Introduction
Colorectal cancer is one of the most common cancers
in the world with over 1.2 million new cancer cases and over
600,000 deaths annually. The risk factors for colorectal cancer
include smoking tobacco; being physically inactive, overweight, or
obese; and consuming red and processed meat or excessive alcohol.
These factors cause gene mutations and altered gene expression,
which promote uncontrolled cell growth and invasion. Increasing
evidence indicates that cancer stem cells (CSCs) are responsible
for cancer formation, recurrence and drug resistance. Colon CSCs
consist of a group of colorectal epithelial cells with the ability
to self-renew, which drive tumorigenesis and differentiation,
generating the heterogeneous mass within a tumor. These CSCs
persist in the tumor mass as a distinct population and cause tumor
relapse and metastasis; however, most CSCs are in a quiescent
state. In other words, they are in a nondividing state, so they are
much less sensitive to classical anti-proliferative
chemotherapeutic regimens (1–4). To
effectively control colorectal cancer in the clinic, there needs to
be a better understanding of colorectal CSCs and their biological
behavior, such as identification of growth regulation of CSCs in a
tumor mass or cell population and their functions in maintaining
stemness and tumorigenicity in colorectal cancer.
To date, a consistent biomarker to precisely
identify CSCs has not been identified; although the molecules
CD133, CD44, CD166 and EpCAM have been proposed as CSC markers in
various types of human cancers (5,6). For
example, CD44 has been linked to certain breast and prostate
cancers for their CSC properties and has been reported to play a
key role in tumor progression and poor prognosis (7). Similarly, CD166 is expressed in
aggressive melanoma and breast, colorectal and bladder cancers
(8–13). CD166 is also highly expressed within
the endogenous intestinal stem cell niche and has been applied as a
marker of pluripotent mesenchymal stem cells (14,15).
In addition, specificity protein 1 (Sp1), a human
transcription factor involved in gene expression in the early
development of an organism, plays an important role in colorectal
cancer development and progression. Sp1 contains a zinc finger
protein motif that binds to the consensus sequence
5′-(G/T)GGGCGG(G/A)(G/A)(C/T)-3′. Expression of Sp1 protein has
been shown to be elevated in different types of tumors, including
colorectal cancers and is associated with patient prognosis
(16,17). Functionally, Sp1 protein regulates
the expression of various genes that are important in
tumorigenesis, such as genes related to cell proliferation,
differentiation, apoptosis, drug resistance and metastasis
(18,19). In this study, we first enriched
colon CSCs from colorectal cancer cell lines and tissues and then
determined whether Sp1 protein can regulate the growth and gene
expression of these CSCs in order to effectively control colorectal
CSCs for the future treatment of colorectal cancer in the
clinic.
Materials and methods
Antibodies and reagents
Rabbit antibodies against Ki-67, Snail, vimentin,
c-kit and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were
obtained from Abcam (Cambridge, UK). PE-conjugated mouse anti-human
CD166 and IgG1κ and FITC-conjugated mouse anti-human CD44 and
IgG2b1κ were purchased from BD Pharmingen (San Diego, CA, USA).
Goat anti-rabbit IgG-FITC, mouse anti-human Sp1, and rabbit
anti-human E-cadherin antibodies were purchased from Santa Cruz
Biotechnology, Inc. (Santa Cruz, CA, USA). Pyronin Y and the Sp1
inhibitor MIT were products of Sigma (St. Louis, MO, USA).
Cell lines and culture
Colon cancer SW480, HCT116, DLD1, HT29, HCT15, LoVo
and SW1116 cell lines were maintained in the laboratory as
previously described (19). To
enrich colon CSCs, SCM was used for cell culture. It was prepared
from Dulbecco’s modified Eagle’s medium (DMEM), i.e., addition of
Nutrient Mixture F12 (DMEM/F12; Gibco-BRL, Rockville, MD, USA) with
N-2 Plus Medium Supplement (Invitrogen Life Technologies, Carlsbad,
CA, USA), 10 ng/ml recombinant human fibroblast growth factor-basic
(FGF-2), 20 ng/ml recombinant human epidermal growth factor (EGF)
(both from Imgenex Corp. San Diego, CA, USA), 100 μg/ml
streptomycin, and 100 U/ml penicillin, without the addition of
serum. The CSCs were cultured in ultra-low attachment dishes or
plates (Corning Inc., Lowell, MA, USA) that do not allow adherence
to a substratum. The cell culture was maintained in a humidified
incubator at 37°C with an atmosphere of 95% air and 5% carbon
dioxide.
Purification of the stem cell
fraction
The mononuclear cells were isolated from colorectal
cancer cell lines by density gradient centrifugation through
Ficoll-Paque™ PLUS (endotoxin tested, sterile; GE Healthcare). In
brief, colorectal cancer SW480 or HCT116 cells were cultured with
SCM in dishes coated with an ultra-low attachment surface for 1
week and refed with SCM every day. Next, the tumor cells were
carefully loaded into Ficoll media and centrifuged for 30 min at
1080 rpm with 0 deceleration, and the interphase layer of cells was
then collected and washed thoroughly with phosphate-buffered saline
(PBS). These enriched colorectal CSCs were further cultured with
SCM for the following experiments.
Immunofluorescence staining
Colorectal cancer cells and the enriched colorectal
CSCs were collected from the monolayer culture using trypsin,
scattered onto glass cover slips using a Shandon Cytospin 4 (Thermo
Electron Corporation, Waltham, MA, USA), and then fixed with 4%
paraformaldehyde. Next, the cells were incubated with the Ki-67
antibody (1:100) for 2 h followed by the FITC-conjugated secondary
antibody (1:100) for 1 h in the dark. The cell nuclei were
counterstained with 1 μg/ml Hoechst 33258. After mounting, images
were obtained under a Zeiss Axioscop fluorescence microscope. The
proliferation index was expressed as the percentage of CSCs to that
of colorectal cancer cells.
Reverse transcription-polymerase chain
reaction (RT-PCR) and quantitative RT-PCR (qRT-PCR)
Total RNA was isolated using TRIzol reagent
(Gibco-BRL, Grand Island, NY, USA), and complementary DNA (cDNA)
was synthesized by a ThermoScript™ reverse-transcription PCR system
(Invitrogen Life Technologies) according to the manufacturer’s
instructions. PCR was performed using a 25-μl system with Hotstart
DNA polymerase (Qiagen, Hilden, Germany) for 36 cycles of 95°C
denaturation for 30 min for the first cycle and then at 94°C for 30
sec, 53°C annealing for 30 sec, and 72°C elongation for 60 sec plus
72°C for 10 min at the last cycle. The PCR products were separated
on a 1% agarose gel.
qPCR was performed using the Option real-time PCR
system (Bio-Rad, Hercules, CA, USA) with Power SYBR-Green PCR
Master Mix. Reactions were carried out in duplicate in a 15-μl
reaction volume. The qPCR conditions were 5 min at 95°C, followed
by 50 cycles of 95°C for 15 sec, 56°C for 30 sec, and 72°C for 40
sec. A final extension at 72°C for 5 min was included before a
temperature ramp from 72 to 95°C at 0.1°C/sec. Gene expression was
normalized to GAPDH (housekeeping gene) and the cycle threshold
values were calculated using the 2−ΔΔCt method. The
primer sequences used in this study are listed in Table I.
 | Table IPrimer sequences for PCR or qPCR. |
Table I
Primer sequences for PCR or qPCR.
| Gene name | Sequence 5′-3′
(forward and reverse) | Length (bp) |
|---|
| CD133 |
CAAATGTGGTGGAGAAATGC
GTGATTTGCCACAAAACCAT | 132 |
| CD44 |
AGACATCTACCCCAGCAACC
GGTGATCCAGGGACTGTCTT | 142 |
| CD166 |
TCTGCTCTTCTGCCTCTTGA
CGGGCTTTTCATATTTCCAT | 155 |
| Sp1 |
TGGATGAGGCACTTCTGTCA
GAAGGTGCCTGCGTCAGTAG | 134 |
| Snail |
AGACGAGGACAGTGGGAAAG
AGATCCTTGGCCTCAGAGAG | 168 |
| E-cadherin |
GTCAGGTGCCTGAGAACGAG
GCCATCGTTGTTCACTGGAT | 158 |
| Vimentin |
GAACTTTGCCGTTGAAGCTG
TCTCAATGTCAAGGGCCATC | 143 |
| c-kit |
CCGAAGGAGGCACTTACACA
GAATCCTGCTGCCACACATT | 146 |
| GAPDH |
GTCAACGGATTTGGTCGTATTG
CTCCTGGAAGATGGTGATGGG | 216 |
Sp1 siRNA construction and
transfection
The sequences of Sp1 siRNA and a control siRNA were
as follows: 5′-AAAGC GCUUCAUGAGGAGUGA-3′ and 5′-UUCUCCGAACGU
GUCACGUTT-3′, respectively. For these siRNA transfections into
cells, colorectal CSCs were grown to 30–50% confluence and then
transfected with Sp1 siRNA or the control siRNA using Lipofectamine
2000 according to the manufacturer’s instructions. The effect of
gene knockdown was evaluated by qPCR.
Protein extraction and western
blotting
Total cellular protein was extracted using RIPA
buffer and then quantified using a BCA kit. Next, lysates
containing 50 μl of protein in each sample were analyzed by
SDS-PAGE, and proteins were transferred to a polyvinylidene
difluoride membrane (Millipore, Bedford, MA, USA). Residual protein
sites were incubated with different primary antibodies followed by
the appropriate horseradish peroxidase-conjugated secondary
antibody for protein visualization with ECL reagents (Millipore,
Billerica, MA, USA).
Flow cytometry
To assess changes in cell cycle distribution in
colorectal cancer cells and CSCs, the cells were grown and
subjected to flow cytometric analysis. In brief, the cells were
first fixed in 70% ethanol and incubated with PBS containing 2
μg/ml Hoechst 33258 for 15 min. Pyronin Y was then added at 4
μg/ml. Fluorescence of 10,000 cells/sample was measured after 20
min. Apoptotic cells were detected using the FITC-Annexin V
Apoptosis Detection Kit I (BD Biosciences, San Diego, CA, USA).
Cells were harvested and resuspended in 500 μl of 1X binding
buffer. A total of 10 μl of FITC-Annexin V and 2 μg/ml propidium
iodide (PI) were added to the cell mixture and incubated for 15 min
in the dark prior to analysis. To observe the expression of stem
cell surface markers, cells were stained with anti-CD44-FITC,
anti-CD166-PE, or the isotype-matched control IgG for 45 min. After
washing, cells were fixed with 0.5% paraformaldehyde prior to FACS
analysis.
Tissue specimens
A total of 45 pairs of colorectal cancer and distant
normal tissue specimens were collected at the Department of
Gastroenterology, Nanfang Hospital (Guangzhou, China). The paraffin
blocks were prepared for immunohistochemistry. This study was
approved by the Medical Ethics Committee of Nanfang Hospital, and
all patients provided written informed consent.
Tissue specimens and
immunohistochemistry
For immunohistochemistry experiments,
paraffin-embedded tissue sections were prepared and then
deparaffinized and rehydrated. The endogenous peroxidase activity
was blocked by incubation with hydrogen peroxide, and antigen
retrieval was then performed by incubation in a pepsin solution at
37°C. The sections were then incubated with an anti-Sp1 antibody
(1:200) at 4°C overnight, followed by incubation with the
biotin-linked anti-mouse IgG (Dako, Copenhagen, Denmark) and then
with the ABC complex. The staining sections were then reviewed and
scored according to our previous published criteria (17). In particular, the cells with <10%
staining were scored as negative staining (−), while 10–49%
staining as +, 50–74% as ++, and 75–100% as +++.
Cell viability MTT assay and colony
formation assay
To assess the effects of Sp1 knockdown in colorectal
CSCs, we transiently transfected Sp1 siRNA into the enriched CSCs.
Alternatively, we also treated them with 500 nM MIT, a Sp1
inhibitor (16). Next, viable cells
were counted up to 3 days. For the colony formation assay, we
plated the cells (1×104) in 35-mm dishes with SCM
containing 0.3% top agar and 0.6% bottom agar and then grew them
for 14 days with observation every 3 days. At the end of the
experiment, the cell colonies were stained with 0.5% violet blue
and counted. Colonies containing >50 cells were counted.
Animal experiment
The animal experiment was approved by the Committee
on the Ethics of Animal Experiments of Southern Medical University.
Briefly, 5–6-week-old BALB/c nude mice were divided into 4 groups
with 3 mice/group. SW480 or HCT116 cells (2×106) with
>90% viability were subcutaneously injected into the right flank
of each mouse. After the tumors became palpable, 25 mg/kg of MIT or
PBS as a control (n=3) was given intraperitoneally every 3 days for
3 weeks. After the last treatment, tumors were removed and digested
into a single-cell suspension for detection of CD44 and CD166
expression using FACS analysis.
Statistical analysis
All data are presented as means ± standard deviation
(SD), and the differences between groups were evaluated by the
two-sided independent samples t-test. Statistical significance was
set at P≤0.05.
Results
Enrichment and identification of a
colorectal CSC population from colorectal cancer cells
In this study, we performed special cell culture of
colorectal cancer cell lines with stem cell medium (SCM) for Ficoll
isolation. We then collected the middle interface cells from the
Ficoll isolation and observed the enriched CSCs under a microscope.
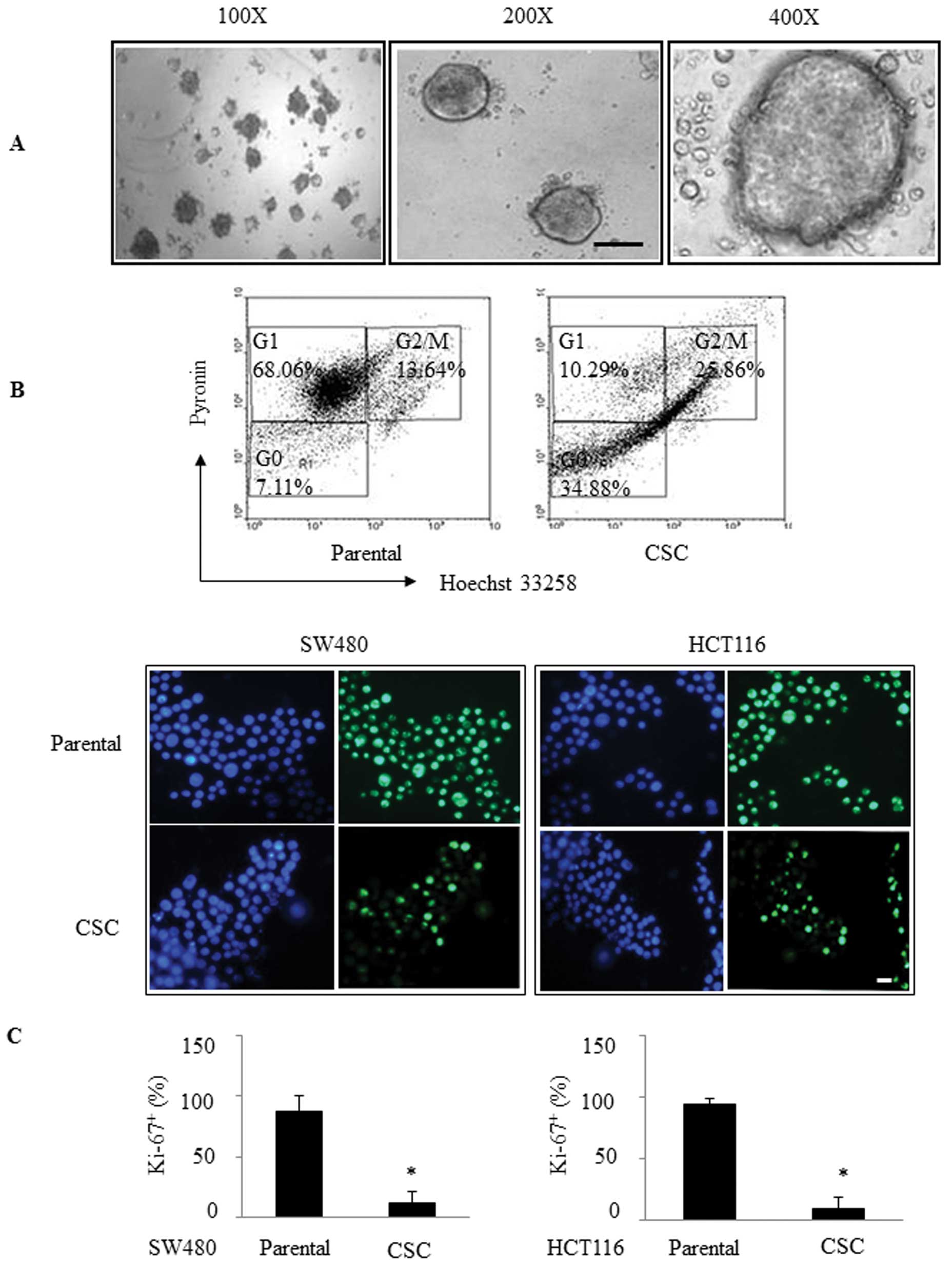
As shown in Fig. 1A, these cells
were large and round in unattached floating spheroid colonies.
Fluorescence-activated cell sorting (FACS) analysis showed that the
percentage of quiescent cells in the stem spheres was greater
compared with that in the parental cells (34.88 vs. 7.11%; Fig. 1B). Moreover, we immunofluorescently
stained them for Ki-67 (a cell-division marker) and found that the
percentage of Ki-67-positive cells was 12.29% in the SW480 CSC
spheres compared to 87.06% in the parental SW480 cells (P<0.05),
while the percentage of Ki-67-positive cells was 9.9% in the HCT116
CSC spheres compared to 94% in the parental HCT116 cells
(P<0.05; Fig. 1C). These
findings suggest that these CSCs grew much more slowly than the
parental cells.
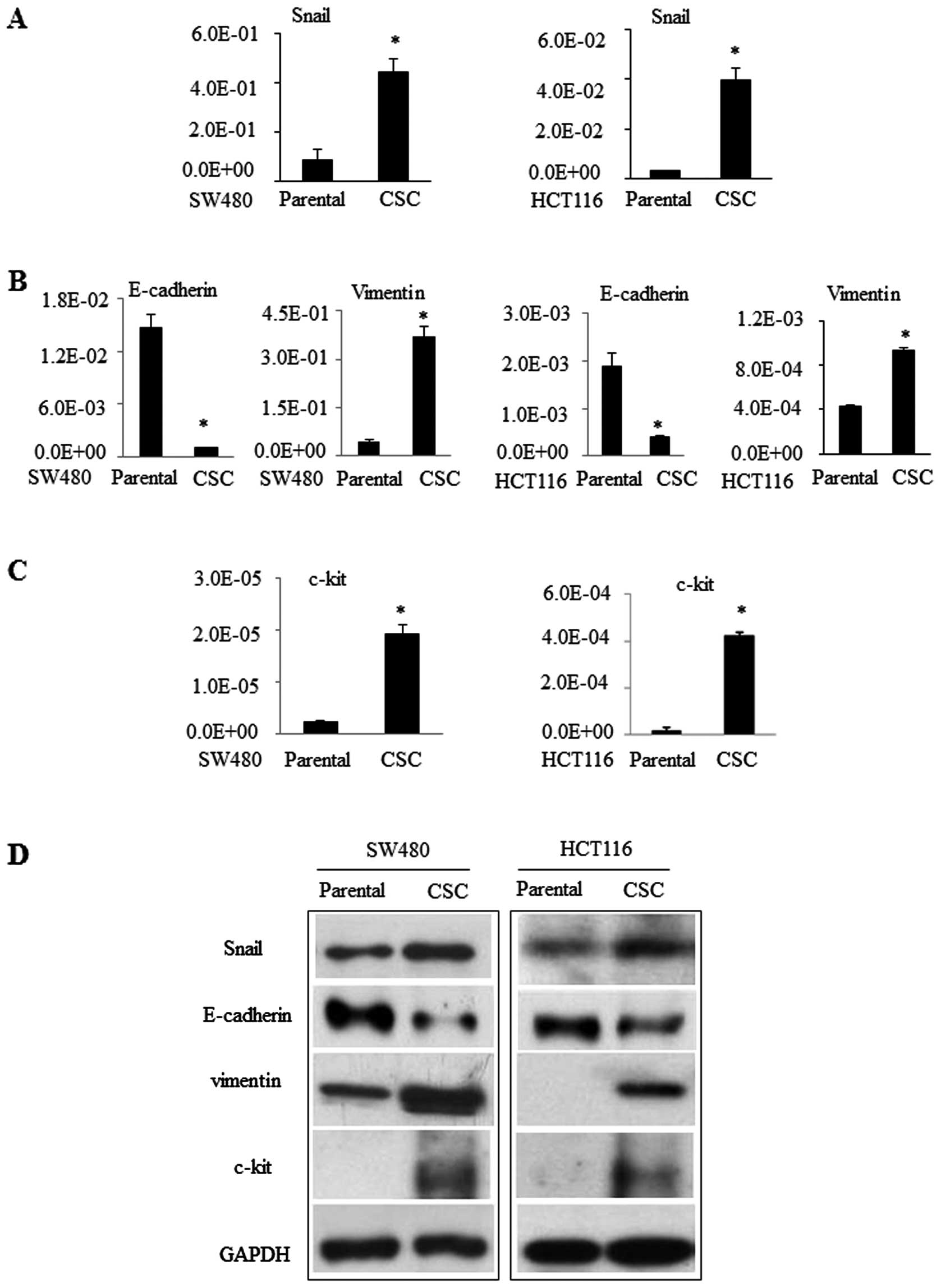
In addition, we detected the expression of
additional genes in these CSCs. For example, expression of
pluripotency genes and acquisition of mesenchymal markers have been
regarded as stem cell phenotypes (20). We assessed expression of Snail [an
epithelial mesenchymal transformation (EMT)-activating
transcription factor], E-cadherin, and vimentin (2 EMT regulatory
proteins) in these CSCs. We found that expression levels of Snail
and vimentin mRNA were significantly greater in CSCs compared to
those in parental SW480 or HCT116 cells. In contrast, E-cadherin
expression was less in CSCs (Fig. 2A
and B). Similarly, CD117 (c-kit) is a stem cell factor receptor
involved in cell signaling transduction in several cell types
(21). CD117 expression in SW480
and HCT116 cells was very low or even undetectable, but it was
significantly greater in CSCs (Fig. 2C
and D). These data suggest that these cells are CSCs.
Differential expression of cell surface
markers in CSCs and parental cells
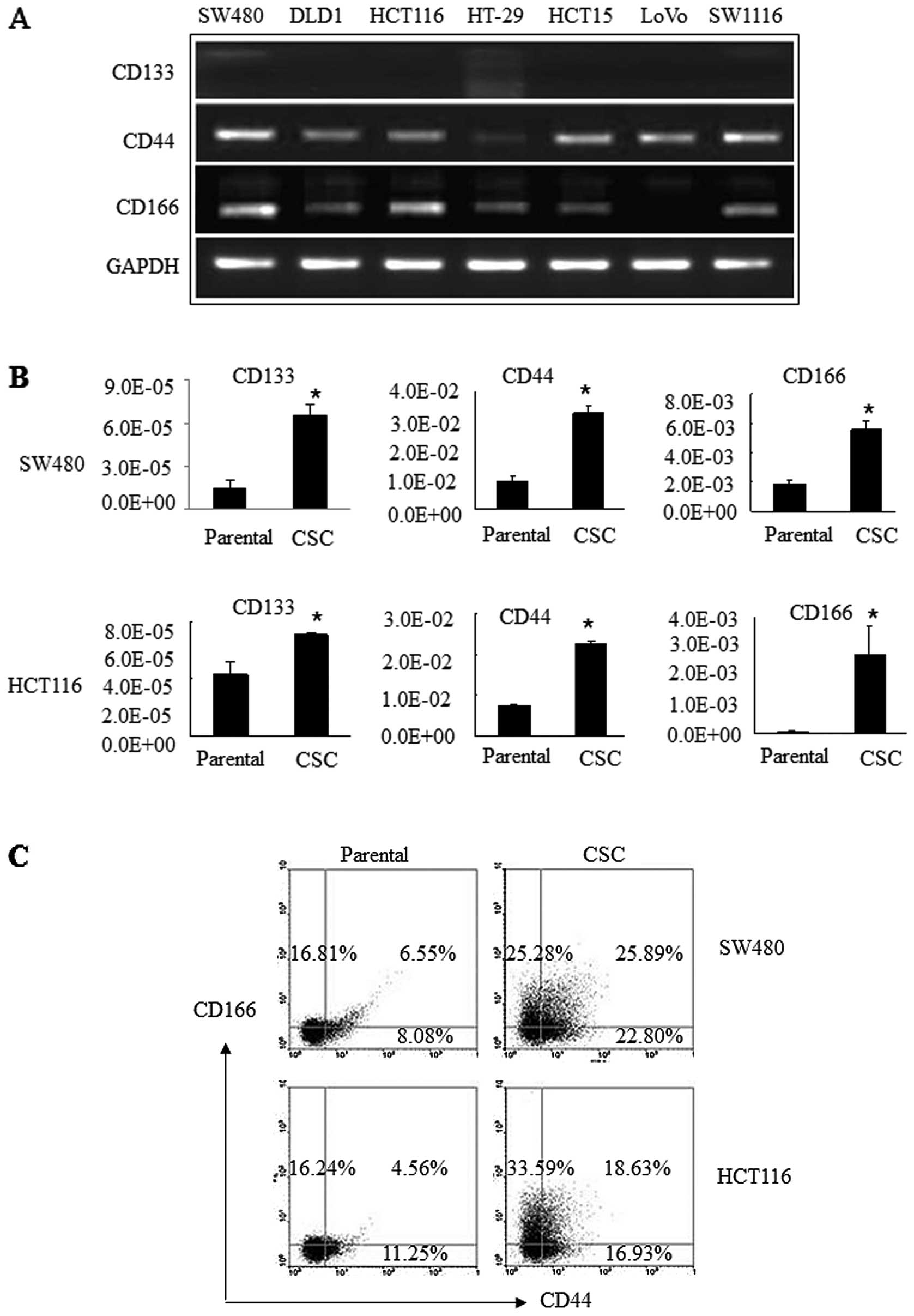
Next, we detected the expression of 3 commonly
accepted stem cell surface markers (CD133, CD44 and CD166) in 7
different colon cancer cell lines. Our data showed that CD44 was
expressed in all 7 colorectal cancer cell lines, i.e., SW480, DLD1,
HCT116, HT29, HCT15, LoVo and SW1116, while CD166 was expressed in
all cell lines except LoVo. In contrast, CD133 was expressed at
very low levels or was even undetectable in these 7 cell lines
(Fig. 3A). We then assessed their
expression levels in the CSCs derived from SW480 and HCT116 cells
and found that all 3 of these surface markers were significantly
upregulated in CSCs compared to the parental cells. In particular,
CD44 and CD166 were expressed at a level of
10−2–10−3/cell, while CD133 was
10−5/cell in CSCs from both SW480 and HCT116 cells
(Fig. 3B). Furthermore, the
percentage of CD44+/CD166+ cells was 25.89%
in SW480 CSC spheres compared to 6.55% in SW480 cells. Similarly,
there were 18.63% of CD44+/CD166+ HCT116 CSC
spheres compared to 4.56% in HCT116 cells (Fig. 3C). These data indicate that CD44 and
CD166 may be useful to define CSCs from colon cancer.
Overexpression of Sp1 protein in colon
cancer tissues
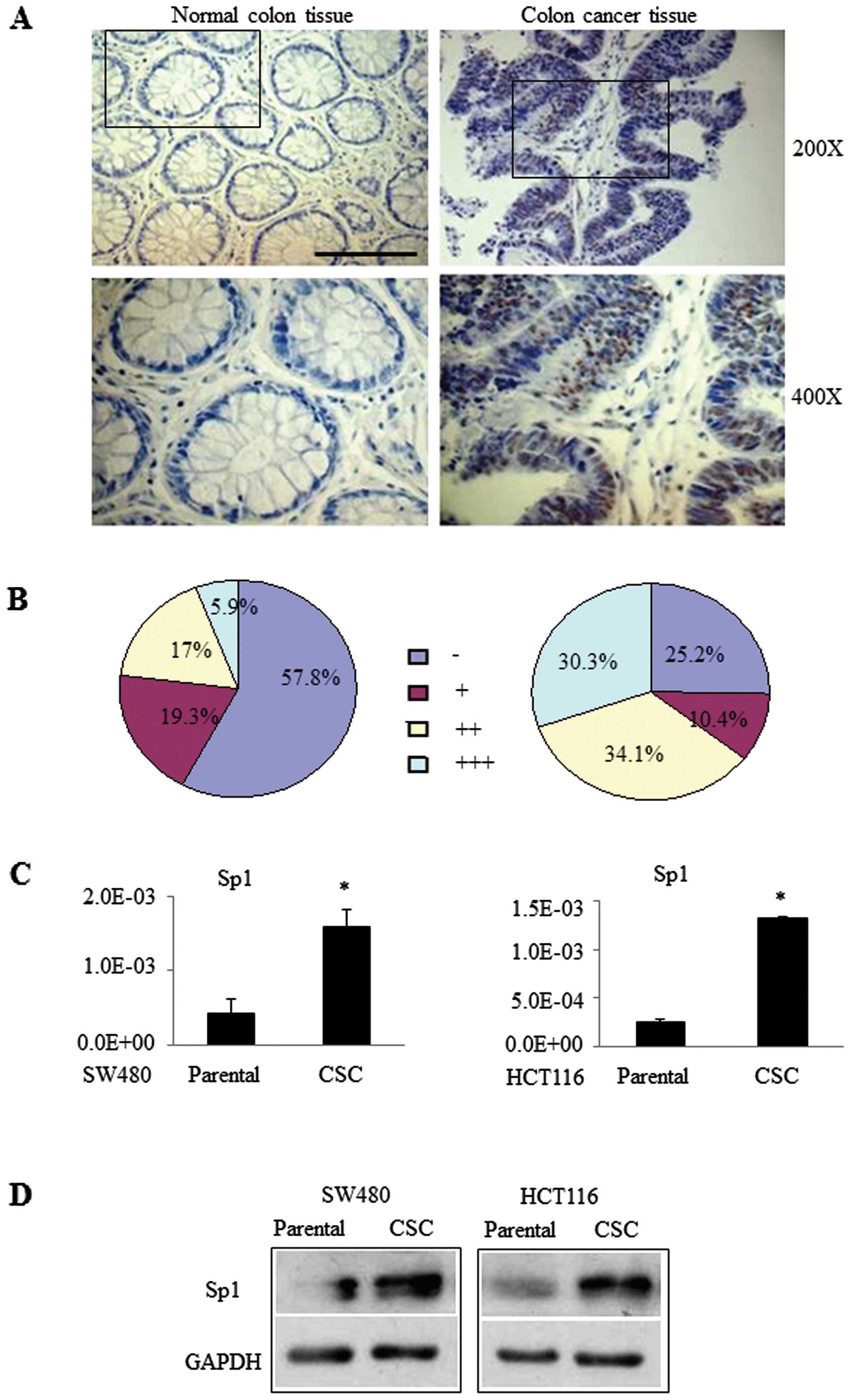
We further confirmed the overexpression of Sp1 in 45
pairs of human distant normal and cancerous colon tissue specimens.
Fig. 4A illustrates the typical
staining of representative tumor and normal mucosa specimens. The
quantitative data on all 45 patients revealed that normal colon
tissues had 42.2% Sp1-positive and only 5.9% strong Sp1-positive
cells, whereas colon cancer tissues had 74.8% Sp1-positive and
30.3% strong Sp1-positive cells (10.4, +; 34.1, ++; 30.3%, +++;
Fig. 4B). Similar results were
found with CSCs and their parental cells (Fig. 4C and D). These data demonstrated
that Sp1 was expressed at a greater level in human colon cancer
specimens than in adjacent normal colon tissues, suggesting that
Sp1 protein may be useful to identify a colon CSC population in
cells and tissues.
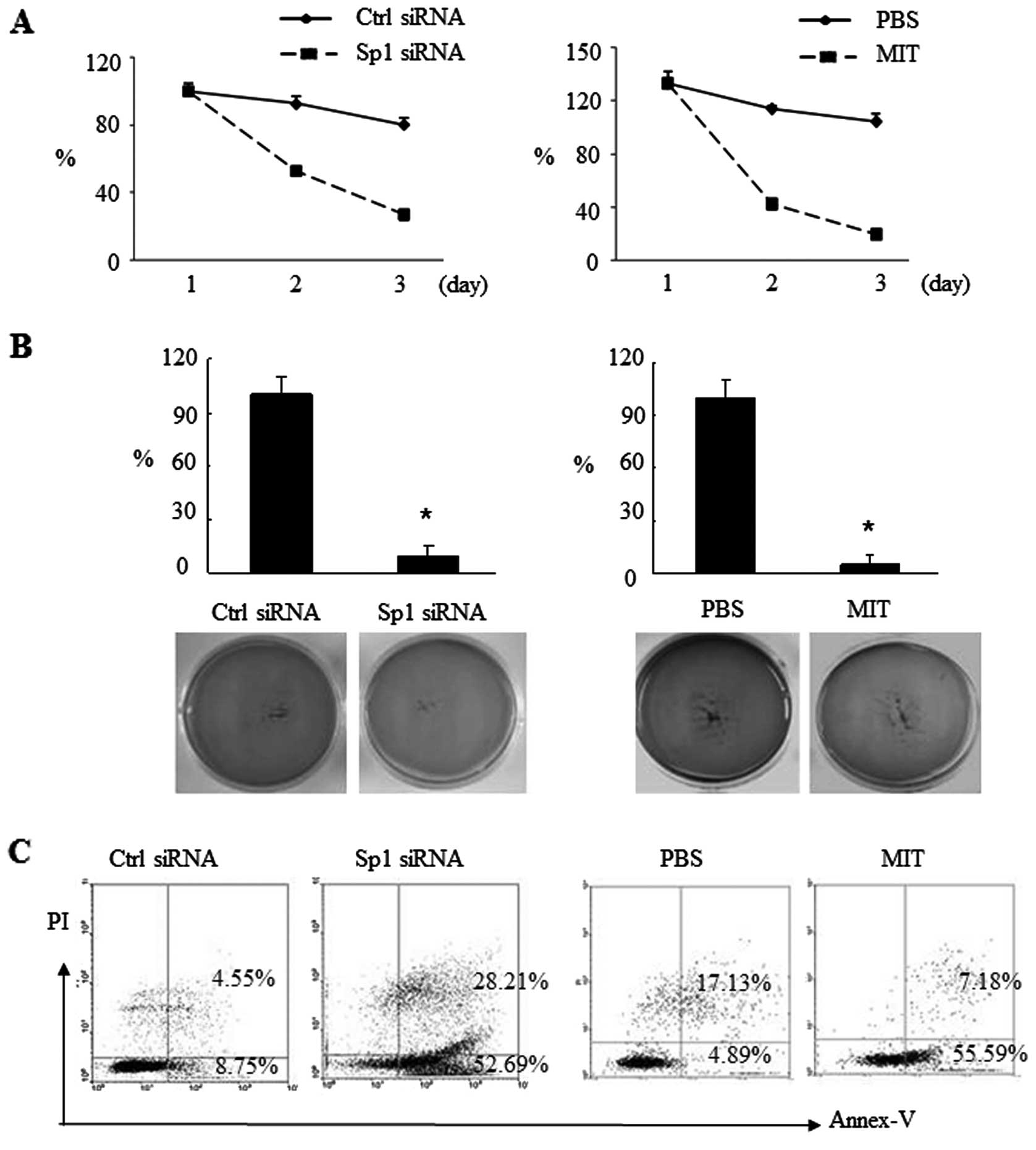
Effects of Sp1 inhibition on suppression
of CSC growth and induction of CSC apoptosis
Next, we determined the effects of Sp1 knockdown on
the regulation of CSC growth and apoptosis by transient Sp1 siRNA
transfection or treatment with the Sp1 inhibitor mithramycin A
(MIT). We found that Sp1 siRNA transfection into CSCs prevented
growth of these CSCs (Fig. 5A). The
soft agar assay showed that Sp1 inhibition sharply reduced colony
formation from 100 to 9.3% in the siRNA group and 4.65% in
MIT-treated cells (Fig. 5B). In
parallel, CSC apoptosis was increased after Sp1 inhibition, i.e.,
Sp1 siRNA transfection induced CSC apoptosis by 43.94% and MIT
treatment induced CSC apoptosis by 50.7% (Fig. 5C).
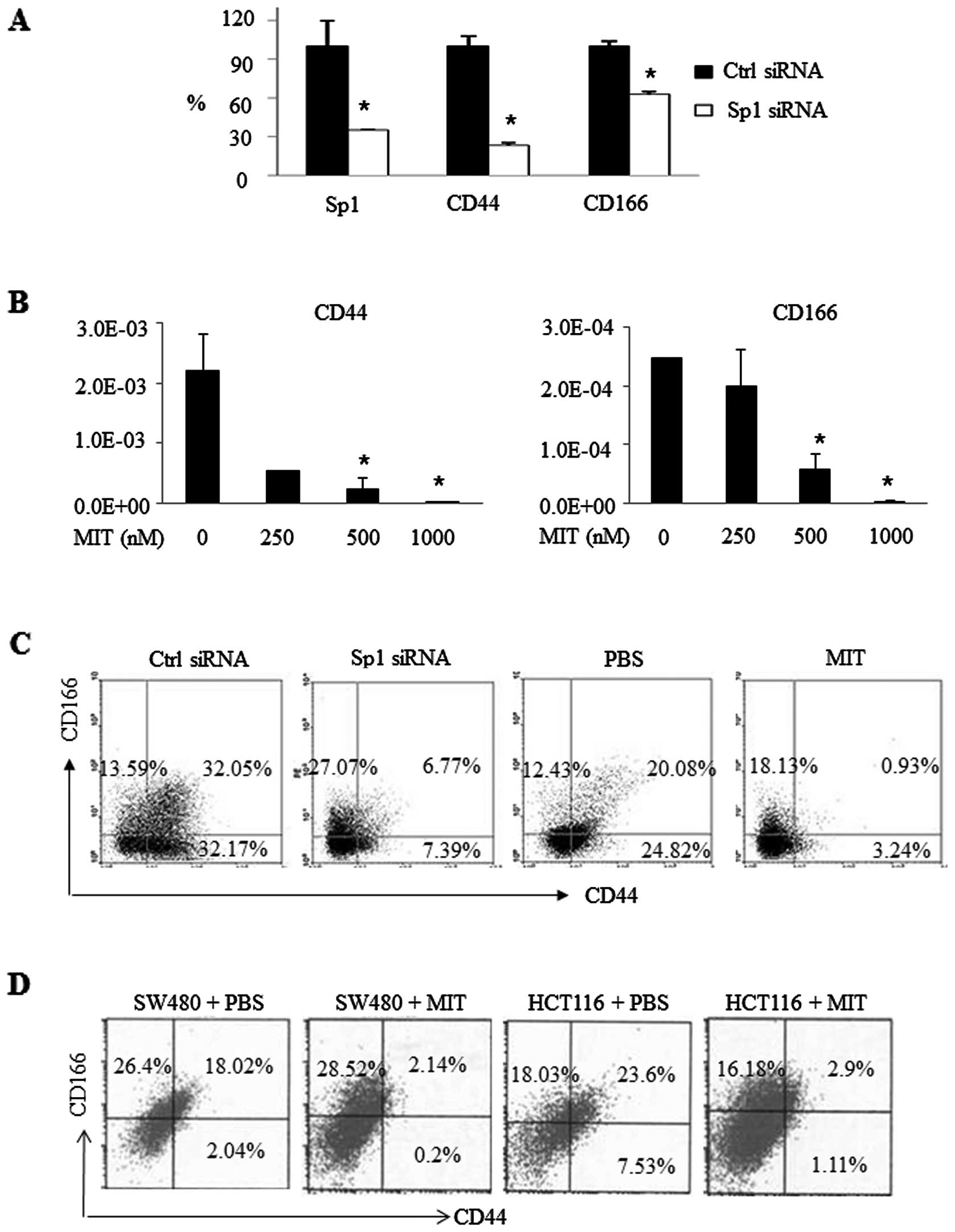
Effects of Sp1 inhibition on the
regulation of CD44 and CD166 expression in vitro and in nude
mice
Next, we explored the effect of Sp1 inhibition on
suppressing the expression of CD44 and CD166 in vitro and in
nude mice. As shown in Fig. 6A,
CD44 expression was inhibited by 76.89%, and CD166 expression was
reduced by 37.05% after Sp1 knockdown by Sp1 siRNA in SW480 CSC
spheres. Consistently, MIT treatment also inhibited CD44 and CD166
expression in a dose-dependent manner (Fig. 6B). FACS data showed that Sp1 siRNA
transfection resulted in 6.77% CD44+/CD166+
cells compared to 32.05% in the control siRNA group, while MIT
treatment decreased CD44+/CD166+ expression
from 20.08 to 0.93% (Fig. 6C).
Furthermore, we also performed nude mouse
experiments by injecting SW480 and HCT116 cells into mice. After
the tumors became palpable, we treated these mice with 25 mg/kg of
MIT or PBS as a control. At the end of the experiment, tumor tissue
was resected and subjected to FACS analysis. The data showed that
the percentage of CD44+/CD166+ cells in the
tumors of MIT-treated mice was 2.14% compared to 18.02% in the
tumors of the SW480 cell-injected mice. Consistently, there were
2.9 and 23.6% of CD44+/CD166+ cells,
respectively, with or without MIT treatment in the tumors of the
HCT116 cell-injected mice (Fig.
6D). Of note, MIT treatment had a greater effect on CD44
inhibition than on CD166 inhibition.
Discussion
In this study, we successfully enriched CSCs from
colorectal cancer SW480 or HCT116 cell lines by using a special
serum-free cultivation method. These enriched CSCs displayed
typical stem cell properties; for example, cell quiescence with
slow growth and expression of stemness markers (e.g., CD44, CD166
and c-kit), mesenchymal markers (vimentin and Snail), and Sp1.
Furthermore, Sp1 inhibition suppressed the growth of these CSCs but
promoted apoptosis. These data suggest that targeting Sp1 protein
could be useful for the development of a novel therapeutic strategy
to control colon cancer.
To date, serum-free cell culture and cancer spheroid
cell formation are the main methods with which to enrich or isolate
CSCs (22). However, most studies
focus on isolation from clinical specimens with mechanical
dissociation and collagenase digestion (23–25).
In the present study, we enriched CSCs from 2 different colon
cancer cell lines, which excluded any mesenchymal cells or stromal
tissues. Furthermore, our current data showed that expression of
CD133, CD44 and CD166 proteins was cell line-specific. Although the
hypothesis that a tumor may originate from a single CSC has been
raised for many years, not all colon cancer cell lines expressed
equally high levels of stemness markers. This result may be due to
the long-term self-renewal potential of CSCs and their ability to
generate heterogeneous progeny.
Recent studies have demonstrated close associations
among tumor EMT, cancer metastasis and CSCs (26–28).
Vimentin is an important mesenchymal marker, and Snail is a
significant transcription factor of EMT. In our study, the
expression of both vimentin and Snail was upregulated in these
CSCs. These results are consistent with previous studies that
showed a direct link between EMT and a gain of epithelial stem cell
properties (26,27). Moreover, c-kit protein plays a
critical role in the growth and differentiation of various types of
cells, including hematopoietic stem cells. A previous study found
that in salivary adenoid cystic carcinoma, c-kit overexpression was
associated with tumor cell invasion and metastasis (29). Most importantly, Sp1 is not only
required for EMT, but it also is able to bind to the c-kit
promoter, thereby inhibiting c-kit gene transcription (30,31).
Thus, the present study linked Sp1 and c-kit together, which may
suggest that Sp1 confers not only EMT but also CSC activity.
Furthermore, CD133 was initially regarded as a
marker of tumor-initiating cells and has been used to isolate CSCs
from fresh lung cancers. However, its role as a marker of colon
CSCs has been subsequently challenged. In the present study, almost
all of the 7 cell lines studied showed low levels of CD133
expression. Similar results were found by Chen et
al(32) (only 0.7% CD133
expression in H1299 cells) and Leung et al(20) (no CD133 expression in tumor cells).
These studies indicate that CD133 may not be a useful marker for
CSCs in certain types of cancer.
In the present study, CD44 and CD166 were
differentially expressed in various colon cancer cell lines. These
results may indicate that expression of stem cell surface markers
are both tissue-specific and cell line-specific. Similar discrepant
observations have also been shown in ovarian and liver cancer
cells. The inconsistent expression may be due to different potency
states and compositional or functional characteristics of the CSC
or progenitor populations (20).
Interestingly, CD44 expression was significantly decreased after
Sp1 inhibition using siRNA knockdown or MIT treatment, whereas such
an inhibitory effect on CD166 expression was not obvious.
Nevertheless, the underlying molecular mechanisms need to be
further elucidated.
In addition, the expression level of Sp1 protein was
much greater in colon cancer tissues than in normal colon tissues.
Similar results were found in CSCs, i.e., Sp1 expression was
significantly greater in colon CSCs than in parental colon cancer
cells. These data suggest that Sp1 expression is a potential marker
associated with colon disease progression. Sp1 siRNA and MIT
treatment suppressed CSC sphere growth and induced apoptosis in
vitro; Sp1 suppression also attenuated CD44 and CD166
expressions in vivo, suggesting that Sp1 expression also has
a close relationship with CSCs. Sp1 knockdown may not only
attenuate the malignant phenotype of colon cancer, but it also
decreases the survival of colon CSCs.
In conclusion, this study demonstrated that Sp1 was
overexpressed in colon CSCs. Inhibition of Sp1 suppressed the
traits of CSCs and promoted cell apoptosis. Hence, the self-renewal
ability, drug resistance, and metastasis potential of colon CSCs
may be partially due to preferentially high expression of Sp1
protein. Our findings link the transcription factor Sp1 to colon
CSCs for the first time and indicate that Sp1 suppression may be a
potential therapeutic strategy for colon cancer.
Acknowledgements
This study was supported by NSFC grants (30973404,
81172057 and 81272761), President Foundation of Nanfang Hospital
(2012B009), high-level topic matching funds of Nanfang Hospital
(2010036, G201227) and Twelfth Five-year-plan (2011BAZ03191) from
the National Technology Support Program.
Abbreviations:
|
Sp1
|
specificity protein 1
|
|
CSCs
|
cancer stem cells
|
References
|
1
|
Vermeulen L, Sprick MR, Kemper K, Stassi G
and Medema JP: Cancer stem cells - old concepts, new insights. Cell
Death Differ. 15:947–958. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gupta PB, Chaffer CL and Weinberg RA:
Cancer stem cells: mirage or reality? Nat Med. 15:1010–1012. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
O’Brien CA, Kreso A and Dick JE: Cancer
stem cells in solid tumors: an overview. Semin Radiat Oncol.
19:71–77. 2009.
|
|
4
|
Lonardo E, Hermann PC, Mueller MT, et al:
Nodal/Activin signaling drives self-renewal and tumorigenicity of
pancreatic cancer stem cells and provides a target for combined
drug therapy. Cell Stem Cell. 9:433–446. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Anderson EC, Hessman C, Levin TG, Monroe
MM and Wong MH: The role of colorectal cancer stem cells in
metastatic disease and therapeutic response. Cancers. 3:319–339.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Horst D, Kriegl L, Engel J, Kirchner T and
Jung A: Prognostic significance of the cancer stem cell markers
CD133, CD44, and CD166 in colorectal cancer. Cancer Invest.
27:844–850. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thorne RF, Legg JW and Isacke CM: The role
of the CD44 transmembrane and cytoplasmic domains in co-ordinating
adhesive and signalling events. J Cell Sci. 117:373–380. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ofori-Acquah SF and King JA: Activated
leukocyte cell adhesion molecule: a new paradox in cancer. Transl
Res. 151:122–128. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van Kempen LC, van den Oord JJ, van Muijen
GN, Weidle UH, Bloemers HP and Swart GW: Activated leukocyte cell
adhesion molecule/CD166, a marker of tumor progression in primary
malignant melanoma of the skin. Am J Pathol. 156:769–774.
2000.PubMed/NCBI
|
|
10
|
King JA, Ofori-Acquah SF, Stevens T,
Al-Mehdi AB, Fodstad O and Jiang WG: Activated leukocyte cell
adhesion molecule in breast cancer: prognostic indicator. Breast
Cancer Res. 6:R478–R487. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Burkhardt M, Mayordomo E, Winzer KJ, et
al: Cytoplasmic overexpression of ALCAM is prognostic of disease
progression in breast cancer. J Clin Pathol. 59:403–409. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Weichert W, Knösel T, Bellach J, Dietel M
and Kristiansen G: ALCAM/CD166 is overexpressed in colorectal
carcinoma and correlates with shortened patient survival. J Clin
Pathol. 57:1160–1164. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tachezy M, Zander H, Gebauer F, et al:
Activated leukocyte cell adhesion molecule (CD166) - its prognostic
power for colorectal cancer patients. J Surg Res. 177:e15–e20.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Levin TG, Powell AE, Davies PS, et al:
Characterization of the intestinal cancer stem cell marker CD166 in
the human and mouse gastrointestinal tract. Gastroenterology.
139:2072–2082. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
van Kilsdonk JW, Wilting RH, Bergers M, et
al: Attenuation of melanoma invasion by a secreted variant of
activated leukocyte cell adhesion molecule. Cancer Res.
68:3671–3679. 2008.PubMed/NCBI
|
|
16
|
Campbell VW, Davin D, Thomas S, et al: The
G-C specific DNA binding drug, mithramycin, selectively inhibits
transcription of the C-MYC and C-HA-RAS genes in regenerating
liver. Am J Med Sci. 307:167–172. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guo Z, Zhang W, Xia G, et al: Sp1
upregulates the four and half lim 2 (FHL2) expression in
gastrointestinal cancers through transcription regulation. Mol
Carcinog. 49:826–836. 2010.PubMed/NCBI
|
|
18
|
Tian H, Qian GW, Li W, et al: A critical
role of Sp1 transcription factor in regulating the human Ki-67 gene
expression. Tumour Biol. 32:273–283. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jungert K, Buck A, von Wichert G, et al:
Sp1 is required for transforming growth factor-β-induced
mesenchymal transition and migration in pancreatic cancer cells.
Cancer Res. 67:1563–1570. 2007.
|
|
20
|
Leung EL, Fiscus RR, Tung JW, et al:
Non-small cell lung cancer cells expressing CD44 are enriched for
stem cell-like properties. PLoS One. 5:e140622010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sperling C, Schwartz S, Buchner T, Thiel E
and Ludwig WD: Expression of the stem cell factor receptor C-KIT
(CD117) in acute leukemias. Haematologica. 82:617–621.
1997.PubMed/NCBI
|
|
22
|
Lee J, Kotliarova S, Kotliarov Y, et al:
Tumor stem cells derived from glioblastomas cultured in bFGF and
EGF more closely mirror the phenotype and genotype of primary
tumors than do serum-cultured cell lines. Cancer Cell. 9:391–403.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu S, Dontu G, Mantle ID, et al: Hedgehog
signaling and Bmi-1 regulate self-renewal of normal and malignant
human mammary stem cells. Cancer Res. 66:6063–6071. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dey D, Saxena M, Paranjape AN, et al:
Phenotypic and functional characterization of human mammary
stem/progenitor cells in long term culture. PLoS One. 4:e53292009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tomuleasa C, Soritau O, Rus-Ciuca D, et
al: Isolation and characterization of hepatic cancer cells with
stem-like properties from hepatocellular carcinoma. J
Gastrointestin Liver Dis. 19:61–67. 2010.PubMed/NCBI
|
|
26
|
Sigurdsson V, Hilmarsdottir B,
Sigmundsdottir H, et al: Endothelial induced EMT in breast
epithelial cells with stem cell properties. PLoS One. 6:e238332011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tellez CS, Juri DE, Do K, et al: EMT and
stem cell-like properties associated with miR-205 and miR-200
epigenetic silencing are early manifestations during
carcinogen-induced transformation of human lung epithelial cells.
Cancer Res. 71:3087–3097. 2011. View Article : Google Scholar
|
|
28
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: an emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang Y, Liang X, Zheng M, et al:
Expression of c-kit and Slug correlates with invasion and
metastasis of salivary adenoid cystic carcinoma. Oral Oncol.
46:311–316. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Maeda K, Nishiyama C, Ogawa H and Okumura
K: GATA2 and Sp1 positively regulate the c-kit promoter in mast
cells. J Immunol. 185:4252–4260. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lecuyer E, Herblot S, Saint-Denis M, et
al: The SCL complex regulates c-kit expression in hematopoietic
cells through functional interaction with Sp1. Blood.
100:2430–2440. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen YC, Hsu HS, Chen YW, et al: Oct-4
expression maintained cancer stem-like properties in lung
cancer-derived CD133-positive cells. PLoS One. 3:e26372008.
View Article : Google Scholar : PubMed/NCBI
|