Introduction
Gastric cancer is one of the most common malignant
tumors in the world. According to data from the National Cancer
Institute (NCI), it is estimated that more than 24,000 patients are
diagnosed with gastric cancer each year in the United States
(1). Patients with advanced gastric
cancer still face a poor prognosis. Identification of genes
responsible for the development and progression of gastric cancer
and a clear understanding of the clinical significance of these
genes are both important for the diagnosis and adequate treatment
of this disease.
Chromatin remodeling is a key mechanism regulating
gene expression. The chromodomain helicase DNA-binding (CHD) family
comprises a group of chromatin remodeling enzymes that use the
energy from ATP hydrolysis to alter the structure or position of
the nucleosome (2–5). In humans, 9 CHD family proteins have
been identified to date (2,5). These proteins share common tandem
chromodomains in the N-terminal region and an ATPase-helicase
domain in the central region (2,5).
Several studies have demonstrated that chromatin remodeling by CHD
family members is involved in the pathogenesis of different types
of cancers. For example, the CHD7 gene is known to be
mutated in small cell lung cancer (SCLC) tissues and SCLC cell
lines express the PVT1-CHD7 fusion gene (6). Moreover, CHD5 has been shown to
control proliferation and apoptosis via the p19–p53 pathway,
functioning as a tumor suppressor (7). In humans, CHD5 is inactivated
not only by deletion, but also by hypermethylation in several types
of cancer (7).
CHD8 was originally isolated as a negative regulator
of the Wnt/β-catenin pathway when it was found to suppress
β-catenin function (8,9). CHD8 has also been suggested to
regulate the expression of various genes, including cyclin
E2 and HOXA2(10–14).
In contrast, CHD8 can also bind to p53 and suppress its
transactivation activity by recruiting histone H1 during
embryogenesis (15). However, the
role of CHD8 in solid malignant tumors has not yet been
elucidated.
In the present study, we analyzed CHD8 mRNA
expression using clinical samples from 101 patients diagnosed with
primary gastric cancer. We then examined the relationship between
CHD8 mRNA expression and clinicopathological factors and
determined the clinical significance of aberrant CHD8
expression. Moreover, we investigated the functional role of CHD8
in gastric cancer by analyzing expression array data in
silico and confirmed the biological significance of CHD8 in
gastric cancer cells in vitro.
Materials and methods
Clinical samples and cell lines
A total of 101 gastric cancer patients were enrolled
in this study. All patients underwent surgery without preoperative
treatments such as chemotherapy and radiotherapy. Tumor and
adjacent normal tissues were obtained. Total RNA was extracted
using the QIAamp DNA Micro Kit (Qiagen) following the
manufacturer’s protocol. Patients were closely observed each month
after surgery and the mean postoperative follow-up period was 2.8
years. Histopathological evaluations were assessed according to the
Japanese Classification of Gastric Cancer, 3rd English edition.
MKN-45 cell lines were provided by the American Type
Culture Collection and were maintained in RPMI-1640 containing 10%
FBS with 100 U/ml penicillin and 100 mg/ml streptomycin. Cells were
cultured in a humidified 5% CO2 incubator at 37°C.
Real-time quantitative reverse
transcription (RT)-PCR
Real-time quantitative RT-PCR was performed using a
LightCycler® System and a LightCycler® 480
Probes Master kit (both from Roche Applied Science, Indianapolis,
IN, USA) following the manufacturer’s protocol with the following
specific CHD8 primers: forward, 5′-AGTGGTGTCTACGTT TGGTGTG-3′ and
reverse, 5′-GATGGGCTCAATGAACAG GT-3′. CHD8 levels were normalized
to GAPDH (primers: forward, 5′-GTCAACGGATTTGGTCTGTATT-3′ and
reverse, 5′-AGTCTTCTGGGTGGCAGTGAT-3′).
siRNA transfections and proliferation
assays
For siRNA knockdown studies, double-stranded RNA
duplexes targeting human CHD8 (5′-AGGAGCGUCCAGUAGAUGAACACG
C-3′/5′-GCGUGUUCAUCUACUGGACGCUCCU-3′; 5′-UU
CAAAUGCUUAAACUUUGGGAUUG-3′/5′-CAAUCCCAA AGUUUAAGCAUUUGAA-3′) were
purchased from Invitrogen (Carlsbad, CA, USA) (Stealth RNAi).
Negative control siRNA (NC) was also purchased from Invitrogen.
MKN-45 and NUGC4 cells were transfected with siRNA at a
concentration of 20 μmol/l using Lipofectamine reagent (RNAiMax) in
glucose-free Opti-MEM (both from Invitrogen). For proliferation
assays, siRNA-, NC- or mock-transfected MKN-45 cells were seeded at
8.0×103 cells/well in 96-well flat-bottomed microtiter
plates in a final volume of 100 μl of culture medium per well.
Cells were incubated in a humidified atmosphere (37°C and 5%
CO2) for 24, 48, 72 or 96 h after initiation of
transfection. The
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay (Roche Diagnostics Corp.) was then used to measure cell
growth inhibition according to standard protocols. Briefly, after
incubation, 10 μl of MTT labeling reagent (final concentration of
0.5 mg/ml) was added to each well, and the plate was incubated for
an additional 4 h in a humidified atmosphere. Solubilization
solution (100 μl) was added to each well, and the plate was
incubated overnight in a humidified atmosphere. After confirming
that the purple formazan crystals were completely solubilized, the
absorbance of each well was measured by a Model 550 Series
Microplate Reader (Bio-Rad Laboratories, Hercules, CA, USA) at a
wavelength of 570 nm corrected to 655 nm. Each sample was run with
6 replicates.
Gene set enrichment analysis (GSEA) of
gastric cancer expression
To investigate CHD8 function in gastric cancer, we
obtained gastric cancer expression profiles from the National
Center for Biotechnology Information (NCBI) Gene Expression Omnibus
(GEO) database (accession code GSE22377) and analyzed these
profiles using GSEA.
Expression profiles were normalized with the ‘affy’
R/BioConductor package (http://www.bioconductor.org/packages/release/bioc/html/affy.html).
We applied a continuous-type CLS file with the CHD8 profile to
phenotype labels in GSEA. The metric for ranking genes was set as
‘Pearson’ and all other parameters were set to their default
values.
Statistical analysis
The significance of differences between 2 groups was
estimated with the Student’s t-test and Chi-square test. Overall
survival curves were plotted according to the Kaplan-Meier method,
with the log-rank test applied for comparison. Variables with a
P-value of <0.05 by univariate analysis were used in subsequent
multivariate analysis on the basis of the Cox proportional hazards
model. All differences were considered statistically significant at
the level of P<0.05. Statistical analyses were conducted using
JMP 5 Software (SAS Institute).
Results
CHD8 expression in gastric cancer
tissues
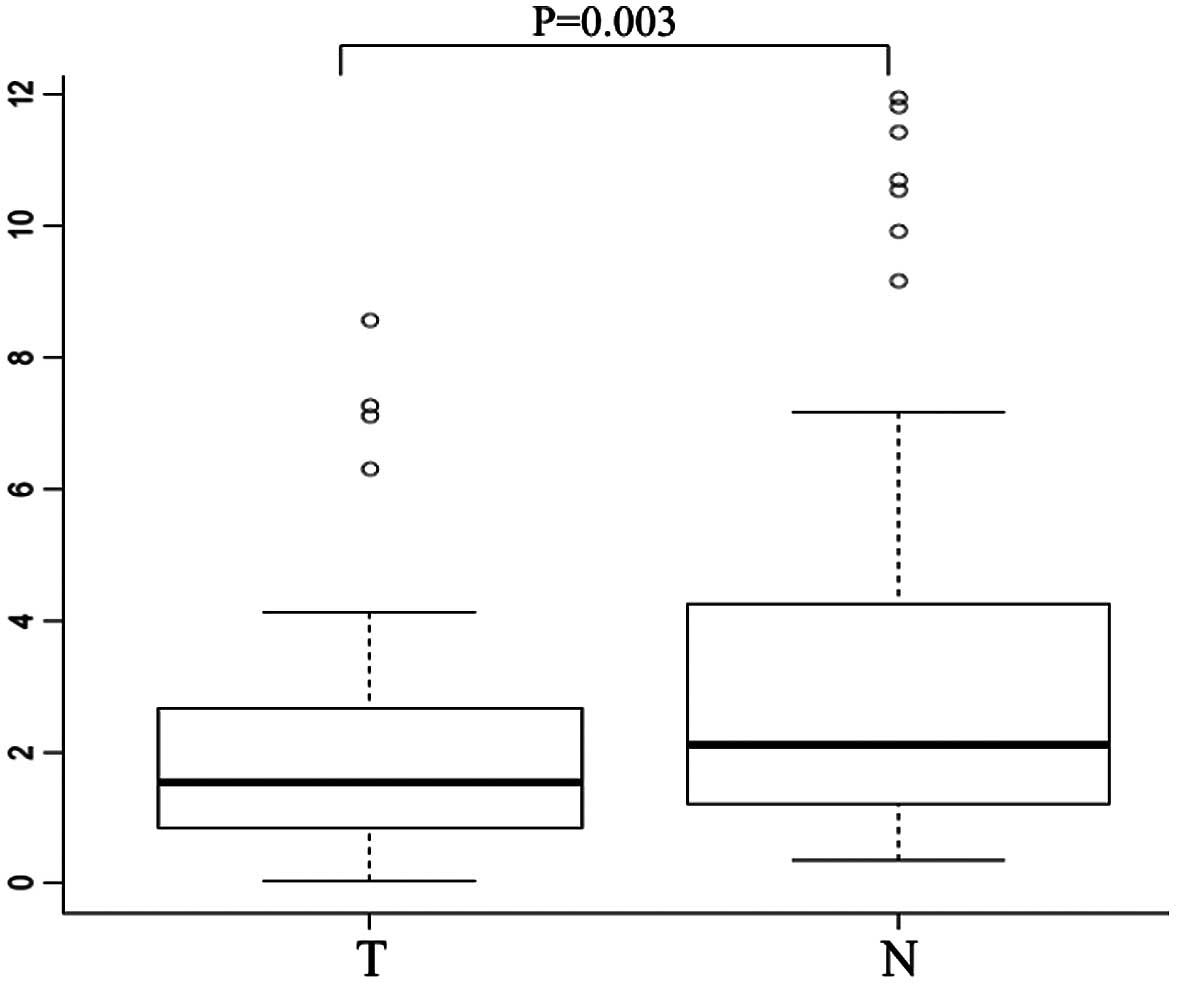
We examined CHD8 mRNA expression in tumor
tissues and the corresponding normal mucosa collected from 101
gastric cancer patients by quantitative real-time PCR. Notably,
CHD8 was significantly downregulated in tumor tissues when
compared to the level in the corresponding normal mucosa (median
CHD8/GAPDH ratio, 1.58 vs. 2.26, respectively; P=0.003)
(Fig. 1).
Association between CHD8 mRNA expression
and clinicopathological factors
As shown in Table I,
we divided the patient population into a high (n=60) and a low
CHD8 expression group (n=41), with a cutoff value of 1.29
for the CHD8/GAPDH ratio in the cancerous tissues. The low
CHD8 expression group was significantly associated with an
increased depth of tumor invasion (P=0.036) and lymph node
metastasis (P=0.028). Relative to the high CHD8 expression
group, the low CHD8 expression group showed an increased
tendency to be associated with peritoneal dissemination (Table I). No significant differences were
observed regarding histological type, lymphatic invasion, or venous
invasion.
 | Table IAssociation between CHD8 mRNA
expression and clinicopathologic factors. |
Table I
Association between CHD8 mRNA
expression and clinicopathologic factors.
| Factors | Low expression
(n=41) | High expression
(n=60) | P-value |
|---|
| Age (mean ± SD) | 63.9±11.3 | 67.2±10.6 | |
| Gender | | | 0.013 |
| Male | 31 | 31 | |
| Female | 10 | 29 | |
| Histological
grade | | | 0.44 |
| Well and mod | 18 | 31 | |
| Por and sig | 23 | 29 | |
| Depth of
invasion | | | 0.036 |
| T1–2 | 12 | 30 | |
| T3–4 | 29 | 30 | |
| Lymph node
metastasis | | | 0.028 |
| Negative | 11 | 29 | |
| Positive | 30 | 31 | |
| Lymphatic
invasion | | | 0.23 |
| Negative | 28 | 34 | |
| Positive | 13 | 26 | |
| Venous
invasion | | | 0.71 |
| Negative | 8 | 10 | |
| Positive | 33 | 50 | |
| Peritoneal
dissemination | | | 0.051 |
| Negative | 32 | 55 | |
| Positive | 9 | 5 | |
Association between CHD8 expression and
prognosis
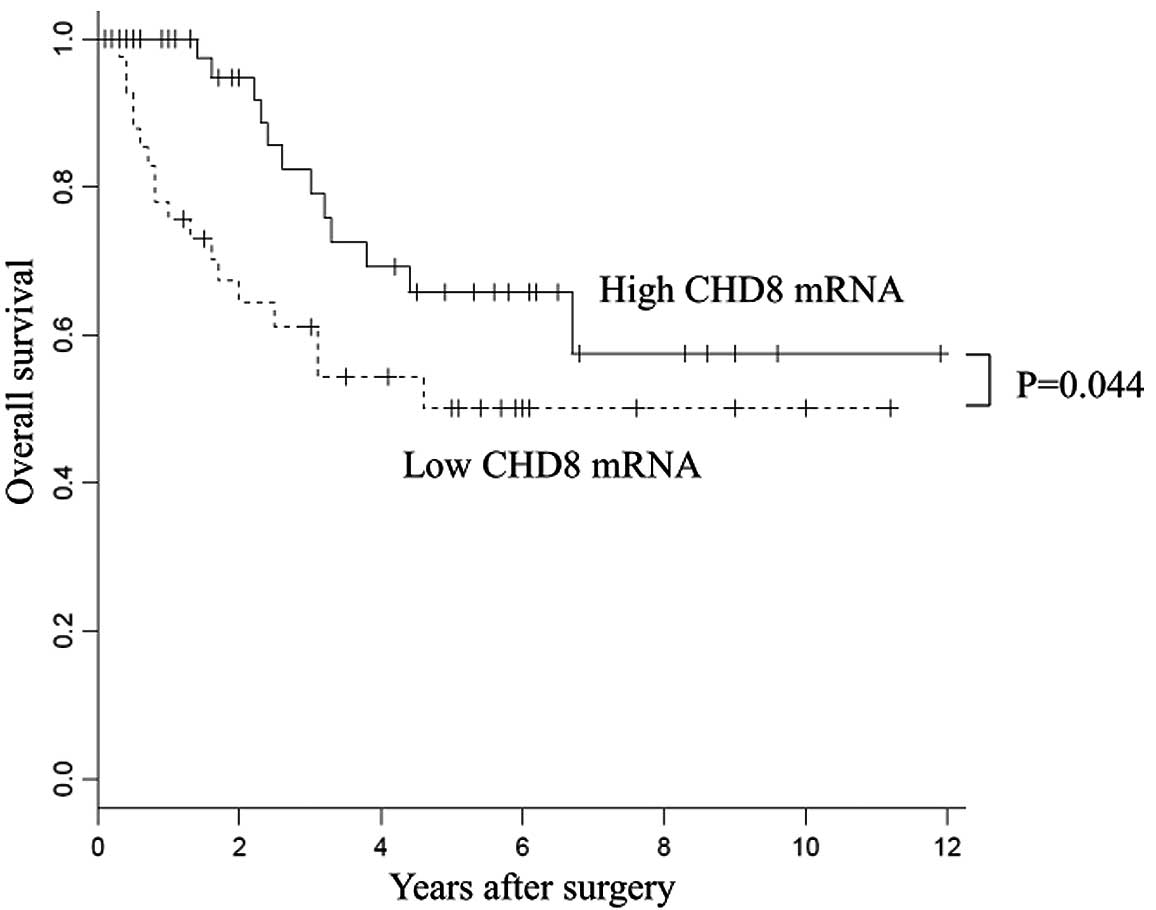
With regard to overall survival, patients with high
CHD8 expression had a significantly better prognosis than
those with low CHD8 expression (P=0.044) (Fig. 2). Univariate analysis revealed that
the level of CHD8 expression, depth of tumor invasion and
presence of lymph node metastasis, lymphatic invasion, or venous
invasion were significantly correlated with prognosis in gastric
cancer patients (Table II). These
factors identified by univariate analysis were then applied to
multivariate analysis, and CHD8 expression was found to be
an independent prognostic indicator for overall survival in
patients with gastric cancer (P=0.048; Table II).
 | Table IIUnivariate and multivariate analyses
for overall survival using the Cox proportional hazards regression
model. |
Table II
Univariate and multivariate analyses
for overall survival using the Cox proportional hazards regression
model.
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
| Factor | RR | 95% CI | P-value | RR | 95% CI | P-value |
|---|
| Age (>65
years) | 0.83 | 0.58–1.20 | 0.336 | - | - | - |
| Gender | 0.97 | 0.67–1.43 | 0.89 | - | - | - |
| Histology grade
(well and mod/por and sig) | 1.13 | 0.79–1.66 | 0.48 | - | - | - |
| Tumor stage
(T1/T2–4) | 2.39 | 1.49–4.41 | 0.00001 | 1.08 | 0.57–2.23 | 0.81 |
| Lymph node
metastasis (negative/positive) | 3.46 | 1.89–8.63 | 0 | 2.17 | 1.04–5.74 | 0.037 |
| Lymphatic invasion
(negative/positive) | 2.39 | 1.42–4.90 | 0.0004 | 1.74 | 0.94–3.78 | 0.076 |
| Venous invasion
(negative/positive) | 2.11 | 1.43–3.04 | 0.0004 | 1.8 | 1.13–2.84 | 0.012 |
| CHD8 mRNA
expression (low/high) | 1.44 | 1.00–2.10 | 0.047 | 1.55 | 1.00–2.45 | 0.048 |
GSEA for analyzing CHD8 function in
gastric cancer
We used 2,996 gene sets, obtained from GSEA, Qiagen
and Sabioscience websites (GSEA: http://www.broadinstitute.org/gsea/index.jsp; Qiagen:
https://www.qiagen.com/geneglobe/pathways.aspx;
Sabioscience: http://www.sabiosciences.com/pathwaycentral.php). The
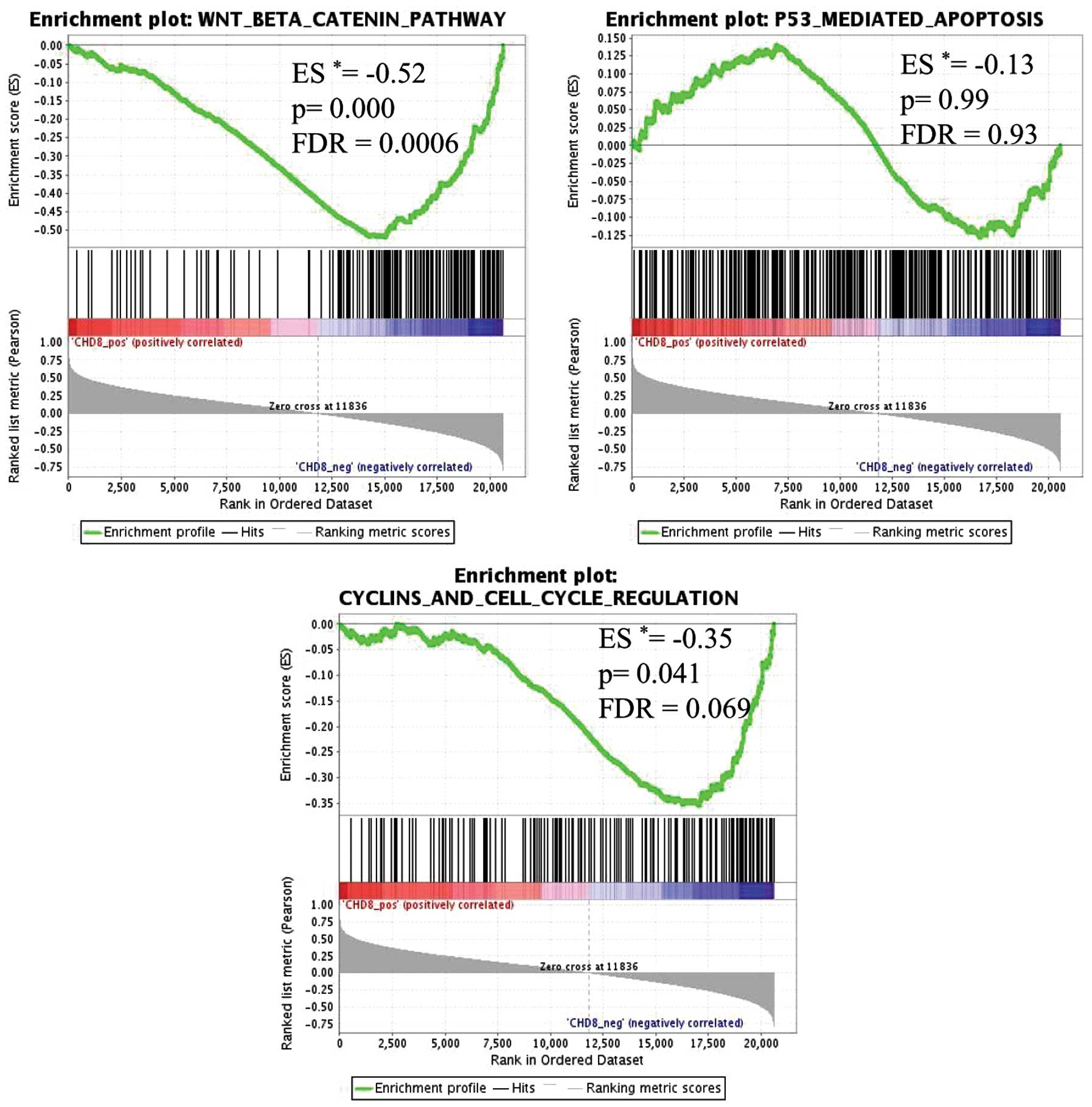
results of GSEA showed that 5 gene sets were significantly
enriched, with a false discovery rate (FDR) of <10% (Table III). In gastric cancer tissues
with low CHD8 expression, genes in the Wnt/β-catenin pathway
signature were the most highly enriched gene set (P=0.000,
FDR=0.000) (Fig. 3). Similarly,
genes within the cell cycle signature were also significantly
enriched (P=0.041, FDR=0.069) (Fig.
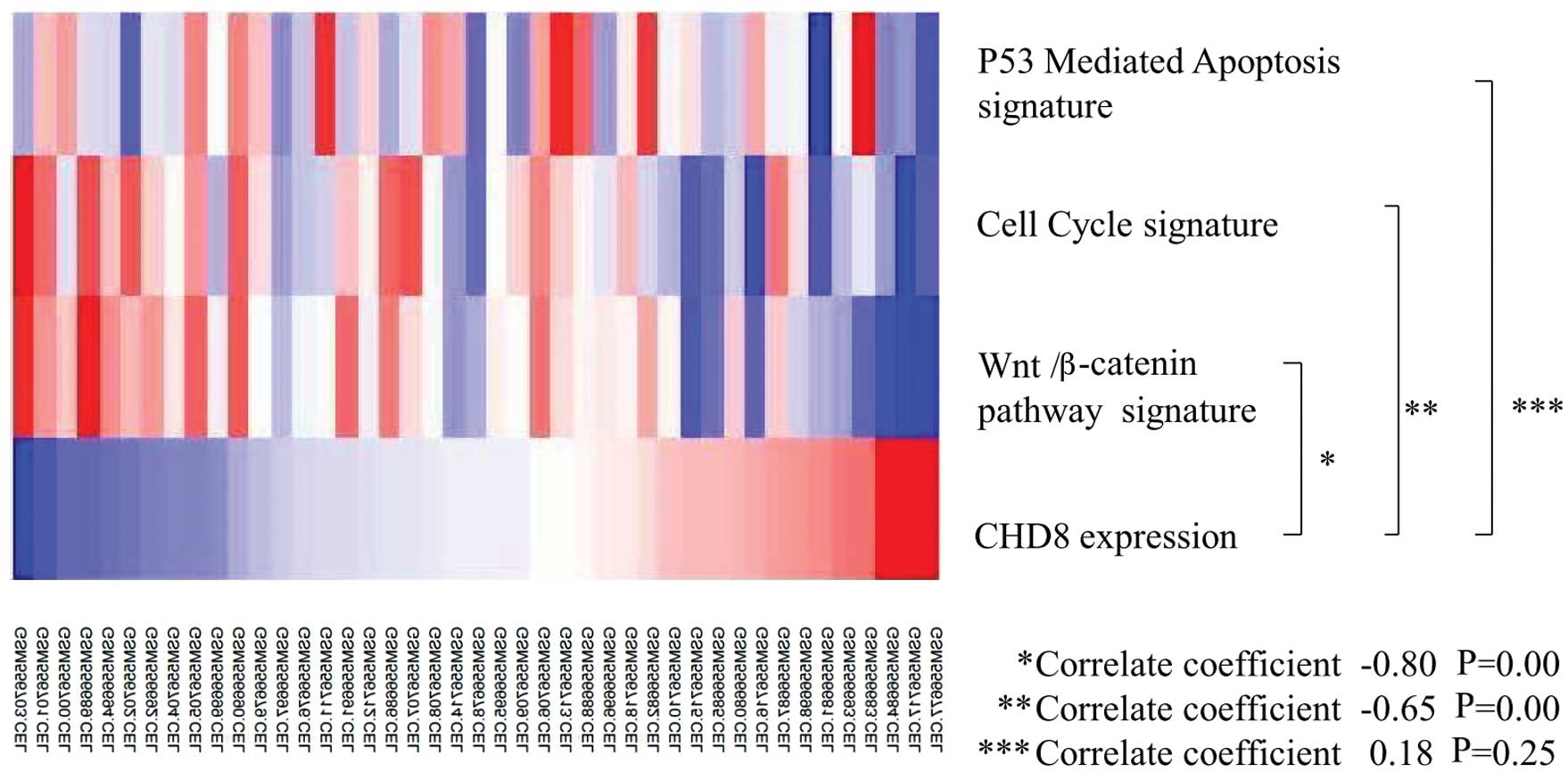
3). Thus, CHD8 expression was inversely correlated with
the Wnt/β-catenin gene signature and the cell cycle gene signature
(Fig. 4).
 | Table IIIResult of GSEA for CHD8
expression in gastric cancer. |
Table III
Result of GSEA for CHD8
expression in gastric cancer.
| Gene set | ES | Nominal
P-value | FDR q-value |
|---|
|
WNT_BETACATENIN_PATHWAY | −0.52 | 0.000 | 0.0006 |
|
GUTIERREZ_MULTIPLE_MYELOMA_DN | −0.71 | 0.000 | 0.050 |
|
ALONSO_METASTASIS_UP | −0.55 | 0.000 | 0.069 |
|
CYCLIN_AND_CELL_CYCLE_REGULATION | −0.35 | 0.041 | 0.069 |
|
CHIANG_LIVER_CANCER_SUBCLASS_UNANNOTATED_DN | −0.72 | 0.000 | 0.074 |
In contrast, p53-mediated apoptosis, which has been
shown to be associated with CHD8 expression in embryonic cells
(15), was not significantly
associated with CHD8 in gastric cancer (Figs. 3 and 4).
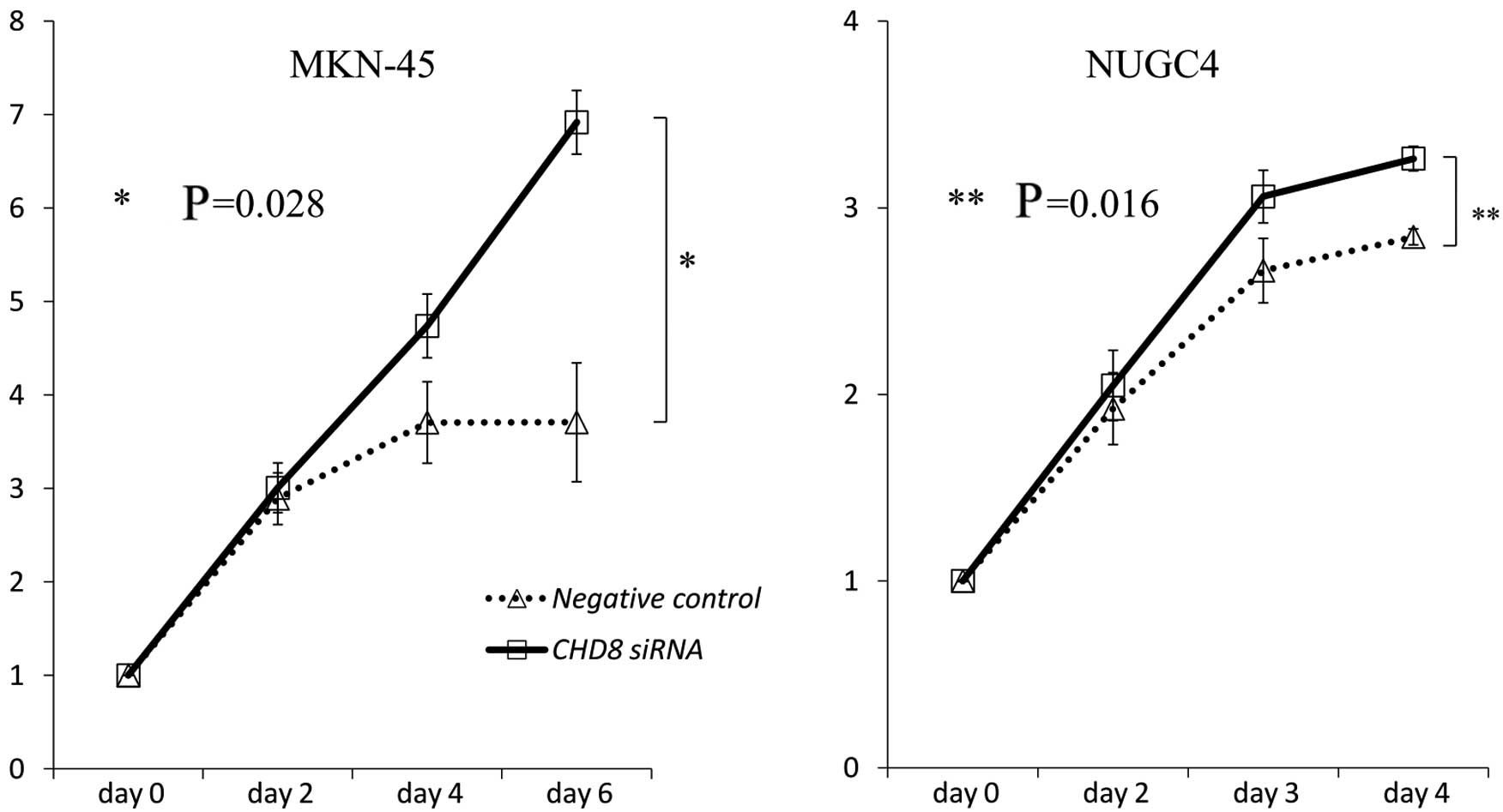
In vitro assessment of CHD8
knockdown
To investigate the role of CHD8 in gastric
cancer progression, CHD8 expression was suppressed by
transient siRNA transfection into MKN-45 and NUGC4 cells. The
reduction in CHD8 expression was confirmed by quantitative
read-time RT-PCR. As revealed by MTT assays, siRNA-mediated
knockdown of CHD8 promoted the proliferation of MKN-45 and
NUGC4 cells (MKN-45: P=0.028; NUGC4: P=0.016 vs. control cells)
(Fig. 5).
Discussion
Our present study revealed that CHD8 mRNA
expression was frequently downregulated in gastric cancer tissues
compared with that in the adjacent normal mucosa. In addition, we
found that CHD8 played a key role as a tumor suppressor in
gastric cancer, i.e. high expression of CHD8 was associated
with a better prognosis. By in vitro analysis, we confirmed
that knockdown of CHD8 mRNA in gastric cancer cell lines
resulted in more aggressive proliferation. Moreover, GSEA based on
GEO expression array data showed that CHD8 expression was
significantly associated with downregulation of genes involved in
the Wnt/β-catenin pathway and the cell cycle. Thus, loss of
CHD8 expression was expected to enhance activation of the
Wnt/β-catenin pathway and promote cell cycle progression in gastric
cancer. Taken together, these data suggest that CHD8 exerts a
suppressive effect on cell proliferation through negative
regulation of the Wnt/β-catenin pathway and cell cycle progression
in gastric cancer.
From the GSEA study, we found significant
associations between CHD8 expression and Wnt/β-catenin
signaling and the cell cycle; this result is consistent with
several studies that have described CHD8 as a negative regulator of
the Wnt/β-catenin pathway. For example, Thompson et al
showed that CHD8 is an ATP-dependent chromatin remodeling
factor that regulates β-catenin target genes in colon cancer cells
(HCT116) (8). Additionally,
Nishiyama et al(16)
suggested that CHD8 negatively regulates β-catenin function by
recruiting histone H1 to the promoters of Wnt target genes.
Together with our current data, these studies suggest that CHD8 is
an essential mediator of Wnt/β-catenin signaling in several
different cellular contexts.
Studies have also suggested that CHD8 controls the
expression of cyclin E2 (CCNE2) and thymidylate synthetase
(TYMS), two genes expressed during G1/S
transition of the cell cycle (11).
Indeed, in the present study, both genes were upregulated in
gastric cancer tissues with low CHD8 expression (data not
shown). Moreover, a previous study also found that p53-dependent
apoptosis was suppressed by histone H1, which is recruited by CHD8
during embryogenesis (15);
however, to date, no studies have investigated the involvement of
CHD8 in p53 activation and signaling in malignancies. Our study
demonstrated that CHD8 expression was not associated with
the p53 pathway in gastric cancer.
The Wnt/β-catenin signaling pathway is an important
functional pathway in development, specification of cell fate and
adult stem cell proliferation (17–20).
Abnormal Wnt signaling has been demonstrated in a variety of human
cancers. For example, Ooi et al(21) demonstrated that 3 oncogenic pathways
(proliferation/stem cell, NF-κB and Wnt/β-catenin) were deregulated
in the majority of gastric cancers and that increased activation of
the Wnt/β-catenin pathway was associated with poor patient survival
in gastric cancer. The present study is the first report to show
that CHD8 interacts with the Wnt/β-catenin pathway in gastric
cancer, with low CHD8 mRNA expression contributing to a poor
prognosis.
In conclusion, our data demonstrate that CHD8
functions as a tumor suppressor by regulating Wnt/β-catenin
signaling and the cell cycle. Moreover, loss of CHD8
expression, commonly observed in gastric cancer, may represent a
novel indicator for the biological aggressiveness of gastric
cancer.
Acknowledgements
We would like to thank T. Shimooka and M. Kasagi for
their technical assistance. This study was funded in part by the
Funding Program for Next Generation World-Leading Researchers
(LS094).
References
|
1
|
Jemal A, Tiwari RC, Murray T, et al:
Cancer statistics, 2004. CA Cancer J Clin. 54:8–29. 2004.
View Article : Google Scholar
|
|
2
|
Marfella CG and Imbalzano AN: The Chd
family of chromatin remodelers. Mutat Res. 618:30–40. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lusser A and Kadonaga JT: Chromatin
remodeling by ATP-dependent molecular machines. Bioessays.
25:1192–1200. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tsukiyama T: The in vivo functions of
ATP-dependent chromatin-remodelling factors. Nat Rev Mol Cell Biol.
3:422–429. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hall JA and Georgel PT: CHD proteins: a
diverse family with Strong Ties. Biochem Cell Biol. 85:463–476.
2007.PubMed/NCBI
|
|
6
|
Pleasance ED, Stephens PJ, O’Meara S, et
al: A small-cell lung cancer genome with complex signatures of
tobacco exposure. Nature. 463:184–190. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bagchi A, Papazoglu C, Wu Y, et al: CHD5
is a tumor suppressor at human 1p36. Cell. 128:459–475. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Thompson BA, Tremblay V, Lin G and Bochar
DA: CHD8 is an ATP-dependent chromatin remodeling factor that
regulates β-catenin target genes. Mol Cell Biol. 28:3894–3904.
2008.PubMed/NCBI
|
|
9
|
Sakamoto I, Kishida S, Fukui A, et al: A
novel β-catenin-binding protein inhibits β-catenin-dependent Tcf
activation and axis formation. J Biol Chem. 275:32871–32878.
2000.
|
|
10
|
Yates JA, Menon T, Thompson BA and Bochar
DA: Regulation of HOXA2 gene expression by the ATP-dependent
chromatin remodeling enzyme CHD8. FEBS Lett. 584:689–693. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rodríguez-Paredes M, Ceballos-Chávez M,
Esteller M, García-Domínguez M and Reyes JC: The chromatin
remodeling factor CHD8 interacts with elongating RNA polymerase II
and controls expression of the cyclin E2 gene. Nucleic Acids Res.
37:2449–2460. 2009.PubMed/NCBI
|
|
12
|
Rodenberg JM, Hoggatt AM, Chen M, Touw K,
Jones R and Herring BP: Regulation of serum response factor
activity and smooth muscle cell apoptosis by chromodomain helicase
DNA-binding protein 8. Am J Physiol Cell Physiol. 299:C1058–C1067.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yuan CC, Zhao X, Florens L, Swanson SK,
Washburn MP and Hernandez N: CHD8 associates with human Staf and
contributes to efficient U6 RNA polymerase III transcription. Mol
Cell Biol. 27:8729–8738. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Menon T, Yates JA and Bochar DA:
Regulation of androgen-responsive transcription by the chromatin
remodeling factor CHD8. Mol Endocrinol. 24:1165–1174. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nishiyama M, Oshikawa K, Tsukada Y, et al:
CHD8 suppresses p53-mediated apoptosis through histone H1
recruitment during early embryogenesis. Nat Cell Biol. 11:172–182.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nishiyama M, Skoultchi AI and Nakayama KI:
Histone H1 recruitment by CHD8 is essential for suppression of the
Wnt-β-catenin signaling pathway. Mol Cell Biol. 32:501–512.
2012.PubMed/NCBI
|
|
17
|
Bienz M and Clevers H: Linking colorectal
cancer to Wnt signaling. Cell. 103:311–320. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moon RT, Kohn AD, De Ferrari GV and Kaykas
A: WNT and β-catenin signalling: diseases and therapies. Nat Rev
Genet. 5:691–701. 2004.
|
|
19
|
Nelson WJ and Nusse R: Convergence of Wnt,
β-catenin, and cadherin pathways. Science. 303:1483–1487. 2004.
|
|
20
|
Willert K and Jones KA: Wnt signaling: is
the party in the nucleus? Genes Dev. 20:1394–1404. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ooi CH, Ivanova T, Wu J, et al: Oncogenic
pathway combinations predict clinical prognosis in gastric cancer.
PLoS Genet. 5:e10006762009. View Article : Google Scholar : PubMed/NCBI
|