Introduction
SGEF was initially identified in a screen for
androgen-responsive genes in human prostate cancer cells (1). SGEF contains N-terminal proline-rich
domain, DH domain in tandem with a PH domain and C-terminal SH3
binding domain. The DH-PH module is essential for GEF activity of
most DBL family members. Proline-rich domain and SH3 binding domain
are assumed to be associated with protein-protein interactions and
modulation of exchange activity of SGEF. SGEF supports the exchange
of GDP for GTP on Rho G and is the first identified mammalian
RhoGEF that promotes macropinocytosis (2). SGEF has been reported to be involved
in the uptake of Salmonella by epithelial cells by stimulating the
formation of the surfaces phagocytic cups (3). It is also reported that SGEF can
promote leukocyte trans-endothelial migration by regulating
endothelial apical cup assembly (4). All the data reported have suggested
that SGEF play a role on various physiological and pathological
situations in association with phagocytosis or the uptake of
particulate material. In contrast, the functions of SGEF in human
cancer still remain unknown.
Prostate cancer (PCa) remains the most commonly
diagnosed and the second leading cause of cancer-related death in
men in the western world (5).
Although early stage PCa is responsive to androgen withdrawal, it
ultimately progresses to AI and there is no curative therapy
available at present. Improved understanding of the molecular
events of prostate cancer progression will contribute to developing
new therapeutic approaches.
In this study, we explored the expression and
potential roles of SGEF in human prostate cancer. Our studies
revealed that SGEF was overexpressed in human specimens and AI
prostate cancer cells. Moreover, the growth of AI prostate cancer
cell lines C4-2 and C4-2B was suppressed by knockdown of SGEF and
this inhibitory effect can be enhanced by bicalutamide, which is
known as AR antagonist. Collectively, these data identify SGEF as a
novel potential promoter of human prostate cancer progression.
Materials and methods
Cell culture
C4-2 and C4-2B cell lines were generous gifts from
L.W. Chung (Emory University, USA), cultured in RPMI-1640 (Gibco,
Paisley, UK) containing 10% fetal bovine serum (FBS; Hyclone,
Logan, UT, USA) at 37°C in 5% CO2. Where indicated,
cells were treated with 1 nM R1881 (methyltrienolone) in fresh
phenol red-free RPMI-1640 with 5% dextran/charcoal absorbed fetal
bovine serum (cFBS; Hyclone) or 10 μM bicalutamide in culture
medium with 10% FBS. For shRNA experiments, C4-2 or C4-2B cell
lines were infected with lentivirus derived from pU6-vshRNA-CMV-GFP
(GeneChem Co., Shanghai, China), containing the shRNA human SGEF
sequence 5′-GGAATCTTGTGACAATGAAGA-3′.
Western blot analysis
Western blot analysis was performed as described
previously (6). Rabbit anti-SGEF
(Sigma, St. Louis, MO, USA), mouse actin antibody (Santa Cruz
Biotechnology, Inc., Heidelberg, Germany), rabbit anti-AKT and
rabbit anti-AKT/phosphor Ser473 (Cell Signaling Technology, Inc.,
Danvers, MA, USA) were used as primary antibodies. Proteins were
visualized by using the enhanced chemiluminescence kit (Pierce)
after incubation with anti-mouse or anti-rabbit HRP conjugated
secondary antibodies (Zhongshan Golden Bridge Biotechnology Co.,
Ltd.).
RT-PCR
Total RNA was extracted from cells using TRIzol
reagent (Invitrogen Life Technologies, CA, USA). Reverse
transcription was performed with 1 μg RNA using the First Strand
Synthesis System kit (Toyobo, Japan) according to the
manufacturer’s instruction. The following primers were used for PCR
amplification: 5′-ACTCGGTGTTGCTCCTCCC-3′ and
5′-GGCTCCTATGTACCGTCCTG-3′ for SGEF; 5′-GAGCTACGAGCGCCTGACG-3′ and
5′-CCTAGAAGCTTTGCGGTGG-3′ for β-actin.
Growth rate assay
To obtain growth curves, 1,500 cells were seeded in
96-well plates. Cell growth was examined at indicated time-points
using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
(MTT) assays according to the manufacturer’s directions (Amresco).
Each experiment was repeated independently four times.
Colony formation assay
For the colony-formation assay, 2,000–6,000 cells
were seeded in 6-well plates. After 12 days, cells were washed in
PBS and colonies were fixed by methanol and stained with 0.1%
crystal violet (Sigma). The colonies containing >50 cells were
counted.
Soft agar assay
A total of 2,000–6,000 cells were suspended in 2 ml
of 0.17% low melting agarose (Difco Laboratories) dissolved in
RPMI-1640/10% FBS and plated on the top of 3 ml underlayer of 0.5%
agarose in the same medium in 6-well culture plates. After 3 weeks
of incubation at 37°C in 5% CO2, the colonies containing
>15 cells were counted.
Luciferase reporter assays
C4-2 and C4-2B cells were seeded in 24-well plates
(5−104 cells per well) and transfected with 300 ng
PSA-luc reporter gene plasmids, 100 ng pCMV-2B or pCMV-2B-SGEF
(PGPU6/GFP/Neo-si-NC or PGPU6/GFP/Neo-si-SGEF) plasmids and 30 ng
of pCMV-β-gal plasmids (internal control) using the Lipofectamine
2000 reagent (Invitrogen). After transfection, cells were treated
with 1 nM R1881 or vehicle for 24 h. Then luciferase activity was
measured using luciferase assay system (Promega Corp., Madison, WI,
USA) normalized by β-galactosidase activity. All transfection
experiments were carried out in triplicate wells and repeated three
times.
Immunohistochemistry
Immunohistochemistry was performed as described
previously using TMA (Chaoying Biotechnology Co., Xi’an, China)
including 75 prostate specimens (10 benign prostate tissues, 19 BPH
tissues and 46 tumor tissues) (6).
SGEF expression was scored blindly by staining intensity in each
biopsy core as being weak, moderate or strong.
Statistical analysis
Statistical evaluation was performed using the
statistical software SAS/STAT. p<0.01 was considered
statistically significant.
Results
Expression analysis of SGEF in human
cancer cells
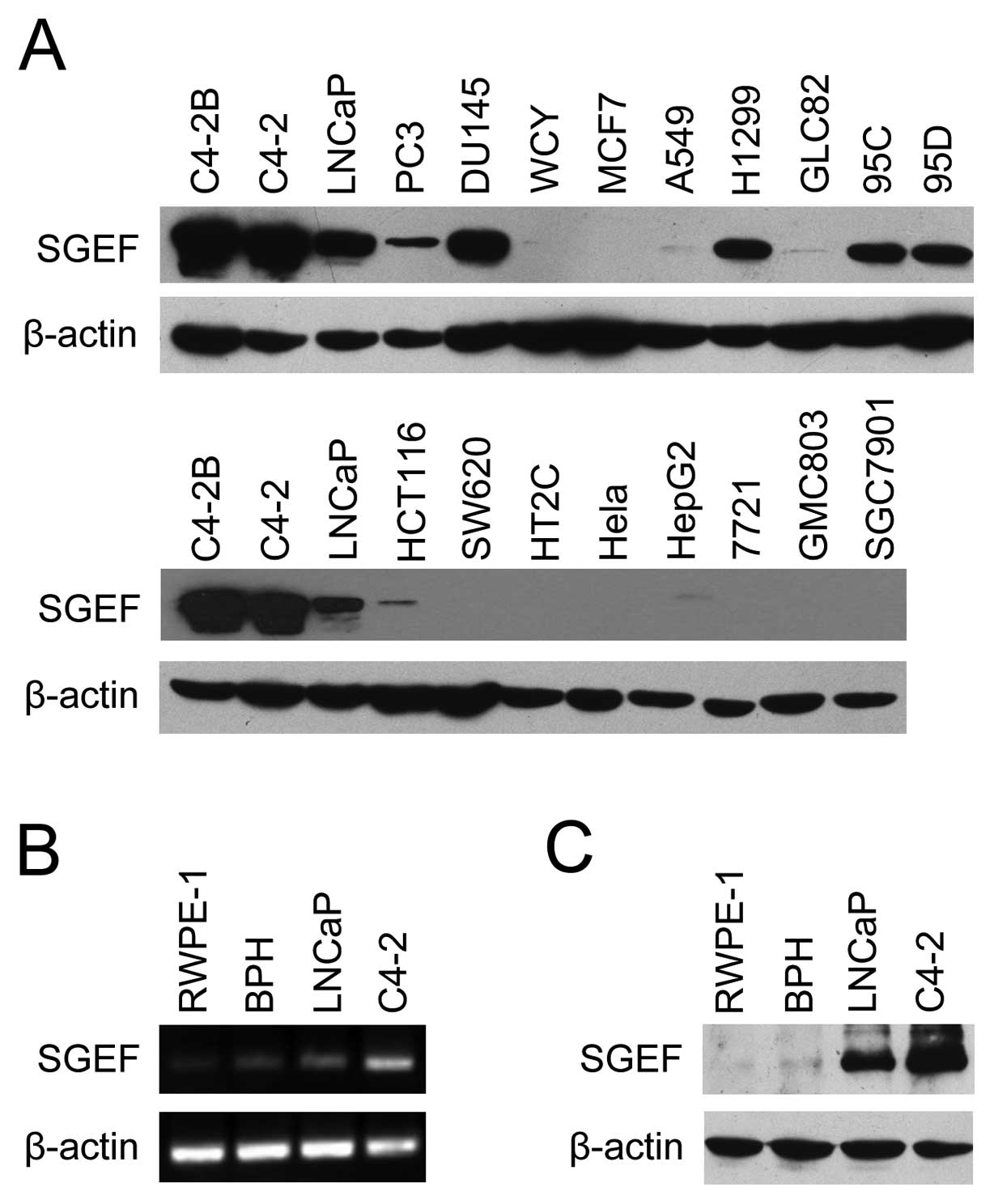
We first examined the expression pattern of SGEF
protein in various cancer cell lines by western blot analyses. As
shown in Fig. 1A, the expression of
SGEF was restricted to the prostate cancer cell lines (C4-2B, C4-2,
LNCaP, PC3 and DU145) and part of the lung cancer cell lines (A549,
H1299, GLC82, 95C and 95D), and absent or very low levels in breast
cancer (MCF10 and WCY), colon cancer (HCT116, SW620 and HT29),
cervical cancer (HeLa), liver cancer (HEPG2 and 7721) and gastric
cancer (GMC803 and SGC7901). Moreover, all prostate cancer cell
lines, except for PC3, had a higher level of SGEF than that of lung
cancer. These results indicated that SGEF may be closely related to
prostate cancer. Then RT-PCR and western blot analyses were
performed to further analyze the expression patterns of SGEF in
prostate cancer cells. As shown in Fig.
1C, SGEF protein was found to be absent or at a very low level
in normal prostate epithelial cell line RWPE-1 and BPH cell line,
moderate level in androgen-dependent prostate cancer cell line
LNCaP, and of the highest level in androgen-independent cell line
C4-2. The results of RT-PCR (Fig.
1B) were consistent with that of western blotting. These
results further showed that SGEF expressions were positively
associated with prostate cancer malignant progression.
SGEF expression is elevated in human
prostate cancer specimens
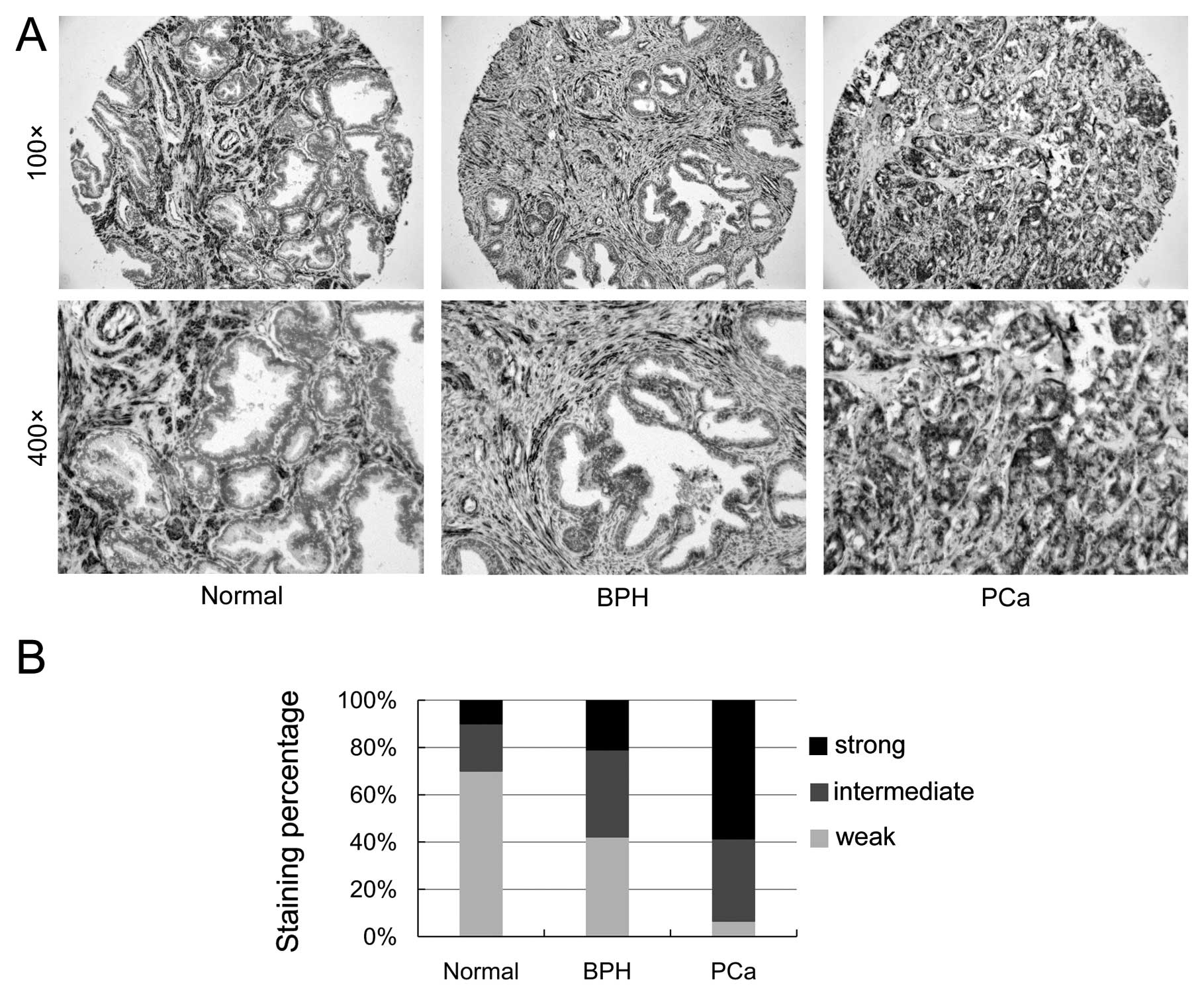
To validate the clinical relevance of SGEF
expression with human prostate cancer progression, we determined
the expression of SGEF in human PCa specimens utilizing TMA
containing 10 normal tissues, 19 BPH tissues and 46 tumor tissues
through immunohistochemistry. Fig.
2A shows the representative images of SGEF expression from
human prostate TMA samples. As shown in Fig. 2B, the strong staining frequency of
SGEF significantly increased in the epithelial cells of tumor
samples (58.7%) in comparison to normal prostate samples (10%) and
BPH samples (21.1%). Compared with normal prostate samples (70%)
and BPH samples (42.1%), the weak staining frequency of SGEF
notably decreased in the epithelial cells of tumor samples (6.5%).
The TMA data suggested that SGEF expression was elevated and
associated with the progression of prostate cancer.
Reducing the expression of SGEF inhibits
the growth of prostate cancer cells
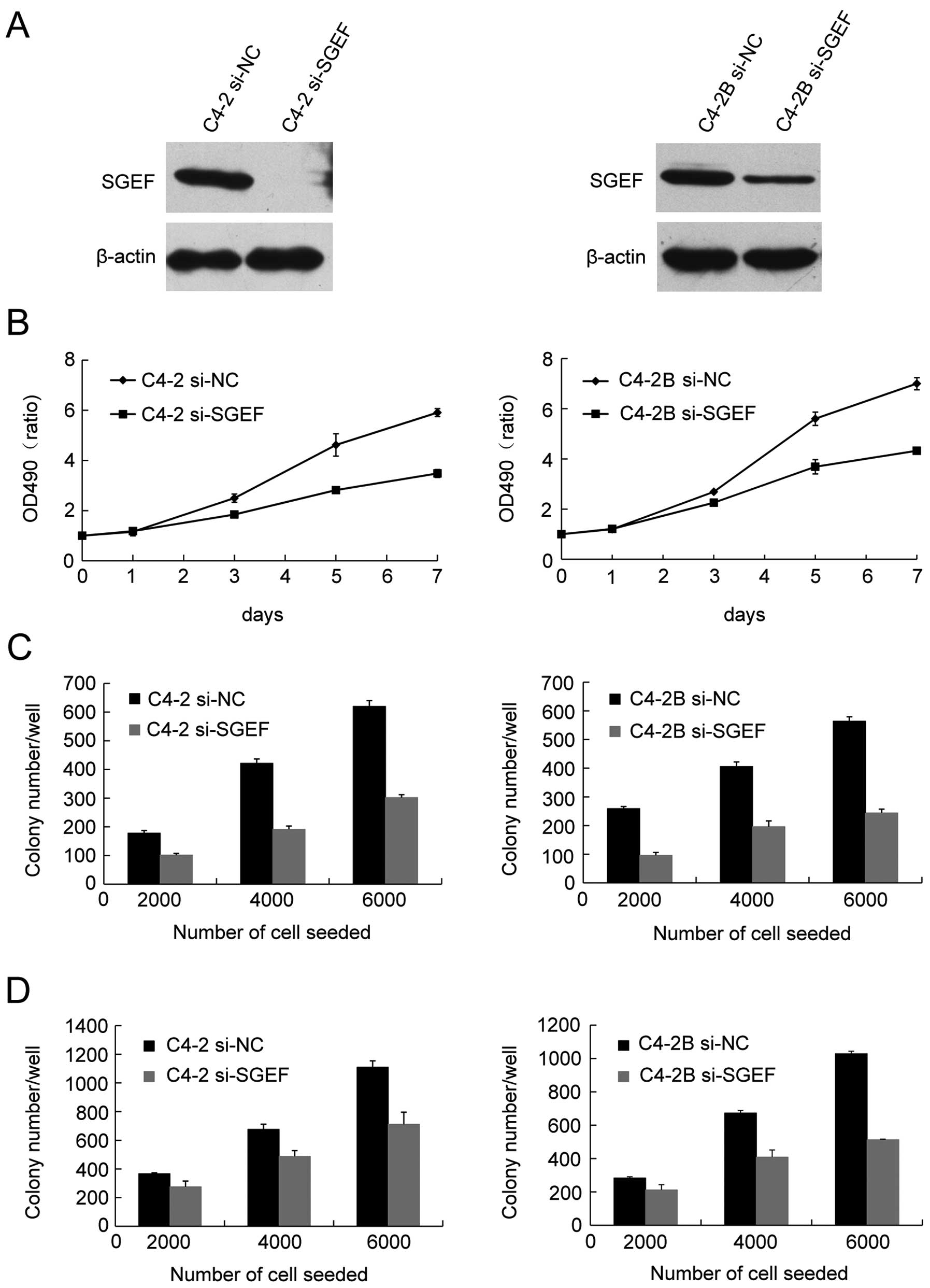
In order to investigate the potential role of SGEF
in human prostate cancer cells growth, C4-2 and C4-2B cells were
infected with lentivirus expressing an SGEF-targeted siRNA. Western
blot assays showed that SGEF protein was sharply reduced (Fig. 3A). MTT assays were used to examine
the correlation between knockdown of SGEF and the growth ability of
C4-2 and C4-2B. The cell growth curves showed that silencing the
expression of SGEF suppressed the growth of C4-2 and C4-2B cells
(Fig. 3B). Then, colony formation
assays and soft agar assays were respectively performed to
determine the effect of SGEF reduction on the viability and
anchorage-independent growth ability of prostate cancer cell. As
data showed, C4-2 and C4-2B cells of the reduced SGEF displayed
significantly decreased clonogenecity and anchorage-independent
growth ability respectively, regardless of the cell number seeded
(Fig. 3C and D). These results
suggested that endogenous SGEF had positive roles to human prostate
cancer cells growth.
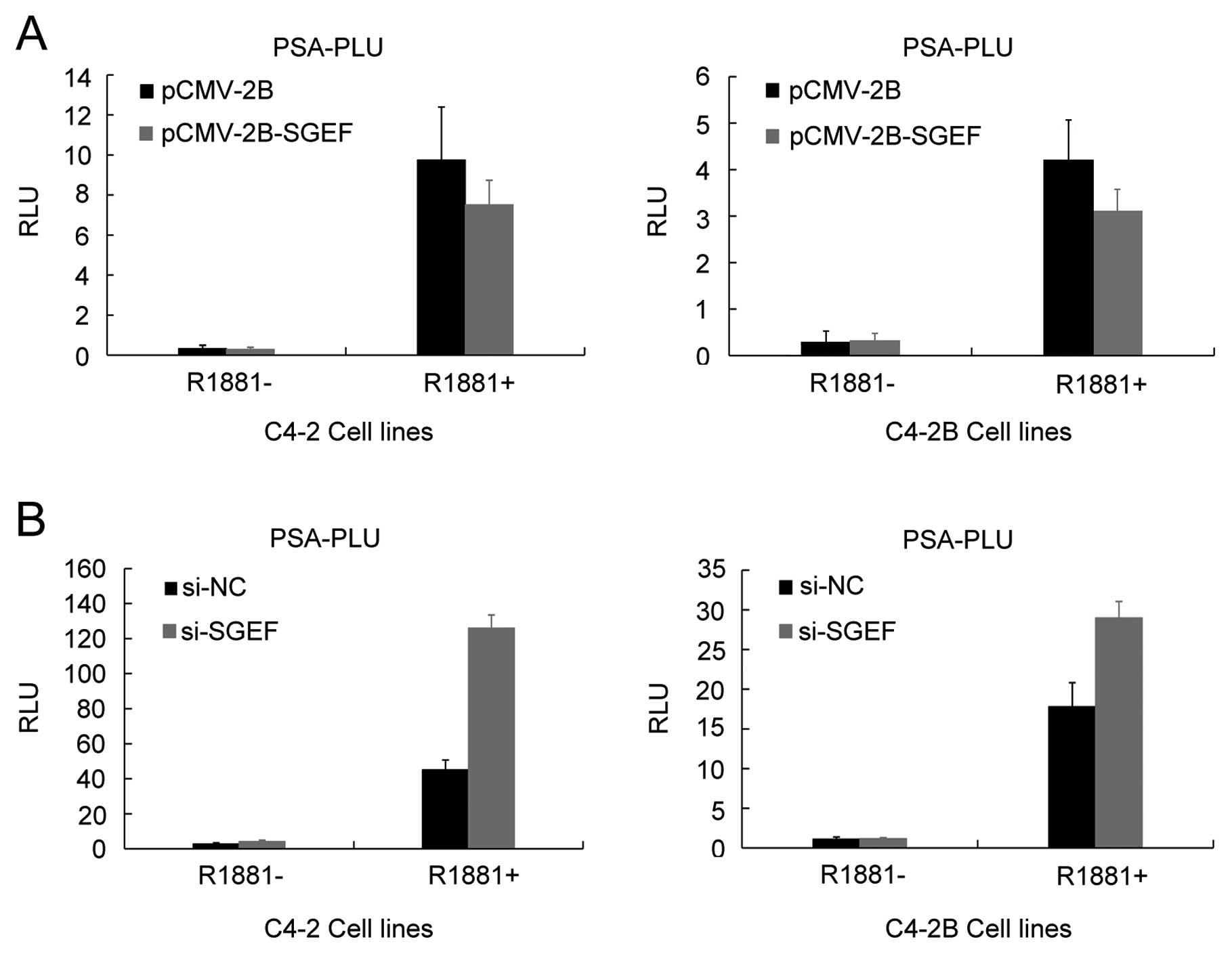
SGEF suppress AR transactivation
It was reported that Vav3 protein, another Rho
GTPase GEF similar to SGEF, contributed to prostate cancer cell
growth through enhancing AR transactivation. Hence, we assumed that
SGEF might function in the same way. To verify this supposition,
FLAG-tagged SGEF or SGEF siRNA plasmid was transiently transfected
into C4-2 and C4-2B cells along with PSA-Luc reporter construct in
the presence of R1881 or not to examine the potential effects of
SGEF on the AR transactivation. As shown in Fig. 4A, overexpression of SGEF decreases
AR transactivation in an androgen-dependent manner. Consistent with
the results of the SGEF overexpression, knockdown of SGEF increases
AR transactivation in two cell lines (Fig. 4B). Taken together, these results
indicated that SGEF could suppress AR transcriptional activity, but
not increase its transcriptional activity.
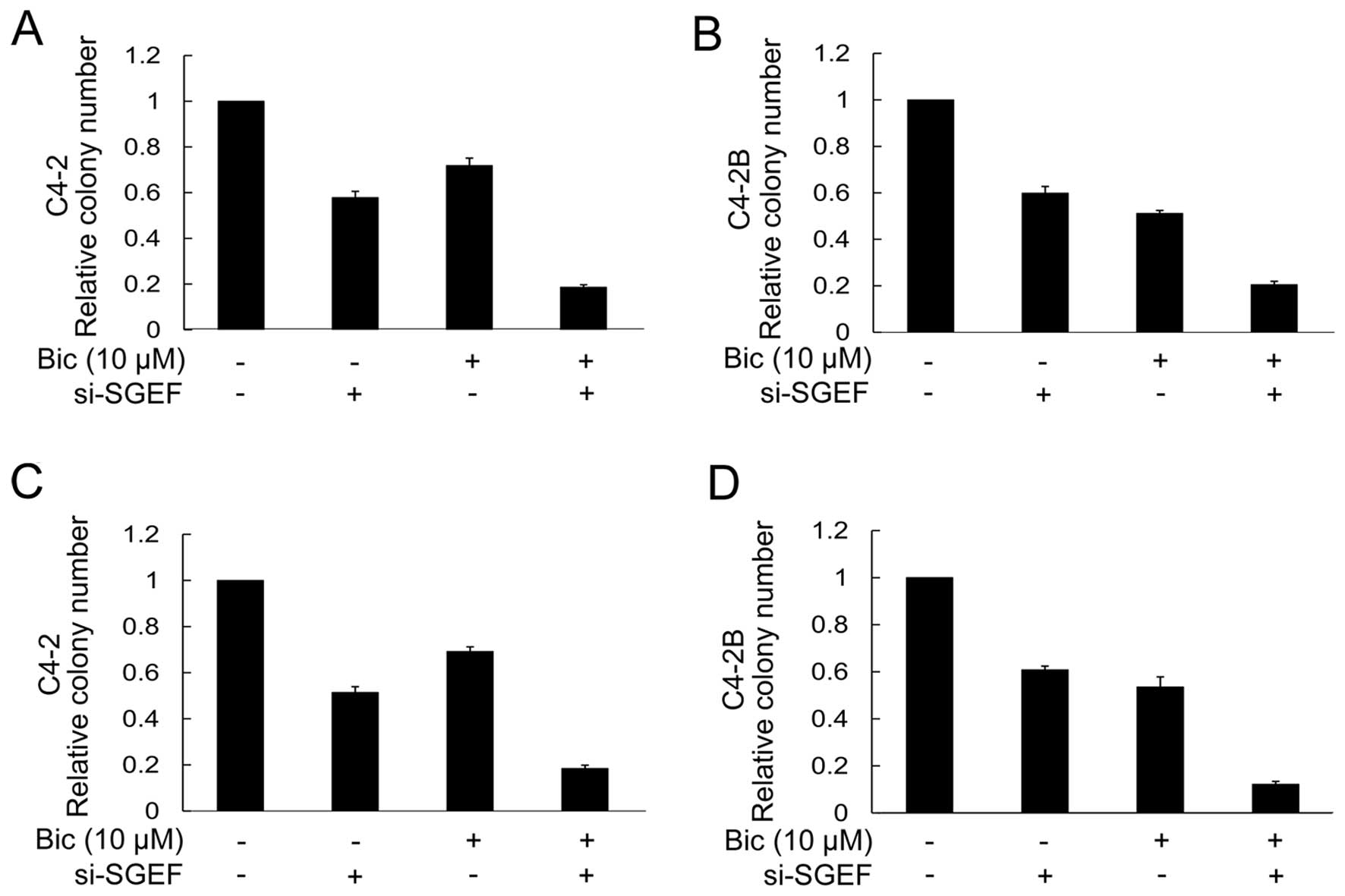
AR antagonist bicalutamide potentiates
inhibitory effects of reduced SGEF on the growth of prostate cancer
cells
The previous data indicated that knockdown of SGEF
inhibited the growth of C4-2 and C4-2B, but augmented the
transcriptional activity of AR. Hence, it is believed that the
inhibitory effect of decreasing the SGEF expression on the growth
of C4-2 and C4-2B cells could be further strengthened by the AR
antagonist, which could repress the AR transactivation. As
expected, bicalutamide, an AR antagonist, further enhanced the
suppression of the clonogenicity and anchorage-independent growth
of C4-2 and C4-2B cells by silencing the SGEF expression. For
example, compared with control cells, the colony-formation rate of
SGEF-knockdown C4-2 or C4-2B cells was decreased by 40 or 42%, but
in the conditions of bicalutamide, the inhibition rate reached up
to 81 or 80%, respectively (Fig. 5A and
B). Similarly, the soft-agar colony formation rate of C4-2 or
C4-2B cells was decreased by 49 or 39% as a result of decreasing
the SGEF expression, whereas the rate was up to 82 or 88% when
treated with bicalutamide (Fig. 5C and
D). These results suggested that bicalutamide facilitated the
inhibitory effect of reduced SGEF on the growth of C4-2 and C4-2B
cells.
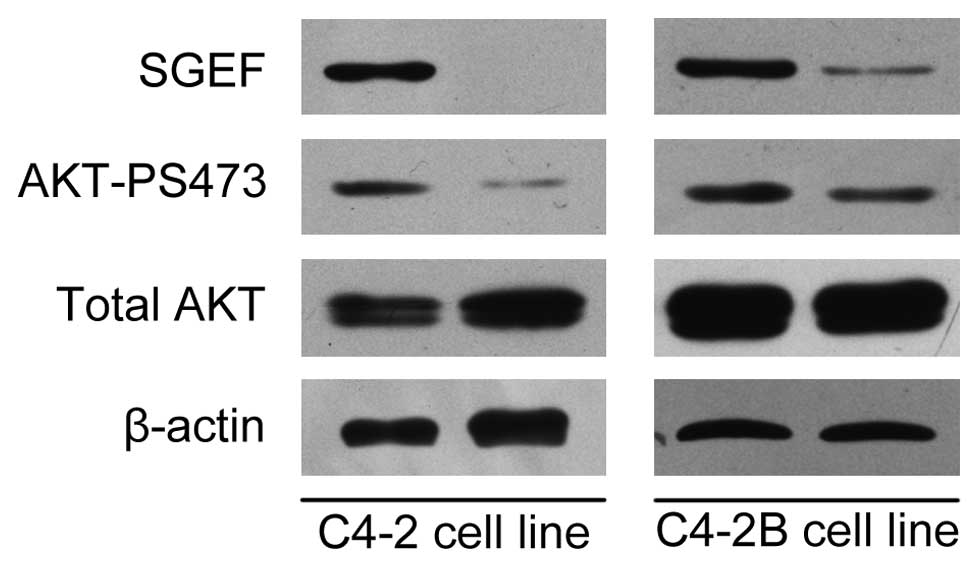
Silencing the expression of SGEF inhibits
Akt signaling pathway
Abnormal activation of Akt/PKB is significantly
associated with the development of prostate cancer. To further
elucidate the mechanism mediating SGEF function in prostate cancer
progression, we examined the effect of SGEF on Akt phosphorylation
in C4-2 and C4-2B cells (Fig. 6).
As a result, knockdown of SGEF was observed to suppress the Ser473
phosphorylation of Akt in these two prostate cancer cell lines.
These results suggest that AKT may be a downstream target of SGEF
and SGEF may mediate tumor cell growth through Akt/PKB signaling
pathways.
Discussion
Previous studies focused on the role of SGEF in
cytoskeleton organization, and little attention was given to its
roles in cancer. This study is the first on the roles of SGEF in
cancer. We found that the expression of SGEF was elevated in human
prostate cancer cells and in the epithelial cells of tumor tissues.
Downregulation of SGEF inhibited both anchorage-dependent and
anchorage-independent growth ability of human prostate cancer
cells. Moreover, we demonstrated that the inhibitory effect of
silenced SGEF on prostate cancer cell growth could be enhanced by
the bicalutamide treatment. Taken together, our results indicate
that increased expression of SGEF is involved in human prostate
cancer progression.
Rho family GTPases are activated in response to a
multitude of stimuli and regulate numerous cellular processes
including gene expression, cytoskeleton organization, cell
proliferation and survival. Because of their central role in
regulating diverse signaling pathways and cellular processes,
deregulation of the RhoGTPase pathway is assumed to be involved in
the development of cancer. In contrast to Ras genes, the typical
members of the GTPase super-family, few point mutations were
detected in human tumors in RhoGTPases (7). Instead, overexpression and
hyperactivity of Rho proteins appear to play a role in human cancer
initiation and progression. These facts indicate that regulator
proteins of RhoGTPases, especially GEFs, have a crucial role in
cancer development. Consistent with this, many GEFs including Dbl,
LARG, Vav1 and Ect2 were found as genetic alteration or aberrant
expression in several human tumors (8–11). In
prostate cancer, several GEFs including Tiam1, PREX1 and Vav3 were
reported to be overexpressed and associated with carcinoma
progression (12–14). These reports further support our
result that SGEF may act as a novel potential oncogene contributing
to prostate cancer development.
Enhancement of AR signaling has been thought to be
involved in prostate cancer initiation and progression (15,16).
Vav3, another GEF similar to SGEF, has been reported to promote
prostate cancer cells growth though enhancing AR transactivation
(14). To assess whether SGEF
functions in the same way, we evaluated the effect of SGEF on the
AR transcriptional activity. Unexpectedly, luciferase reporter
assays suggested that SGEF repressed AR transactivation. These data
indicate that the positive effect of SGEF expression on prostate
cancer cell growth is not due to elevated AR activity. Furthermore,
these results also provide us an assumption that AR antagonist may
further enhance the inhibition of decreased SGEF on the growth of
prostate cancer cells though reducing the elevated AR
transcriptional activity from silencing SGEF expression. Our
subsequent experimental results confirm our hypothesis. In
addition, we also found that reducing the expression of SGEF and
treatment of the androgen antagonist had a synergistic effect on
the suppression of prostate cancer cell growth. These studies
provide the possibility that increased SGEF expression is involved
in androgen antagonist treatment resistance, which is a critical
stage for poor prognosis in prostate cancer.
The PI3K/Akt signaling pathway has a critical role
in prostate cancer progression and development. Increased Akt
kinase activity correlates with poor prognosis in human prostate
cancer and is associated with a hormone therapy-resistant phenotype
(17–20). Previous studies have shown that SGEF
can activate RhoG, a member of the Rho family of small GTPases
(2–4). In addition, RhoG was reported to
inhibit anoikis through a phosphatidylinositol 3-kinase-dependent
mechanism (21). Overexpression of
Vav3, another GEF similar to SGEF, has been reported to elevate the
phosphorylation of Akt in prostate cancer cells (14). These studies lead us to examine
whether SGEF activate the Akt signaling pathway. Our data show that
knockdown of SGEF decreases levels of phosphorylated Akt. Since Akt
can regulate a variation of substrates involved in multiple
cellular processes, including cell proliferation, cell migration
and cell differentiation, we speculate that the activation of Akt
signaling pathway may be one of the reason that SGEF contributes to
prostate cancer progression. Further studies are necessary to
determine how SGEF regulate Akt activity and which substrate of Akt
affects cell proliferation as a downstream target of SGEF.
Previous reports have demonstrated that inhibition
of PI3K/Akt signaling pathways enhances AR transcriptional activity
(22–25). Additionally, a recent study shows
that PI3K-AKT-mTOR pathway is dominant over AR signaling in
prostate cancer cell growth (26).
Data presented here suggest that decreasing the expression of SGEF
enhance the AR transactivation, but inhibits the PI3K/Akt
signaling. However, in spite of an increase in AR signaling, which
is thought to have proliferative effects, the end result of
knockdown of SGEF is reduced prostate cancer cell growth which can
be further enhanced when AR signaling is blocked. Our results
provide a possibility that reduced SGEF increases AR signaling
though inhibition of the PI3K/AKT pathway and results in an
inhibitory effect on cell growth due to the dominant role of
PI3K/AKT pathway in prostate cancer cell growth. Further
experiments should be performed to check this hypothesis.
In conclusion, this study reveals that SGEF is
overexpressed in human prostate cancer cells and contribute to
prostate cancer progression. These data suggest SGEF could be a new
potential marker and an efficacious therapeutic target for human
prostate cancer.
Acknowledgements
This study was supported by grants to Jianguang Zhou
and Jian Wang from National Natural Science Foundation of China
(nos. 30870961 and 81172445).
Abbreviations:
|
SGEF
|
SH3-containing guanine nucleotide
exchange factor
|
|
PCa
|
prostate cancer
|
|
AI
|
androgen independence
|
|
AR
|
androgen receptor
|
|
BPH
|
benign prostatic hyperplasia
|
|
TMA
|
tissue microarray
|
|
GTPase
|
guanosinetriphosphatase
|
|
GEF
|
guanine nucleotide exchange factor
|
References
|
1
|
Qi H, Fournier A, Grenier J, Fillion C,
Labrie Y and Labrie C: Isolation of the novel human guanine
nucleotide exchange factor Src homology 3 domain-containing guanine
nucleotide exchange factor and of C-terminal SGEF, an N-terminally
truncated form of SGEF, the expression of which is regulated by
androgen in prostate cancer cells. Endocrinology. 144:1742–1752.
2003.
|
|
2
|
Ellerbroek SM, Wennerberg K, Arthur WT, et
al: SGEF, a RhoG guanine nucleotide exchange factor that stimulates
macropinocytosis. Mol Biol Cell. 15:3309–3319. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Patel JC and Galan JE: Differential
activation and function of Rho GTPases during Salmonella-host cell
interactions. J Cell Biol. 175:453–463. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Van Buul JD, Allingham MJ, Samson T, et
al: RhoG regulates endothelial apical cup assembly downstream from
ICAM1 engagement and is involved in leukocyte trans-endothelial
migration. J Cell Biol. 178:1279–1293. 2007.PubMed/NCBI
|
|
5
|
Jemal A, Siegel R, Ward E, Murray, Xu J
and Thun MJ: Cancer statistics. Cancer J Clin. 57:43–66. 2007.
|
|
6
|
Pang B, Zhang H, Wang J, et al: Ubiquitous
mitochondrial creatine kinase is overexpressed in the conditioned
medium and the extract of LNCaP lineaged androgen independent cell
lines and facilitates prostate cancer progression. Prostate.
69:1176–1187. 2009. View Article : Google Scholar
|
|
7
|
Schubbert S, Shannon K and Bollag G:
Hyperactive Ras in developmental disorders and cancer. Nat Rev
Cancer. 7:295–308. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Advani AS and Pendergast AM: Bcr-Abl
variants: biological and clinical aspects. Leuk Res. 26:713–720.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kourlas PJ, Strout MP, Becknell B, et al:
Identification of a gene at 11q23 encoding a guanine nucleotide
exchange factor: evidence for its fusion with MLL in acute myeloid
leukemia. Proc Natl Acad Sci USA. 97:2145–2150. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fernandez-Zapico ME, Gonzalez-Paz NC,
Weiss E, et al: Ectopic expression of VAV1 reveals an unexpected
role in pancreatic cancer tumorigenesis. Cancer Cell. 7:39–49.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hirata D, Yamabuki T and Miki D:
Involvement of epithelial cell transforming sequence-2 oncoantigen
in lung and esophageal cancer progression. Clin Cancer Res.
15:256–266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Engers R, Mueller M, Walter A, Collard JG,
Willers R and Gabbert HE: Prognostic relevance of Tiam1 protein
expression in prostate carcinomas. Br J Cancer. 95:1081–1086. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qin J, Xie Y, Wang B, et al: Upregulation
of PIP3-dependent Rac exchanger 1 (P-Rex1) promotes prostate cancer
metastasis. Oncogene. 28:1853–1863. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dong ZY, Liu Y, Lu S, et al: Vav3 oncogene
is overexpressed and regulates cell growth and androgen receptor
activity in human prostate cancer. Mol Endocrinol. 20:2315–2325.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Linja MJ, Savinainen KJ, Saramaki OR,
Tammela TL, Vessella RL and Visakorpi T: Amplification and
overexpression of androgen receptor gene in hormone-refractory
prostate cancer. Cancer Res. 61:3550–3555. 2001.PubMed/NCBI
|
|
16
|
Ruizeveld de Winter JA, Janssen PJ,
Sleddens HM, et al: Androgen receptor status in localized and
locally progressive hormone refractory human prostate cancer. Am J
Pathol. 144:735–746. 1994.PubMed/NCBI
|
|
17
|
Edwards J, Krishna NS, Witton CJ and
Bartlett JM: Gene amplifications associated with the development of
hormone resistant prostate cancer. Clin Cancer Res. 9:5271–5281.
2003.PubMed/NCBI
|
|
18
|
Zhang H, Wang J, Pang B, et al: PC-1/PrLZ
contributes to malignant progression in prostate cancer. Cancer
Res. 67:8906–8913. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nakatani K, Thompson DA, Barthel A, et al:
Upregulation of Akt3 in estrogen receptor deficient breast cancers
and androgen-independent prostate cancer lines. J Biol Chem.
274:21528–21532. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ayala G, Thompson T, Yang G, et al: High
levels of phosphorylated form of Akt-1 in prostate cancer and
non-neoplastic tissues are strong predictors of biochemical
recurrence. Clin Cancer Res. 10:6572–6578. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yamaki N, Negishi M and Katoh H: RhoG
regulates anoikis through a phosphatidylinositol 3-kinase-dependent
mechanism. Exp Cell Res. 313:2821–2832. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang L, Lin HK, Altuwaijri S, Xie S, Wang
L and Chang C: APPL suppresses androgen receptor transactivation
via potentiating Akt activity. J Biol Chem. 278:16820–16827. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin HK, Yeh S, Kang HY and Chang C: Akt
suppresses androgen-induced apoptosis by phosphorylating and
inhibiting androgen receptor. Proc Natl Acad Sci USA. 98:7200–7205.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang L, Wang L, Lin HK, et al:
Interleukin-6 differentially regulates androgen receptor
transactivation via PI3K-Akt, STAT3, and MAPK, three distinct
signal pathways in prostate cancer cell. Biochem Biophys Res
Commun. 305:462–469. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang L, Xie S, Jamaluddin MS, et al:
Induction of androgen receptor expression by
phosphatidylinositol3-kinase/Akt downstream substrate, FOXO3a, and
their roles in apoptosis of LNCaP prostate cancer cells. J Biol
Chem. 280:33558–33565. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kaarbø M, Mikkelsen OL, Malerød L, et al:
PI3K-AKT-mTOR pathway is dominant over androgen receptor signaling
in prostate cancer cells. Cell Oncol. 32:11–27. 2010.PubMed/NCBI
|