Introduction
Colorectal cancer (CRC) continues to be a leading
malignancy in gastrointestinal tumors. It has been reported that
CRC is the fourth most common cancer and the second leading cause
of cancer-related mortality in the United States (1–3).
Although significant progress has been made in the treatment of
CRC, it is still difficult to treat advanced CRC, which has a poor
prognosis and a 40% overall mortality rate (4,5). Thus,
understanding the molecular mechanisms of CRC progression may help
to develop more efficient therapy strategies for the disease.
The Wnt/β-catenin pathway involved in the early
development process of various types of cancer is also involved in
the development of CRC (6,7). The key step in activating the Wnt
pathway is the stabilization of β-catenin and its translocation
into the nucleus, which may serve as a potential target for CRC
therapy (8). Therefore, clarifying
the mechanism of β-catenin regulation may aid in the diagnosis and
treatment of CRC (9–11). The latest research shows that
pancreatic adenocarcinoma upregulated factor (PAUF), a novel
secretory protein, may be one of the regulatory genes of β-catenin
(12).
PAUF, as a newly discovered oncogene, has been
reported to play its oncogenic role by upregulating the expression
of β-catenin and increasing its transcriptional activity in
pancreatic cancer (13).
Researchers also found that PAUF is highly expressed in pancreatic
cancer tissues as well as in colon, ovary and stomach cancer
tissues (12,13). However, in CRC, whether PAUF also
play its oncogenic role through the increase of β-catenin
expression and upregulation of its transcriptional activity remains
unknown.
In the present study, we detected the expression of
PAUF and β-catenin in CRC and investigated whether PAUF affected
the biological behavior of CRC through the adjustment of the
Wnt/β-catenin pathway by RNA interference.
Materials and methods
Reagents
Rabbit anti-human PAUF antibody, rabbit anti-human
β-catenin antibody, rat anti-human β-actin antibody and goat
anti-rabbit IgG secondary antibody were purchased from Santa Cruz
Biotechnology, Inc. (Santa Cruz, CA, USA). Lipofectamine™ 2000 was
provided by Invitrogen Life Technologies (Carlsbad, CA, USA).
RT-PCR kits were purchased from Fermentas (USA). Small interfering
RNA (siRNA)-PAUF were designed and synthesized by GenePharma Co.,
Ltd. (Shanghai, China). Annexin V apoptosis kit (Keygentec, China)
and Matrigel (BD Biosciences, San Jose, CA, USA) were used in the
present study.
Cell lines and tissue samples
The human CRC cell lines (SW480, LoVo, SW1116, SW620
and HCT116) were purchased from the Chinese Academy of Sciences
(Shanghai, China). The cell lines were seeded in 6-well plates at a
density of 1.5×105/well and maintained in RPMI-1640
(Invitrogen Life Technologies) supplemented with 10% fetal bovine
serum (FBS; Sijiqing Biological Engineering Materials Co.,
Hangzhou, China). All cells were cultured at 37°C in a humidified
atmosphere containing 5% CO2. Surgical specimens of CRC
tissue and their corresponding adjacent non-tumor tissues (NATs)
(at least 2 cm distance from the tumor site) were obtained from 48
patients at the Department of General Surgery, the First Affiliated
Hospital of Soochow University, from 2008–2010. All patients had a
clear histological diagnosis of CRC, based on the
clinicopathological criteria described by the UICC. Informed
consent was obtained from all the patients and research protocols
were approved by the Independent Ethics Committee (IEC) of our
hospital.
Immunohistochemical analysis
For immunohistochemical analysis, samples were fixed
in 10% neutral formaldehyde, embedded in paraffin and sliced.
Briefly, the paraffin-embedded tissues were serially cut into 4 μm
sections, dewaxed and rehydrated. Sections were then blocked with
peroxide and non-immune animal serum and incubated sequentially
with rat anti-human PAUF and β-catenin (1:1,000), and
biotin-labeled goat anti-rabbit IgG (1:1,000). Finally, the
sections were stained with DBA, counterstained with hematoxylin,
dehydrated, cleared in xylene, and fixed. Histological assessment
was performed as previously described (2). Immunostaining was independently
examined by two clinical pathologists. Five high-power fields (x400
magnification) were randomly counted for each section. The brown
staining on the cytoplasm was read as positive reactivity for PAUF
and β-catenin. The presence of brown colored granules on the
cytoplasm was considered a positive signal, and was divided by
color intensity into not colored, light yellow, brown, tan, and was
recorded as 0, 1, 2, 3, respectively. We also selected five
high-power fields from each slice and scored them. A positive cell
rate of <25% was a score of 1, a positive cell rate of 25–50%
was a score of 2, a positive cell rate of 51–75% was a score of 3,
and a positive cell rate of >75% was a score of 4. The final
score was determined by multiplying positive cell rate and score
values: 0 was equal to negative (−), 1–4 was equal to weakly
positive (+), 5–8 was equal to moderate positive (++), 9–12 was
equal to strongly positive (+++).
Transfection of cells with
PAUF-siRNA
PAUF-siRNA sequences were designed and synthesized
by GenePharma Co., Ltd. (Table I),
which included 3 sets of 25-nucleotide stealth RNAi targeting PAUF,
and a fluorescently labeled siRNA oligos segment which was used to
detect the transfection efficacy by flow cytometric analysis
(FACS). HCT116 cells (1.5×105) were divided into 6
groups: (i) PAUF-siRNA1, (ii) PAUF-siRNA2, (iii) PAUF-siRNA3, (iv)
scrambled siRNA and (v) non-siRNA control. Cells were transfected
with PAUF-siRNA or negative control siRNA using Lipofectamine 2000
(Invitrogen Life Technologies), according to the manufacturer’s
protocol. Cells were exposed to siRNA in Dulbecco’s modified
Eagle’s medium (DMEM) for 6 h, after which the medium was replaced
with DMEM containing 10% FBS and the cells were incubated for 48 h.
Our preliminary study confirmed that the maximal transfection
efficacy is obtained when the ratio of Lipofectamine 2000 to siRNA
is 4 μl:4 μl. At 48 h post-transfecion, cells were harvested and
subjected to total RNA extraction and adhesion, migration and
invasion assay. Secreted protein was prepared from culture medium
at 48 h post-transfection.
 | Table ISynthesized oligonucleotide sequences
for siRNA. |
Table I
Synthesized oligonucleotide sequences
for siRNA.
| Primer | Sequence |
|---|
| PAUF-siRNA1 |
| Sense |
5′-UCCCUGGGUAUUCCCACCUAAGGCU-3′ |
| Antisense |
5′-AGCCUUAGGUGGGAAUACCCAGGAA-3′ |
| PAUF-siRNA |
| Sense |
5′-AAAUAGAAAUAGCGGUCCUUGCUGG-3′ |
| Antisense |
5′-CCAGCAAGGACCGCUAUUUCUAUUU-3′ |
| PAUF-siRNA |
| Sense |
5′-GAGUAUGUGAGAUUAACUGGUGGC-3′ |
| Antisense |
5′-GCCACCAGUUAAUCUCACAUACUCA-3′ |
| FAM-negative-
control |
| Sense |
5′-UUCUCCGAACGUGUCACGUTT-3′ |
| Antisense |
5′-ACGUGACACGUUCGGAGAATT-3′ |
| Negative-control |
| Sense |
5′-UUCUCCGAACGUGUCACGUTT-3′ |
| Antisense |
5′-ACGUACACGUUCGGAGAATT-3′ |
RT-PCR and real-time PCR gene expression
analysis
The mRNA expression of PAUF and β-catenin in HCT116
cells following PAUF-siRNA transfection were quantified by RT-PCR.
Total RNA was isolated from cells using TRIzol Reagent (Invitrogen
Life Technologies) and quantified. cDNA was synthesized from 5 μg
of RNA using AMV reverse transcriptase (Fermentas) according to the
manufacturer’s instructions. PAUF and β-catenin were amplified from
the cDNA by RT-PCR. The PCR conditions consisted of 5 min at 95°C 1
cycle, 30 sec at 95°C, 30 sec at 55°C, 30 sec at 72°C and 7 min at
72°C 35 cycles. The primer sequences were: PAUF forward,
5′-CACCTGGGCAGGGAAGATGTA-3′ and reverse,
5′-GCTCAGTGGTCGGCTCCTCT-3′; β-catenin forward,
5′-TGCCAAGTGGGTGGTATAGAG-3′ and reverse,
5′-TGG GATGGTGGGTGTAAGAG-3′; β-actin forward,
5′-AACTC CATCATGAAGGGTTGTGA-3′ and reverse,
5′-ACTCCTG CTTGCTGATCCAC-3′.
The mRNA expression of PAUF and β-catenin in 12 CRC
samples was quantified by real-time PCR. A total of 200 mg of tumor
tissues or NATs was homogenized in liquid nitrogen. Total RNA was
extracted and reverse transcribed into cDNA, which was then used
for amplification of PAUF and β-catenin. The real-time PCR
conditions consisted of 1 cycle at 95°C for 10 min followed by 35
cycles at 95°C for 30 sec, at 55°C for 30 sec and at 72°C for 30
sec. GAPDH was employed as an internal standard. The primer
sequences were: PAUF forward,
5′-CCTGGAGGAGGCAAGTATTTCA-3′ and reverse,
5′-GACCTACAGACACCCGCAGC-3′; β-catenin forward,
5′-TGCCAAGTGGGTGGTATAGAG-3′ and reverse,
5′-TGGGATGGTGGGTGTAAGAG-3′; GAPDH forward,
5′-AGGGGCCATCCACAGTCTTC-3′ and reverse,
5′-AGAA GGCTGGGGCTCATTTG-3′. The 2−ΔΔCT method
was applied to analyze the relative changes in gene expression.
Western blot analysis
After 72 h of transfection, protein was extracted
from HCT116 cells as previously described by Kim et
al(12), and then subjected to
SDS-PAGE. Protein concentrations were transferred onto PVDF
membranes and the membranes were then blocked and incubated with
rabbit anti-human PAUF (1:1,000) or β-catenin antibody (1:1,000) at
4°C overnight. After 3 washes with TBST solution for 10 min, the
membranes underwent hybridization with a goat anti-rabbit IgG
secondary antibody (1:1,000) at 37°C for 1 h. After further
washing, PAUF and β-catenin levels were visualized using an ECL
chemiluminescence kit.
MTT assay
HCT116 cells were digested, re-suspended and seeded
in a 96-well culture plate 6 h after transfection. After 24, 48 and
72 h of incubation, cells were stained with 20 μl MTT solution (5
mg/ml) at 37°C for 4 h and subsequently made soluble in 150 μl
DMSO. The reaction product was quantified by measuring the
absorbance (A) of 490 nm at room temperature.
Flow cytometric analysis
Transfection efficiency was estimated with the use
of fluorescein phosphoramidite (FAM)-antisense
oligodeoxynucleotides by fluorescence-activated cell sorter (FACS).
Cells were transfected with the mixture of Lipofectamine 2000 and
FAM-NC-siRNA according to a pre-set mixing ratio. After successful
transfection, cells were harvested and washed with phosphate
buffered saline (PBS) twice. The maximal transfection efficacy
could then be tested by FACS.
Cells were trypsinized and centrifuged at 1,500
rpm/min for 5 min at 48 h after transfection. Cells were harvested
and washed with PBS twice. Reagents for apoptosis detection were
added and then cells were incubated in the dark for 30 min and
subjected to FACS. Cells were collected, washed with PBS, fixed
with 75% ethanol at 20°C overnight, and centrifuged at 1,500
rpm/min for 5 min. Then, ethanol was removed and cells were washed
with PBS twice. Propidium iodide (PI) and 500 μl of RNase were
added and the cells were incubated in the dark at 4°C for 60 min.
Lastly, cells were subjected to cell cycle analysis by FACS.
Cell invasion assay
Invasion capability in vitro was measured in
Transwell chambers (CoStar Group Inc., Washington, DC, USA)
according to the manufacturer’s protocol. Briefly, the upper
chambers of the Transwell inserts were coated with 100 ml diluted
Matrigel solution at 37°C for 4 h, and then pretreated with
serum-free RPMI-1640 medium at 37°C for 1 h before seeding the
cells at a density of 1×105/well in 100 μl medium with
1% FBS. The lower chambers were filled with 500 ml RPMI-1640 with
10% FBS and HCT116 contained medium as a chemoattractant. The
Transwells were then incubated at 37°C with 5% CO2 for
48 h to allow cells to migrate. At the end of the incubation, the
cells on the upper side of the insert filter were completely
removed by wiping with a cotton swab, and the cells that had
invaded through the Matrigel-coated filter were fixed in ethanol
and stained with crystal violet. For quantification, the cells were
counted under a microscope on 5 random fields at ×100. Dye bound to
the cells was solubilized with 0.1% SDS, and absorbance at 560 nm
was measured. The A560 value represents the number of cells that
had invaded through the Matrigel-coated filter.
Cell adhesion assay
Subsequently, 96-well plates were coated with
Matrigel (2.5 mg/ml) 100 μl, 4°C overnight. Cells exposed to siRNA
for 48 h were seeded at a density of 1×104/100 μl and
then incubated for 80 min. Five duplicate wells were set up for
each group. At the end of the experiment, cells were washed twice
with PBS to remove non-adherent cells. Cells that adhered to the
substrate were stained with crystal violet. Dye bound to adhered
cells was solubilized with 0.1% SDS, and absorbance at 560 nm was
measured.
Cell migration assay
Cell migration assay was performed by the
wound-healing method. Cells (1×105) exposed to
adenovirus or siRNA for 48 h were plated in 6-well plates and grown
to confluence. The monolayer was wounded by scratching with a
sterile pipette tip lengthwise along the chamber. After wounding,
cells were washed twice with PBS and cultured at 37°C for 24 h.
Images were captured immediately after cell wounding (0 h) and 24 h
after cell wounding. Wound width (μm) was measured using OpenLab
software. Wound healing rate = (0 h scratch width - 24 h scratch
width)/0 h scratch width × 100%.
Statistical analysis
Statistical analysis was performed with SPSS17.0
software (SPSS Inc, Chicago, IL, USA). Data are expressed as the
means ± standard deviation (SD). A Student’s t-test was used for
comparisons between groups, and F-test was applied for correlation
analyses. A value of P<0.05 was considered to indicate a
statistically significant difference.
Results
Increased expression of PAUF and
β-catenin in CRC tissues and NATs
To investigate the expression of PAUF and β-catenin
in CRC, we detected the expression of PAUF and β-catenin in fresh
CRC tissues and NATs from the same patient. Real-time PCR analysis
showed that mRNA expression levels of PAUF and β-catenin in tumor
tissues were significantly higher than in NATs (P<0.05)
(Fig. 1B and C). In addition, the
expression of PAUF was correlated with the expression of β-catenin
in both tumor tissues (r=0.658, P<0.05) and NATs (r=0.732,
P<0.05).
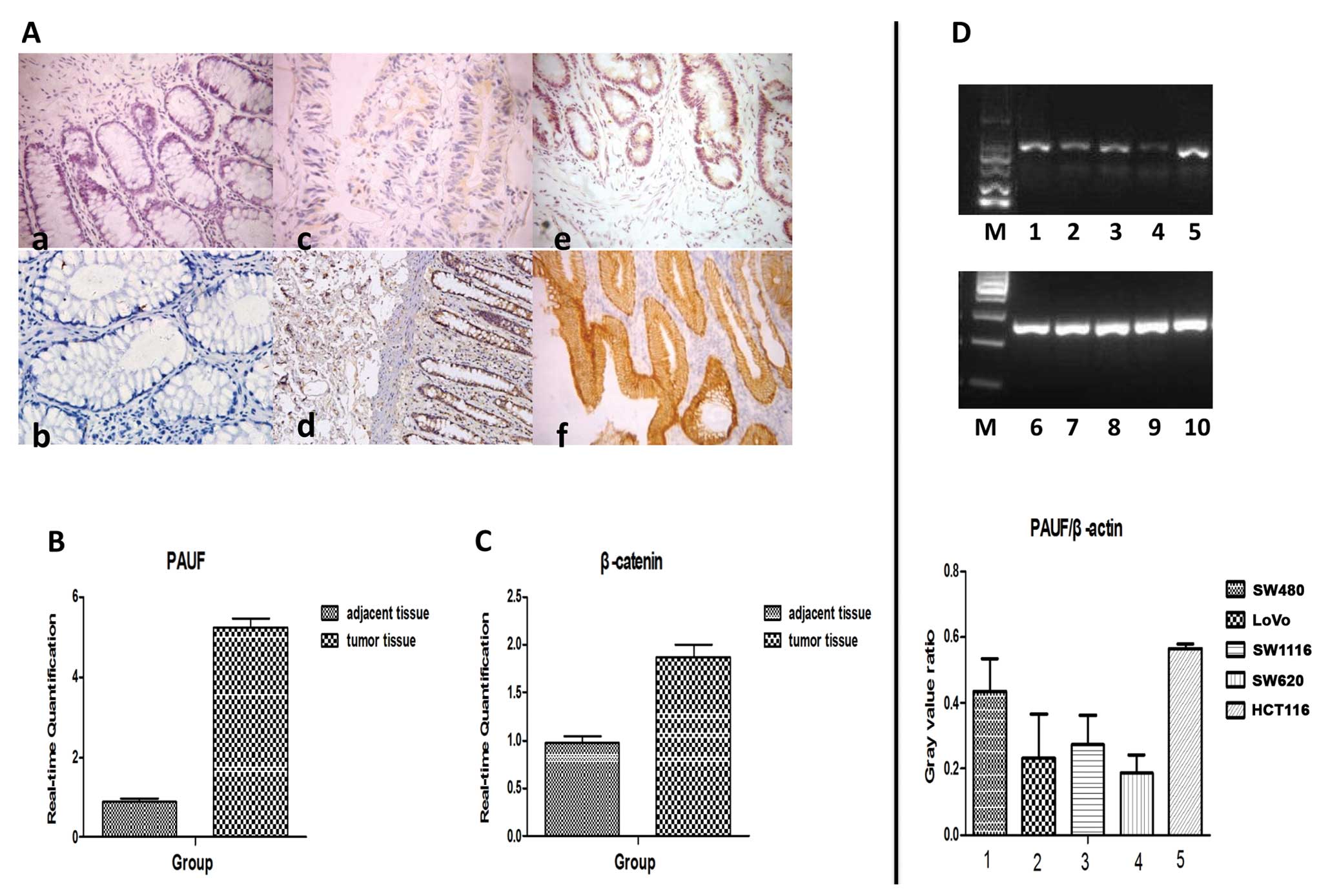 | Figure 1The expression of pancreatic
adenocarcinoma upregulated factor (PAUF) and β-catenin in
colorectal cancer (CRC), adjacent non-tumor tissues (NATs) and CRC
cells. (A) Immunohistochemical analysis of CRC and NATs for
expression of PAUF and β-catenin protein. The expression of PAUF
(a,c,e) and β-catenin (b,d,f) was stained brown and was present in
cancer tissue and NATs. Representative sites with negative (a,
×400), moderate positive (c, ×400), strongly positive (e, ×400)
expression of PAUF and corresponding weakly positive (b, ×400),
moderate positive (d, ×400), strongly positive (f, ×400) expression
of β-catenin. (B and C) Relative mRNA expression of PAUF and
β-catenin in CRC and NATs detected by real-time PCR. (D) Relative
expression of PAUF mRNA in CRC cells detected by RT-PCR: Lanes
1,2,3,4,5 represent PAUF mRNA expression of SW480, LoVo, SW1116,
SW620 and HCT116, respectively. Lanes 6–10 represent corresponding
internal reference. M, marker. Results are given as average value
of the gray in 3 target genes and internal controls from 3
independent experiments. |
Consistent with these results, immunohistochemical
analysis showed strong cytoplasmic overexpression of PAUF and
β-catenin in cancer cells was clearly observed; however, NATs did
not react with the antibody (Fig.
1A). Of the 48 CRCs, the positive rate of PAUF and β-catenin
was 62.5% (30/48) and 91.7% (44/48). However, in NATs, the positive
rate was 16.7% (8/48) and 81.3% (39/48), respectively. The
difference between them was statistically significant (P<0.05).
As anticipated, the expression of PAUF was also correlated with the
expression of β-catenin in the tumor tissues (r=0.355, P<0.05)
and NATs (r=0.416, P<0.05).
Transfection of CRC cells with PAUF-siRNA
inhibits mRNA and protein expression of PAUF and β-catenin
RT-PCR results showed that PAUF was expressed in all
5 CRC cell lines (SW480, LoVo, SW1116, SW620 and HCT116) at a high
frequency, and the HCT116 cell line showed the highest elevation of
PAUF mRNA among the 5 cell lines (Fig.
1D). In order to achieve a significant interference effect, we
used the HCT116 cell line for the follow-up experiment.
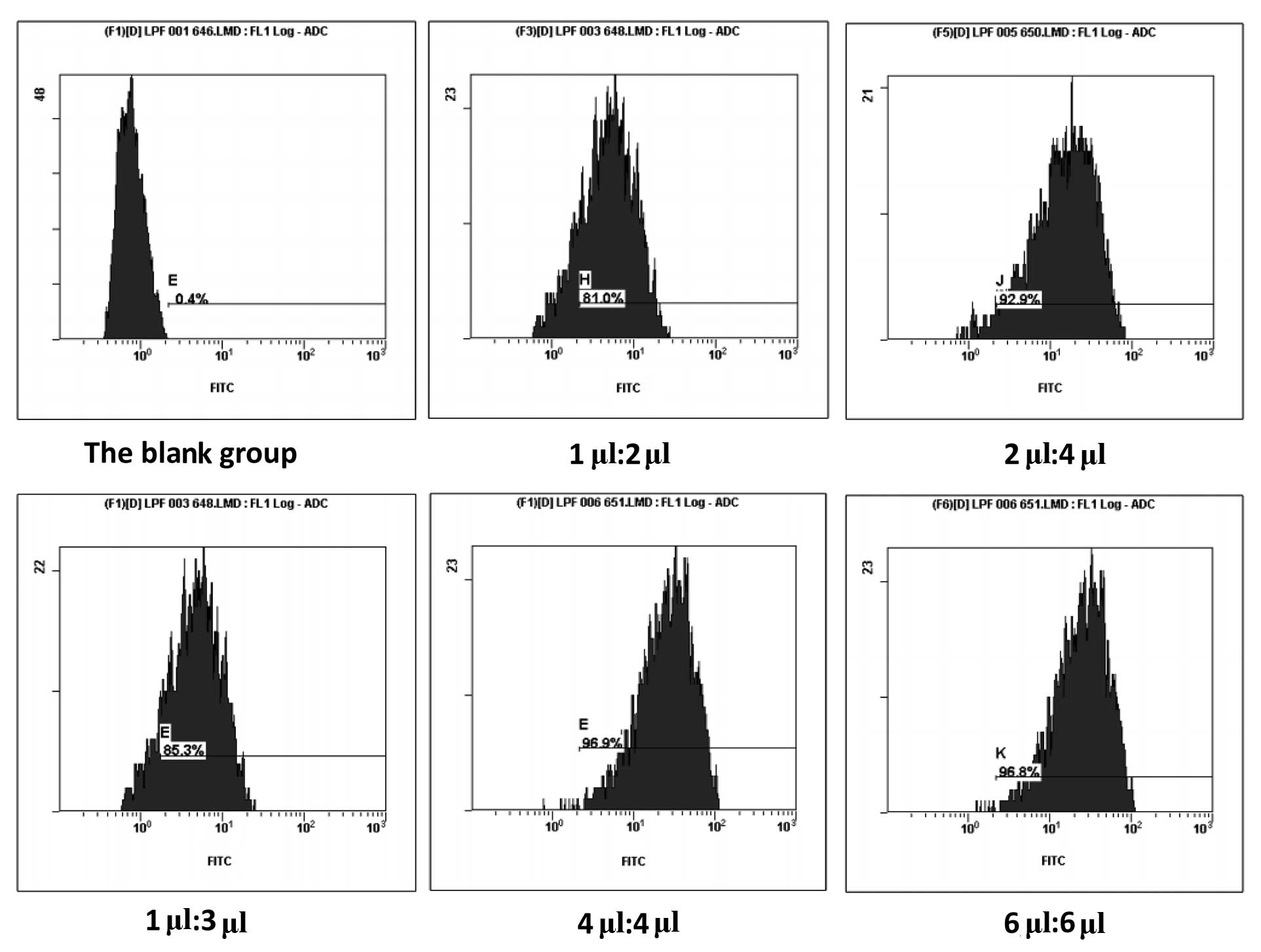
A fluorescently labeled siRNA oligos segment was
used to detect the transfection efficacy by FACS. Our study
confirmed that the maximal transfection efficacy could be obtained
when the ratio of Lipofectamine 2000 to siRNA was 4 μl:4 μl.
Increase of the reagents did not improve transfection efficiency
(Fig. 2). Three siRNA oligos
segments targeting PAUF were designed to knock down PAUF (Table I).
To test the efficacy of PAUF siRNA in downregulating
the expression of PAUF, we evaluated the PAUF expression in
transfected cells at the transcript and protein level. The results
demonstrated that mRNA and protein expression of PAUF were reduced
by the 3 siRNA oligos segments to varying degrees, compared with
the scrambled siRNA group and the non-siRNA group (P<0.05) and
the no. 2 siRNA oligos segment (PAUF-siRNA2 group) was the most
efficient. However, there was no significant difference between the
scrambled siRNA group and the non-siRNA group (Fig. 3A and C). In addition, PAUF-siRNA
transfection downregulated the expression of PAUF in HCT116 cells,
which was also associated with a decrease in the expression of
β-catenin, and the PAUF-siRNA2 group was also the most efficient
(Fig. 3B and D). In order to
achieve a significant interference effect, PAUF-siRNA2 was selected
for subsequent experiments.
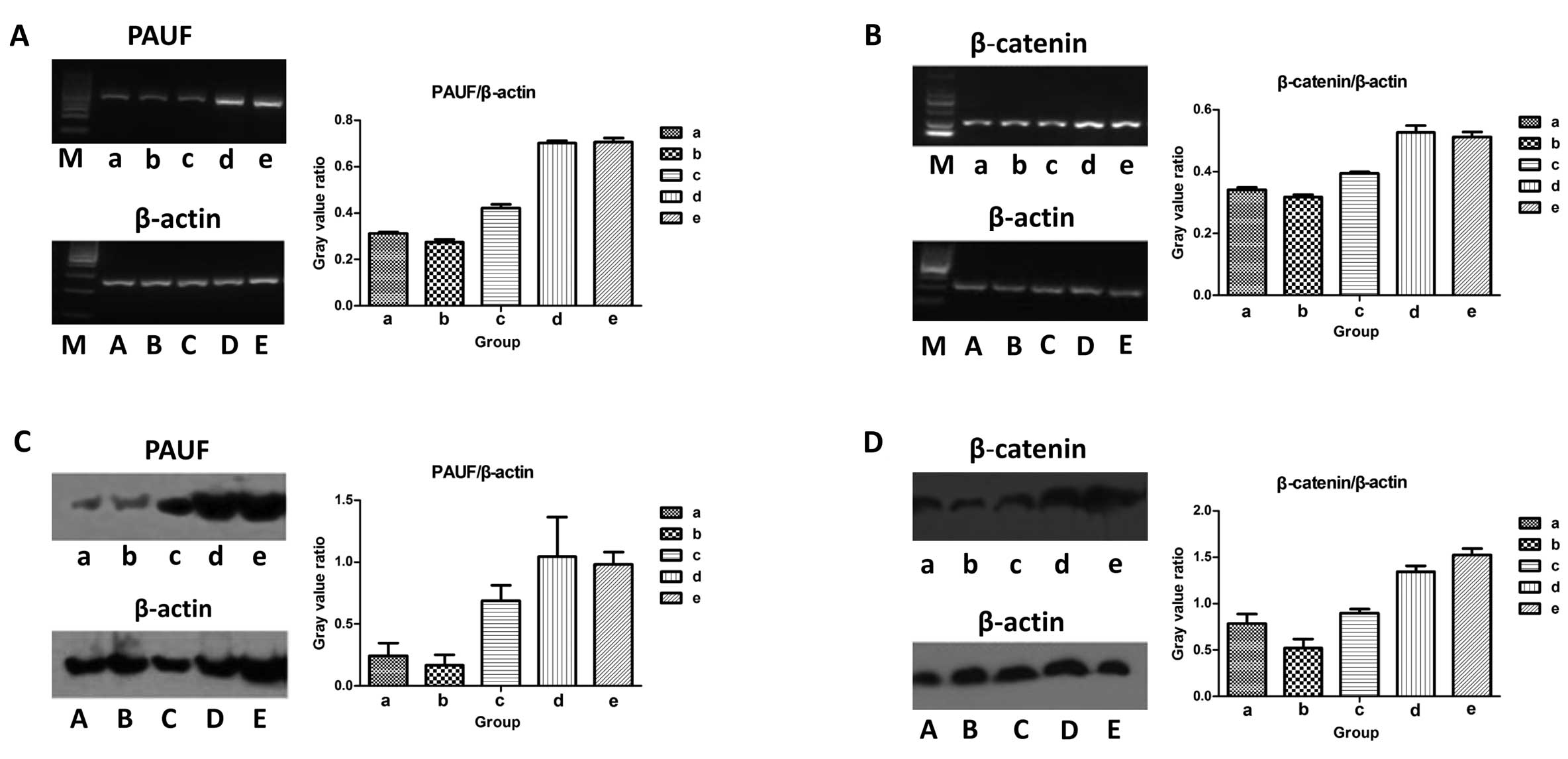 | Figure 3Pancreatic adenocarcinoma upregulated
factor (PAUF) and β-catenin expression level in PAUF-small
interfering RNA (siRNA) transfected HCT116 cells. (A and B) PAUF
and β-catenin mRNA expression of PAUF-siRNA transfected HCT116
cells detected by RT-PCR. After 48 h of PAUF-siRNA transfection,
RT-PCR was performed to detect PAUF (A) and β-catenin (B) mRNA
expression. (a) PAUF-siRNA1 group; (b) PAUF-siRNA2 group; (c)
PAUF-siRNA3 group; (d) scrambled siRNA group; (e) non-siRNA group.
Lanes A,B,C,D,E represent corresponding internal reference. M,
marker. Results are given as average value of the gray in 3 target
genes and internal controls from 3 independent experiments. (C and
D) Representative western blot analysis of PAUF and β-catenin
expression level in PAUF-siRNA transfected HCT116 cells. After 48 h
of PAUF-siRNA transfection of HCT116 cells, protein expression of
PAUF (C) and β-catenin (D) was determined by western blot assay.
(a) PAUF-siRNA1 group; (b) PAUF-siRNA2 group; (c) PAUF-siRNA3
group; (d) scrambled siRNA group; (e) non-siRNA group. Lanes
A,B,C,D,E represent corresponding internal reference. Results are
given as average value of the gray in 3 target genes and internal
controls from 3 independent experiments. |
Effect of PAUF-siRNA transfection on the
growth of HCT116 cells
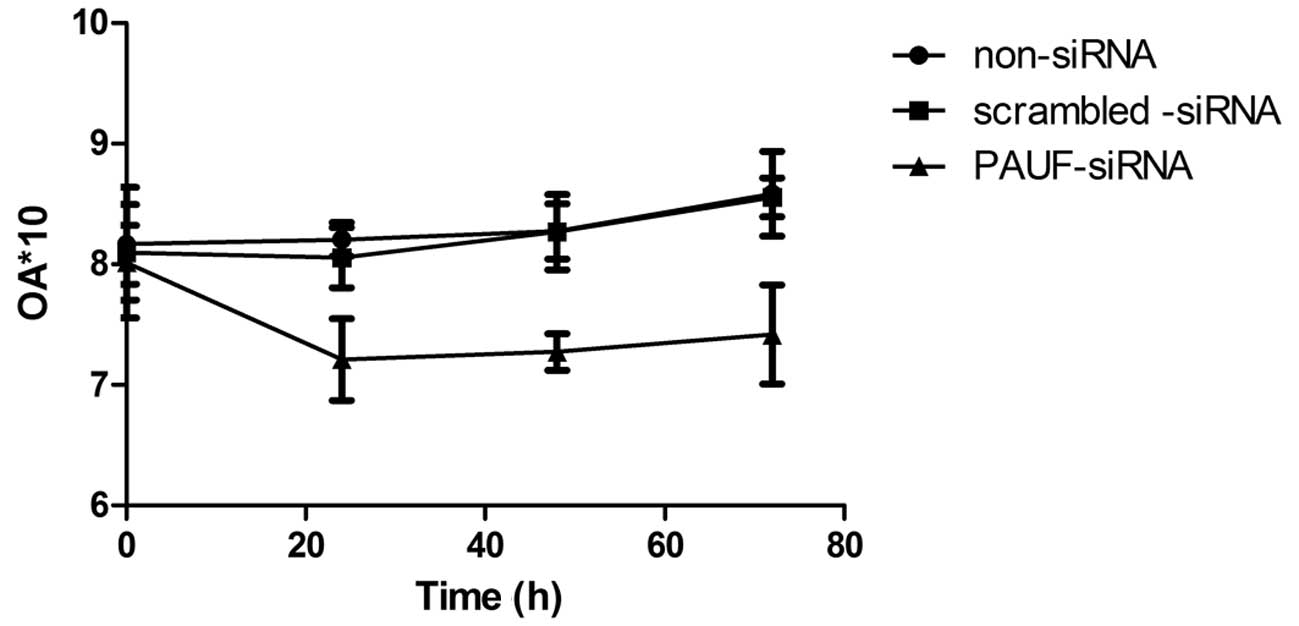
In phenotype analysis, we investigated the effect of
PAUF on cell growth of HCT116 cells. The cell viability was reduced
significantly after treatment with PAUF siRNA at 24, 48 and 72 h
compared with other control groups (P<0.05) (Fig. 4). The results indicated that RNA
interference mediated specific downregulation of PAUF induced
strong inhibition of CRC cell growth.
Effect of PAUF-siRNA transfection on the
apoptosis and cell cycle of HCT116 cells
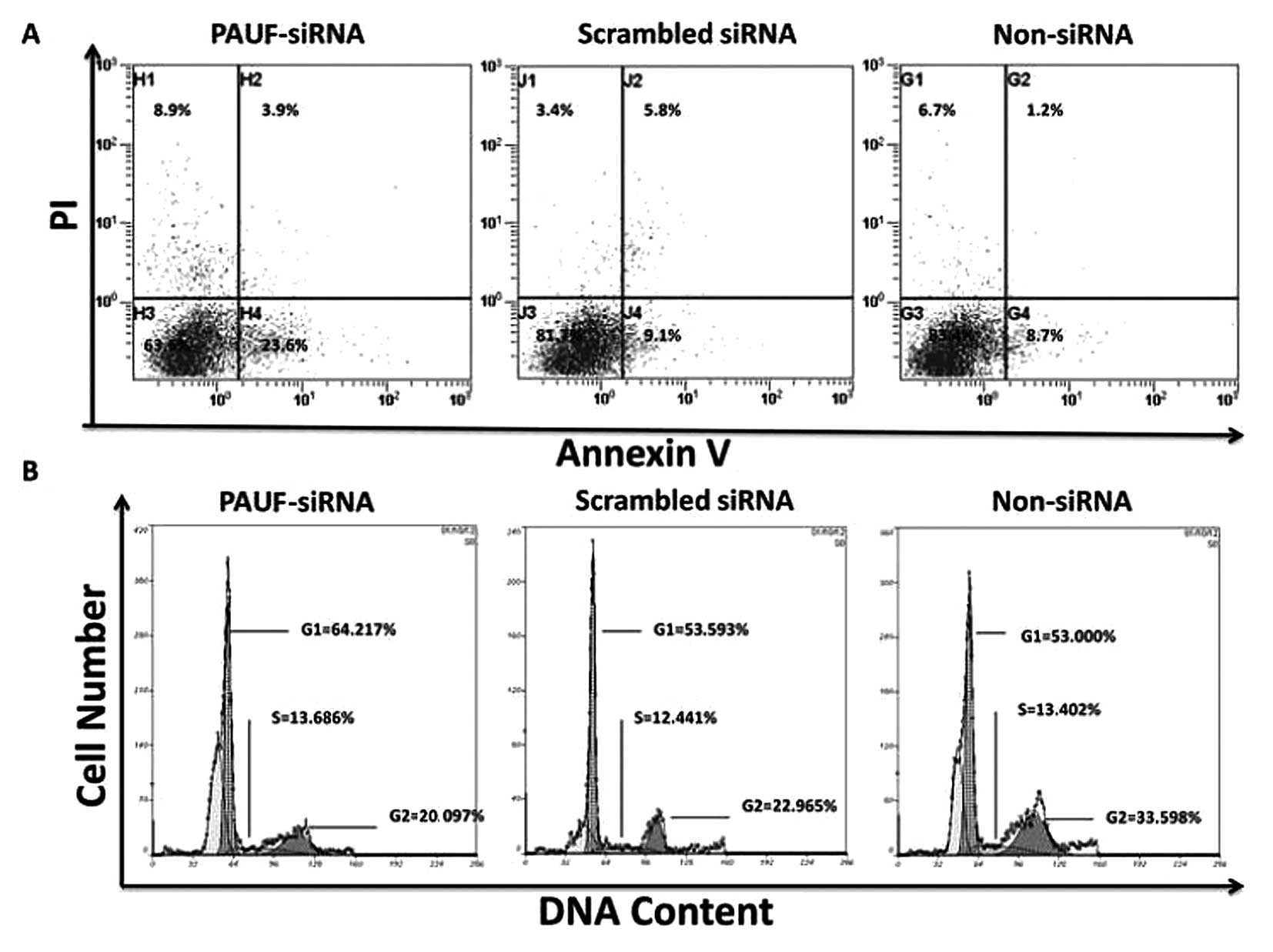
We performed experiments to evaluate the apoptosis
and cell cycle distribution of HCT116 cells with PAUF-siRNA by PI
staining. The results showed that following 48 h of transfection,
the rate of apoptosis in the PAUF-siRNA group (36.4±1.24%) was
significantly higher than that of the scrambled siRNA (18.3±1.74%)
and non-siRNA groups (16.6±1.84%) (P<0.01) (Fig. 5A). On the other hand, the cell cycle
analysis in the PAUF-siRNA group showed increased G0/G1 phase cells
(66.59±4.15%, P<0.05) and decreased percentage of S and G2/M
(20.97±2.15%, P<0.05) phase cells.
Effect of PAUF-siRNA transfection on the
invasion, adhesion and migration of HCT116 cells
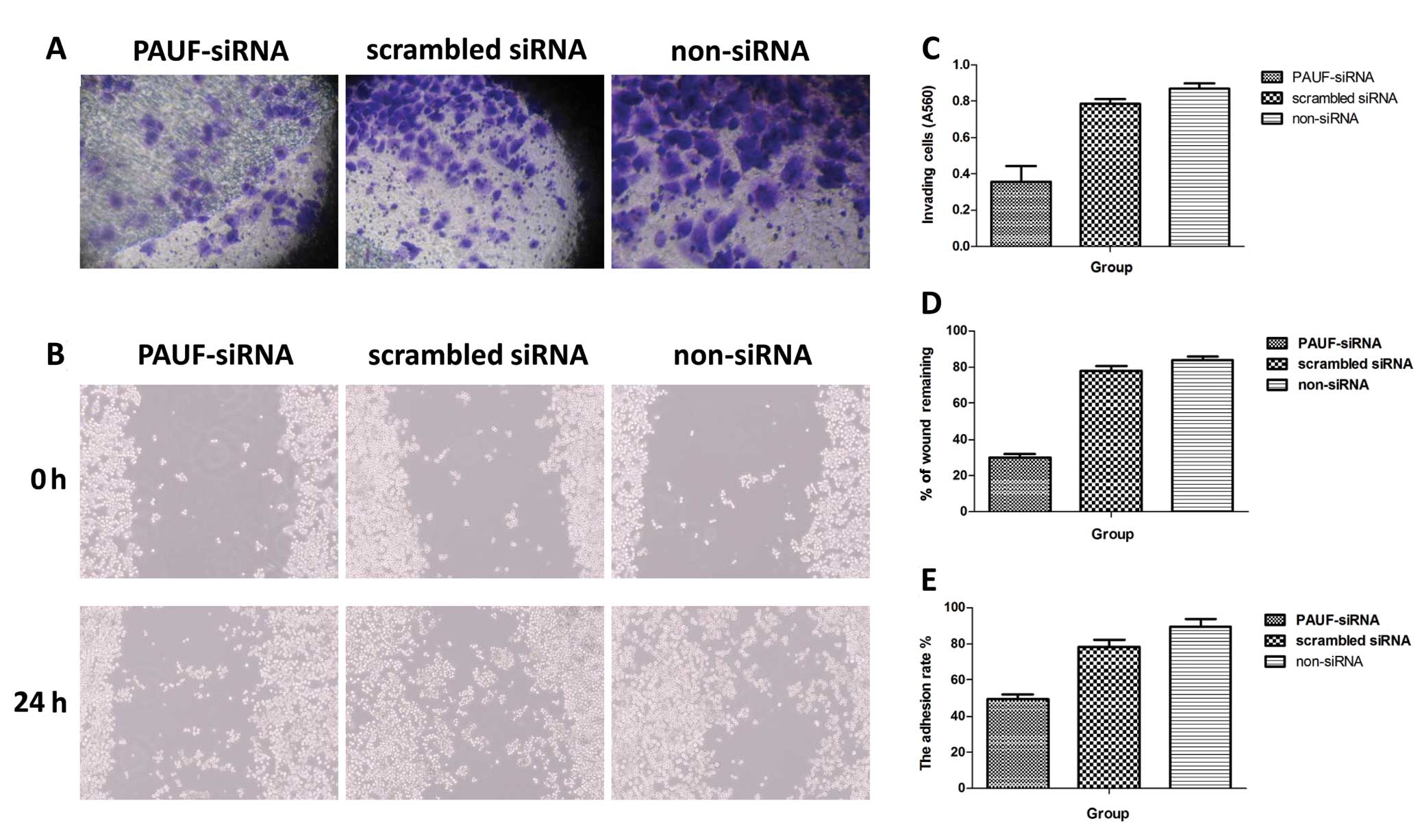
Invasion capability was measured in Transwell
chambers (CoStar Group Inc.) according to the manufacturer’s
protocol. The cells that had invaded through the Matrigel-coated
filter were fixed in ethanol and stained with crystal violet. The
cells were counted under a microscope on 5 random fields at ×100
(Fig. 6A). Dye bound to the cells
was solubilized with 0.1% SDS, and absorbance at 560 nm was
measured. The A560 value represents the number of cells that had
invaded through the Matrigel-coated filter. The histograms
represent the quantification of cells that invaded through Matrigel
(Fig. 6C).
Changes in adhesion were evaluated by cell adhesion
assay. The A560 value represents the number of cells that adhered
to the Matrigel. The adhesion rate = the number of adhered
cells/the number of total cells × 100%. Results showed that the
adhesion rate of the PAUF-siRNA transfected group was
(49.52±2.45%), which was significantly lower than that of the
scrambled siRNA (78.23±3.86%) and the non-siRNA group (89.59±4.23%)
(P<0.05) (Fig. 6E).
Cell migration assay was performed by the
wound-healing method. Images were captured immediately after cell
wounding (0 h) and at 24 h after cell wounding (Fig. 6B). Wound healing rate = (0 h scratch
width - 24 h scratch width)/0 h scratch width × 100%. The results
showed that the wound healing rate of the PAUF-siRNA transfected
group was 30.54±3.35%, which was significantly lower than that of
the scrambled siRNA (78.67±2.81%) and the non-siRNA group
(84.79±2.63%) (P<0.01) (Fig.
6D).
Discussion
In the present study, we examined the expression
pattern of PAUF transcript and protein in CRC tissues and cell
lines, and our findings demonstrated that PAUF is highly expressed
in CRC at the transcript and protein level when compared with NATs.
Wee also evaluated the role of PAUF in the biological behavior of
CRC through the adjustment of the Wnt/β-catenin pathway.
The disorder of several genes, and cell signaling
pathways involved in the evolution of CRC, has been extensively
investigated and mutations of key genes in the Wnt/β-catenin
signaling pathway play an important role in the occurrence and
development of CRC (1,14). Furthermore, β-catenin plays a
critical role in this signaling pathway (15,16).
Previous studies have shown that the regulation of β-catenin is
multifactorial (17), such as CDK8
(2,17,18) of
E2F1 (19). Whether other genes
play a decisive role in regulating the stability oβ-catenin remains
to be examined. In the current study, we focused on gene silencing
techniques to detect whether PAUF, a newly discovered oncogene, is
involved in the regulation of β-catenin and whether PAUF
participates in the evolution of CRC.
Several studies have demonstrated that the new
oncogene PAUF is highly expressed in pancreatic cancer, CRC,
ovarian cancer and gastric cancer tissues (12,13),
and there is evidence to show that in pancreatic cancer, PAUF can
contribute to the oncogenesis of pancreatic cells by upregulating
the expression and transcriptional activity of β-catenin (13). PAUF also plays important roles in
cancer progression, including cell proliferation, adhesion,
migration and invasion (20–23).
In our present study, we detected the expression levels of PAUF in
48 cases of CRC by using real-time RT-PCR. Our data demonstrated
that PAUF expression levels were significantly upregulated in
cancer tissues compared to NATs, and the expression of PAUF was
related to the expression of β-catenin in both tumor tissues and
NATs. These data shows that PAUF may be involved in the evolution
of CRC by regulating the Wnt/β-catenin pathway.
In order to obtain a deeper understanding of the
functional mechanism of PAUF in the initiation and progression of
CRC, we employed the RNA interference technique for knockdown of
PAUF expression in HCT116 cells. The RNA interference allows
inexpensive and rapid analysis of gene function in mammals and
represents an effective approach that could be exploited for gene
therapy (24,25). In our study, we observed that PAUF
inhibitor can markedly inhibit the cell proliferation in HCT116
cells. Flow cytometry analysis found that the cell apoptotic rate
in the PAUF inhibitor group was significantly higher than in the NC
group. The cell cycle data also revealed that the cells of S and
G2/M phase in the PAUF inhibitor group were markedly lower than in
the NC group, while cells of the G0/G1 phase were significantly
increased. This suggests that PAUF downregulation may induce G0/G1
cell cycle arrest, more cell apoptosis and the inhibition of cell
proliferation in HCT116 cells. After silencing the expression of
PAUF, we found that PAUF-siRNA not only inhibited the proliferation
of CRC cells, promoted their apoptosis and arrested these cells in
the G0/G1 phase, but also the invasion, adhesion and migration of
the tumor cells were inhibited to varying degrees.
Although the precise molecular mechanisms of PAUF in
CRC have not been fully clarified, our study, for the first time,
illustrated that PAUF could effectively influence the malignant
phenotypes of CRC through the Wnt/β-catenin pathway. A deeper
understanding of its clinical implications and targeted therapeutic
interventions in CRC requires further investigation.
In summary, the current study showed that PAUF is
involved in the progression of CRC development by regulating the
Wnt/β-catenin pathway, and PAUF downregulation with RNA
interference can inhibit proliferation, apoptosis, cell cycle
progression, invasion, adhesion and migration of cancer cells.
These results also suggested that PAUF may serve as an efficient
biomarker for diagnosis and is a novel prognostic indicator in CRC.
Our study represents a potential new approach to understanding the
role of this gene function in cancer and provides a novel strategy
for CRC therapy.
Acknowledgements
This work was supported by Project of Medical
Science and Technology Development Foundation of Jiangsu Province
of PR. China (H201209), Project of Nature Science Foundation of
P.R. China (81201905), Nature Science Research Grants in University
of Jiangsu Province of P.R. China (12KJB320009), Shanghai
Postdoctoral Scientific Program of P.R. China (13R21415200) and the
Science and Technology Research Project of in Science and
Technology Bureau of Suzhou city of P.R. China (SYS201220).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
He SB, Yuan Y, Wang L, Yu MJ, Zhu YB and
Zhu XG: Effects of cyclin-dependent kinase 8 specific siRNA on the
proliferation and apoptosis of colon cancer cells. J Exp Clin
Cancer Res. 30:1092011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wan D, He S, Xie B, Xu G, Gu W, Shen C, Hu
Y, Wang X, Zhi Q and Wang L: Aberrant expression of miR-199a-3p and
its clinical significance in colorectal cancers. Med Oncol.
30:3782013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Markowitz SD and Bertagnolli MM: Molecular
origins of cancer: molecular basis of colorectal cancer. N Engl J
Med. 361:2449–2460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Walther A, Johnstone E, Swanton C, Midgley
R, Tomlinson I and Kerr D: Genetic prognostic and predictive
markers in colorectal cancer. Nat Rev Cancer. 9:489–499. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen HJ, Hsu LS, Shia YT, Lin MW and Lin
CM: The β-catenin/TCF complex as a novel target of resveratrol in
the Wnt/β-catenin signaling pathway. Biochem Pharmacol.
84:1143–1153. 2012.
|
|
8
|
Firestein R and Hahn WC: Revving the
Throttle on an oncogene: CDK8 takes the driver seat. Cancer Res.
69:7899–7901. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luu HH, Zhang R, Haydon RC, Rayburn E,
Kang Q, Si W, Park JK, Wang H, Peng Y, Jiang W and He TC:
Wnt/β-catenin signaling pathway as a novel cancer drug target. Curr
Cancer Drug Targets. 4:653–671. 2004.
|
|
10
|
Dihlmann S and von Knebel Doeberitz M:
Wnt/β-catenin-pathway as a molecular target for future anti-cancer
therapeutics. Int J Cancer. 113:515–524. 2005.
|
|
11
|
Kundu JK, Choi KY and Surh YJ:
β-catenin-mediated signaling: a novel molecular target for
chemoprevention with anti-inflammatory substances. Biochim Biophys
Acta. 1765:14–24. 2006.
|
|
12
|
Kim SA, Lee Y, Jung DE, Park KH, Park JY,
Gang J, Jeon SB, Park EC, Kim YG, Lee B, Liu Q, Zeng W, Yeramilli
S, Lee S, Koh SS and Song SY: Pancreatic adenocarcinoma
up-regulated factor (PAUF), a novel up-regulated secretory protein
in pancreatic ductal adenocarcinoma. Cancer Sci. 100:828–836. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cho IR, Koh SS, Min HJ, Kim SJ, Lee Y,
Park EH, Ratakorn S, Jhun BH, Oh S, Johnston RN and Chung YH:
Pancreatic adenocarcinoma up-regulated factor (PAUF) enhances the
expression of β-catenin, leading to a rapid proliferation of
pancreatic cells. Exp Mol Med. 43:82–90. 2011.PubMed/NCBI
|
|
14
|
Bienz M and Clevers H: Linking colorectal
cancer to Wnt signaling. Cell. 103:311–320. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nelson WJ and Nusse R: Convergence of Wnt,
β-catenin, and cadherin pathways. Science. 303:1483–1487. 2004.
|
|
16
|
Coluccia AM, Benati D, Dekhil H, De
Filippo A, Lan C and Gambacorti-Passerini C: SKI-606 decreases
growth and motility of colorectal cancer cells by preventing
pp60(c-Src)-dependent tyrosine phosphorylation of β-catenin and its
nuclear signaling. Cancer Res. 66:2279–2286. 2006.PubMed/NCBI
|
|
17
|
Seo JO, Han SI and Lim SC: Role of CDK8
and β-catenin in colorectal adenocarcinoma. Oncol Rep. 24:285–291.
2010.
|
|
18
|
Firestein R, Bass AJ, Kim SY, Dunn IF,
Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, Chheda MG,
Tamayo P, Finn S, Shrestha Y, Boehm JS, Jain S, Bojarski E, Mermel
C, Barretina J, Chan JA, Baselga J, Tabernero J, Root DE, Fuchs CS,
Loda M, Shivdasani RA, Meyerson M and Hahn WC: CDK8 is a colorectal
cancer oncogene that regulates β-catenin activity. Nature.
455:547–551. 2008.PubMed/NCBI
|
|
19
|
Morris EJ, Ji JY, Yang F, Di Stefano L,
Herr A, Moon NS, Kwon EJ, Haigis KM, Näär AM and Dyson NJ: E2F1
represses β-catenin transcription and is antagonized by both pRB
and CDK8. Nature. 455:552–556. 2008.
|
|
20
|
Lee Y, Kim SJ, Park HD, Park EH, Huang SM,
Jeon SB, Kim JM, Lim DS and Koh SS: PAUF functions in the
metastasis of human pancreatic cancer cells and upregulates CXCR4
expression. Oncogene. 29:56–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Park HD, Lee Y, Oh YK, Jung JG, Park YW,
Myung K, Kim KH, Koh SS and Lim DS: Pancreatic adenocarcinoma
upregulated factor promotes metastasis by regulating TLR/CXCR4
activation. Oncogene. 30:201–211. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee YS, Kim SJ, Min HJ, Jo JY, Park EH and
Koh SS: PAUF promotes adhesiveness of pancreatic cancer cells by
modulating focal adhesion kinase. Exp Mol Med. 43:291–297. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim YH, Sung HJ, Kim S, Kim EO, Lee JW,
Moon JY, Choi K, Jung JE, Lee Y, Koh SS, Rhee SG, Heo K and Kim IH:
An RNA aptamer that specifically binds pancreatic adenocarcinoma
up-regulated factor inhibits migration and growth of pancreatic
cancer cells. Cancer Lett. 313:76–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Merritt WM, Bar-Eli M and Sood AK: The
dicey role of dicer: implications for RNAi therapy. Cancer Res.
70:2571–2574. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brummelkamp TR, Bemards R and Agami R:
Stable suppression of tumorigenicity by virus-mediated RNA
interference. Cancer Cell. 2:243–247. 2002. View Article : Google Scholar : PubMed/NCBI
|