gene expression in
hepatocellular carcinoma promotes cell dedifferentiation and
resistance to chemotherapy. Cancer Res. 69:1358–1367. 2009.
|
|
36
|
Tatsumi N, Oji Y, Tsuji N, Tsuda A,
Higashio M, Aoyagi S, et al: Wilms’ tumor gene WT1-shRNA as a
potent apoptosis-inducing agent for solid tumors. Int J Oncol.
32:701–711. 2008.
|
|
37
|
McCarty G, Awad O and Loeb DM: WT1 protein
directly regulates expression of vascular endothelial growth factor
and is a mediator of tumor response to hypoxia. J Biol Chem.
286:43634–43643. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jomgeow T, Oji Y, Tsuji N, Ikeda Y, Ito K,
Tsuda A, et al: Wilms’ tumor gene WT1 17AA(−)/KTS(−) isoform
induces morphological changes and promotes cell migration and
invasion in vitro. Cancer Sci. 97:259–270. 2006.
|