1. Introduction
Liver cancer is one of the most common cancers
worldwide and is a main cause of cancer-related death. There are
many risk factors related to hepatocellular carcinoma (HCC), such
as hepatitis B virus (HBV) infection, hepatitis C virus (HCV)
infection, alcohol abuse, obesity-related fatty liver disease,
aflatoxin and various carcinogens (1–5).
Effective treatments for localized HCC include partial liver
resection, liver transplantation and local ablation, such as
radiofrequency ablation (RFA), interstitial laser coagulation,
percutaneous ethanol injection (PEI) and percutaneous acetic acid
injection (PAI) (6–8). These treatments result in a cure for
cancer only for early stage tumors. Systemic therapy is the
conventional treatment for advanced HCC, but the outcomes are not
satisfactory. Therefore, the mechanisms involved in the formation
and progression of HCC require further investigation to discover
more effective therapies for liver cancer.
Currently, the theory of a ‘cancer stem cell’ may
partially explain the process of HCC formation. According to the
theory, there is a rare population of stem-like cells in tumor
tissue, called liver cancer stem cells (LCSCs), which are
responsible for the self-renewal, malignant transformation,
metastasis and chemoresistance of HCC. Dysregulation of signaling
pathways, including the transforming growth factor β (TGF-β), Wnt,
Notch and Hedgehog pathways, has been found to be involved in the
process of hepatocarcinogenesis. In order to isolate LCSCs from
tumor tissue, biomarkers need to be defined. At present, several
markers that identify LCSCs have been reported. These include
CD133, epithelial cell adhesion molecule (EpCAM), ABCG2 and
CD90.
2. Cancer stem cells
Currently, all of the cancer cells in a tumor are
thought to be responsible for tumor growth. However, recently
emerging evidence suggests that there is a rare population of
stem-like cells in tumors that determine cancer characteristics.
Reya et al(9) proposed a
theory of cancer stem cells (CSCs). They stated that ‘tumors may
often originate from the transformation of normal stem cells,
similar signaling pathways may regulate self-renewal in stem cells
and cancer cells, and cancer cells may include ‘cancer stem cells’
- rare cells with indefinite potential for self-renewal that drive
tumorigenesis’.
Bonnet and Dick (10) first reported the existence of CSCs
in acute myeloid leukemia (AML), and CSCs have been subsequently
found in some solid tumors. Al-Hajj et al(11) first successfully isolated CSCs from
breast tumors. Many studies also demonstrated the presence of CSCs
in prostate (12,13), lung (14,15),
colon (16,17), pancreatic (18,19)
and brain tumors (20,21).
At present, the mechanisms responsible for the
formation and features of HCC are not clear, but the CSC theory
suggests that LCSCs may be responsible for HCC. Sun et
al(22) analyzed different
expression patterns of stem-cell markers in HBV-associated
cirrhotic livers and in HCC and demonstrated that the stem-like
cells possessed tumorigenic capacity and that these cells might be
LCSCs.
3. Liver stem/progenitor cells and liver
cancer stem cells
Liver progenitor cells, a type of bipotential cell
in human liver tissue, give rise to both hepatocytes and the
biliary tree. There are two main potential sources of liver stem
cells: adult liver stem/progenitor cells and extrahepatic stem
cells. The adult stem cells reside in the mature liver and can be
activated by certain factors. The oval cells, located in the canal
of Hering, have the ability to differentiate into both hepatocytes
and biliary epithelia and are now generally acknowledged to be
liver stem/progenitor cells (23).
In addition, liver stem cells may also be derived from other
organs, such as bone marrow (24,25).
Increasing evidence shows that bone marrow stem cells participate
in liver regeneration (26,27) and that Thy1-positive bone marrow
stem cells might be the source of these liver stem cells (28).
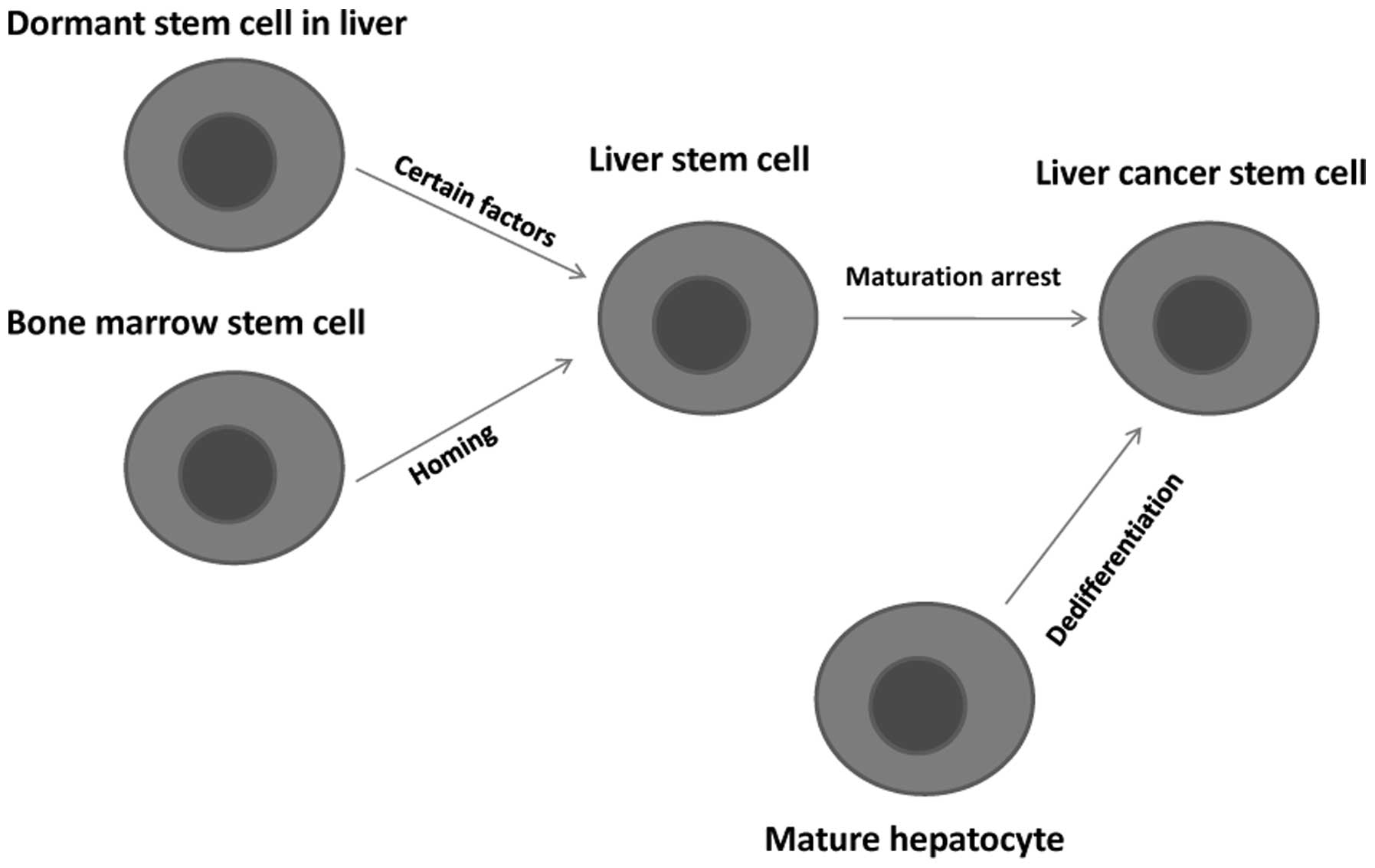
The CSC theory suggests that LCSCs exist, but the
origin of LCSCs is unclear. There are two main hypotheses to
explain the origin of LCSCs: the dedifferentiation of mature
hepatocytes and the maturation arrest of liver stem cells. Early
studies in rat models mainly focused on premalignant foci and
nodules, and the results supported the dedifferentiation hypothesis
(29,30).
However, this hypothesis has been challenged by
subsequent research. At present, it is commonly believed that liver
stem/progenitor cells are the potential source of HCC, intrahepatic
cholangiocarcinoma (ICC), combined hepatocellular
cholangiocarcinoma (CHC) and cholangiolocellular carcinoma (CLC), a
subtype of cholangiocellular carcinoma (CC) (31–36).
To study the effect of oval cells upon tumorigenesis, de Lima et
al(37) established a rat model
of non-alcoholic steatohepatitis (NASH), cirrhosis and HCC, showing
that oval cells could proliferate in this model and that these
cells may be the origin of malignancy. In another model (the
Solt-Farber carcinogenic model), hepatic progenitor cells,
identified by the expression of glypican-3 (GPC3), were shown to
play an important role in hepatic carcinogenesis (38). A summary of the origin of LCSCs is
presented in Fig. 1.
4. Signaling pathways and liver cancer stem
cells
Dysregulation of signaling pathways has been
observed in the process of hepatocarcinogenesis, and the TGF-β,
Wnt, Notch and Hedgehog signaling pathways have been extensively
studied. The signaling pathways involved in LCSCs are presented in
Fig. 2.
 | Figure 2The LCSC signaling pathways. The
TGF-β signaling pathway, the Wnt signaling pathway, the Notch
signaling pathway and the Hedgehog signaling pathway.
Abbreviations: Dhh, desert hedgehog; Hh, hedgehog; Ihh, Indian
hedgehog; NICD, Notch intracellular domain; Ptch, patched homolog;
Shh, sonic the hedgehog homolog; Smo, Smoothened; TGF-β,
transforming growth factor β. |
TGF-β signaling pathway
The TGF-β signaling pathway plays a crucial role in
cell cycle regulation, the immune system and apoptosis. In HCC,
TGF-β signaling inhibits oncogenesis at an early stage by inducing
apoptosis (39). This physiological
phenomenon is involved in TGF-β-induced TRAIL expression and in the
ability of Smad 3 to repress Bcl-2 transcription and p53-dependent
apoptosis, which is mediated by the TGF-β signaling pathway
(40–42). In addition, a recent study
demonstrated that TGF-β activates autophagy in certain HCCs to
suppress tumor formation (43), and
emerging evidence also suggests that dysregulation of TGF-β
signaling is associated with hepatocarcinogenesis (44,45).
In HCC cells, higher TGF-β1, Smad 7 and NF-κB expression and lower
Tβ-RII, Tβ-RIII and Smad 4 expression have been observed (46,47).
Mechanism of escape from TGF-β growth
inhibition in HCC cells
Recent studies have mainly focused on the HCC cell
mechanism of escape from TGF-β growth inhibition since TGF-β
promotes apoptosis in HCC cells and also activates survival
signals, such as AKT (39). The AKT
pathway is also involved in IL-4-transduced signaling pathways,
which are able to protect HCC cells from TGF-β-induced apoptosis
(48). Smad 3 plays a dual role in
carcinogenesis, as it promotes apoptosis and is essential for
TGF-β-mediated immune suppression (49). Smad 7, another member of the TGF-β
signaling pathway, confers resistance to the antiproliferative
effects of TGF-β on HCC cells by inhibiting the formation of the
TGF-β-induced functional Smad-DNA complex (50). Loss of ELF, an embryonic liver
fodrin belonging to the type II β-spectrin adaptor proteins, is
considered an early event in hepatocarcinogenesis and stimulation
of angiogenesis in HCC tissue (51,52).
TGF-β-induced apoptosis requires the participation of NADPH
oxidase, NOX4, and thus, impairing NOX4 upregulation inhibits
TGF-β-induced cell death in HCC (53). Further exploration of the mechanism
showed that NOX4 upregulation was impaired by the overactivation of
the MEK/ERK pathway (54). Disabled
p53, p21Cip1, or Rb genes may also be involved in the escape from
TGF-β growth inhibition in HCC cells (55).
TGF-β signaling pathway and LCSCs
The cooperation between TGF-β and oncogenic RAS
activates the nuclear β-catenin signaling pathway, which causes
neoplastic hepatocyte dedifferentiation to immature progenitor
cells and facilitates HCC recurrence (56). This evidence not only supports the
dedifferentiation theory for the source of LCSCs, but also shows
the relationship between TGF-β signaling and LCSCs. Activation of
IL-6/STAT3, a main signaling pathway in liver stem cells, can
induce malignant transformation in liver stem cells along with
inactivation of the TGF-β signaling pathway (57,58).
In addition, downregulation of Socs1 induced activation of STAT3,
and this process plays a crucial role in malignant transformation
(59).
Wnt signaling pathway
The Wnt signaling pathway plays an important role in
embryogenesis and tumor development. It consists of a large number
of proteins that interact with each other to regulate the signaling
pathway. β-catenin is a key component in the pathway and is
inhibited by a protein complex that includes GSK-3, axin and APC
(60,61). Binding of the Wnt proteins to the
Frizzled receptors activates the Dishevelled (DSH) protein family
(62). Subsequently, DSH inhibits
the axin/GSK-3/APC complex, and β-catenin is able to enter the
nucleus to interact with the TCF/LEF family of transcription
factors to promote expression of specific genes, such as cyclin D1,
Myc and TCF-1 (63–65).
Activation of the canonical Wnt signaling pathway
drives tumor formation in liver stem cells (66,67).
Recent studies have shown that the expression of β-catenin was
higher in HCC than in non-tumor tissues (68), and inhibition of Wnt-1 signaling
caused antitumor effects (69). In
addition, the noncanonical Wnt signaling pathway plays an important
role in HCC. Yuzugullu et al(70) reported that noncanonical Wnt5a
represses noncanonical Wnt signaling. This study suggests that the
Wnt pathway is selectively activated or repressed depending on the
differentiation stages of HCC cells. Wnt signaling is activated in
well-differentiated HCC cells and is repressed in poorly
differentiated cell lines. In a subsequent study, Wnt11, a member
of the noncanonical cascade, was also able to inhibit HCC cell
proliferation and migration (71).
Polycomb-group gene products play a pivotal role in
HCC formation and maintenance by modulating the Wnt pathway.
Polycomb proteins can form two major complexes: polycomb repressive
complex 1 and 2 (PRC1 and PRC2). BMI1, a subunit of PRC1, and EZH2,
a subunit of PRC2, are expressed in large quantities within HCC
tissues and enable in vitro HCC cell growth (72). In addition, a definite link between
high levels of BMI1 or EZH2 expression and the maintenance of
tumor-initiating cells in HCC has been observed (73,74).
Furthermore, the expression of EZH2 activates the Wnt/β-catenin
signaling pathway by silencing the Wnt antagonists, thereby
inducing HCC cell proliferation (75).
The Wnt signaling pathway also plays a crucial role
in promoting liver growth and regulating liver stem cells (76,77).
Yamashita et al(77)
employed a novel prognostic HCC subtype cell line to identify the
relationship between Wnt signaling and EpCAM, a hepatic stem cell
marker. They concluded that EpCAM was a target gene of Wnt
signaling, and that EpCAM(+) HCC cells have the ability to both
self-renew and differentiate, which suggests that EpCAM(+) HCC
cells may be LCSCs (78).
Notch signaling pathway
The Notch signaling pathway involves multiple cell
differentiation processes during embryonic development and
throughout adulthood. It has also been demonstrated that Notch
signaling plays an important role in many types of human cancers,
including T-cell leukemia, lymphoma, medulloblastoma and
colorectal, pancreatic, mammary, ovarian, lung, gastric, cervical
and breast carcinoma (79–82). The involvement of Notch in cancer
development is complex, since Notch can function as an oncogene or
a tumor suppressor depending on the tissue.
Notch signaling was first highlighted in human
T-cell leukemia. Dysregulated Notch signaling can promote
tumorigenesis, and direct Notch inhibition has been found to have
antiproliferative effects on T-cell acute lymphoblastic leukemia
(T-ALL) (83,84). Activated Notch signaling has been
observed in a wide variety of breast carcinomas (85,86).
High Notch1 protein expression is an early event in breast cancer
development and is associated with the HER-2 molecular subtype
(87); there is also a general
increase in the Notch1, Notch2, Notch4, Jagged1, Jagged2 and
Delta-like 4 protein expression in breast carcinoma (88). Emerging evidence suggests that the
Notch signaling pathway may be a potential therapeutic target in
breast carcinoma (89,90).
Low expression levels of Notch1/Jagged1 were
frequently observed, and downregulation of Notch1/Jagged1 signaling
may sustain tumor progression in HCC (68). Upregulation of Notch1 was also shown
to retard hepatocarcinogenesis by arresting the cell cycle and
inducing apoptosis (91). In
addition, high Notch3 and low Notch4 expression levels may be
associated with HCC (92).
In some solid tumors, dysregulation of the Notch
signaling pathway is correlated with tumor initiation (93–95).
These findings suggest that aberrant Notch expression may influence
CSC regulation and induce tumorigenesis (96).
Hedgehog signaling pathway
The Hedgehog signaling pathway plays a key role in
embryonic development and carcinogenesis. The main members of the
Hedgehog signaling pathway include the polypeptide ligands Hh (Shh,
Ihh, Dhh), cell-surface transmembrane receptors (PTCH and SMO) and
a downstream transcription factor (Gli). A large number of
experiments demonstrate that Hedgehog signaling activation is
involved in HCC oncogenesis, proliferation and invasiveness
(97,98). Blocking the Hedgehog signaling
pathway could inhibit HCC formation by restraining proliferation,
inducing apoptosis and repressing c-Myc and cyclin D expression
(99).
Members of the Hedgehog signaling pathway perform
differently in hepatocarcinogenesis. PTCH (PTCH1), the Hedgehog
signaling receptor, is associated with the early stage of HCC
formation (100). Smo is
considered a prognostic factor for HCC formation and it plays a
critical role in hepatocarcinogenesis by mediating c-myc
overexpression (101). The basal
expression of Gli2, which is regulated by p53, Notch and TGF-β
signaling, could prime the Hedgehog signaling pathway and lead to
HCC tumor formation (102).
Activation of the Hedgehog signaling pathway may influence the Wnt
signaling pathway by regulating the transcription of a secreted
frizzled-related protein (sFRP-1), which has the ability to
suppress Wnt signaling (103). In
addition, knockdown of Rab23, an essential negative regulator of
the Hedgehog signaling pathway, is reported to suppress HCC cell
growth (104).
It has been suggested that Hedgehog signaling
pathway activation might be related to LCSCs. In normal liver
tissue, the expression of Hh is low and mature hepatocytes are not
Hh-responsive. Omenetti and Diehl (105) found that high levels of Hh were
expressed after liver injury and that this favored the survival of
Hh-responsive cells, such as myofibroblastic and progenitor cells.
During subsequent differentiation, the original Hh-responsive
population progeny proliferates and this may lead to hepatic
fibrosis and neoplasia. Therefore, the progenitor cells that
survived may initiate hepatocarcinogenesis. A recent report also
demonstrated that HBV/HCV infection induced high Hh ligand
expression levels and Hh-responsive cell proliferation, promoting
liver fibrosis and cancer (106).
In addition, Hedgehog signaling pathway activation may cause
malignant embryonal liver cell transformation in hepatoblastoma
(107). In summary, the Hedgehog
signaling pathway plays an important role in LCSC regulation.
5. Markers for liver cancer stem cells
In order to isolate LCSCs from HCC tissues, several
biomarkers have been identified, including CD133, EpCAM and ABCG2.
These biomarkers and others are discussed below.
CD133
CD133, which is expressed in hematopoietic and
neuronal stem cells, has long been considered an important CSC
marker in HCC. In normal liver tissues, CD133(+) cells are related
to liver regeneration and may also serve as self-renewing bipotent
primitive hepatic cells (108).
Further study showed that CD133(+)CD45(−) cells from chronic liver
disease represented a bipotent liver stem cell population at the
stage of primary carcinoma formation, which had CSC characteristics
(109). Emerging evidence suggests
that CD133 expression is a putative marker for LCSCs as follows: i)
a small population of CD133(+) cells was observed in HCC tissues
(110); ii) CD133(+) HCC cells had
a higher proliferative potential and a greater ability to form
colonies (111); iii) CD133(+) HCC
cells possess the characteristics of progenitor cells (111); iv) the high expression level of
‘stemness’ genes and the low expression level of the mature
hepatocyte markers, glutamine synthetase and cytochrome P450 3A4
were observed in CD133(+) HCC cells as compared with CD133(−) HCC
cells (111,112); v) after injection into SCID mice,
CD133(+) cells from HCC tissue formed tumors, while CD133(−) cells
did not (112); vi) CD133
expression may contribute to HCC survival (113); and vii) knockdown of CD133
expression reduces the ability to form colonies and alter the cell
cycle distribution in HCC (114).
In addition, increased CD133 expression may indicate a poor
prognosis and tumor recurrence in patients with HCC (115).
It has been demonstrated that co-expression of CD133
and other cell surface markers could define CSCs. CD133(+)ALDH(+)
cells represent the CSC population in HCC tissue and there is a
hierarchical organization in HCC bearing tumorigenic capacity in
the following order: CD133(+)ALDH(+) > CD133(+)ALDH(−) >
CD133(−)ALDH(−) (116). Higher
tumorigenic potential was also observed in CD133(+)CD44(+) HCC
cells compared to CD133(+)CD44(−). Therefore, the co-expression of
CD133 and CD44 could be considered markers for LCSCs (117).
The relationship between CD133 expression and
signaling pathways has been studied extensively. TGF-β1 induces
CD133 expression in HCC by inhibiting DNMT1 and DNMT3β expression
and these CD133(+) cells subsequently initiate tumor formation
(118). The Akt/PKB and Bcl-2
pathway is involved in CD133(+)-HCC cell chemoresistance and this
pathway could represent a new target for HCC therapy (119).
A differential analysis between the microRNA
expression profiles of CD133(+) and CD133(−) liver cancer cells
showed a higher miR-130b expression level in CD133(+) cells
(120). In addition, miR-130b
plays a critical role in maintaining the stem-like characteristics
of CD133(+) cancer cells by silencing tumor protein p53-inducible
nuclear protein 1 (TP53INP1) (120).
However, the migratory properties do not differ
between CD133(+) and CD133(−) HCC cells and the amount of CD133(+)
cells is not related to the HCC clinical status (121). Therefore, it is still uncertain
whether or not CD133 can serve as a marker for LCSCs.
EpCAM
EpCAM is expressed during early liver development,
but not in hepatocytes. EpCAM is also observed in hepatic stem
cells and most hepatoblasts (122). Accumulating evidence suggests that
EpCAM may be a potential biomarker for LCSCs, and is presented as
follows: i) high levels of known hepatic stem cell markers are
expressed in EpCAM(+) cells, whereas mature hepatocyte markers are
increased significantly in EpCAM(−) cells (78); ii) compared with EpCAM(−) cells,
EpCAM(+) cells showed a greater colony formation rate (123); iii) EpCAM(+) cells contain a
multipotent cell population, and they can differentiate into both
EpCAM(+) and EpCAM(−) cells (123); iv) after injection into NOD/SCID
mice, EpCAM(+) cells efficiently initiated tumors, while EpCAM(−)
cells could not (78); and v) in
the HuH7 cell line, EpCAM(+) cells are much more invasive than
EpCAM(−) cells (78). Taken
together, this information suggests that EpCAM(+) HCC cells
represent hepatic stem cells and that these cells may also serve as
LCSCs.
EpCAM expression is regulated by the
Wnt/β-catenin signaling pathway
Accumulation of β-catenin induces EpCAM expression
in normal liver tissue and in HCC tissue, while degradation of
β-catenin or inhibition of Tcf/β-catenin complex formation
suppresses EpCAM expression (77).
A novel regulatory relationship between miR-181 and EpCAM(+) HCC
cells has been observed; inhibition of miR-181 reduced the amount
of EpCAM(+) cells and their ability to initiate tumors (124). Therefore, miR-181 may serve as a
potential therapeutic target for HCC. In addition, EpCAM, the
target of β-catenin and miR-181, contributes to the regulation of
several reprogramming genes, including c-MYC, OCT-4, NANOG, SOX2
and KLF4, thereby playing a critical role in the maintenance of HCC
cell ‘stemness’ (125,126).
ABCG2
Goodell et al(127) first described a type of primitive
stem cell, the side population (SP), in the bone marrow; these
cells were distinguished by their ability to exclude Hoechst 33342
dye and they defined this characteristic as a side population
phenotype. Recently, SP cells have been considered to be CSCs in
many types of tumor tissues (128). In HCC, SP cells harbor CSC-like
properties, and they may be related to tumorigenesis, metastasis
and therapeutic resistance (129–131). In addition, a study of the HCC
cell cycle distribution showed that G0 cells were
present in the SP fraction and that they may play a crucial role in
tumor pathogenesis (132,133).
ABCG2, an ATP binding cassette (ABC)
half-transporter that is highly expressed on the SP plasma
membrane, efficiently extrudes a wide variety of compounds such as
anticancer agents across cell membranes, and is considered to be
the determinant of the SP phenotype. Recent evidence suggests that
ABCG2 serves as a CSC biomarker in many types of tumors, such as
lung cancer, pancreatic cancer and retinoblastoma (133–136). In addition, Zen et
al(137) compared ABCG2(+)
with ABCG2(−) subpopulations from HCC tissues. The results showed
that other progenitor cell markers, such as CK19 and AFP, were
mainly located in ABCG2(+) subpopulations and that ABCG2(+) cells
may play an important role in hepatocarcinogenesis through their
ability to generate both ABCG2(+) and ABCG2(−) progenies. Our
previous study also supported the potential for ABCG2 to be a LCSC
marker (138). Further study
explored the mechanism of ABCG2 expression in HCC, demonstrating
that the Akt signaling pathways regulated the SP phenotype activity
by altering the subcellular localization of ABCG2 and by
suppressing Akt signaling that could help overcome ABCG2-induced
chemotherapy resistance (128,139).
Other putative markers
CD90, also named Thy-1, is a conserved cell surface
protein that can be used as a marker for a variety of stem cells.
In precancerous liver tissues, CD90 expression is observed in
proliferating bile ductules and its co-expression with CD34
represents hepatic stem cells (140). Yang et al(141) compared CD90(+) cells with CD90(−)
cells from HCC cell lines and demonstrated that CD45(−)CD90(+)
cells were detected in all of the tumor specimens and in 90% of the
blood samples from HCC patients. These researchers also
demonstrated that CD90 expression increased during tumor formation
and that CD90(+) cells formed tumor nodules in immunodeficient
mice; CD90(+) cells generated tumor nodules after serial
transplantation in a second and then in a third group of
immunodeficient mice. Therefore, CD90 may be considered to be a
marker for LCSCs.
Co-expression of CD90 and CD13 was found to play an
important role in hepatocarcinogenesis, and combination of a CD13
inhibitor and a CD90 inhibitor drastically reduced tumor volume
compared with either agent alone. In addition, CD13(+) cells
demonstrated CSC characteristics such as proliferation, formation
of cellular clusters in cancer foci and the ability to survive
during treatment (142). Given
these results, CD13 is a potential marker for LCSCs.
Cytokeratin 19 (CK19), a member of the keratin
family, is a stemness-related marker. CK19 is expressed in normal
human liver bile duct cells and is also observed scattered in the
parenchyma of cirrhotic livers and within HCCs (143,144). Compared with CK19(−) cells,
CK19(+) early lesions and advanced HCCs contain genetic changes
consistent with remodeling toward a differentiated phenotype, and
they are an important predictive factor for prognosis, patient
survival and tumor recurrence (145). The expression of
epithelial-mesenchymal transition (EMT)-related proteins, which
play a pivotal role in the tumor-cell invasion process, is
increased in CK19(+) HCC cells; therefore, CK19(+) HCC cells
demonstrate high invasive ability (146).
OV6, a hepatic progenitor cell marker, has recently
been regarded as a putative marker for LCSCs. OV6(+) HCC cells
demonstrate greater chemoresistance and a greater ability to form
tumors in vivo compared to OV6(−) cells (147). Activation of the Wnt pathway tends
to give rise to an increase in the proportion of OV6(+) cells, and
inhibition of β-catenin signaling suppresses OV6 expression within
HCC cells (147); therefore, the
OV6 expression is regulated by the Wnt pathway.
In addition, the expression of other markers, such
as CD44, DLK1, Oct4, Nanog, c-kit and Ezmin, may also be related to
LCSCs (144,148–152). However, the exact pattern of LCSC
marker expression is still unknown. Jabari et al(153) demonstrated that different HCC cell
lines express different stem cell markers. The classical
cholangiocellular type (Huh-7, Huh-7 pcDNA3.1, Hep3B) expressed
CK7/19, β-catenin and CD34; a dedifferentiated
mesenchymal-proliferative type (Huh-7 5–15) was characterized by
CK19, vimentin and Ki-67; a dedifferentiated embryonic-development
type (Hep3B implanted in Matrigel) expressed CK19, β-catenin and
PTC; and a classical HCC type (HepG2) expressed CK18/19 and
β-catenin. In addition, EpCAM(+) cells have a greater capacity to
initiate tumors than do CD133(+) cells in the Huh1 cell line, while
EpCAM(+) and CD133(+) cells showed similar tumorigenic ability in
the Huh7 cell line (78).
Therefore, determination of LCSC markers requires further research.
A summary of putative LCSC markers is provided in Table I.
 | Table IPutative markers of LCSCs. |
Table I
Putative markers of LCSCs.
| Surface
markers | Percentages of
cells expressing markersa | Minimum no. of
cells for tumor formation | Injection site of
mice | Strain | Latency | Ref. |
|---|
|
CD133(+)ALDH(+) | 0.94–55.71% | 500 | s.c. | SCID | 82 days | (116) |
| CD133(+) | 0.1–2.0% | 100 | i.p. | BNX | 10 weeks | (110) |
| CD133(+) | 0.10–93.18% | 100 | s.c. | NOD/SCID | 3 months | (117) |
|
CD133(+)CD44(+) | 0.09–1.88% | 100 | s.c. | NOD/SCID | 2 months | (117) |
| EpCAM(+) | 0.7–99.6% | 100 | s.c. | NOG | 6–7 weeks | (123) |
| ABCG2 (SP
cells) | 0.25–0.80% | 1000 | s.c. | NOD/SCID | 16 weeks | (129) |
| CD90(+) | 0.04–2.34% | 500 | s.c. | SCID/Beige | 3 months | (141) |
| CD90(+)CD44(+) | 0.02–2.53% | 500 | s.c. | SCID/Beige | 3 months | (141) |
6. Discussion
Although the exact mechanism that controls
hepatocarcinogenesis remains unclear, the ‘cancer stem cell’ theory
has been proposed as a potential explanation. LCSCs, a rare
population in HCC cells with stem-like characteristics, are thought
to be responsible for oncogenic cell transformation.
Dysregulation of signaling pathways such as TGF-β,
Wnt, Notch and Hedgehog plays a crucial role in HCC formation and
LCSC maintenance. Recent research has shown that additional factors
also contribute to this progression, especially microRNAs. In
addition to miR-130b and miR-181 mentioned above, other microRNAs,
which participate in cancer cell ‘stemness’, have also been
identified. LIN28, a miRNA-binding protein, is expressed without
restriction in embryonic stem cells and various human cancer cells.
LIN28 was found to be one of the reprogramming factors, which are
able to reprogram somatic cells to pluripotent stem cells (154). Under physiological conditions, the
miRNAs let-7, mir-125, mir-9 and mir-30 negatively regulate LIN28
expression and the downregulation of these miRNAs may lead to LIN28
overexpression in tumors (155).
In addition, high expression levels of LIN28 can promote tumor
formation and malignant transformation by repressing the let-7
family miRNA expression (156).
Let-7 is sufficient to negatively regulate LIN28 via a feedback
loop (157). In tumor tissue,
LIN28 upregulation is tightly linked to a high proportion of
ALDH(+) cells, which are regarded as CSC representatives, and LIN28
plays a crucial role in the maintenance of ALDH(+) cancer cells.
Changes in the LIN28/let-7 regulatory loop induce the
‘reprogramming-like’ process in tumors, which may explain the
formation of CSCs (157).
Numerous markers have been used to identify LCSCs,
and these markers may be putative therapeutic targets in HCC.
Immunotherapy may also be an effective way to treat HCC by
targeting these biomarkers. CD133(+) HCC cells have long been
regarded as potential LCSCs and an anti-CD133 antibody conjugated
to a cytotoxic drug is reported to inhibit HCC cell proliferation
in vitro(158); therefore,
CD133(+) cancer cells may be considered a novel target for HCC
therapy. An anti-CD44 antibody-mediated liposomal nanoparticle,
containing a triple fusion gene (herpes simplex virus truncated
thymidine kinase, renilla luciferase and red fluorescent protein),
can be used in gene therapy and in the molecular imaging of HCC
(159). CD13(+) HCC cells are able
to resist regular ROS-inducing chemoradiation therapy as CD13
protects HCC cells from ROS-induced DNA damage. Therefore, a
combination of a CD13 inhibitor and ROS-inducing chemoradiation
therapy may enhance treatment effectiveness (142). A large amount of circulating CSCs,
represented by CD45(+)CD90(+)CD44(+) HCC cells, is tightly linked
to an increased possibility of HCC recurrence after hepatectomy,
which means these CSCs may be a potential target for the prevention
of HCC recurrence (160).
Acknowledgements
This study was supported by the Jiangsu Science
Foundation of China (no. LW201008) and the Nanjing Science
Foundation of China (no. ZKX08025).
References
|
1
|
Yu MC and Yuan JM: Environmental factors
and risk for hepatocellular carcinoma. Gastroenterology. 127(Suppl
1): S72–S78. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bosch FX, Ribes J, Cleries R and Diaz M:
Epidemiology of hepatocellular carcinoma. Clin Liver Dis.
9:191–211. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gomaa AI, Khan SA, Toledano MB, Waked I
and Taylor-Robinson SD: Hepatocellular carcinoma: epidemiology,
risk factors and pathogenesis. World J Gastroenterol. 14:4300–4308.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shariff MI, Cox IJ, Gomaa AI, Khan SA,
Gedroyc W and Taylor-Robinson SD: Hepatocellular carcinoma: current
trends in worldwide epidemiology, risk factors, diagnosis and
therapeutics. Expert Rev Gastroenterol Hepatol. 3:353–367. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kumar M, Kumar R, Hissar SS, et al: Risk
factors analysis for hepatocellular carcinoma in patients with and
without cirrhosis: a case-control study of 213 hepatocellular
carcinoma patients from India. J Gastroenterol Hepatol.
22:1104–1111. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Carr BI: Hepatocellular carcinoma: current
management and future trends. Gastroenterology. 127(Suppl 1):
S218–S224. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kassahun WT, Fangmann J, Harms J, Hauss J
and Bartels M: Liver resection and transplantation in the
management of hepatocellular carcinoma: a review. Exp Clin
Transplant. 4:549–558. 2006.PubMed/NCBI
|
|
8
|
Witjes CD, Verhoef C, Verheul HM and
Eskens FA: Systemic treatment in hepatocellular carcinoma; ‘A small
step for man’. Neth J Med. 67:86–90. 2009.
|
|
9
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bonnet D and Dick JE: Human acute myeloid
leukemia is organized as a hierarchy that originates from a
primitive hematopoietic cell. Nat Med. 3:730–737. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Collins AT, Berry PA, Hyde C, Stower MJ
and Maitland NJ: Prospective identification of tumorigenic prostate
cancer stem cells. Cancer Res. 65:10946–10951. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Klarmann GJ, Hurt EM, Mathews LA, et al:
Invasive prostate cancer cells are tumor initiating cells that have
a stem cell-like genomic signature. Clin Exp Metastasis.
26:433–446. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim CF, Jackson EL, Woolfenden AE, et al:
Identification of bronchioalveolar stem cells in normal lung and
lung cancer. Cell. 121:823–835. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eramo A, Lotti F, Sette G, et al:
Identification and expansion of the tumorigenic lung cancer stem
cell population. Cell Death Differ. 15:504–514. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
O’Brien CA, Pollett A, Gallinger S and
Dick JE: A human colon cancer cell capable of initiating tumour
growth in immunodeficient mice. Nature. 445:106–110.
2007.PubMed/NCBI
|
|
17
|
Chu P, Clanton DJ, Snipas TS, et al:
Characterization of a subpopulation of colon cancer cells with stem
cell-like properties. Int J Cancer. 124:1312–1321. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li C, Heidt DG, Dalerba P, et al:
Identification of pancreatic cancer stem cells. Cancer Res.
67:1030–1037. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li C, Lee CJ and Simeone DM:
Identification of human pancreatic cancer stem cells. Methods Mol
Biol. 568:161–173. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Singh SK, Clarke ID, Terasaki M, et al:
Identification of a cancer stem cell in human brain tumors. Cancer
Res. 63:5821–5828. 2003.PubMed/NCBI
|
|
21
|
Rahman R, Heath R and Grundy R: Cellular
immortality in brain tumours: an integration of the cancer stem
cell paradigm. Biochim Biophys Acta. 1792:280–288. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun YL, Yin SY, Xie HY, et al: Stem-like
cells in hepatitis B virus-associated cirrhotic livers and adjacent
tissue to hepatocellular carcinomas possess the capacity of
tumorigenicity. J Gastroenterol Hepatol. 23:1280–1286. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Roskams TA, Theise ND, Balabaud C, et al:
Nomenclature of the finer branches of the biliary tree: canals,
ductules, and ductular reactions in human livers. Hepatology.
39:1739–1745. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Petersen BE, Bowen WC, Patrene KD, et al:
Bone marrow as a potential source of hepatic oval cells. Science.
284:1168–1170. 1999.PubMed/NCBI
|
|
25
|
Shi XL, Gu JY, Han B, Xu HY, Fang L and
Ding YT: Magnetically labeled mesenchymal stem cells after
autologous transplantation into acutely injured liver. World J
Gastroenterol. 16:3674–3679. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Theise ND, Nimmakayalu M, Gardner R, et
al: Liver from bone marrow in humans. Hepatology. 32:11–16. 2000.
View Article : Google Scholar
|
|
27
|
Rowe PM: Chronic Lyme disease: the debate
goes on. Lancet. 355:14362000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bae SH, Choi JY, Yoon SK, et al:
Thy1-positive bone marrow stem cells express liver-specific genes
in vitro and can mature into hepatocytes in vivo. Hepatol Int.
2:63–71. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gournay J, Auvigne I, Pichard V, Ligeza C,
Bralet MP and Ferry N: In vivo cell lineage analysis during
chemical hepatocarcinogenesis in rats using retroviral-mediated
gene transfer: evidence for dedifferentiation of mature
hepatocytes. Lab Invest. 82:781–788. 2002. View Article : Google Scholar
|
|
30
|
Bralet MP, Pichard V and Ferry N:
Demonstration of direct lineage between hepatocytes and
hepatocellular carcinoma in diethylnitrosamine-treated rats.
Hepatology. 36:623–630. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dumble ML, Croager EJ, Yeoh GC and Quail
EA: Generation and characterization of p53 null transformed hepatic
progenitor cells: oval cells give rise to hepatocellular carcinoma.
Carcinogenesis. 23:435–445. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fujii T, Zen Y, Harada K, et al:
Participation of liver cancer stem/progenitor cells in
tumorigenesis of scirrhous hepatocellular carcinoma - human and
cell culture study. Hum Pathol. 39:1185–1196. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nomoto K, Tsuneyama K, Cheng C, et al:
Intrahepatic cholangiocarcinoma arising in cirrhotic liver
frequently expressed p63-positive basal/stem-cell phenotype. Pathol
Res Pract. 202:71–76. 2006. View Article : Google Scholar
|
|
34
|
Tanaka S, Yamamoto T, Tanaka H, et al:
Potentiality of combined hepatocellular and intrahepatic
cholangiocellular carcinoma originating from a hepatic precursor
cell: immunohistochemical evidence. Hepatol Res. 32:52–57. 2005.
View Article : Google Scholar
|
|
35
|
Zhang F, Chen XP, Zhang W, et al: Combined
hepatocellular cholangiocarcinoma originating from hepatic
progenitor cells: immunohistochemical and double-fluorescence
immunostaining evidence. Histopathology. 52:224–232. 2008.
View Article : Google Scholar
|
|
36
|
Komuta M, Spee B, Vander Borght S, et al:
Clinicopathological study on cholangiolocellular carcinoma
suggesting hepatic progenitor cell origin. Hepatology.
47:1544–1556. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
de Lima VM, Oliveira CP, Alves VA, et al:
A rodent model of NASH with cirrhosis, oval cell proliferation and
hepatocellular carcinoma. J Hepatol. 49:1055–1061. 2008.PubMed/NCBI
|
|
38
|
Grozdanov PN, Yovchev MI and Dabeva MD:
The oncofetal protein glypican-3 is a novel marker of hepatic
progenitor/oval cells. Lab Invest. 86:1272–1284. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Caja L, Ortiz C, Bertran E, et al:
Differential intracellular signalling induced by TGF-beta in rat
adult hepatocytes and hepatoma cells: implications in liver
carcinogenesis. Cell Signal. 19:683–694. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Herzer K, Grosse-Wilde A, Krammer PH,
Galle PR and Kanzler S: Transforming growth factor-beta-mediated
tumor necrosis factor-related apoptosis-inducing ligand expression
and apoptosis in hepatoma cells requires functional cooperation
between Smad proteins and activator protein-1. Mol Cancer Res.
6:1169–1177. 2008. View Article : Google Scholar
|
|
41
|
Wang CL, Wan YL, Liu YC and Huang ZQ:
TGF-beta1/SMAD signaling pathway mediates p53-dependent apoptosis
in hepatoma cell lines. Chin Med Sci J. 21:33–35. 2006.PubMed/NCBI
|
|
42
|
Yang YA, Zhang GM, Feigenbaum L and Zhang
YE: Smad3 reduces susceptibility to hepatocarcinoma by sensitizing
hepatocytes to apoptosis through downregulation of Bcl-2. Cancer
Cell. 9:445–457. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kiyono K, Suzuki HI, Matsuyama H, et al:
Autophagy is activated by TGF-beta and potentiates
TGF-beta-mediated growth inhibition in human hepatocellular
carcinoma cells. Cancer Res. 69:8844–8852. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mazzocca A, Fransvea E, Lavezzari G,
Antonaci S and Giannelli G: Inhibition of transforming growth
factor beta receptor I kinase blocks hepatocellular carcinoma
growth through neo-angiogenesis regulation. Hepatology.
50:1140–1151. 2009. View Article : Google Scholar
|
|
45
|
Mikula M, Proell V, Fischer AN and
Mikulits W: Activated hepatic stellate cells induce tumor
progression of neoplastic hepatocytes in a TGF-beta dependent
fashion. J Cell Physiol. 209:560–567. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bae HJ, Eun JW, Noh JH, et al:
Down-regulation of transforming growth factor beta receptor type
III in hepatocellular carcinoma is not directly associated with
genetic alterations or loss of heterozygosity. Oncol Rep.
22:475–480. 2009.PubMed/NCBI
|
|
47
|
Ji GZ, Wang XH, Miao L, et al: Role of
transforming growth factor-beta1-smad signal transduction pathway
in patients with hepatocellular carcinoma. World J Gastroenterol.
12:644–648. 2006.PubMed/NCBI
|
|
48
|
Lin SJ, Chang C, Ng AK, Wang SH, Li JJ and
Hu CP: Prevention of TGF-beta-induced apoptosis by interleukin-4
through Akt activation and p70S6K survival signaling pathways.
Apoptosis. 12:1659–1670. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Millet C and Zhang YE: Roles of Smad3 in
TGF-beta signaling during carcinogenesis. Crit Rev Eukaryot Gene
Expr. 17:281–293. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang S, Fei T, Zhang L, et al: Smad7
antagonizes transforming growth factor beta signaling in the
nucleus by interfering with functional Smad-DNA complex formation.
Mol Cell Biol. 27:4488–4499. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kitisin K, Ganesan N, Tang Y, et al:
Disruption of transforming growth factor-beta signaling through
beta-spectrin ELF leads to hepatocellular cancer through cyclin D1
activation. Oncogene. 26:7103–7110. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Baek HJ, Lim SC, Kitisin K, et al:
Hepatocellular cancer arises from loss of transforming growth
factor beta signaling adaptor protein embryonic liver fodrin
through abnormal angiogenesis. Hepatology. 48:1128–1137. 2008.
View Article : Google Scholar
|
|
53
|
Carmona-Cuenca I, Roncero C, Sancho P, et
al: Upregulation of the NADPH oxidase NOX4 by TGF-beta in
hepatocytes is required for its pro-apoptotic activity. J Hepatol.
49:965–976. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Caja L, Sancho P, Bertran E,
Iglesias-Serret D, Gil J and Fabregat I: Overactivation of the
MEK/ERK pathway in liver tumor cells confers resistance to
TGF-{beta}-induced cell death through impairing up-regulation of
the NADPH oxidase NOX4. Cancer Res. 69:7595–7602. 2009.PubMed/NCBI
|
|
55
|
Sheahan S, Bellamy CO, Dunbar DR, Harrison
DJ and Prost S: Deficiency of G1 regulators P53, P21Cip1 and/or pRb
decreases hepatocyte sensitivity to TGFbeta cell cycle arrest. BMC
Cancer. 7:2152007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zulehner G, Mikula M, Schneller D, et al:
Nuclear beta-catenin induces an early liver progenitor phenotype in
hepatocellular carcinoma and promotes tumor recurrence. Am J
Pathol. 176:472–481. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tang Y, Kitisin K, Jogunoori W, et al:
Progenitor/stem cells give rise to liver cancer due to aberrant
TGF-beta and IL-6 signaling. Proc Natl Acad Sci USA. 105:2445–2450.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lin L, Amin R, Gallicano GI, et al: The
STAT3 inhibitor NSC 74859 is effective in hepatocellular cancers
with disrupted TGF-beta signaling. Oncogene. 28:961–972. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Bagnyukova TV, Tryndyak VP, Muskhelishvili
L, Ross SA, Beland FA and Pogribny IP: Epigenetic downregulation of
the suppressor of cytokine signaling 1 (Socs1) gene is associated
with the STAT3 activation and development of hepatocellular
carcinoma induced by methyl-deficiency in rats. Cell Cycle.
7:3202–3210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Dajani R, Fraser E, Roe SM, et al:
Structural basis for recruitment of glycogen synthase kinase 3beta
to the axin-APC scaffold complex. EMBO J. 22:494–501. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ha NC, Tonozuka T, Stamos JL, Choi HJ and
Weis WI: Mechanism of phosphorylation-dependent binding of APC to
beta-catenin and its role in beta-catenin degradation. Mol Cell.
15:511–521. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Tauriello DV, Jordens I, Kirchner K, et
al: Wnt/β-catenin signaling requires interaction of the Dishevelled
DEP domain and C terminus with a discontinuous motif in Frizzled.
Proc Natl Acad Sci USA. 109:E812–E820. 2012.
|
|
63
|
Tetsu O and McCormick F: Beta-catenin
regulates expression of cyclin D1 in colon carcinoma cells. Nature.
398:422–426. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yochum GS, Sherrick CM, Macpartlin M and
Goodman RH: A beta-catenin/TCF-coordinated chromatin loop at MYC
integrates 5′ and 3′ Wnt responsive enhancers. Proc Natl Acad Sci
USA. 107:145–150. 2010.PubMed/NCBI
|
|
65
|
Staal FJ, Meeldijk J, Moerer P, et al: Wnt
signaling is required for thymocyte development and activates Tcf-1
mediated transcription. Eur J Immunol. 31:285–293. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Taniguchi H and Chiba T: Stem cells and
cancer in the liver. Dis Markers. 24:223–229. 2008. View Article : Google Scholar
|
|
67
|
Takigawa Y and Brown AM: Wnt signaling in
liver cancer. Curr Drug Targets. 9:1013–1024. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wang M, Xue L, Cao Q, et al: Expression of
Notch1, Jagged1 and beta-catenin and their clinicopathological
significance in hepatocellular carcinoma. Neoplasma. 56:533–541.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wei W, Chua MS, Grepper S and So SK:
Blockade of Wnt-1 signaling leads to anti-tumor effects in
hepatocellular carcinoma cells. Mol Cancer. 8:762009. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yuzugullu H, Benhaj K, Ozturk N, et al:
Canonical Wnt signaling is antagonized by noncanonical Wnt5a in
hepatocellular carcinoma cells. Mol Cancer. 8:902009. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Toyama T, Lee HC, Koga H, Wands JR and Kim
M: Noncanonical Wnt11 inhibits hepatocellular carcinoma cell
proliferation and migration. Mol Cancer Res. 8:254–265. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yonemitsu Y, Imazeki F, Chiba T, et al:
Distinct expression of polycomb group proteins EZH2 and BMI1 in
hepatocellular carcinoma. Hum Pathol. 40:1304–1311. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Chiba T, Miyagi S, Saraya A, et al: The
polycomb gene product BMI1 contributes to the maintenance of
tumor-initiating side population cells in hepatocellular carcinoma.
Cancer Res. 68:7742–7749. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chiba T, Suzuki E, Negishi M, et al:
3-Deazaneplanocin A is a promising therapeutic agent for the
eradication of tumor-initiating hepatocellular carcinoma cells. Int
J Cancer. 130:2557–2567. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Cheng AS, Lau SS, Chen Y, et al:
EZH2-mediated concordant repression of Wnt antagonists promotes
β-catenin-dependent hepatocarcinogenesis. Cancer Res. 71:4028–4039.
2011.PubMed/NCBI
|
|
76
|
Zaret KS: Genetic programming of liver and
pancreas progenitors: lessons for stem-cell differentiation. Nat
Rev Genet. 9:329–340. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yamashita T, Budhu A, Forgues M and Wang
XW: Activation of hepatic stem cell marker EpCAM by
Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res.
67:10831–10839. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yamashita T, Ji J, Budhu A, et al:
EpCAM-positive hepatocellular carcinoma cells are tumor-initiating
cells with stem/progenitor cell features. Gastroenterology.
136:1012–1024. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Yin L, Velazquez OC and Liu ZJ: Notch
signaling: emerging molecular targets for cancer therapy. Biochem
Pharmacol. 80:690–701. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Sharma VM, Draheim KM and Kelliher MA: The
Notch1/c-Myc pathway in T cell leukemia. Cell Cycle. 6:927–930.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Moserle L, Ghisi M, Amadori A and
Indraccolo S: Side population and cancer stem cells: therapeutic
implications. Cancer Lett. 288:1–9. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wu WK, Cho CH, Lee CW, et al:
Dysregulation of cellular signaling in gastric cancer. Cancer Lett.
295:144–153. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Moellering RE, Cornejo M, Davis TN, et al:
Direct inhibition of the NOTCH transcription factor complex.
Nature. 462:182–188. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Arora PS and Ansari AZ: Chemical biology:
A Notch above other inhibitors. Nature. 462:171–173. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Farnie G and Clarke RB: Mammary stem cells
and breast cancer - role of Notch signalling. Stem Cell Rev.
3:169–175. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Stylianou S, Clarke RB and Brennan K:
Aberrant activation of notch signaling in human breast cancer.
Cancer Res. 66:1517–1525. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zardawi SJ, Zardawi I, McNeil CM, et al:
High Notch1 protein expression is an early event in breast cancer
development and is associated with the HER-2 molecular subtype.
Histopathology. 56:286–296. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Mittal S, Subramanyam D, Dey D, Kumar RV
and Rangarajan A: Cooperation of Notch and Ras/MAPK signaling
pathways in human breast carcinogenesis. Mol Cancer. 8:1282009.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Hirose H, Ishii H, Mimori K, et al: Notch
pathway as candidate therapeutic target in Her2/Neu/ErbB2
receptor-negative breast tumors. Oncol Rep. 23:35–43.
2010.PubMed/NCBI
|
|
90
|
Korkaya H and Wicha MS: HER-2, notch, and
breast cancer stem cells: targeting an axis of evil. Clin Cancer
Res. 15:1845–1847. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Wang C, Qi R, Li N, et al: Notch1
signaling sensitizes tumor necrosis factor-related
apoptosis-inducing ligand-induced apoptosis in human hepatocellular
carcinoma cells by inhibiting Akt/Hdm2-mediated p53 degradation and
up-regulating p53-dependent DR5 expression. J Biol Chem.
284:16183–16190. 2009. View Article : Google Scholar
|
|
92
|
Gramantieri L, Giovannini C, Lanzi A, et
al: Aberrant Notch3 and Notch4 expression in human hepatocellular
carcinoma. Liver Int. 27:997–1007. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Sikandar SS, Pate KT, Anderson S, et al:
NOTCH signaling is required for formation and self-renewal of
tumor-initiating cells and for repression of secretory cell
differentiation in colon cancer. Cancer Res. 70:1469–1478. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Harrison H, Farnie G, Howell SJ, et al:
Regulation of breast cancer stem cell activity by signaling through
the Notch4 receptor. Cancer Res. 70:709–718. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhen Y, Zhao S, Li Q, Li Y and Kawamoto K:
Arsenic trioxide-mediated Notch pathway inhibition depletes the
cancer stem-like cell population in gliomas. Cancer Lett.
292:64–72. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Hambardzumyan D, Becher OJ and Holland EC:
Cancer stem cells and survival pathways. Cell Cycle. 7:1371–1378.
2008. View Article : Google Scholar
|
|
97
|
Huang S, He J, Zhang X, et al: Activation
of the hedgehog pathway in human hepatocellular carcinomas.
Carcinogenesis. 27:1334–1340. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Cheng WT, Xu K, Tian DY, Zhang ZG, Liu LJ
and Chen Y: Role of Hedgehog signaling pathway in proliferation and
invasiveness of hepatocellular carcinoma cells. Int J Oncol.
34:829–836. 2009.PubMed/NCBI
|
|
99
|
Patil MA, Zhang J, Ho C, Cheung ST, Fan ST
and Chen X: Hedgehog signaling in human hepatocellular carcinoma.
Cancer Biol Ther. 5:111–117. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Fu X, Wang Q, Chen X, et al: Expression
patterns and polymorphisms of PTCH in Chinese hepatocellular
carcinoma patients. Exp Mol Pathol. 84:195–199. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Sicklick JK, Li YX, Jayaraman A, et al:
Dysregulation of the Hedgehog pathway in human
hepatocarcinogenesis. Carcinogenesis. 27:748–757. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Katoh Y and Katoh M: Integrative genomic
analyses on GLI2: mechanism of Hedgehog priming through basal GLI2
expression, and interaction map of stem cell signaling network with
P53. Int J Oncol. 33:881–886. 2008.PubMed/NCBI
|
|
103
|
He J, Sheng T, Stelter AA, et al:
Suppressing Wnt signaling by the hedgehog pathway through sFRP-1. J
Biol Chem. 281:35598–35602. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Liu YJ, Wang Q, Li W, et al: Rab23 is a
potential biological target for treating hepatocellular carcinoma.
World J Gastroenterol. 13:1010–1017. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Omenetti A and Diehl AM: The adventures of
sonic hedgehog in development and repair. II Sonic hedgehog and
liver development, inflammation, and cancer. Am J Physiol
Gastrointest Liver Physiol. 294:G595–G598. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
de Pereira TA, Witek RP, Syn WK, et al:
Viral factors induce Hedgehog pathway activation in humans with
viral hepatitis, cirrhosis, and hepatocellular carcinoma. Lab
Invest. 90:1690–1703. 2010.PubMed/NCBI
|
|
107
|
Eichenmuller M, Gruner I, Hagl B, et al:
Blocking the hedgehog pathway inhibits hepatoblastoma growth.
Hepatology. 49:482–490. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Suzuki A, Sekiya S, Onishi M, et al: Flow
cytometric isolation and clonal identification of self-renewing
bipotent hepatic progenitor cells in adult mouse liver. Hepatology.
48:1964–1978. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Rountree CB, Ding W, He L and Stiles B:
Expansion of CD133-expressing liver cancer stem cells in
liver-specific phosphatase and tensin homolog deleted on chromosome
10-deleted mice. Stem Cells. 27:290–299. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Yin S, Li J, Hu C, et al: CD133 positive
hepatocellular carcinoma cells possess high capacity for
tumorigenicity. Int J Cancer. 120:1444–1450. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Ma S, Chan KW, Hu L, et al: Identification
and characterization of tumorigenic liver cancer stem/progenitor
cells. Gastroenterology. 132:2542–2556. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Suetsugu A, Nagaki M, Aoki H, Motohashi T,
Kunisada T and Moriwaki H: Characterization of CD133+
hepatocellular carcinoma cells as cancer stem/progenitor cells.
Biochem Biophys Res Commun. 351:820–824. 2006.
|
|
113
|
Kohga K, Tatsumi T, Takehara T, et al:
Expression of CD133 confers malignant potential by regulating
metalloproteinases in human hepatocellular carcinoma. J Hepatol.
52:872–879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Yao J, Zhang T, Ren J, Yu M and Wu G:
Effect of CD133/prominin-1 antisense oligodeoxynucleotide on in
vitro growth characteristics of Huh-7 human hepatocarcinoma
cells and U251 human glioma cells. Oncol Rep. 22:781–787.
2009.PubMed/NCBI
|
|
115
|
Song W, Li H, Tao K, et al: Expression and
clinical significance of the stem cell marker CD133 in
hepatocellular carcinoma. Int J Clin Pract. 62:1212–1218. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Ma S, Chan KW, Lee TK, et al: Aldehyde
dehydrogenase discriminates the CD133 liver cancer stem cell
populations. Mol Cancer Res. 6:1146–1153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Zhu Z, Hao X, Yan M, et al: Cancer
stem/progenitor cells are highly enriched in
CD133+CD44+ population in hepatocellular
carcinoma. Int J Cancer. 126:2067–2078. 2010.PubMed/NCBI
|
|
118
|
You H, Ding W and Rountree CB: Epigenetic
regulation of cancer stem cell marker CD133 by transforming growth
factor-beta. Hepatology. 51:1635–1644. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Ma S, Lee TK, Zheng BJ, Chan KW and Guan
XY: CD133+ HCC cancer stem cells confer chemoresistance
by preferential expression of the Akt/PKB survival pathway.
Oncogene. 27:1749–1758. 2008.
|
|
120
|
Ma S, Tang KH, Chan YP, et al: miR-130b
promotes CD133(+) liver tumor-initiating cell growth and
self-renewal via tumor protein 53-induced nuclear protein 1. Cell
Stem Cell. 7:694–707. 2010.PubMed/NCBI
|
|
121
|
Salnikov AV, Kusumawidjaja G, Rausch V, et
al: Cancer stem cell marker expression in hepatocellular carcinoma
and liver metastases is not sufficient as single prognostic
parameter. Cancer Lett. 275:185–193. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Schmelzer E and Reid LM: EpCAM expression
in normal, non-pathological tissues. Front Biosci. 13:3096–3100.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Kimura O, Takahashi T, Ishii N, et al:
Characterization of the epithelial cell adhesion molecule
(EpCAM)+ cell population in hepatocellular carcinoma
cell lines. Cancer Sci. 101:2145–2155. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Ji J, Yamashita T, Budhu A, et al:
Identification of microRNA-181 by genome-wide screening as a
critical player in EpCAM-positive hepatic cancer stem cells.
Hepatology. 50:472–480. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Arzumanyan A, Friedman T, Ng IO, Clayton
MM, Lian Z and Feitelson MA: Does the hepatitis B antigen HBx
promote the appearance of liver cancer stem cells? Cancer Res.
71:3701–3708. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Lu TY, Lu RM, Liao MY, et al: Epithelial
cell adhesion molecule regulation is associated with the
maintenance of the undifferentiated phenotype of human embryonic
stem cells. J Biol Chem. 285:8719–8732. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Goodell MA, Brose K, Paradis G, Conner AS
and Mulligan RC: Isolation and functional properties of murine
hematopoietic stem cells that are replicating in vivo. J Exp Med.
183:1797–1806. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Ding XW, Wu JH and Jiang CP: ABCG2: a
potential marker of stem cells and novel target in stem cell and
cancer therapy. Life Sci. 86:631–637. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Chiba T, Kita K, Zheng YW, et al: Side
population purified from hepatocellular carcinoma cells harbors
cancer stem cell-like properties. Hepatology. 44:240–251. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Shi GM, Xu Y, Fan J, et al: Identification
of side population cells in human hepatocellular carcinoma cell
lines with stepwise metastatic potentials. J Cancer Res Clin Oncol.
134:1155–1163. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Zhang N, Li R, Tao KS, et al:
Characterization of a stem-like population in hepatocellular
carcinoma MHCC97 cells. Oncol Rep. 23:827–831. 2010.PubMed/NCBI
|
|
132
|
Kamohara Y, Haraguchi N, Mimori K, et al:
The search for cancer stem cells in hepatocellular carcinoma.
Surgery. 144:119–124. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Qiang GH, Yu DC and Jiang CP: Side
population cells and liver cancer stem cells. World Chin J
Digestol. 18:971–974. 2010.(In Chinese).
|
|
134
|
Polgar O, Robey RW and Bates SE: ABCG2:
structure, function and role in drug response. Expert Opin Drug
Metab Toxicol. 4:1–15. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Sarkadi B, Ozvegy-Laczka C, Nemet K and
Varadi A: ABCG2 - a transporter for all seasons. FEBS Lett.
567:116–120. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Han B and Zhang JT: Multidrug resistance
in cancer chemotherapy and xenobiotic protection mediated by the
half ATP-binding cassette transporter ABCG2. Curr Med Chem
Anticancer Agents. 4:31–42. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Zen Y, Fujii T, Yoshikawa S, et al:
Histological and culture studies with respect to ABCG2 expression
support the existence of a cancer cell hierarchy in human
hepatocellular carcinoma. Am J Pathol. 170:1750–1762. 2007.
View Article : Google Scholar
|
|
138
|
Xi Z, Jiang CP and Ding YT: Expression of
stem cell marker ABCG2 and its significance in hepatocellular
carcinoma tissue and cell lines. World Chin J Digestol. 17:247–252.
2009.
|
|
139
|
Hu C, Li H, Li J, et al: Analysis of ABCG2
expression and side population identifies intrinsic drug efflux in
the HCC cell line MHCC-97L and its modulation by Akt signaling.
Carcinogenesis. 29:2289–2297. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Yang ZF, Ngai P, Ho DW, et al:
Identification of local and circulating cancer stem cells in human
liver cancer. Hepatology. 47:919–928. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Yang ZF, Ho DW, Ng MN, et al: Significance
of CD90+ cancer stem cells in human liver cancer. Cancer
Cell. 13:153–166. 2008.
|
|
142
|
Haraguchi N, Ishii H, Mimori K, et al:
CD13 is a therapeutic target in human liver cancer stem cells. J
Clin Invest. 120:3326–3339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Oliva J, French BA, Qing X and French SW:
The identification of stem cells in human liver diseases and
hepatocellular carcinoma. Exp Mol Pathol. 88:331–340. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Martinez-Chantar ML, Lu SC, Mato JM, et
al: The role of stem cells/progenitor cells in liver carcinogenesis
in glycine N-methyltransferase deficient mice. Exp Mol Pathol.
88:234–237. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Andersen JB, Loi R, Perra A, et al:
Progenitor-derived hepatocellular carcinoma model in the rat.
Hepatology. 51:1401–1409. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Kim H, Choi GH, Na DC, et al: Human
hepatocellular carcinomas with ‘stemness’-related marker
expression: keratin 19 expression and a poor prognosis. Hepatology.
54:1707–1717. 2011.
|
|
147
|
Yang W, Yan HX, Chen L, et al:
Wnt/beta-catenin signaling contributes to activation of normal and
tumorigenic liver progenitor cells. Cancer Res. 68:4287–4295. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Xie Z, Choong PF, Poon LF, et al:
Inhibition of CD44 expression in hepatocellular carcinoma cells
enhances apoptosis, chemosensitivity, and reduces tumorigenesis and
invasion. Cancer Chemother Pharmacol. 62:949–957. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Yu F, Hao X, Zhao H, et al: Delta-like 1
contributes to cell growth by increasing the interferon-inducible
protein 16 expression in hepatocellular carcinoma. Liver Int.
30:703–714. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Machida K, Tsukamoto H, Mkrtchyan H, et
al: Toll-like receptor 4 mediates synergism between alcohol and HCV
in hepatic oncogenesis involving stem cell marker Nanog. Proc Natl
Acad Sci USA. 106:1548–1553. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Knight B, Tirnitz-Parker JE and Olynyk JK:
C-kit inhibition by imatinib mesylate attenuates progenitor cell
expansion and inhibits liver tumor formation in mice.
Gastroenterology. 135:969–979. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Okamura D, Ohtsuka M, Kimura F, et al:
Ezrin expression is associated with hepatocellular carcinoma
possibly derived from progenitor cells and early recurrence after
surgical resection. Mod Pathol. 21:847–855. 2008. View Article : Google Scholar
|
|
153
|
Jabari S, Meissnitzer M, Quint K, et al:
Cellular plasticity of trans- and dedifferentiation markers in
human hepatoma cells in vitro and in vivo. Int J
Oncol. 35:69–80. 2009.PubMed/NCBI
|
|
154
|
Yu J, Vodyanik MA, Smuga-Otto K, et al:
Induced pluripotent stem cell lines derived from human somatic
cells. Science. 318:1917–1920. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Zhong X, Li N, Liang S, Huang Q, Coukos G
and Zhang L: Identification of microRNAs regulating reprogramming
factor LIN28 in embryonic stem cells and cancer cells. J Biol Chem.
285:41961–41971. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Viswanathan SR, Powers JT, Einhorn W, et
al: Lin28 promotes transformation and is associated with advanced
human malignancies. Nat Genet. 41:843–848. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Yang X, Lin X, Zhong X, et al:
Double-negative feedback loop between reprogramming factor LIN28
and microRNA let-7 regulates aldehyde dehydrogenase 1-positive
cancer stem cells. Cancer Res. 70:9463–9472. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Smith LM, Nesterova A, Ryan MC, et al:
CD133/prominin-1 is a potential therapeutic target for
antibody-drug conjugates in hepatocellular and gastric cancers. Br
J Cancer. 99:100–109. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Wang L, Su W, Liu Z, et al: CD44
antibody-targeted liposomal nanoparticles for molecular imaging and
therapy of hepatocellular carcinoma. Biomaterials. 33:5107–5114.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Fan ST, Yang ZF, Ho DW, Ng MN, Yu WC and
Wong J: Prediction of posthepatectomy recurrence of hepatocellular
carcinoma by circulating cancer stem cells: a prospective study.
Ann Surg. 254:569–576. 2011. View Article : Google Scholar : PubMed/NCBI
|