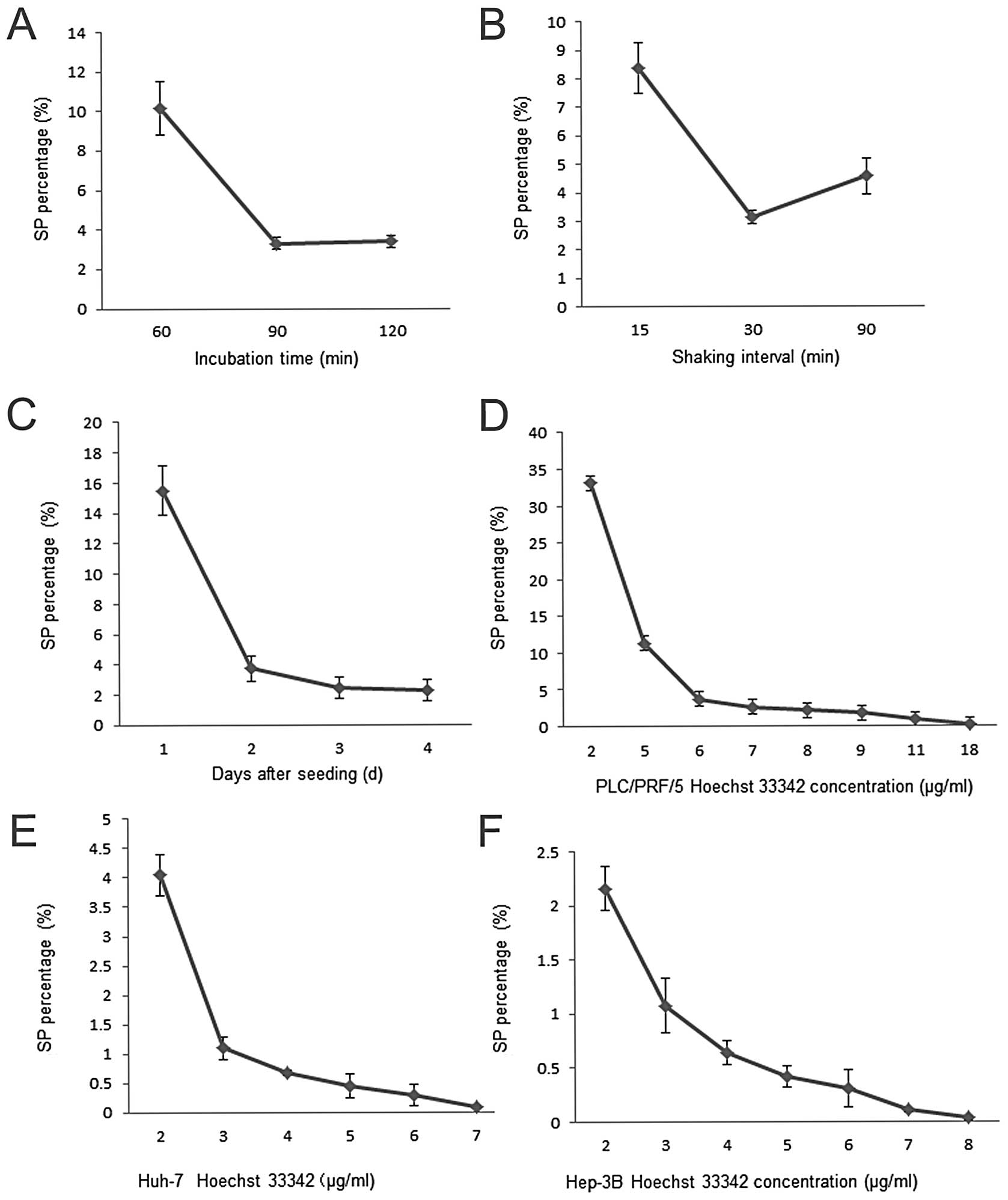
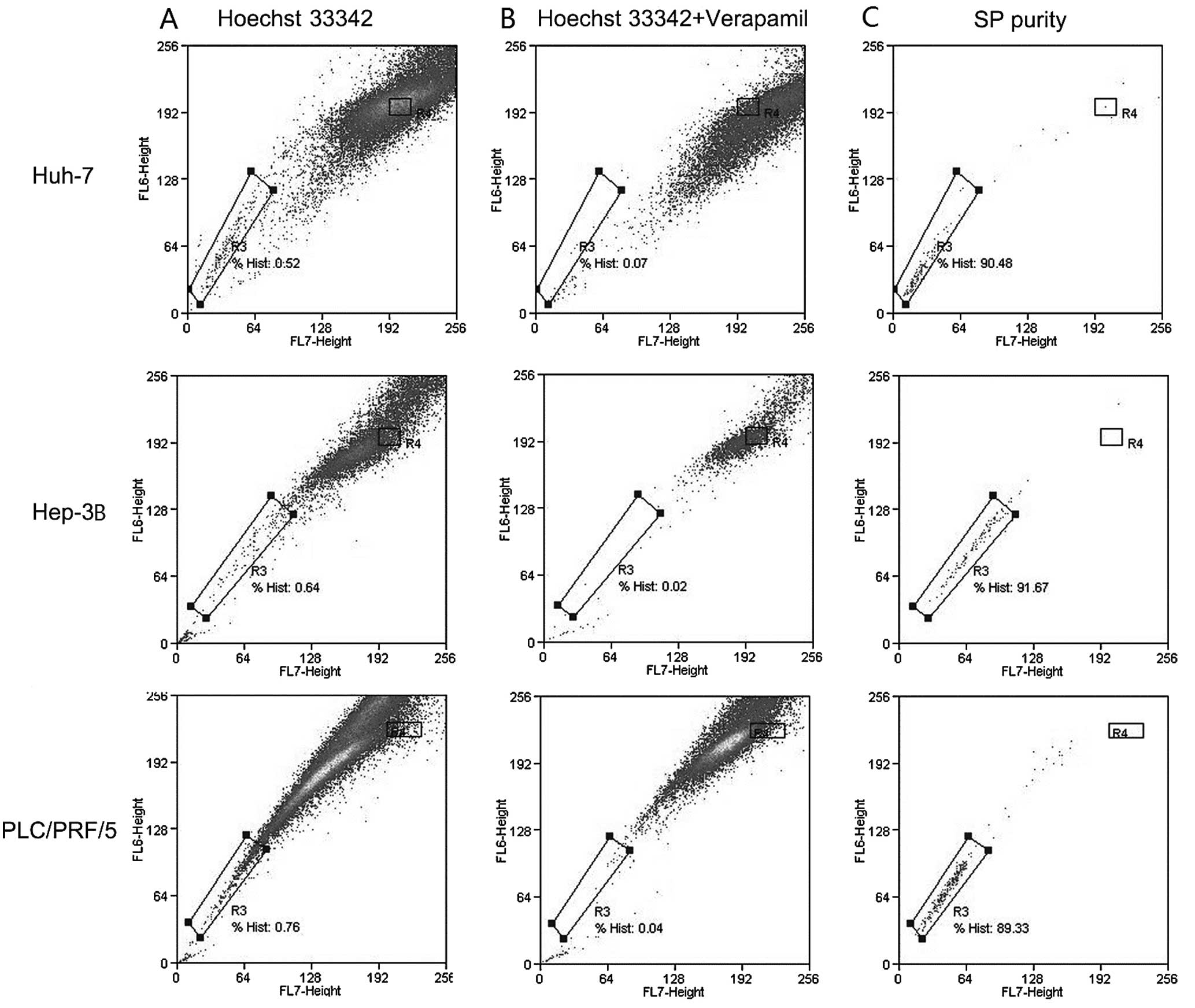
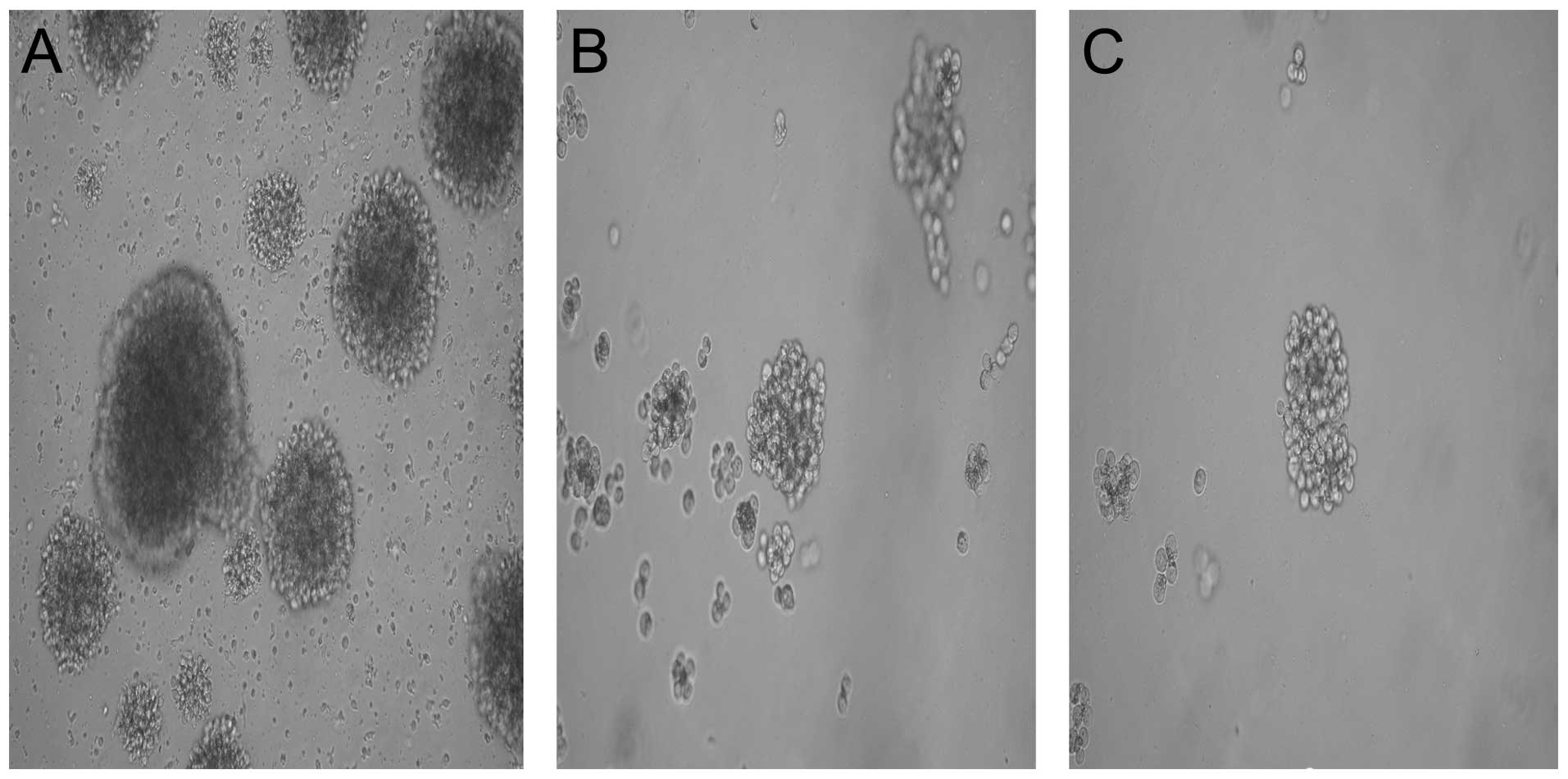
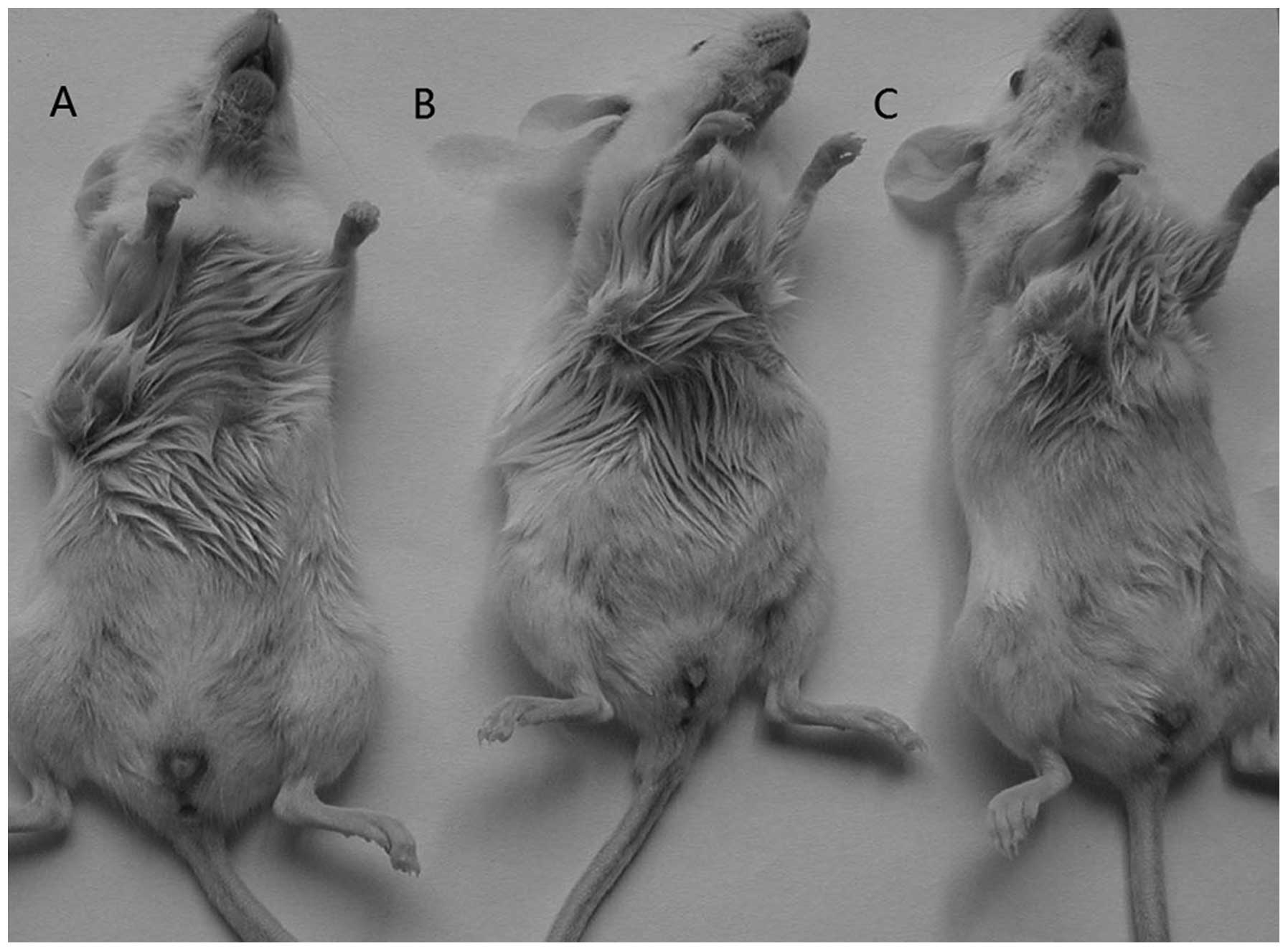
|
1
|
Haraguchi N, Ishii H, Mimori K, et al:
CD13 is a therapeutic target in human liver cancer stem cells. J
Clin Invest. 120:3326–3339. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim HM, Haraguchi N, Ishii H, et al:
Increased CD13 expression reduces reactive oxygen species,
promoting survival of liver cancer stem cells via an
epithelial-mesenchymal transition-like phenomenon. Ann Surg Oncol.
19:539–548. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang W, Wang C, Lin Y, et al: OV6(+)
tumor-initiating cells contribute to tumor progression and invasion
in human hepatocellular carcinoma. J Hepatol. 57:613–620. 2012.
|
|
4
|
Piao LS, Hur W, Kim TK, et al:
CD133+ liver cancer stem cells modulate radioresistance
in human hepatocellular carcinoma. Cancer Lett. 315:129–137.
2011.
|
|
5
|
Lingala S, Cui YY, Chen X, et al:
Immunohistochemical staining of cancer stem cell markers in
hepatocellular carcinoma. Exp Mol Pathol. 89:27–35. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sukowati CH, Rosso N, Pascut D, et al:
Gene and functional up-regulation of the BCRP/ABCG2 transporter in
hepatocellular carcinoma. BMC Gastroenterol. 12:1602012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yamashita T, Ji J, Budhu A, et al:
EpCAM-positive hepatocellular carcinoma cells are tumor-initiating
cells with stem/progenitor cell features. Gastroenterology.
136:1012–1024. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi GM, Xu Y, Fan J, et al: Identification
of side population cells in human hepatocellular carcinoma cell
lines with stepwise metastatic potentials. J Cancer Res Clin Oncol.
134:1155–1163. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma S, Chan KW, Hu L, et al: Identification
and characterization of tumorigenic liver cancer stem/progenitor
cells. Gastroenterology. 132:2542–2556. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chiba T, Miyagi S, Saraya A, et al: The
polycomb gene product BMI1 contributes to the maintenance of
tumor-initiating side population cells in hepatocellular carcinoma.
Cancer Res. 68:7742–7749. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chiba T, Kita K, Zheng YW, et al: Side
population purified from hepatocellular carcinoma cells harbors
cancer stem cell-like properties. Hepatology. 44:240–251. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Golebiewska A, Brons NH, Bjerkvig R and
Niclou SP: Critical appraisal of the side population assay in stem
cell and cancer stem cell research. Cell Stem Cell. 8:136–147.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pastrana E, Silva-Vargas V and Doetsch F:
Eyes wide open: a critical review of sphere-formation as an assay
for stem cells. Cell Stem Cell. 8:486–498. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Uchida Y, Tanaka S, Aihara A, et al:
Analogy between sphere forming ability and stemness of human
hepatoma cells. Oncol Rep. 24:1147–1151. 2010.PubMed/NCBI
|
|
15
|
Zimmerman AL and Wu S: MicroRNAs, cancer
and cancer stem cells. Cancer Lett. 300:10–19. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ji J, Yamashita T, Budhu A, et al:
Identification of microRNA-181 by genome-wide screening as a
critical player in EpCAM-positive hepatic cancer stem cells.
Hepatology. 50:472–480. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li R, Qian N, Tao K, et al: MicroRNAs
involved in neoplastic transformation of liver cancer stem cells. J
Exp Clin Cancer Res. 29:1692012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
DiMeo TA, Anderson K, Phadke P, et al: A
novel lung metastasis signature links Wnt signaling with cancer
cell self-renewal and epithelial-mesenchymal-transition in
basal-like breast cancer. Cancer Res. 69:5364–5373. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Meng Z, Fu X, Chen X, et al: miR-194 is a
marker of hepatic epithelial cells and suppresses metastasis of
liver cancer cells in mice. Hepatology. 52:2148–2157. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dong P, Kaneuchi M, Watari H, et al:
MicroRNA-194 inhibits epithelial to mesenchymal transition of
endometrial cancer cells by targeting oncogene BMI-1. Mol Cancer.
10:992011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi L, Zhang J, Pan T, et al: MiR-125b is
critical for the suppression of human U251 glioma stem cell
proliferation. Brain Res. 1312:120–126. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang L, Wong CM, Ying Q, et al:
MicroRNA-125b suppressesed human liver cancer cell proliferation
and metastasis by directly targeting oncogene LIN28B2. Hepatology.
52:1731–1740. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tirino V, Desiderio V, Paino F, et al:
Cancer stem cells in solid tumors: an overview and new approaches
for their isolation and characterization. FASEB J. 27:13–24. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mo SL, Li J, Loh YS, et al: Factors
influencing the abundance of the side population in a human myeloma
cell line. Bone Marrow Res. 52:2148–2157. 2011.
|
|
25
|
Mott JL: MicroRNAs involved in tumor
suppressor and oncogene pathways: implications for hepatobiliary
neoplasia. Hepatology. 50:630–637. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jeon HM, Sohn YW, Oh SY, et al: ID4
imparts chemoresistance and cancer stemness to glioma cells by
derepressing miR-9*-mediated suppression of SOX2. Cancer
Res. 71:3410–3421. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu F, Yao H, Zhu P, et al: let-7 regulates
self- renewal and tumorigenicity of breast cancer cells. Cell.
131:1109–1123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hurst DR, Edmonds MD, Scott GK, et al:
Breast cancer metastasis suppressor 1 up-regulates miR-146, which
suppresses breast cancer metastasis. Cancer Res. 69:1279–1283.
2009. View Article : Google Scholar : PubMed/NCBI
|