Introduction
MicroRNAs (miRNAs) are short single-stranded
nucleotide RNA molecules, which function as master regulators of
gene expression by post-transcriptional modifications of target
mRNAs (1). The pattern of
regulation of gene expression is sequence-specific. MiRNAs bind to
3′ untranslated regions (3′-UTRs) of mRNAs and then reduce the
translation and/or stability of that mRNA, leading to a reduction
in protein levels. Based on the unique feature of their targeting,
miRNAs could have many targets (2),
and, thus, control a large number of proteins.
MiRNAs may serve as either oncogenes or tumor
suppressors (3,4). In our study, the miR-145 is a
tumor-suppressive miRNA (5) and its
expression is low in different kinds of cancers. Mir-145 inhibits
proliferation by targeting c-myc (6) and MUC1 (7). In addition, miR-145 can also promote
differentiation and repress cell growth by repressing OCT4
(8).
ADAMs are best known as ectodomain sheddases, and
their domains function as metalloproteases. The ADAM family is a
Zn-dependent metalloproteinase (9,10).
ADAM17 is an important member of the ADAM family and involved in
proteolysis of collagen IV of the extracellular matrix and also the
release from the cell surface of several integrins, suggesting that
ADAM17 influences the invasive activity of different cells
including glioma cells (11).
ADAM17 is a primary upstream component for multiple EGFR
pro-ligands (12,13). EGFR binding with ligands
subsequently activates MEK/ERK and PI3K/Akt pathways, which
contribute to invasiveness and other malignant phenotypes (14).
In this study, the expression of miR-145 in glioma
cells compared with normal brain tissue was studied by real-time
RT-PCR. Proliferation, migration and invasion were examined to
confirm the effects of miR-145 in glioma cells. The suppression of
ADAM17 by miR-145 was confirmed by experiments using luciferase
analysis and western blotting. We found that ectopic expression of
miR-145 in U87 and U251n glioma cells caused decreased
proliferation, migration and invasion with accompanying low protein
expression of ADAM17 and EGFR. Our study provides direct evidence
that miR-145 functions as an anti-oncogene in glioma cells and may
be the target of therapies for glioma patients.
Materials and methods
Cells and miRNA transfection
U87-MG, U251n and T98G cells were obtained from the
American Type Culture Collection (Manassas, VA, USA). HF66 cells
were established by the Neurosurgical Department of Henry Ford
Hospital. Cells and were maintained in Dulbecco’s modified Eagle’s
medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Total
RNA from human frontal cortex brain tissue were obtained from
Agilent Technologies (Santa Clara, CA, USA). Transfection of the
miRIDIAN hsa-miR-145 miRNA mimic (Applied Biosystems), inactive
(scrambled) control cel-mir-67 (Thermo Scientific Dharmacon, IL,
USA), or pMIR-Report vectors was performed using Lipofectamine 2000
transfection reagent (Invitrogen, CA, USA) with 300 nmol of miRNA
or 1 μg/ml DNA plasmid, respectively.
Western blot analysis and real-time
RT-PCR
Western blot analysis was performed to detect ADAM17
(Abcam, MA, USA), EGFR, p-EGFR (Santa Cruz Biotechnology, Santa
Cruz, CA, USA), and β-actin (Santa Cruz). Protein expression was
measured 72 h after transfection. Protein concentration was
quantified using a BCA protein assay kit (Pierce, Rockford, IL,
USA). For quantitative analysis, densitometric measurements of
three blots per group were averaged. Densitometry was performed
with the MCID image analysis program (Cambridge, UK). The image
density (1/intensity) of each band was normalized to β-actin, and
experimental group band density was normalized to the band density
of control. RT-PCR for miRNAs was performed using a miR-145 TaqMan
MicroRNA assay. U6 miRNA was used as a house-keeping control. Each
sample was tested in triplicate and relative gene expression was
determined.
Invasion assay
Matrigel chambers (BD Biosciences) were used to
determine the effect of miR-145 on invasiveness according to the
manufacturer’s instructions. Cells (5×104), after being
transfected with miR-145 for 72 h, were re-suspended in 500 μl of
serum-free medium and added to the upper chamber while the lower
chamber was filled with 0.5 ml of complete medium that served as a
chemo-attractant. Cells were then incubated for 24 h at 37°C. After
removal of cells on the upper surface of the membrane, cells on the
lower surface of the membrane were stained with CellTracker™ Green
(Molecular Probes, Eugene, OR, USA) for 45 min and fixed in 4%
formaldehyde. Nine fields of cells were counted randomly in each
well under a fluorescent microscope at magnification ×100. All the
experiments were done in duplicate and results were expressed as
mean ± SEM of three independent experiments.
Migration assay
After transfection, cells were seeded in 12-well
plates in complete medium. When the cell confluence reached ~90%,
at 72 h post-transfection, an artificial circle wound was made onto
the monolayer with a 1-ml micropipette tip. The wound area was then
examined after 24 h of incubation under an optical microscope at
magnification ×100. Photographs were taken and the cell migration
ability was expressed by the gap closure.
Proliferation assay
Cell proliferation was measured using an ELISA kit
(Roche Applied Science, Germany) according to the manufacturer’s
instructions. Briefly, 4×103 cells were plated to each
well of a 96-well plate after transfection, and then cells were
incubated with bromodeoxyuridine for 2 h. The cells were
subsequently fixed and incubated with 100 μl of detection antibody
conjugated with peroxidase and substrate, respectively. Light
emission of the samples was measured in a microplate
luminometer.
MirRNA luciferase assay
Position 416–422 of the ADAM17 3′-UTR is a predicted
interaction position of miR-145. We cloned an 80-bp sequence
containing the predicted binding site or a scrambled sequence
downstream of the pMIR luciferase reporter to generate pMIR-ADAM17
and pMIR-mut-ADAM17 vectors, respectively. Luciferase assays were
carried out in U87 cells. First, cells were co-transfected with
appropriate plasmids with either hsa-miR-145 miRNA mimic or control
cel-mir-67 mimic in 12-well plates. Then, the cells were harvested
and lysed for luciferase assay 24 h after transfection. Luciferase
assays were performed using a luciferase assay kit (Promega)
according to the manufacturer’s instructions.
Statistical analysis
Data are presented as mean and standard error (mean
± SEM). Statistical significance was analyzed by one-way ANOVA
using the GraphPad Prism software (version 4.0). P-value smaller
than 0.05 (P<0.05) was considered significant.
Results
Expression profile of miR-145 in glioma
cells
A growing body of studies report that miR-145 is
downregulated in cancers (5,15–19).
However, the expression of miR-145 in glioma has not been well
documented (20,21). We sought to identify the role of
miR-145 in human glioma cells. We compared the expression levels of
miR-145 in glioma cell lines with total RNA from frontal cortex
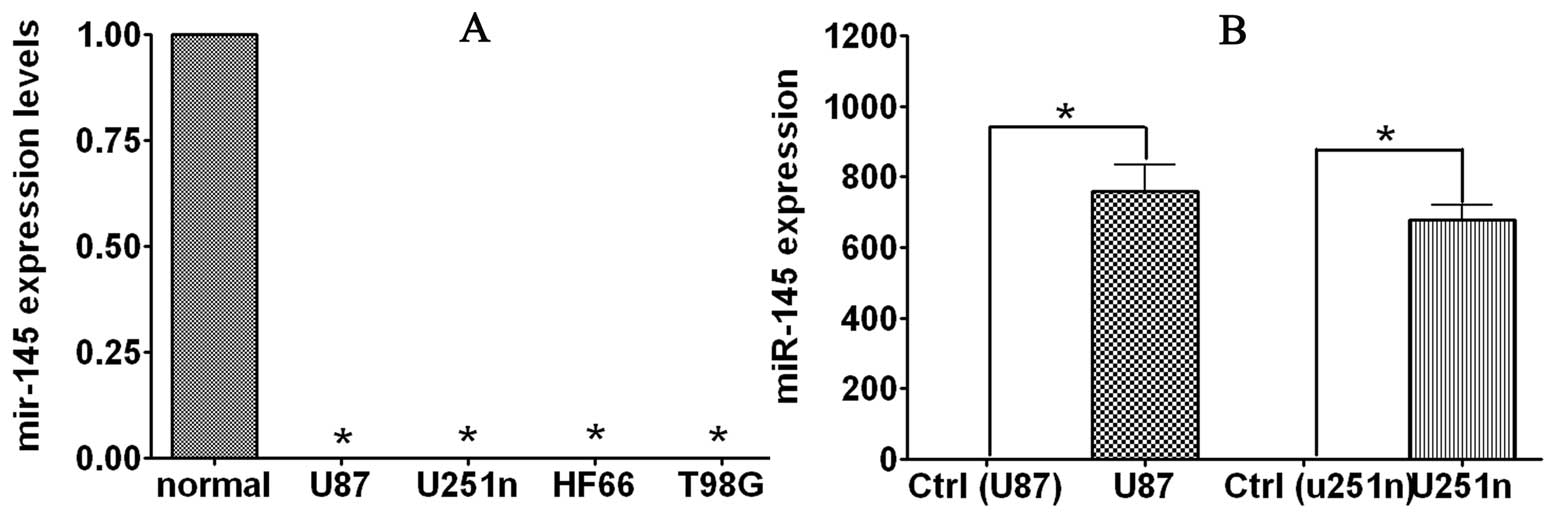
using real-time RT-PCR. As shown in the Fig. 1A, miR-145 expression levels were
significantly decreased in tumor cell lines compared to total RNA
from normal brain tissues. Expression of miR-145 in U87, U251n,
T98G and HF66 glioma cell lines 0.113, 0.002, 0.045 and 0.002% of
normal brain tissue. The reduced expression of miR-145 in glioma
cells suggests that miR-145 is a potential target in glioma
therapy.
MiR-145 overexpression inhibits
proliferation of glioma cells
To test the effect of miR-145 on cell growth, we
used miR-145 precursor microRNA to infect human glioma U251n and
U87 cells. After transfection, miR-145 levels were increased in
both cell lines, indicating that enhancement was due to the
introduction of precursor miR-145 (Fig.
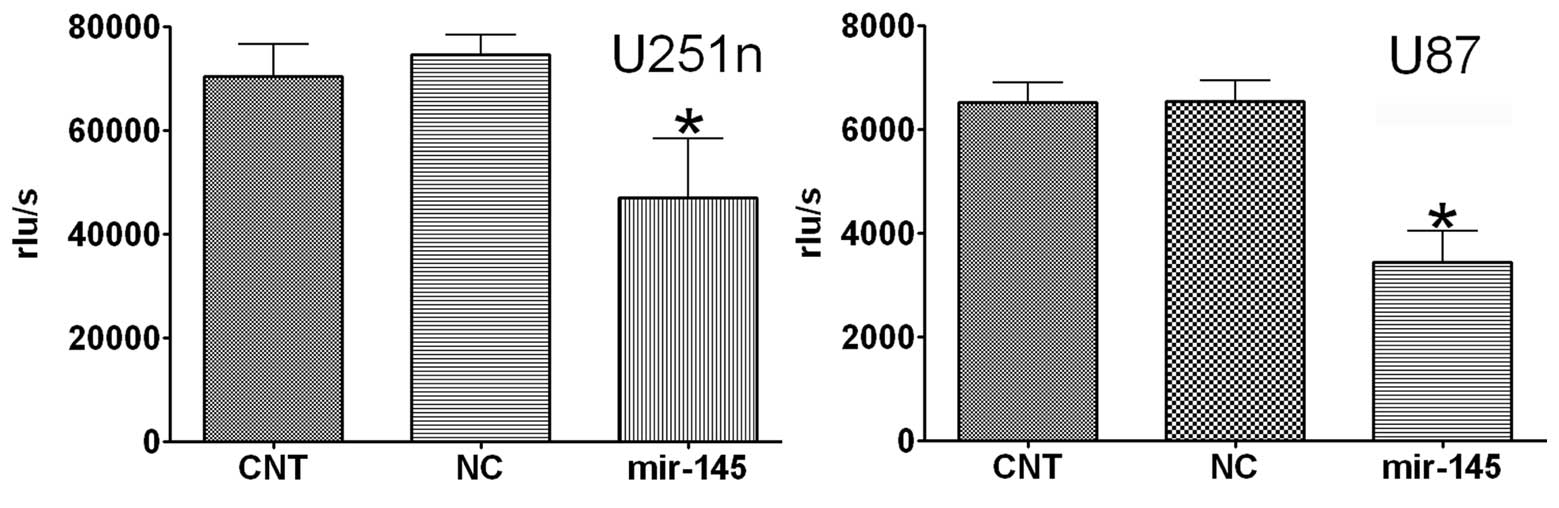
1B). As demonstrated by BrdU assay, overexpression of miR-145
significantly reduced cell proliferation by >40% in U87 and
U251n as compared to cells transfected with control (Fig. 2). These data suggest miR-145 has
anti-proliferative effects on U87 and U251n in vitro.
Ectopic expression of miR-145 inhibits
glioma cell migration and invasion
Previous studies reported that miR-145 decreases
invasion and migration of breast cancer cells in vitro and
in vivo(7). ADAM17 and EGFR
have been reported to play important roles in glioma, and
overexpression of ADAM17 promotes glioma invasiveness (22,23).
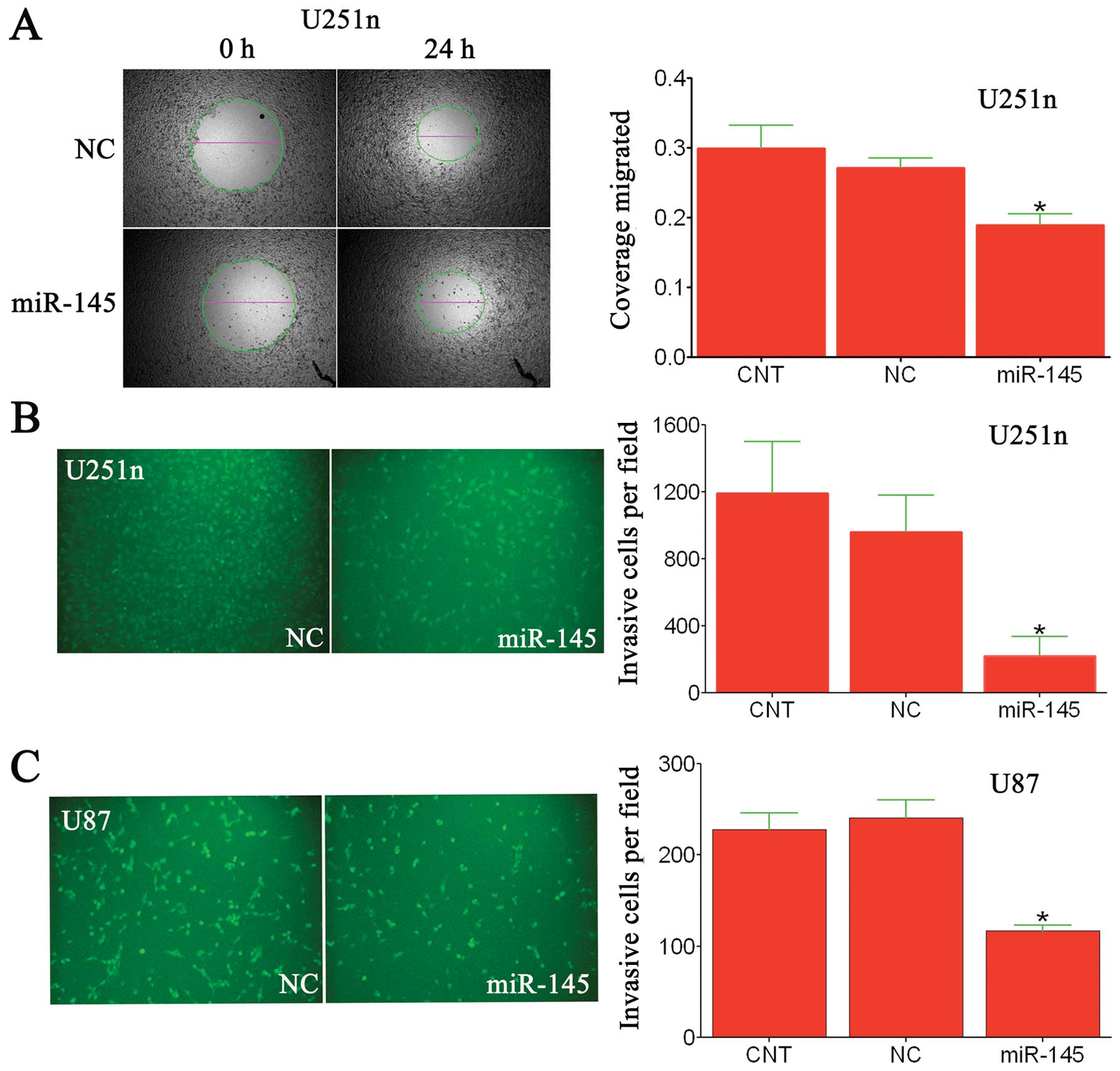
To determine if miR-145 had a similar effect on glioma cells, we
employed a wound healing migration assay and a Matrigel invasion
chamber assay. Seventy-two hours after transfection with miR-145 or
scrambled-miRNA, U87 and U251n cells were seeded into the upper
chamber, and then cells that invaded through the extracellular
matrix after 24 h were imaged and counted (Fig. 3B and C). In both cell lines, miR-145
significantly decreased the number of cells that invaded compared
to controls. Similarly, using a wound healing assay, we examined
the effect of miR-145 on U251n cell migration (Fig. 3A). Here, we found miR-145 inhibited
migration of U251n glioma cell migration, as compared with
scrambled-miRNA transfected control groups. Taken together, these
data indicate that miR-145 serves as a regulatory molecule involved
in cell migration and invasion in vitro.
ADAM17 and EGFR as targets of miR-145 for
post-transcriptional repression
Given that miR-145 reduces migration and invasion of
U251n and U87 glioma cells, we then sought to identify the target
genes of miR-145 using the online miRNA target prediction programs
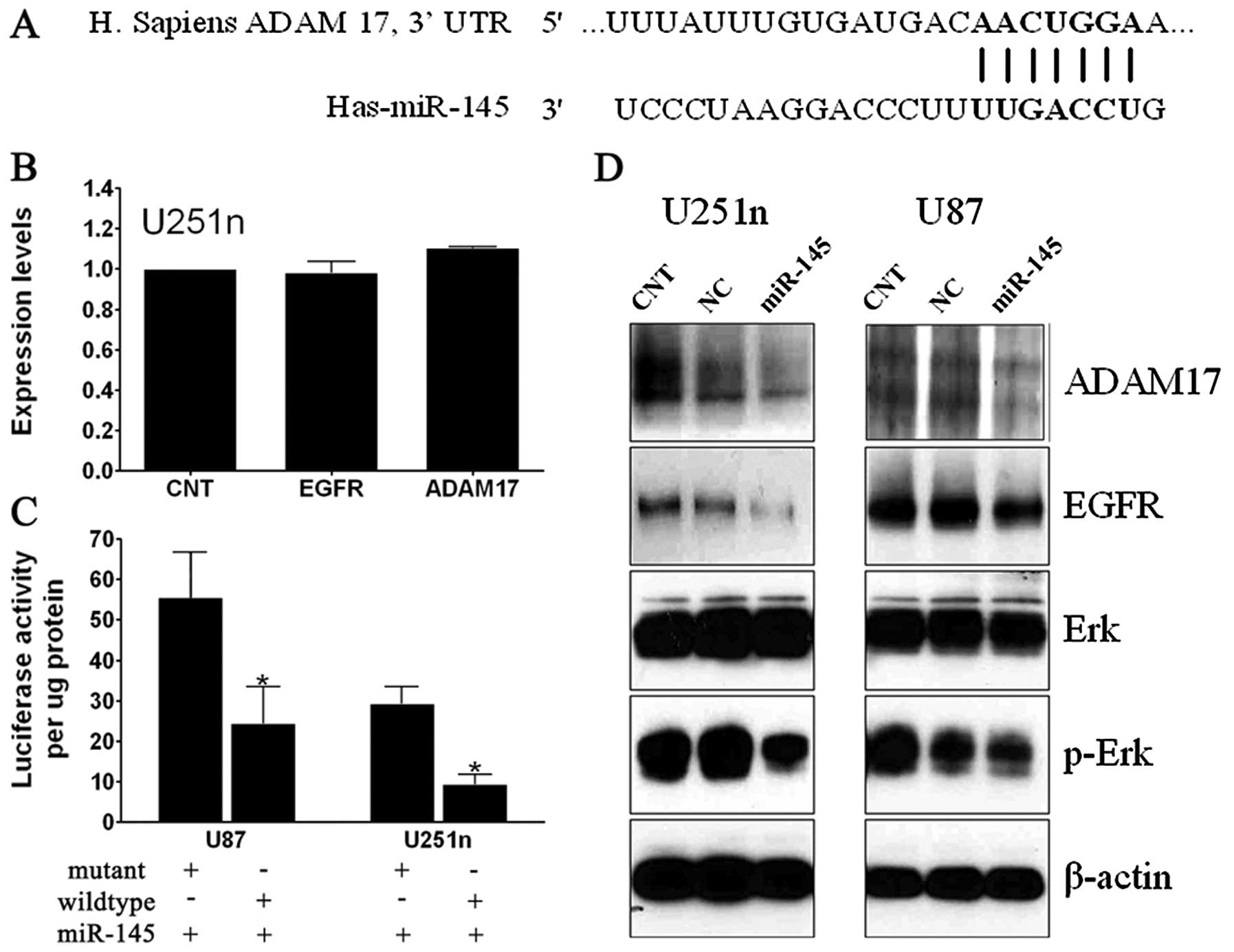
Targetscan and Pictar (Fig. 4A).
Approximately 150 targets of miR-145 were predicted from these
programs. ADAM17 was of particular interest because our previous
studies revealed that its expression level was upregulated in
glioma specimens. ADAM17 overexpression contributes to glioma
progression (23). It was recently
reported that miR-145 inhibits expression of EGFR and reduces cell
growth of lung cancer cells (24).
To test whether miR-145 suppresses the expression of
ADAM17 and EGFR at protein level in glioma, we performed Western
blot analysis. Employing a ‘scrambled’ cel-mir-67 or miR-145 mimic,
we found that ADAM17 and EGFR protein expression were significantly
reduced by miR-145 compared to miRNA controls in both cell lines
(Fig. 4D). However, overexpression
of miR-145 did not change mRNA expression of ADAM17 and EGFR
(Fig. 4B). These data suggest that
miR-145 targets ADAM17 and EGFR by post-transcriptional
repression.
ADAM17 is a primary sheddase for multiple EGFR
pro-ligands (25,26). EGFR is an important mediator
responsible for the invasiveness of malignant gliomas (14,27).
EGFR ligand binding results in receptor self-dimerization,
autophosphorylation and subsequent activation of downstream
Ras/MAPK/ERK signaling pathways, which contribute to the malignant
phenotype (28). As reduced protein
expression of ADAM17 and EGFR by miR-145, Western blot analysis was
also employed to determine the expression of downstream signaling
protein Erk/p-Erk. Compared to controls, miR-145 has no effect on
Erk, but significantly decreased p-Erk expression. These data
indicate that miR-145 may reduce migration and invasion through the
ADAM17/EGFR/Erk/p-Erk signaling pathway.
To further confirm that the effects observed above
are ascribed to the specific interaction between miR-145 and the
binding sites for miR-145 in the 3′-UTR of ADAM17, we performed a
Luciferase activity assay. As shown in Fig. 4C, miR-145 suppressed >40 and 50%
activity, respectively, of Luc-ADAM17-UTR (wild-type) in U87 and
U251n compared with mutant ADAM17 3′-UTR, which suggest that
miR-145 directly targets the 3′-UTR of ADAM17 mRNA.
Discussion
In the present study we detected the miR-145
expression level in U87, U251n, T98G and HF66 human glioma cell
lines, and found a significant reduction of miR-145 in human glioma
cell lines compared to total RNA of frontal cortex. The expression
level of miR-145 negatively regulates glioma malignancy. Ectopic
expression of miR-145 decreased proliferation, migration, and
invasion of glioma cells. These data suggest that miR-145 plays a
critical role in glioma development, and it may function as an
anti-tumor factor in glioma cells.
Our studies indicate that ADAM17 is a target gene of
miR-145, and several lines of evidence support the direct
interaction between miR-145 and ADAM17: i) the 3′-UTR of ADAM17
contains a binding site for miR-145 with significant seed match;
ii) miR-145 suppresses the activity of a luciferase reporter with
the 3′-UTR of ADAM17 mRNA; iii) miR-145 represses ADAM17 expression
at protein level; (4) our previous
studies have shown that ADAM17 knockdown by siRNAs resulted in a
decreased glioma cell proliferation, migration and invasiveness. We
therefore, the first time, identify miR-145 as directly regulating
ADAM17.
ADAM17 expression is elevated in different cancers
and is associated with cancer progression (29,30).
High levels of ADAM17 have been reported in a variety of cell lines
and clinical tissue samples, including breast, colon, prostate and
glioblastoma (23,31–33).
Our previous study showed that high level expression of ADAM17 was
found in high grade glioma samples. Knockdown of ADAM17 inhibits
breast cancer and glioma cell proliferation and invasion (23,31).
Ectopic expression of ADAM17 induced tumorigenicity of cortical
astrocyte cell line (34) and
promotes both breast and glioma cell malignant phenotypes (23,31).
In our present study, we demonstrated that miR-145 is an important
regulator of ADAM17, and directly binds the 3′-UTR of ADAM17 mRNA
in glioma cells. Thus, the reduction of miR-145 in glioma cells
yields an increased expression of ADAM17.
One of the characteristics of glioma is
invasiveness. The EGFR is over-expressed in ~50–60% of gliomas
(35), and EGFR increases with
malignancy grade, and is required for maintenance of glioma growth
(36). EGFR serves as an aggressive
agonist of glioma invasion (37).
In our study, transfection of glioma cells with miR-145 resulted in
decreased protein expression of EGFR, which may be partly
responsible for the miR-145 induced decrease of glioma cell
invasion, and consistent with a recent report that miR-145
decreased EGFR expression and proliferation in lung cancers
(38). These data suggest that
miR-145 may also target the EGFR gene. ADAM17 is involved in the
ectodomain shedding of multiple membrane-bound ligands and
cytokines, implicated in diverse biological processes, including
growth and inflammation (23,39).
Interestingly, ADAM17 has recently been identified as the primary
sheddase for multiple EGFR pro-ligands. EGFR ligand-binding results
in receptor self-dimerization, autophosphorylation and subsequent
activation of downstream Ras/MAPK/ERK signaling pathways (28,40).
Taken together with reduced p-Erk expression, miR-145 transfection
may decrease glioma cell migration and invasion through
ADAM17/EGFR/ERK signaling pathway.
In conclusion, our data suggested that miR-145 plays
a key role in the malignancy of glioma cells possibly by direct
regulation of ADAM17 protein expression, which affects glioma cell
proliferation, migration and invasion. However, further study is
needed to determine if ADAM17 activity is influenced by miR-145
in vivo in glioma.
Acknowledgements
This work was supported by NIH grant RO1
CA129446.
References
|
1
|
Yi R and Fuchs E: MicroRNAs and their
roles in mammalian stem cells. J Cell Sci. 124:1775–1783. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gusev Y: Computational methods for
analysis of cellular functions and pathways collectively targeted
by differentially expressed microRNA. Methods. 44:61–72. 2008.
View Article : Google Scholar
|
|
3
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar
|
|
4
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Akao Y, Nakagawa Y and Naoe T:
MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 26:311–320.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen Z, Zeng H, Guo Y, et al: miRNA-145
inhibits non-small cell lung cancer cell proliferation by targeting
c-Myc. J Exp Clin Cancer Res. 29:1512010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sachdeva M and Mo YY: MicroRNA-145
suppresses cell invasion and metastasis by directly targeting mucin
1. Cancer Res. 70:378–387. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu Y, Liu S, Xin H, et al: Up-regulation
of microRNA-145 promotes differentiation by repressing OCT4 in
human endometrial adenocarcinoma cells. Cancer. 117:3989–3998.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu X, Lu D, Scully M and Kakkar V: ADAM
proteins - therapeutic potential in cancer. Curr Cancer Drug
Targets. 8:720–732. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kheradmand F and Werb Z: Shedding light on
sheddases: role in growth and development. Bioessays. 24:8–12.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wildeboer D, Naus S, Amy Sang QX, Bartsch
JW and Pagenstecher A: Metalloproteinase disintegrins ADAM8 and
ADAM19 are highly regulated in human primary brain tumors and their
expression levels and activities are associated with invasiveness.
J Neuropathol Exp Neurol. 65:516–527. 2006. View Article : Google Scholar
|
|
12
|
Zheng X, Jiang F, Katakowski M, et al:
Inhibition of ADAM17 reduces hypoxia-induced brain tumor cell
invasiveness. Cancer Sci. 98:674–684. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zheng X, Jiang F, Katakowski M, et al:
Sensitization of cerebral tissue in nude mice with photodynamic
therapy induces ADAM17/TACE and promotes glioma cell invasion.
Cancer Lett. 265:177–187. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tsatas D, Kanagasundaram V, Kaye A and
Novak U: EGF receptor modifies cellular responses to hyaluronan in
glioblastoma cell lines. J Clin Neurosci. 9:282–288. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gregersen LH, Jacobsen AB, Frankel LB, Wen
J, Krogh A and Lund AH: MicroRNA-145 targets YES and STAT1 in colon
cancer cells. PLoS One. 5. pp. e88362010, View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ozen M, Creighton CJ, Ozdemir M and
Ittmann M: Widespread deregulation of microRNA expression in human
prostate cancer. Oncogene. 27:1788–1793. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Takagi T, Iio A, Nakagawa Y, Naoe T,
Tanigawa N and Akao Y: Decreased expression of microRNA-143 and
-145 in human gastric cancers. Oncology. 77:12–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Akao Y, Nakagawa Y, Kitade Y, Kinoshita T
and Naoe T: Downregulation of microRNAs-143 and -145 in B-cell
malignancies. Cancer Sci. 98:1914–1920. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ichimi T, Enokida H, Okuno Y, et al:
Identification of novel microRNA targets based on microRNA
signatures in bladder cancer. Int J Cancer. 125:345–352. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fischer L, Hummel M, Korfel A, Lenze D,
Joehrens K and Thiel E: Differential micro-RNA expression in
primary CNS and nodal diffuse large B-cell lymphomas.
Neurooncology. 13:1090–1098. 2011.PubMed/NCBI
|
|
21
|
Fang X, Yoon JG, Li L, et al: The SOX2
response program in glioblastoma multiforme: an integrated
ChIP-seq, expression microarray, and microRNA analysis. BMC
Genomics. 12:112011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lo HW: EGFR-targeted therapy in malignant
glioma: novel aspects and mechanisms of drug resistance. Curr Mol
Pharmacol. 3:37–52. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zheng X, Jiang F, Katakowski M, Lu Y and
Chopp M: ADAM17 promotes glioma cell malignant phenotype. Mol
Carcinog. 51:150–164. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cho WC, Chow AS and Au JS: Restoration of
tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung
adenocarcinoma patients with epidermal growth factor receptor
mutation. Eur J Cancer. 45:2197–2206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hart S, Fischer OM and Ullrich A:
Cannabinoids induce cancer cell proliferation via tumor necrosis
factor alpha-converting enzyme (TACE/ADAM17)-mediated
transactivation of the epidermal growth factor receptor. Cancer
Res. 64:1943–1950. 2004. View Article : Google Scholar
|
|
26
|
Lee DC, Sunnarborg SW, Hinkle CL, et al:
TACE/ADAM17 processing of EGFR ligands indicates a role as a
physiological convertase. Ann NY Acad Sci. 995:22–38. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Laerum OD, Nygaar SJ, Steine S, et al:
Invasiveness in vitro and biological markers in human primary
glioblastomas. J Neurooncol. 54:1–8. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wells A: EGF receptor. Int J Biochem Cell
Biol. 31:637–643. 1999. View Article : Google Scholar
|
|
29
|
Kornfeld JW, Meder S, Wohlberg M, et al:
Overexpression of TACE and TIMP3 mRNA in head and neck cancer:
association with tumour development and progression. Br J Cancer.
104:138–145. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Narita D, Seclaman E, Ursoniu S and Anghel
A: Increased expression of ADAM12 and ADAM17 genes in laser-capture
microdissected breast cancers and correlations with clinical and
pathological characteristics. Acta Histochem. 114:131–139. 2012.
View Article : Google Scholar
|
|
31
|
Zheng X, Jiang F, Katakowski M, Zhang ZG,
Lu QE and Chopp M: ADAM17 promotes breast cancer cell malignant
phenotype through EGFR-PI3K-AKT activation. Cancer Biol Ther.
8:1045–1054. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Blanchot-Jossic F, Jarry A, Masson D, et
al: Up-regulated expression of ADAM17 in human colon carcinoma:
co-expression with EGFR in neoplastic and endothelial cells. J
Pathol. 207:156–163. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin P, Sun X, Feng T, et al: ADAM17
regulates prostate cancer cell proliferation through mediating cell
cycle progression by EGFR/PI3K/AKT pathway. Mol Cell Biochem.
359:235–243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Katakowski M, Jiang F, Zheng X, Gutierrez
JA, Szalad A and Chopp M: Tumorigenicity of cortical astrocyte cell
line induced by the protease ADAM17. Cancer Sci. 100:1597–1604.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Heimberger AB, Hlatky R, Suki D, et al:
Prognostic effect of epidermal growth factor receptor and EGFRvIII
in glioblastoma multiforme patients. Clin Cancer Res. 11:1462–1466.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mukasa A, Wykosky J, Ligon KL, Chin L,
Cavenee WK and Furnari F: Mutant EGFR is required for maintenance
of glioma growth in vivo, and its ablation leads to escape from
receptor dependence. Proc Natl Acad Sci USA. 107:2616–2621. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tysnes BB, Haugland HK and Bjerkvig R:
Epidermal growth factor and laminin receptors contribute to
migratory and invasive properties of gliomas. Invasion Metastasis.
17:270–280. 1997.PubMed/NCBI
|
|
38
|
Cho WC, Chow AS and Au JS: MiR-145
inhibits cell proliferation of human lung adenocarcinoma by
targeting EGFR and NUDT1. RNA Biol. 8:125–131. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Franovic A, Robert I, Smith K, et al:
Multiple acquired renal carcinoma tumor capabilities abolished upon
silencing of ADAM17. Cancer Res. 66:8083–8090. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lal A, Glazer CA, Martinson HM, et al:
Mutant epidermal growth factor receptor up-regulates molecular
effectors of tumor invasion. Cancer Res. 62:3335–3339.
2002.PubMed/NCBI
|