Introduction
Colorectal cancer is a significant health burden
worldwide, accounting for over 1,2 million new cancer cases and
more than 600,000 cases of cancer-related mortality annually
(1). However, incidence of this
type of cancer has recently decreased in several Western countries,
largely due to increased awareness, early detection and prevention
measures (2). In addition,
advancements in treatments have improved the survival of patients
with this disease. To date, surgery is curative for the early
stages of colorectal cancer, while chemotherapy plus surgery is
used to treat the later stages of the disease. However, most
patients eventually develop metastatic disease and succumb to
colorectal cancer. Thus, novel approaches for the early detection,
treatment and suppression of tumor progression are required to
effectively control this disease.
microRNAs (miRNAs) are a group of
naturally-occurring, small non-coding RNAs, 18–22 nucleotides long,
that regulate target gene expression by specifically inhibiting the
translation of mRNA or inducing mRNA degradation (2,3).
Accumulating evidence has demonstrated that miRNAs play a critical
role in regulating a variety of physiological and pathological
processes in the human body, such as cell differentiation,
morphogenesis and tumorigenesis (4,5). In
human cancer, miRNAs can function either as oncogenes or tumor
suppressor genes in regulating tumor development and progression
(6), depending on the target gene.
Thus, identifying and understanding the molecular targets of miRNAs
by using computational in silico analysis with a specific
algorithm, such as TargetScan (7),
could elucidate the signaling cascades involved in colorectal
cancer progression.
It was recently shown that miR-22 is dysregulated
and functions as a tumor suppressor gene in several types of
cancer. In addition, its altered expression is associated with the
poor prognosis of colon cancer. Furthermore, previous studies
showed that miR-22 expression inhibited the proliferation of A539
and H1299 lung cancer cells by post-transcriptional regulation of
ErB3 expression (8), while in
breast cancer, miR-22 inhibited estrogen receptor α (ER-α) and
ectopic viral integration site-1 (EVI-1) expression to suppress
tumor cell proliferation and metastatic potential (9–11).
miR-22 expression also inhibited hepatocellular carcinoma cell
proliferation by targeting HDAC4 (12), while inhibiting ovarian cancer cell
migration and invasion by targeting T-cell lymphoma invasion and
metastasis 1 (TIAM1) expression (13,14).
Furthermore, it is showed that TNFAIP8-rs11064 variant G allele
weaken the binding affinity of miR-22 to TNFAIP8 3′-UTR
contributing to cervical cancer risk (38). Similar data were shown in colon
cancer cells, where PTEN expression was downregulated by miR-22,
which contributed to sensitizing paclitaxel-induced chemoresistance
(15,16). miR-22 expression was also shown to
regulate hypoxia signaling in colon cancer cells by suppressing the
expression of hypoxia inducible factor-1α (HIF-1α) (17), while miR-22 expression was induced
by vitamin D treatment to have anti-proliferative and
anti-migratory functions in colon cancer cells (18). These data indicate that there may be
multiple genes regulated by miR-22 in different types of
cancer.
In the present study, we investigated the effect of
miR-22 expression on colorectal cancer cell viability, migration
and invasion. In addition, computational in silico analysis
with TargetScan revealed TIAM1 as a potential miR-22 target gene;
we subsequently performed qRT-PCR, western blotting and
immunocytochemistry to confirm this finding. TIAM1 is a guanine
nucleotide exchange factor protein that modulates the activity of
Rho GTP-binding protein Rac1; in turn, Rac1 regulates the
reorganization of cytoskeletal structure and promotes cell adhesion
and movement, thereby contributing to the invasion and metastasis
of tumor cells (19). In normal
tissues, with the exception of the brain and testis, TIAM1
expression is absent or at low levels. By contrast, TIAM1 is highly
expressed in different types of cancer (particularly metastatic
cancer), including colon (20),
liver (21–23), kidney (24), esophagus (25), nasopharynx (26), ovary (13), breast (27) and lung cancer (28).
Materials and methods
Cell culture and transfection
The HCT-116 human colorectal cancer cell line was
purchased from the American Type Culture Collection (Manassas, VA,
USA) and routinely cultured in McCoy’s 5A modified medium
supplemented with 10% fetal calf serum (FCS), 100 U/ml penicillin
and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA, USA) in a
37°C humidified incubator containing 5% CO2. hsa-miR-22
precursor sequences (5′-AGUUCUUCAGUGGCAAGCUUUA-3′) were cloned into
the SD13 expression vector (GeneChem, Shanghai, China), amplified
and sequence-confirmed prior to use. The plasmid encoding the
miR-22 precursor was labeled SD13-hsa-miR-22.
To induce miR-22 expression in HCT-116 cells, we
transfected SD13-hsa-miR-22 or control vector into HCT-116 cells
using Lipofectamine 2000 (Invitrogen) according to the
manufacturer’s instructions. Briefly, cells plated at 70%
confluence in 24-well plates were transfected with 0.8 μg
SD13-hsa-miR-22 or control plasmid and re-fed with fresh medium 4 h
after transfection. The expression of green fluorescent protein
(GFP) was monitored in cells as an indicator of transfection
efficiency and total RNA was isolated 48 h after transfection to
verify gene expression.
Online search of GenBank databases
To predict miR-22 target genes, we performed online
searches of two different Genbank databases; i.e., miRNA database;
http://www.sanger.ac.uk/Software/Rfam/mirna and
TargetScan algorithm (http://www.targetscan.org).
qRT-PCR
Total RNA was isolated using RNAiso for Small RNA
(Takara, Japan), according to the manufacturer’s instructions, and
then reverse transcribed into cDNA using One Step
PrimeScript® miRNA cDNA Synthesis kit (Perfect Real
Time, Takara, Japan). The newly synthesized cDNA was then used as a
template for detection of miR-22 and other genes. Specifically, 100
ng cDNA was mixed with SYBR Premix Ex TaqTMII (Perfect
Real Time) in a 20-μl reaction. All reactions were run in
triplicate with the ABI PRISM® 7300 Real-Time PCR system
(Applied Biosystems, Foster City, CA, USA) using primers specific
for miR-22, β-actin, matrix metalloproteinases 2 and 9 (MMP-2 and
MMP-9), TIAM1 and vascular endothelial growth factor (VEGF)
(Table I). Reaction conditions
were: 50°C for 2 min and 95°C for 1 min, followed by 40 cycles of
95°C for 5 sec and 60°C for 1 min. The relative expression of mRNA
was analyzed using 7300 System SDS software. U6 small nuclear RNA
was used as an internal control.
 | Table IPrimers for qPCR. |
Table I
Primers for qPCR.
| Primer | Sequence | Size of PCR product
(bp) |
|---|
| hsa-miR-22 | Purchased from
RiboBio (Guangzhou, China) | 88 |
| U6 | Purchased from
RiboBio (Guangzhou, China) | 96 |
| β-actin |
5′-CTGGAACGGTGAAGGTGACA-3′ | 140 |
|
5′-AAGGGACTTCCTGTAACAACGCA-3′ | |
| MMP-2 |
5′-GCTGGAGACAAATTCTGGAGATACA-3′ | 281 |
|
5′-GTATCGAAGGCAGTGGAGAGGA-3′ | |
| MMP-9 |
5′-GCTGGCAGAGGAATACCTGTAC-3′ | 112 |
|
5′-CAGGGACAGTTGCTTCTGGA-3′ | |
| TIAM1 |
5′-AAGACGTACTCAGGCCATGTCC-3′ | 253 |
|
5′-GACCCAAATGTCGCAGTCAG-3′ | |
| VEGF | 5′-AAGATCCGCAGAC
GTGTAAATGTT-3′ | 101 |
|
5′-CGGCTTGTCACATCTGCAAGTA-3′ | |
MTT cell viability assay
To determine the effect of miR-22 on colorectal cell
viability, we performed the MTT assay. Briefly, HCT-116 colon
cancer cells transfected with control vector (NC) or
SD13-hsa-miR-22 were plated in a 96-well plate at a density of
1×104 cells per well and grown for up to 7 days in
complete cell culture media. Thereafter, 20 μl of 10X MTT reagent
(250 mg of 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium
bromide; Sigma, St. Louis, MO, USA) was added and incubated with
the cells for 4 h. The media was subsequently aspirated and
replaced with DMSO and the cells were incubated at room temperature
for 15 min with vigorous shaking. Optical absorbance was measured
spectrophotometrically at 490 nm daily for 7 days to quantify cell
proliferation. Cell proliferation curves were plotted based on
absorbance.
Immunocytochemistry
To detect changes in gene expression in HCT-116
cells in the absence or presence of miR-22 transfection, we
performed immunocytochemistry using a streptavidin peroxidase
method. The protein expression of MMP-2, MMP-9, VEGF and TIAM1 was
assessed using the corresponding antibodies in colon cancer cells.
Following transfection, the cells were fixed in 10% formalin for 10
min and then treated with 0.5% Triton X-100 for 10 min. After
blocking in 20% normal serum, the cells were incubated with a
primary antibody for 1 h and a biotinylated secondary antibody for
10 min, followed by incubation with streptavidin-conjugated
horseradish peroxidase for 15 min at 37°C. The immune reaction was
visualized with DAB (3,3′-diaminobenzidine) and images were
captured with Image-Pro Plus (Media Cybernetics, Bethesda, MD,
USA).
In vitro tumor cell migration and
invasion assay
To determine the effects of miR-22 on colorectal
cell migration and invasion, we performed cell migration and
invasion assays. In brief, Matrigel was thawed on ice overnight at
4°C and diluted with cold basal medium. Diluted Matrigel (30 μl)
was added to the upper chamber of the two-chamber Transwell system
(8-μm pore size and 6.5-mm diameter; Corning, Chelmsford St.
Lowell, MA, USA) and the plate was allowed to sit at room
temperature at 37°C for 30 min. Thereafter, 200 μl of cell
suspension (1×106 cells/ml) in McCoy’s 5A medium
supplemented with 0.1% FCS was plated on the top chamber of the
coated transwell. The bottom chamber was filled with 600 μl McCoy’s
5A medium containing 30% FBS and served as a chemo-attractant.
After a 48-h incubation at 37°C, cells on the upper surface of the
filter were carefully removed with a cotton swab. Cells that had
traversed the membrane were fixed in methanol at 4°C for 30 min and
then stained with 0.1% crystal violet for 20 min. To quantify the
migrated cells microscopically, cells were counted in five random
fields/filter (magnification, ×400). The migration assay was
performed in a similar fashion in a transwell (Corning) without
Matrigel coating.
Protein extraction and western blot
analysis
To determine the effects of miR-22 on the regulation
of expression of different genes, in colorectal cells, HCT-116
cells transfected with miR-22 or NC expression vectors were washed
twice with cold phosphate-buffered saline (PBS), harvested by
scraping and lysed in a lysis buffer (Takara). Following
centrifugation, the supernatant was collected and protein
concentration was determined using a Protein Assay kit II (Bio-Rad,
Hercules, CA, USA). For western blotting, 50 mg of each sample was
mixed with sodium dodecyl sulfate (SDS) loading buffer, heated at
95°C for 5 min, resolved on 12% SDS-polyacrylamide gel
electrophoresis (SDS-PAGE) gels and transferred onto polyvinylidene
difluoride (PVDF) membranes (Perkin-Elmer Life Sciences, Waltham,
MA, USA). Following incubation for 1 h with 5% skim milk in PBS,
the membranes were incubated overnight at 4°C with primary
antibodies against MMP-2, MMP-9, VEGF and TIAM1. Protein bands of
interest were visualized using enhanced chemiluminescence (ECL),
followed by exposure to X-ray film following incubation with
horseradish peroxidase-conjugated secondary antibodies. The film
was scanned and analyzed with Gel Image System software (Simon,
ProteinSimple, Santa Clara, CA, USA).
Statistical analysis
Results are expressed as the means ± SD (standard
deviation) unless otherwise specified. Statistical analyses were
performed between two groups with the Student’s t-test and between
multiple groups by ANOVA. P-values ≤0.05 were considered to
indicate statistically significant differences.
Results
Expression of miR-22 in colon cancer
cells
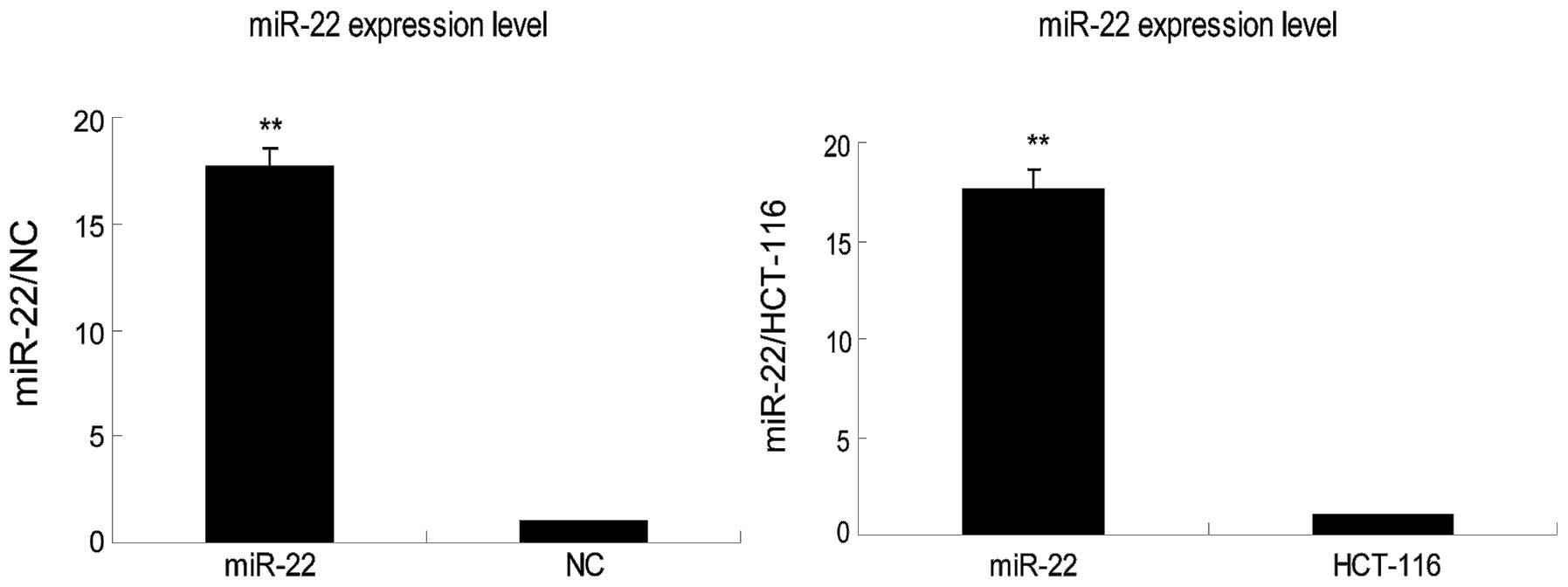
In this study, we used qRT-PCR to examine miR-22
expression in HCT-116 colorectal cancer cells and found that it was
weakly expressed (Fig. 1). We
therefore transfected a miR-22 expression vector into this cell
line. Our data showed that the expression level of transfected
miR-22 was 17.65±0.98-fold higher than that of NC or parental cells
(Fig. 1).
Effect of miR-22 on colon cancer cell
proliferation
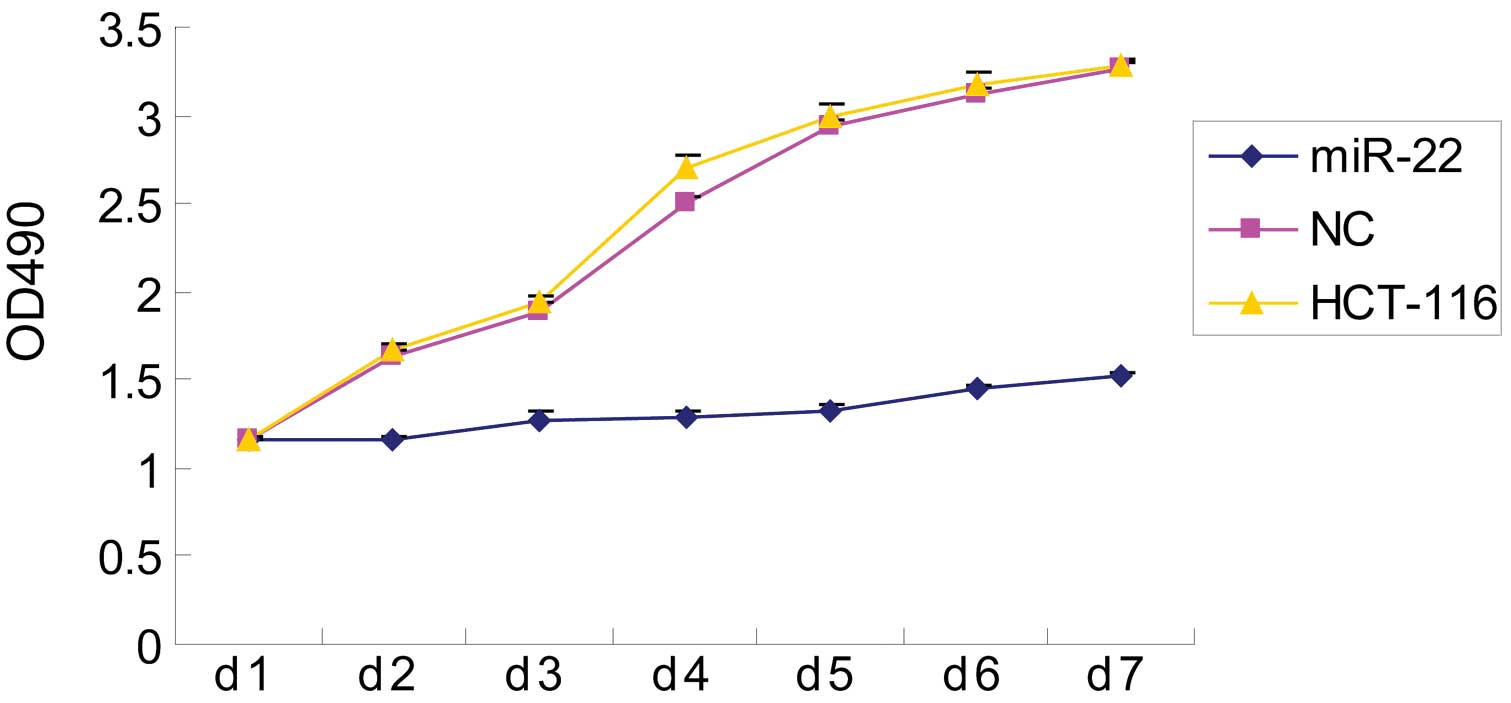
Next, we assessed the effect of miR-22 on colon
cancer cell proliferation and found that miR-22 significantly
inhibited cell viability (Fig. 2).
Specifically, the optical absorbance rates of parental and NC cells
were >2-fold higher than those of miR-22-transfected HCT-116
cells (P<0.05). By contrast, there was no significant difference
between parental and NC control cells during the 7 days of
culture.
Effects of miR-22 on colon cancer cell
migration and invasion
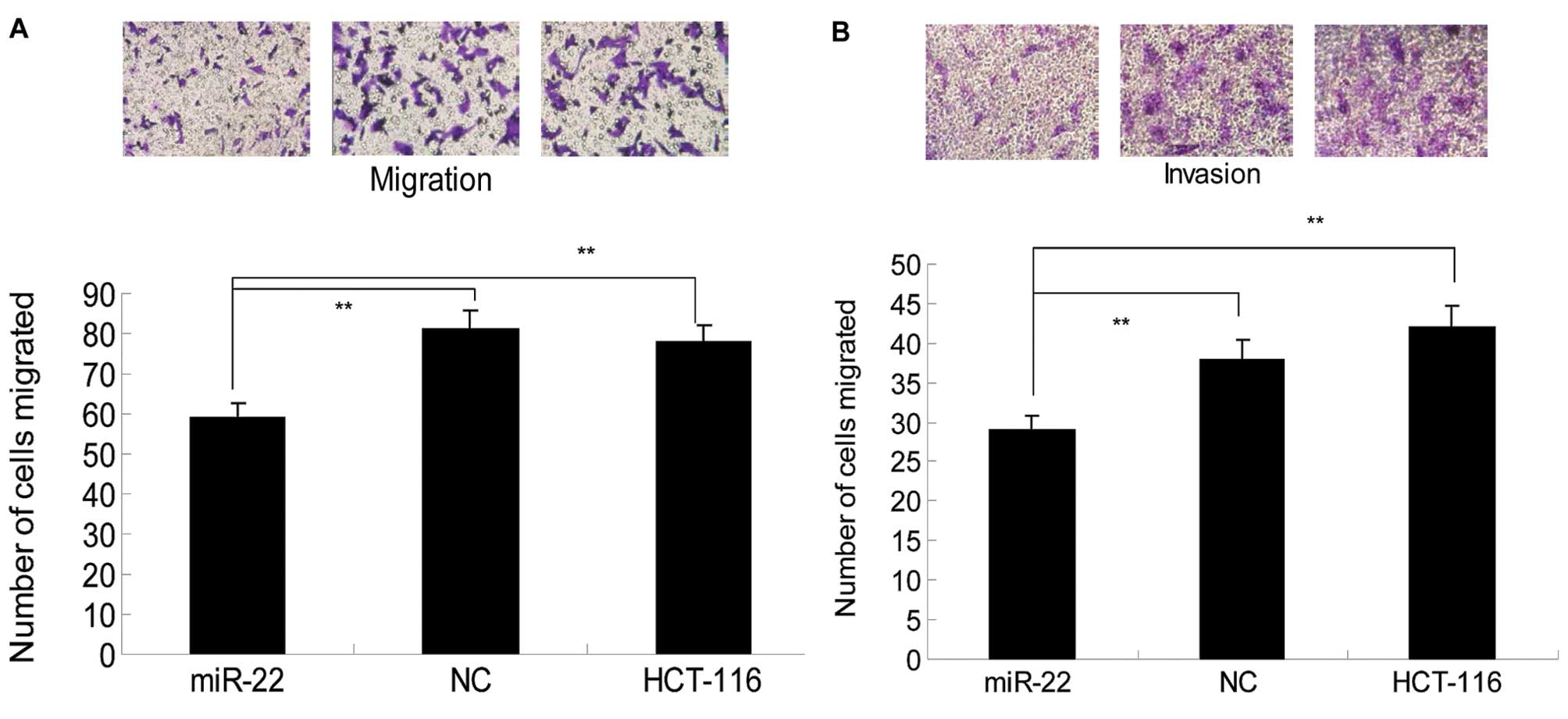
Previous studies demonstrated that miR-22 was able
to inhibit the invasion and metastasis of lung (8), breast (22) and ovarian cancer cells (15). This study investigated whether
miR-22 is also able to suppress colon cancer cell invasion.
Therefore, parental, NC and miR-22-expressing HCT-116 cells were
subjected to transwell migration and invasion assays. We found that
the number of cells that migrated through the pores in
miR-22-expressing cells was 59±3.75, which was significantly lower
than that of NC (81±4.65) or parental cells (78±3.97) (P<0.05;
Fig. 3A), suggesting that miR-22
expression suppressed the migration of colon cancer cells.
Similarly, the invasion assay showed that the number of cells that
invaded the Matrigel and migrated through the pore in the membrane
was markedly lower in tumor cells expressing miR-22 (29±1.75)
compared to parental (42±2.65) or NC cells (38±2.32) (Fig. 3B). Collectively, these results
suggest that induction of miR-22 expression significantly inhibits
the migration and invasion of colon cancer cells.
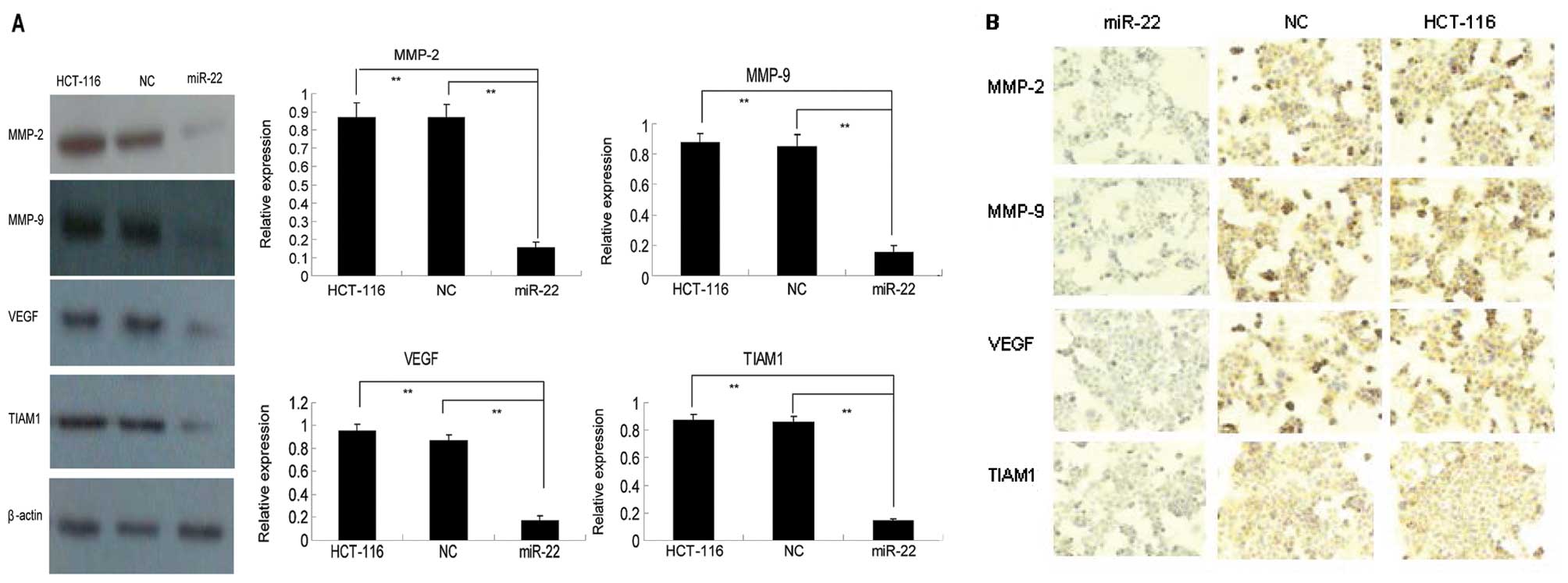
Effects of miR-22 on TIAM1, MMP-2, MMP-9
and VEGF expression in colon cancer cells
To elucidate the underlying molecular events
responsible for miR-22-mediated inhibition of colon cancer cell
migration and invasion, we first performed an in silico
analysis using TargetScan to search potential miR-22 target genes
and found that TIAM1, a Rho GTPase that was shown to be associated
with colon cancer metastasis (29,30),
is a target of miR-22. We then verified our finding in
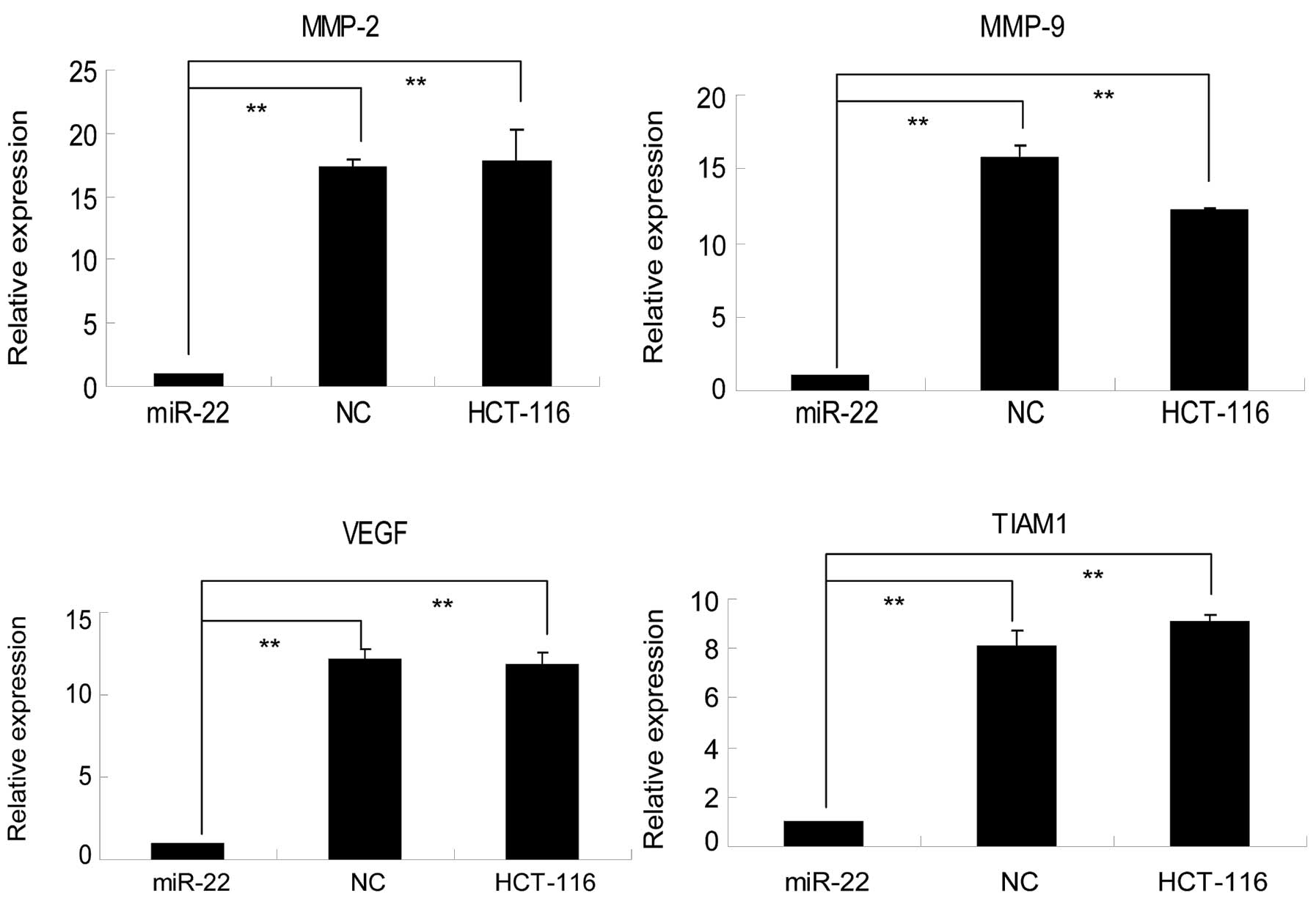
miR-22-transfected HCT-116 cells. As shown in Fig. 4, levels of TIAM1 mRNA in negative
control shRNA-transfected HCT-116 cells were 8.05±0.64-fold higher
than those of miR-22-transfected cells. Western blot analysis
showed that TIAM1 protein expression was >4-fold higher in both
the parental or NC cells than in miR-22-transfected cells (Fig. 5A). Immunocytochemistry confirmed our
western blotting data (Fig. 5B).
Collectively, these data demonstrate that miR-22 is able to
suppress TIAM1 mRNA and protein expression and, consequently, to
contribute to the suppression of colon cancer cell migration and
invasion.
Furthermore, we analyzed expression of matrix MMP-2
and MMP-9, which play critical roles in cancer cell invasion and
metastasis (31), as well as the
expression of pro-angiogenic protein VEGF (32) in these cells. As shown in Figs. 4 and 5, the expression of MMP-2, MMP-9 and VEGF
was well associated with miR-22 expression at both the
transcriptional and translational levels. In particular, in
parental and NC cells, the level of MMP-2 mRNA expression was
17.27±0.64 and 17.74±2.53-fold higher, respectively, than that in
miR-22-transfected cells, while levels of MMP-9 mRNA in parental
and NC cells were 15.76±0.85 and 16.04±0.23-fold higher,
respectively, than those in miR-22-transfected cells. Similarly,
VEGF expression levels in parental and NC cells were 12.14±0.61 and
11.78±0.82-fold higher, respectively, than those in
miR-22-transfected cells (Fig. 4).
A similar trend was observed at the protein level (Fig. 5).
Discussion
Emerging evidence has demonstrated that the altered
expression of miR-22 plays a role in the progression of different
types of cancer; however, the precise role of miR-22 in colon
cancer cells remains elusive. In the present study, we
overexpressed miR-22 in HCT-116 human colon cancer cells. Compared
to the negative control, overexpression of miR-22 significantly
inhibited the proliferation of colon cancer cells in vitro.
Moreover, expression of miR-22 also exerted inhibitory effects on
the suppression of HCT-116 cell migration and invasion.
Molecularly, we evaluated and verified that miR-22 expression
downregulated TIAM1 expression which, in turn, downregulated MMP-2,
MMP-9 and VEGF expression in HCT-116 cells. This study indicated
that the inhibitory effects of miR-22 on colorectal cancer cell
viability and invasion merit further investigation as a novel
strategy for the treatment of colorectal cancer and may thus be
used to prevent colorectal cancer metastasis.
miR-22 has been extensively studied in different
human types of cancer in vitro and ex vivo, but very
few studies have reported its role in colorectal cancer. miR-22 is
highly conserved across several vertebrate species. Early studies
showed that miR-22 plays a role in erythrocyte maturation (33). More recent data has demonstrated
that miR-22 expression is altered in various types of cancer and
plays a role in tumorigenesis; thus, it is thought to function as a
tumor suppressor in human cancer. It should be pointed out that the
relative expression levels of miR-22 were significantly lower in
colorectal cancer tissues than those in the normal adjacent mucosa
(39). However, the role of miR-22
in colorectal cancer remains unknown. In the present study, we
showed that overexpression of miR-22 reduced colorectal cancer cell
viability and migration and invasion capacity, indicating that
miR-22 may be useful in the control of colorectal cancer
progression in future clinic.
Furthermore, in terms of miR-22-targeting genes,
previous studies showed that miR-22 is able to target a variety of
genes, such as HDAC4 (11) and
c-Myc (34) and HIF-1α (17). Nevertheless, in the present study,
TIAM1 was predicted to be the target gene of miR-22 based on
computational in silico analysis. TIAM1 has been shown to
regulate migration, invasion and apoptosis of colon cancer cells
and TIAM1 expression contributed to the metastatic phenotype of
colon cancer in nude mice (32).
This finding is in accordance with our current observation that
miR-22 expression inhibited colorectal cancer cell viability and
invasion, possibly via downregulation of TIAM1 expression. However,
further confirmation is required to determine whether these changes
that occur in colorectal HCT-116 cells upon miR-22 transfection are
due to decreases in TIAM1 expression. Thus, in future studies, we
will manipulate TIAM1 expression in miR-22-transfected tumor cells
to demonstrate that TIAM1 mediates miR-22 effects on colon cancer
cell viability and invasion.
Aside from TIAM1, the present study also showed that
gene expression of MMP-2, MMP-9 and VEGF was significantly reduced
upon transfection with miR-22 expression in colon cancer cells. To
date, there are no studies showing the association between TIAM1
and MMP-2 and MMP-9 expression; however, a previous study
demonstrated that TIAM1 promoted lymphangiogenesis by regulating
Rac/COX-2/VEGF-C signaling (35).
In turn, Rac-1 promotes homophilic complex formation of MT-MMP at
the lamellipodia and promotes cell migration (36,37).
Therefore, miR-22 might target TIAM1, which in turn leads to
defective MMP-2 and MMP-9 activation and cellular invasion of colon
cancer cells. Since both TIAM1 and VEGF expression were compromised
upon expression of miR-22, we proposed that the inhibitory action
of miR-22 on colon cancer proliferation is mediated via TIAM1,
which then regulates VEGF expression. Further studies are required
to confirm this hypothesis.
In conclusion, our data suggest that the
pharmacological manipulation of miR-22 expression may be a tool for
the diagnosis and treatment of colon cancer. However, as this study
is proof-of-principle, further investigations are required to
firmly establish the role of miR-22 in human colon cancer.
Acknowledgements
The authors thank Medjaden Bioscience Limited for
assisting in the preparation of this manuscript.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nature reviews Genetics.
9:102–114. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nature reviews Cancer.
6:259–269. 2006. View
Article : Google Scholar
|
|
5
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Slack FJ and Weidhaas JB: MicroRNA in
cancer prognosis. N Engl J Med. 359:2720–2722. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Grimson A, Farh KK, Johnston WK,
Garrett-Engele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: determinants beyond seed pairing. Mol Cell.
27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ling B, Wang GX, Long G, Qiu JH and Hu ZL:
Tumor suppressor miR-22 suppresses lung cancer cell progression
through post-transcriptional regulation of ErbB3. J Cancer Res Clin
Oncol. 138:1355–1361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pandey DP and Picard D: miR-22 inhibits
estrogen signaling by directly targeting the estrogen receptor
alpha mRNA. Mol Cell Biol. 29:3783–3790. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiong J, Yu D, Wei N, et al: An estrogen
receptor alpha suppressor, microRNA-22, is downregulated in
estrogen receptor alpha-positive human breast cancer cell lines and
clinical samples. FEBS J. 277:1684–1694. 2010. View Article : Google Scholar
|
|
11
|
Patel JB, Appaiah HN, Burnett RM, et al:
Control of EVI-1 oncogene expression in metastatic breast cancer
cells through microRNA miR-22. Oncogene. 30:1290–1301. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang J, Yang Y, Yang T, et al:
microRNA-22, downregulated in hepatocellular carcinoma and
correlated with prognosis, suppresses cell proliferation and
tumourigenicity. Br J Cancer. 103:1215–1220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li J, Liang S, Jin H, Xu C, Ma D and Lu X:
Tiam1, negatively regulated by miR-22, miR-183 and miR-31, is
involved in migration, invasion and viability of ovarian cancer
cells. Oncol Rep. 27:1835–1842. 2012.PubMed/NCBI
|
|
14
|
Li J, Liang S, Yu H, Zhang J, Ma D and Lu
X: An inhibitory effect of miR-22 on cell migration and invasion in
ovarian cancer. Gynecol Oncol. 119:543–548. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li J, Zhang Y, Zhao J, Kong F and Chen Y:
Overexpression of miR-22 reverses paclitaxel-induced
chemoresistance through activation of PTEN signaling in p53-mutated
colon cancer cells. Mol Cell Biochem. 357:31–38. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bar N and Dikstein R: miR-22 forms a
regulatory loop in PTEN/AKT pathway and modulates signaling
kinetics. PloS One. 5:e108592010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yamakuchi M, Yagi S, Ito T and Lowenstein
CJ: MicroRNA-22 regulates hypoxia signaling in colon cancer cells.
PloS One. 6:e202912011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Alvarez-Diaz S, Valle N, Ferrer-Mayorga G,
et al: MicroRNA-22 is induced by vitamin D and contributes to its
antiproliferative, antimigratory and gene regulatory effects in
colon cancer cells. Hum Mol Genet. 21:2157–2165. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Strumane K, Rygiel TP and Collard JG: The
Rac activator Tiam1 and Ras-induced oncogenesis. Methods Enzymol.
407:269–281. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jin H, Li T, Ding Y, et al: Methylation
status of T-lymphoma invasion and metastasis 1 promoter and its
overexpression in colorectal cancer. Hum Pathol. 42:541–551. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang J, Ye X, Guan J, et al: Tiam1 is
associated with hepatocellular carcinoma metastasis. Int J Cancer.
132:90–100. 2012. View Article : Google Scholar
|
|
22
|
Yang W, Lv S, Liu X, Liu H and Hu F:
Up-regulation of Tiam1 and Rac1 correlates with poor prognosis in
hepatocellular carcinoma. Jpn J Clin Oncol. 40:1053–1059. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ding Y, Chen B, Wang S, et al:
Overexpression of Tiam1 in hepatocellular carcinomas predicts poor
prognosis of HCC patients. Int J Cancer. 124:653–658. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao L, Liu Y, Sun X, He M and Ding Y:
Overexpression of T lymphoma invasion and metastasis 1 predict
renal cell carcinoma metastasis and overall patient survival. J
Cancer Res Clin Oncol. 137:393–398. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu H, Shi G, Liu X, Wu H, Fan Q and Wang
X: Overexpression of Tiam1 predicts poor prognosis in patients with
esophageal squamous cell carcinoma. Oncol Rep. 25:841–848.
2011.PubMed/NCBI
|
|
26
|
Qi Y, Huang B, Yu L, Wang Q, Lan G and
Zhang Q: Prognostic value of Tiam1 and Rac1 overexpression in
nasopharyngeal carcinoma. ORL J Otorhinolaryngol Relat Spec.
71:163–171. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Minard ME, Kim LS, Price JE and Gallick
GE: The role of the guanine nucleotide exchange factor Tiam1 in
cellular migration, invasion, adhesion and tumor progression.
Breast Cancer Res Treat. 84:21–32. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang HM and Wang J: Expression of Tiam1 in
lung cancer and its clinical significance. Asian Pac J Cancer Prev.
13:613–615. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Moriarty CH, Pursell B and Mercurio AM:
miR-10b targets Tiam1: implications for Rac activation and
carcinoma migration. J Biol Chem. 285:20541–20546. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cottonham CL, Kaneko S and Xu L: miR-21
and miR-31 converge on TIAM1 to regulate migration and invasion of
colon carcinoma cells. J Biol Chem. 285:35293–35302. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Deryugina EI and Quigley JP: Matrix
metalloproteinases and tumor metastasis. Cancer Metastasis Rev.
25:9–34. 2006. View Article : Google Scholar
|
|
32
|
Patan S: Vasculogenesis and angiogenesis.
Cancer Treat Res. 117:3–32. 2004. View Article : Google Scholar
|
|
33
|
Choong ML, Yang HH and McNiece I: MicroRNA
expression profiling during human cord blood-derived CD34 cell
erythropoiesis. Exp Hematol. 35:551–564. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xiong J, Du Q and Liang Z:
Tumor-suppressive microRNA-22 inhibits the transcription of
E-box-containing c-Myc target genes by silencing c-Myc binding
protein. Oncogene. 29:4980–4988. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhong D, Li Y, Peng Q, et al: Expression
of Tiam1 and VEGF-C correlates with lymphangiogenesis in human
colorectal carcinoma. Cancer Biol Ther. 8:689–695. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhuge Y and Xu J: Rac1 mediates type I
collagen-dependent MMP-2 activation. role in cell invasion across
collagen barrier. J Biol Chem. 276:16248–16256. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Itoh Y, Takamura A, Ito N, et al:
Homophilic complex formation of MT1-MMP facilitates proMMP-2
activation on the cell surface and promotes tumor cell invasion.
EMBO J. 20:4782–4793. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shi TY, Cheng X, Yu KD, et al: Functional
variants in TNFAIP8 associated with cervical cancer susceptibility
and clinical outcomes. Carcinogenesis. January 8–2013.(Epub ahead
of print).
|
|
39
|
Zhang G, Xia S, Tian H, et al: Clinical
significance of miR-22 expression in patients with colorectal
cancer. Med Oncol. 29:3108–3112. 2012. View Article : Google Scholar : PubMed/NCBI
|