Introduction
Gastric cancer (GC), is the fourth most common type
of malignancy and the second most common cause of cancer death in
the world (1), over 70% of the
gastric cancer cases occur in the developing countries, and half of
the total cases occur in Eastern Asia (mainly in China) (2). Gastric cancer is a biologically and
genetically heterogeneous carcinoma (3), and accumulating evidence has suggested
that various genetic and epigenetic alterations are related to
human gastric cancer (4), including
overexpression of oncogenes such as c-met and c-erbB2
(5–7), inactivation of tumor suppressor genes
such as p53, β-catenin and PTEN (8–10), as
well as alterations of cell cycle regulators, cell adhesion
molecules and DNA repair genes (4).
Sakata et al have reported that methylation of HACE1
(HECT domain and ankyrin repeat containing E3 ubiquitin-protein
ligase 1) and downregulation of EGFL8 (epidermal growth
factor-like domain 8) were intimately related to gastric cancer
(11,12). The majority of gastric cancer cases
are diagnosed at advanced stages which are generally resistant to
chemotherapy or radiotherapy, and the current 5-year survival rate
of gastric cancer is <20% (13,14).
Nevertheless, if gastric cancer could be diagnosed at an early
stage, it is a curative disease. Therefore, it is crucial to
identify clinically useful biomarkers that can diagnose gastric
cancer at an early stage (15).
Thus, further investigations to identify genetic changes as new
parameters for assessing the progression of gastric cancer are
necessary.
Carboxyl terminus of heat shock cognate 70
interacting protein (CHIP) is a cytoplasmic protein containing a
34-amino-acid tetratricopeptide repeat (TPR) domain (16), which is referred to in
protein-protein interactions (17),
an intervening charged domain and a ‘U-box’ domain (18). The U-box domain contains an E3
ubiquitin ligase activity and can induce ubiquitylation and
subsequent proteasome-dependent degradation of tumor-related
proteins (19,20). Therefore, many studies have focused
on the relationship between CHIP and carcinomas. For instance, CHIP
acts as an upstream regulator of oncogenic pathways and inhibits
cell growth and metastatic potential by degrading oncogenic
proteins including SRC-3 in breast cancer (21). And a recent report demonstrated CHIP
contributes to the oncogenesis of glioma (22). Moreover, a present study found that
CHIP interacts with endogenous Met in lung cancer cells (H358
cells) via inducing the ubiquitination and degradation of Met
receptor and CHIP inhibits the tumor growth by decreasing Met in
vivo (23). However, the
expression of CHIP in human gastric cancer remains unknown.
Therefore, the current study was carried out to evaluate the
expression of CHIP in gastric cancer and to explore the
correlations between CHIP expression and clinicopathological
characteristics of gastric cancer. In our present study, we found
the decreased expression of CHIP is associated with the clinically
aggressive phenotype in gastric cancer.
Materials and methods
Clinical patient samples
Fifty-three patients (median age, 56.0 years; range,
16–77 years; 32 males, 21 females) with primary gastric cancer were
included in this study. A total of 53 paired cancerous samples and
matched adjacent normal mucosa located at least 6 cm away from the
tumor site were collected from patients who underwent initial
surgical resection at Tongji Hospital, Tongji Medical College
(Wuhan, China) between April 2011 and January 2012. The
non-cancerous samples were confirmed to be without any tumor cell
infiltration by histological examination. All patients were
pathologically diagnosed as stomach carcinoma, without any
metastatic diseases or any other tumors. Informed written consent
was obtained from all the patients and the study was approved by
the local ethics committee. For each sample, a portion of the
lesion was frozen in liquid nitrogen immediately after surgical
resection and then stored at −80°C, while another portion was fixed
in 10% formalin-buffered and paraffin-embedded.
Total RNA extraction and first strand
cDNA synthesis
RNAiso Plus extraction of total RNA was carried out
essentially according to the manufacturer's instructions (Takara,
Dalian, China). The RNA pellets were dissolved in 40 μl of
RNase-free water and stored at −80°C. RNA integrity was assessed
prior to cDNA synthesis. The concentration of total RNA was
measured by UNICO UV-2800 spectrophotometric readings (Shanghai,
China) and the OD260/OD280 ratio of all RNA samples were up to 2.0.
The first strand cDNA was synthesized using the RevertAid™ First
Strand cDNA Synthesis kit (Fermentas, MBI, Lithuania) according to
the manufacturer's protocol.
Polymerase chain reaction and
quantitative real-time PCR
Polymerase chain reactions (PCR) were performed in a
total volume of 20 μl, containing 10 μl 2X Taq PCR MasterMix, 0.5
μl of each primer (10 pM each), 1 μl cDNA template and 8 μl sterile
water. The amplification protocol consisted of an initial
denaturation at 94°C for 5 min, followed by 35 cycles of
denaturation for 30 sec at 94°C, annealing for 45 sec at 64°C and
extension for 30 sec at 72°C, followed by a final extension at 72°C
for 10 min. The PCR products were verified by 1.5% agarose gel
electrophoresis and analyzed using the Gel Doc™ XR Imaging System
(Bio-Rad, Foster City, CA, USA). The PCR and real-time PCR primers
for CHIP (151 bp): forward, 5′-GAGGCCAAGCACGACAAGTAC-3′;
reverse, 5′-TGATGCCACTGGGCGTGATGC-3′. GAPDH (218 bp):
forward, 5′-GGTCGGAGTCAACGGATTTG-3′; reverse,
5′-GGAAGATGGTGATGGGATTTC-3′. The primers of CHIP and
GAPDH genes were designed by Primer Premier 5.0 software
(Premier Biosoft International, Palo Alto, CA, USA). Quantitative
real-time PCR was performed with a continuous fluorescence detector
- StepOne machine (Applied Biosystems, Forster City, CA, USA).
Quantitative real-time PCR reaction was carried out using SuperReal
PreMix SYBR-Green kit (Tiangen Biotech Co., Ltd., Beijing, China).
The cycling parameters were: initial denaturation at 95°C for 15
min, followed by 40 cycles at 95°C for 10 sec, 64°C for 30 sec and
72°C for 30 sec. The cycling was followed by melting curve analysis
to distinguish specificity of the PCR products. CHIP
expression was normalized with glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) as an internal control in the same
sample. Each sample was run three times. No template controls (no
cDNA in PCR reaction) were run to detect unspecific or genomic
amplification and primer dimerization. The average threshold cycle
(Ct) for three replicates per sample was used to calculate ΔCt.
Relative quantification of CHIP expression was calculated with the
2−ΔΔCt method.
Tissue immunohistochemistry and
immunoblotting
Tissue immunohistochemistry (IHC) was performed
using a standard peroxidase-based staining method. Tissue sections
(4 μm) were dewaxed in xylene, hydrated with graded ethanol. Then
antigen retrieval was performed by pretreatment of the slides in
0.01 M citrate buffer (pH 6.0) using a microwave oven.
Subsequently, the sections were treated with 3% hydrogen peroxide
(H2O2) for 10 min in order to block
endogenous peroxidase. The sections were washed with 0.01 M
phosphate-buffered saline (PBS) (pH 7.4), and were incubated with
rabbit anti-CHIP antibody (dilution 1:250; Abcam Co., USA)
overnight at 4°C. The sections were then washed with 0.01 M PBS and
incubated with biotinylated goat anti-rabbit IgG (SP9000, Zhongshan
Goldenbridge Biotechnology Co., Ltd., Beijing, China). For each
sample, the omission of primary antibody was used as a negative
control. In addition, total protein was extracted only with a
tissue lysis buffer containing protease and phosphatase inhibitors
(50 mM Tris-base pH 7.4, 100 mM NaCl, 1% NP-40, 10 mM EDTA, 20 mM
NaF, 1 mM PMSF, 3 mM Na3VO4, protease
inhibitor mixture), the concentration of protein for each sample
was determined using the Enhanced BCA Protein Assay kit (Beyotime
Institute of Biotechnology, Shanghai, China). Protein samples (20
μg) were separated by 10% SDS-polyacrylamide gel electrophoresis
and then transferred to nitrocellulose membranes (transfer buffer:
25 mM Tris, 190 mM glycine, 20% methanol, 0.5% sodium dodecyl
sulfate). The membranes were washed in Tris-buffered saline (TBS)
(20 mM Tris-HCl, pH 7.6, 140 mM NaCl) and blocked with 5% bovine
serum albumin (BSA) in TBS containing 0.5% Tween-20 (TBS-T). The
membranes were incubated overnight at 4°C with the primary antibody
rabbit anti-CHIP (dilution 1:1000; Cell Signaling Technology, Inc.,
USA). Membranes were washed with TBS-T solution, incubated for 60
min with horseradish peroxidase (HRP)-conjugated mouse anti-rabbit
IgG (dilution 1:3000; Upstate Biotechnology, Lake Placid, NY),
washed with TBS-T, rinsed with double deionized water and immersed
in enhanced chemiluminescence (ECL)-detecting substrate
(SuperSignalWest Pico; Pierce Chemical Co., Rockford, IL, USA).
Images were captured with Micro Chemi (DNR Bio-Imaging Systems,
Israel), the pictures were scanned and the optical density of the
bands was determined using NIH ImageJ software (National Institutes
of Health, Bethesda, MD) and was standardized to GAPDH detected
using mouse anti-GAPDH monoclonal antibody (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA). Each case of gastric
cancer and the matched normal mucosa was repeated at least 3
times.
Statistical analysis
The non-parametric Mann-Whitney U-test was used to
analyze the mRNA expression levels of CHIP in the gastric cancerous
samples and the matched normal samples of human gastric cancers.
The significance of correlations between CHIP expression and
clinicopathological characteristics was analyzed by Student's
t-test and Pearson's χ2 test (Tables I and II). The continuous data were expressed as
mean ± SEM. All statistical analyses were two-sided and performed
by the SPSS 13.0 software package (SPSS Inc., Chicago, IL, USA).
The level of statistical significance was set at P<0.05.
 | Table IAssociation between the mRNA
expression of CHIP with histopathological features of
gastric cancer patients. |
Table I
Association between the mRNA
expression of CHIP with histopathological features of
gastric cancer patients.
| Total
(n=53)
No. | CHIP mRNA
levels | P-value |
|---|
|
|---|
| Not decreaseda (n=16) | Decreaseda (n=37) |
|---|
|
|
|---|
| No. | (%) | No. | (%) |
|---|
| Gender |
| Male | 32 | 8 | 25 | 24 | 75 | 0.310 |
| Female | 21 | 8 | 38 | 13 | 62 | |
| Age (years) |
| ≤55 | 26 | 10 | 38 | 16 | 62 | 0.059 |
| >55 | 27 | 6 | 22 | 21 | 78 | |
| TNM stage |
| T1+T2 | 11 | 6 | 55 | 5 | 45 | 0.048 |
| T3+T4 | 42 | 10 | 24 | 32 | 76 | |
| Lymph node
metastasis |
| Negative | 7 | 5 | 71 | 2 | 29 | 0.010 |
| Positive | 46 | 11 | 24 | 35 | 76 | |
|
Differentiation |
| Poor | 45 | 12 | 27 | 33 | 73 | 0.185 |
| Well and
moderated | 8 | 4 | 50 | 4 | 50 | |
 | Table IIAssociation between the protein
levels of CHIP with clinicopathological data in gastric cancer
patients. |
Table II
Association between the protein
levels of CHIP with clinicopathological data in gastric cancer
patients.
| Total
(n=53)
No. | CHIP
expression | P-value |
|---|
|
|---|
| Not
decreaseda (n=24) |
Decreaseda (n=29) |
|---|
|
|
|---|
| No. | (%) | No. | (%) |
|---|
| Gender |
| Male | 32 | 15 | 47 | 17 | 53 | 0.774 |
| Female | 21 | 9 | 43 | 12 | 57 | |
| Age (years) |
| ≤55 | 26 | 15 | 58 | 11 | 42 | 0.075 |
| >55 | 27 | 9 | 33 | 18 | 67 | |
| TNM stages |
| T1+T2 | 11 | 7 | 64 | 4 | 36 | 0.170 |
| T3+T4 | 42 | 17 | 40 | 25 | 60 | |
| Lymph node
metastasis |
| Negative | 7 | 6 | 86 | 1 | 14 | 0.021 |
| Positive | 46 | 18 | 39 | 28 | 61 | |
|
Differentiation |
| Poor | 45 | 17 | 38 | 28 | 62 | 0.009 |
| Well and
moderated | 8 | 7 | 87.5 | 1 | 12.5 | |
Results
Fifty-three patients suffered from gastric cancer
were involved in this research. The gastric cancerous tissues and
the matched normal non-cancerous tissues from each patient were
detected to determine the expression of CHIP at both mRNA and
protein levels. The clinicopathological characteristics including
gender, age, TNM stage, lymph node metastasis and tumor
differentiation of each patient were evaluated in this study.
CHIP mRNA levels were decreased in
gastric cancer and the relationship with histopathologic
features
The matched normal mucosa and cancerous tissue
samples which were normalized to GAPDH levels were detected
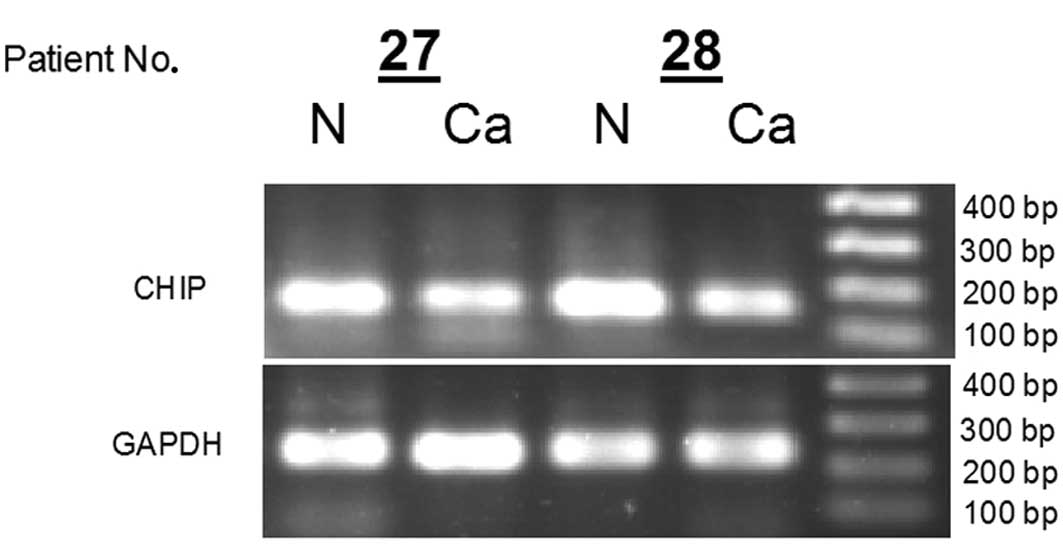
from each GC patients (n=2) by PCR (Fig. 1). The results showed mRNA levels of
CHIP in cancerous tissues were decreased compared with
normal tissues. Furthermore, the expression of CHIP mRNA was
detected in 53 gastric cancer samples and the corresponding normal
samples by real-time PCR analysis. The relative mRNA expression of
CHIP in the gastric cancer samples was significantly lower
than that in the corresponding normal samples (3.44±1.33 vs.
11.40±2.87, 2−ΔCt, P=0.022, paired t-test). As shown in
Table I, the downregulation of CHIP
expression occurred in 70% (37 of 53) of gastric cancer patients.
Furthermore, the clinical significance of decreased CHIP expression
correlated with the clinicopathological data was also explored.
There were remarkable differences in CHIP mRNA expression in pT1/T2
stage tumors vs. pT3/T4 stage tumors (P=0.048, χ2 test),
and lymph node non-invasive tumors vs. lymph node invasive tumors
(P=0.01, χ2 test) (Table
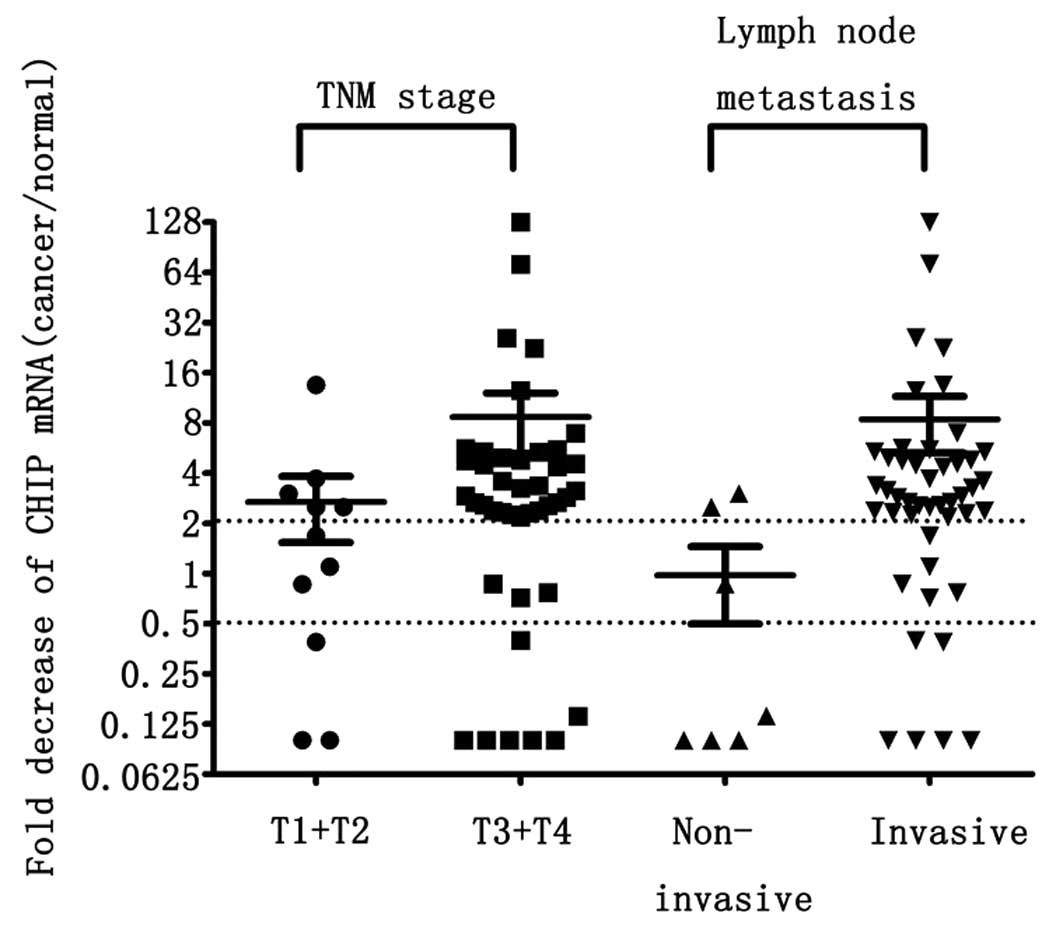
I). CHIP mRNA was reduced 2.68±1.14-fold in 11 pT1/T2 stage
tumors and 8.67±3.42-fold in 42 pT3/T4 stage tumors (P=0.048,
Z=−1.974, Mann-Whitney U-test), respectively. In addition, CHIP
mRNA was decreased 0.97±0.48-fold in 7 lymph node non-invasive
cancers and 8.41±3.13-fold in 46 lymph node invasive cancers
(P=0.008, Z=−2.67, Mann-Whitney U-test) (Fig. 2).
Protein expression of CHIP was
downregulated in gastric cancers and the correlation with
clinicopathological para-meters
In this study, the protein levels of CHIP were also
examined using western blot analysis. The presence of CHIP in the
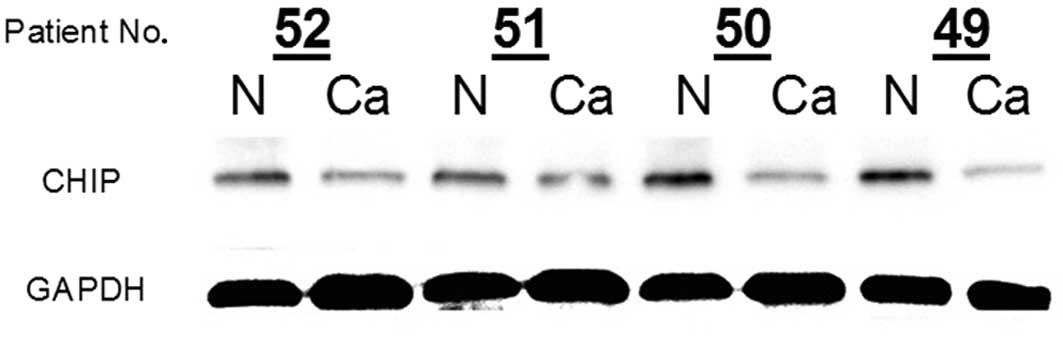
normal gastric mucosa was confirmed (Fig. 3). However, CHIP protein expression
was notably reduced in cancerous samples compared with the matched
normal mucosa in 4 cases of gastric cancer (Fig. 3). An immunohistochemical assay was
used to estimate the endosomatic status of CHIP expression in
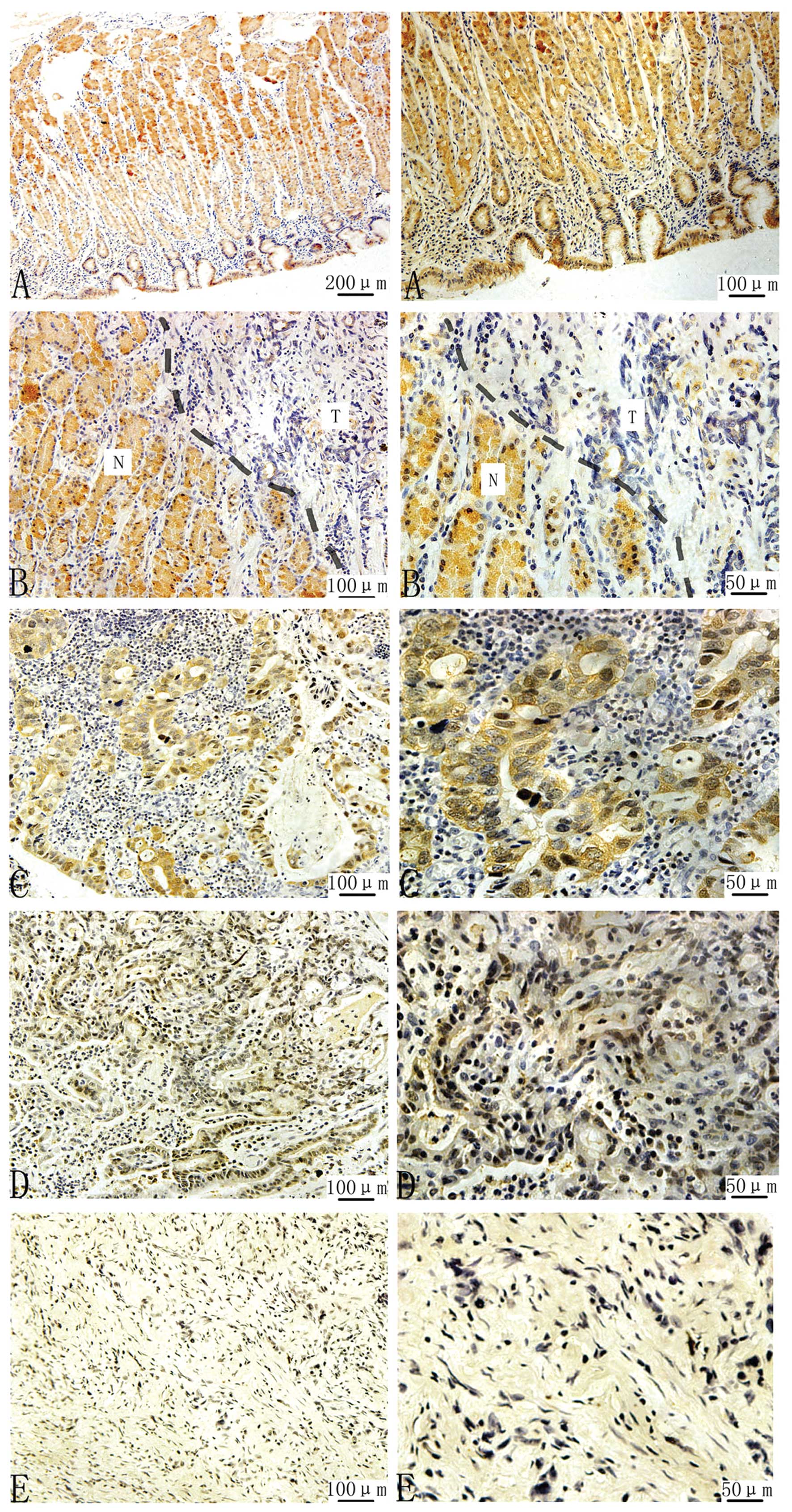
normal stomach. High levels of CHIP expression occurred in
non-cancerous gastric epithelial cells but not in adjacent stromal
or inflammatory cells (Fig. 4A).
The immunohistochemical staining was not observed in the
superficial gastric foveolar cells, but was remarkable in the
epithelium from the neck region to deeper glands (Fig. 4A). Therefore, CHIP was shown to
align from the basal to middle portions of the gastric mucosa. CHIP
staining was diffuse throughout the cytoplasm of the gastric
epithelial cells (Fig. 4A). In
Fig. 4B, immunohistochemical
staining was remarkably decreased at the protein level of CHIP
expression in cancerous tissue compared with normal tissue.
However, in the cancerous sample, CHIP expression was significantly
reduced in well-/moderated-/poor-differentiated gastric cancer
(Fig. 4C-E), and with the
differentiation turning poor, the staining was gradually less
strong. In addition, CHIP downregulation was found in 55% (29 of
53) of gastric cancer patients (Table
II). We further assessed the correlations between downregulated
CHIP expression and clinicopathological features (Table II). Statistical data showed that
downregulated CHIP expression was associated with lymph node
metastasis and tumor differentiation. CHIP expression was decreased
in 14% (1 of 7) of lymph node non-invasive gastric cancer and in
61% (28 of 46) of lymph node invasive gastric cancer (P=0.021,
χ2 test) (Table II).
CHIP expression was decreased in 12.5% (1 of 8) of
well-/moderated-differentiated gastric cancer and in 62% (28 of 45)
of poor-differentiated gastric cancer (P=0.009, χ2 test)
(Table II).
Discussion
Increasing amounts of evidence strongly suggest that
E3 ubiquitin ligases are involved in cancer proliferation and
tumorigenesis. Furthermore, E3 ubiquitin ligases, such as murine
double minute 2 (MDM2), S-phase-kinase-associated protein
(Skp)-Cullin-F-Box (SCF), inhibitor of apoptosis protein have
emerged as prognostic biomarkers and potential cancer drug targets
(24). As a member of the E3
ubiquitin ligases, CHIP has been demonstrated to be involved in
tumorigenesis, proliferation and invasion in several malignancies
(21). CHIP is an E3 ubiquitin
ligase that induces the ubiquitination and proteasomal degradation
of its substrates. CHIP interacts with Hsp/Hsc70 and Hsp90 through
its TPR domain and negatively regulates chaperone functions. The
U-box domain at the carboxyl terminus of CHIP contains its E3
ubiquitin ligase activity, and was able to promote ubiquitylation
and degradation of many tumor-related proteins, such as ErbB2 in
breast cancer and ovarian cancer (25,26).
ErbB2 overexpression contributes to the evolution of a substantial
group of human cancers and signifies a poor clinical prognosis
(25). Previous study suggests that
ErbB2 is a target of CHIP and wild-type CHIP induces ErbB2
ubiquitination and downregulation in vivo (25). CHIP overexpression results in
decreased levels of endogenous ERα in ERα-positive breast cancer
MCF7 cells (27). In addition, CHIP
interacted with Met receptor leading to proteasomal degradation of
the receptor in vitro and CHIP overexpression inhibited
Met-mediated lung cancer cell growth and invasion (23). Other tumor-related proteins such as
p53 (28,29), FOXO1 (30,31)
and hypoxia-inducible factor (HIF)-1-α (32) can also be regulated by CHIP.
Because CHIP can regulate these tumor-related
proteins through ubiquitylation and degradation, it might play an
important role in cancers. Kajiro et al showed that CHIP
suppresses tumor progression by inhibiting oncogenic pathways in
human breast cancer. Knockdown of CHIP (shCHIP) significantly
enhanced the metastatic potential of the cancer cells due to
increased expression of Bcl2, Akt1, Smad and Twist. These
observations demonstrated that CHIP inhibits anchorage-independent
cell growth and metastatic potential by degrading oncogenic
proteins including SRC-3 (21).
Interestingly, the roles of CHIP in gliomas were totally opposite
to those in breast cancer. Xu et al showed that CHIP
expressed stronger in high-grade gliomas than in low-grade gliomas.
Glioma cells proliferation and colony formation were enhanced due
to overexpression of CHIP, while knockdown of CHIP suppressed
proliferation and colony formation. Notably, CHIP RNAi lentivirus
significantly delayed tumor growth. In contrast, overexpression of
CHIP resulted in enhanced tumor growth in a nude mouse xenograft
model. This study demonstrated that CHIP contributes to oncogenesis
of glioma (22). These results
indicate that CHIP might play different roles in different human
cancers. However, the role of CHIP in the progression of gastric
cancer has not been investigated.
In the current study, we presented some primary data
that CHIP was frequently downregulated in gastric cancer using
RT-PCR, real-time PCR, western blot and immunohistochemical assays.
We showed CHIP was expressed in the neck and deeper glands of
gastric mucosa in normal tissues. However, CHIP expression was
significantly decreased in the cancerous tissues. Notably, it was
almost disappeared in some highly lymph node invasive gastric
cancer patients. Meanwhile, the well-differentiated and
moderate-differentiated samples showed higher expression of CHIP
than the poorly-differentiated gastric cancer samples. Therefore,
it seems that a negative correlation exists between CHIP expression
and tumor malignancy in human gastric cancer.
Invasion and metastasis of tumor cells are major
causes of mortality in cancer patients. In the present study, we
found that CHIP expression was almost absent in the advanced
gastric cancer, such as lymph node invasive gastric cancer and
poorly-differentiated gastric cancer. Therefore, CHIP may play a
significant role in the progression of gastric cancer.
Thus, further investigation on the molecular
mechanism between CHIP expression and lymph node metastasis would
provide some useful insight into the understanding of
carcinogenesis of gastric cancer. However, our study only reported
the primary data on the relationships between CHIP downregulation
and clinically aggressive phenotype of gastric cancer. We showed
that the decreased CHIP expression was associated with lymph node
metastasis, TNM stage and tumor differentiation. Such information
indicates that CHIP may be a potential diagnostic biomarker and
therapeutic target for gastric cancer. However, our study only
investigated the correlations between CHIP and clinicopathological
characteristics of gastric cancer, and a further prospective
analysis to elucidate the molecular mechanism of the downregulated
CHIP in gastric cancer could be informative.
Acknowledgements
The authors express their sincere appreciation to
the Department of Sugery of Tongji Hospital, Tongji Medical
College, for supplying resection samples.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vogiatzi P, Vindigni C, Roviello F,
Renieri A and Giordano A: Deciphering the underlying genetic and
epigenetic events leading to gastric carcinogenesis. J Cell
Physiol. 211:287–295. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sasui W, Yokozaki H, Fujimoto J, Naka K,
Kuniyasu H and Tahara E: Genetic and epigenetic alterations in
multistep carcinogenesis of the stomach. J Gastroenterol.
35:111–115. 2000.PubMed/NCBI
|
|
5
|
Lee JH, Han SU, Cho H, et al: A novel germ
line juxtamembrane Met mutation in human gastric cancer. Oncogene.
19:4947–4953. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Allgayer H, Babic R, Gruetzner KU,
Tarabichi A, Schildberg FW and Heiss MM: c-erbB-2 is of independent
prognostic relevance in gastric cancer and is associated with the
expression of tumor-associated protease systems. J Clin Oncol.
18:2201–2209. 2000.PubMed/NCBI
|
|
7
|
Drebber U, Baldus SE, Nolden B, et al: The
overexpression of c-met as a prognostic indicator for gastric
carcinoma compared to p53 and p21 nuclear accumulation. Oncol Rep.
19:1477–1483. 2008.PubMed/NCBI
|
|
8
|
Shiao YH, Rugge M, Correa P, Lehmann HP
and Scheer WD: p53 alteration in gastric precancerous lesions. Am J
Pathol. 144:511–517. 1994.PubMed/NCBI
|
|
9
|
Ebert MP, Yu J, Hoffmann J, et al: Loss of
beta-catenin expression in metastatic gastric cancer. J Clin Oncol.
21:1708–1714. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wen YG, Wang Q, Zhou CZ, Qiu GQ, Peng ZH
and Tang HM: Mutation analysis of tumor suppressor gene PTEN in
patients with gastric carcinomas and its impact on PI3K/AKT
pathway. Oncol Rep. 24:89–95. 2010.PubMed/NCBI
|
|
11
|
Sakata M, Kitamura YH, Sakuraba K, et al:
Methylation of HACE1 in gastric carcinoma. Anticancer Res.
29:2231–2233. 2009.PubMed/NCBI
|
|
12
|
Wu F, Shirahata A, Sakuraba K, et al:
Down-regulation of EGFL8: a novel biomarker for advanced gastric
cancer. Anticancer Res. 31:3377–3380. 2011.PubMed/NCBI
|
|
13
|
Paoletti X, Oba K, Burzykowski T, et al:
Benefit of adjuvant chemotherapy for resectable gastric cancer: a
meta-analysis. JAMA. 303:1729–1737. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Crew KD and Neugut AI: Epidemiology of
gastric cancer. World J Gastroenterol. 12:354–362. 2006.
|
|
15
|
Anderson WF, Camargo MC, Fraumeni JF Jr,
Correa P, Rosenberg PS and Rabkin CS: Age-specific trends in
incidence of non-cardia gastric cancer in US adults. JAMA.
303:1723–1728. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ballinger CA, Connell P, Wu Y, Hu Z,
Thompson LJ, Yin LY and Patterson C: Identification of CHIP, a
novel tetratricopeptide repeat-containing protein that interacts
with heat shock proteins and negatively regulates chaperone
functions. Mol Cell Biol. 19:4535–4545. 1999.
|
|
17
|
Lamb JR, Tugendreich S and Hieter P:
Tetratricopeptide repeat interactions: to TPR or not to TPR? Trends
Biochem Sci. 20:257–259. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Murata S, Minami Y, Minami M, Chiba T and
Tanaka K: CHIP is a chaperone-dependent E3 ligase that
ubiquitylates unfolded protein. EMBO Rep. 2:1133–1138. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Connell P, Ballinger CA, Jiang J, Wu Y,
Thompson LJ, Hohfeld J and Patterson C: The co-chaperone CHIP
regulates protein triage decisions mediated by heat-shock proteins.
Nat Cell Biol. 3:93–96. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meacham GC, Patterson C, Zhang W, Younger
JM and Cyr DM: The Hsc70 co-chaperone CHIP targets immature CFTR
for proteasomal degradation. Nat Cell Biol. 3:100–105. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kajiro M, Hirota R, Nakajima Y, et al: The
ubiquitin ligase CHIP acts as an upstream regulator of oncogenic
pathways. Nat Cell Biol. 11:312–319. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu T, Zhou Q, Zhou J, et al: Carboxyl
terminus of Hsp70-interacting protein (CHIP) contributes to human
glioma oncogenesis. Cancer Sci. 102:959–966. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jang KW, Lee JE, Kim SY, et al: The
C-terminus of Hsp70-interacting protein promotes Met receptor
degradation. J Thorac Oncol. 6:679–687. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lakshmanan M, Bughani U, Duraisamy S,
Diwan M, Dastidar S and Ray A: Molecular targeting of E3 ligases -
a therapeutic approach for cancer. Expert Opin Ther Targets.
12:855–870. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou P, Fernandes N, Dodge IL, et al:
ErbB2 degradation mediated by the co-chaperone protein CHIP. J Biol
Chem. 278:13829–13837. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
McDonough H and Patterson C: CHIP: a link
between the chaperone and proteasome systems. Cell Stress
Chaperones. 8:303–308. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fan M, Park A and Nephew KP: CHIP
(carboxyl terminus of Hsc70-interacting protein) promotes basal and
geldanamycin-induced degradation of estrogen receptor-α. Mol
Endocrinol. 19:2901–2914. 2005.PubMed/NCBI
|
|
28
|
Esser C, Scheffner M and Hohfeld J: The
chaperone-associated ubiquitin ligase CHIP is able to target p53
for proteasomal degradation. J Biol Chem. 280:27443–27448. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
McDonough H, Charles PC, Hilliard EG, et
al: Stress-dependent Daxx-CHIP interaction suppresses the p53
apoptotic program. J Biol Chem. 284:20649–20659. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang H and Tindall DJ: Dynamic FoxO
transcription factors. J Cell Sci. 120:2479–2487. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li F, Xie P, Fan Y, et al: C terminus of
Hsc70-interacting protein promotes smooth muscle cell proliferation
and survival through ubiquitin-mediated degradation of FoxO1. J
Biol Chem. 284:20090–20098. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Luo W, Zhong J, Chang R, Hu H, Pandey A
and Semenza G: Hsp70 and CHIP selectively mediate ubiquitination
and degradation of hypoxia-inducible factor (HIF)-1α but not
HIF-2α. J Biol Chem. 285:3651–3663. 2010.PubMed/NCBI
|