Introduction
microRNAs (miRNAs) are an endogenous conserved class
of non-coding 20–24 nucleotide small RNAs that regulate gene
expression at post-transcriptional level by mainly binding to
3′-UTR of target mRNAs, leading to mRNA degradation or translation
inhibition (1,2). Bioinformatic analyses have predicted
that 30% of human genes may be regulated by miRNA (3,4).
miRNAs have been shown to regulate a variety of biological
processes, including developmental timing, cell proliferation, cell
differentiation and cell death (5,6).
Several reports have elucidated the role of certain miRNAs as a
class of oncogenes or suppressors of tumors, depending on their
targeted genes (6–9). Moreover, accumulating evidence shows
that miRNAs can influence multiple steps of metastasis, such as
tumor cell migration, invasion and colonization and play an
important role in tumor metastasis (10).
Brain metastases (BMs) generally tend to occur late
in the progression of multiple types of cancer and are associated
with poor patient survival. The BMs are the most common
malignancies and occur in 20–40% of patients, an incidence 10 times
greater than primary brain tumors, with a rising incidence in all
countries (11,12). The most common tumors to metastasize
to the brain originate in the lung, breast and skin (melanomas)
(13,14). Lung cancer has the highest incidence
for metastasis to the brain. Epidemiological studies have shown
that approximately 15–30% of lung cancer patients develop BMs. The
types of lung cancer with higher incidence of BMs are small-cell
lung carcinomas (SCLC) and adenocarcinomas, and in more advanced
cancer stages (15).
Metastasis, the spread of cancer cells from the site
of primary tumor growth to distant organs, is a leading cause of
cancer morbidity and mortality. The metastatic process is complex,
requiring invasion from the primary tumor, intravasation, survival,
arrest and extravasation of the circulatory system and colonization
of a distant site (16,17). When tumor cells arrive at a new
metastasis site, the vast majority of tumor cells that have
undergone extravasation are still not able to effectively colonize
the new site. For several types of carcinomas, solitary tumor cells
can be detected in the bone marrow years before the development of
overt metastasis (18). Thus, a
better understanding of molecular factors that contribute to the
growth and colonization of tumor cells in secondary sites is key in
treating metastatic disease.
Molecules such as parathyroid hormone-related
protein and transforming growth factor-β (TGF-β) that are produced
by the cancer cells or are present in the bone microenvironment may
mediate this growth (19). Some
molecular factors that influence the ability of colon and other
cancer cells to grow in the liver have also been identified, for
example, the expression of the epidermal growth-factor receptor
coupled with expression of growth factors in the tissue, such as
transforming growth factor-α (TGF-α) (20–22).
Reactive glia are recruited by highly proliferative BMs of breast
cancer and promote tumor cell colonization (23). Expression of miR-200, which promotes
a mesenchymal to epithelial cell transition (MET) by inhibiting
Zeb2 expression, unexpectedly enhances macroscopic metastases in
mouse breast cancer cell lines, enhances mouse breast cancer cell
colonization to form distant metastases (24). These studies identified that some
molecular and miRNAs can regulate growth and colonization of cancer
cells in specific secondary sites.
To date, relatively little is known about mechanisms
that enable BM and colonization of tumor cells, and even less is
known about what causes it, such as miRNAs. To address miRNAs that
regulate BM in lung adenocarcinoma, we compared the miRNA
expression profiles between clinical human primary lung
adenocarcinoma samples and BM samples from lung adenocarcinoma
using Agilent Human miRNA Microarrays (25). In the present study, we showed that
specific miRNAs may be involved in BM from primary lung
adenocarcinoma. Furthermore, we demonstrated that the upregulation
of miR-145 can suppress proliferation of human lung adenocarcinoma
cells, but does not affect cell invasion and migration in
vitro. These results suggest that miR-145 may play important
roles for effective colonization of distant brain tissue sites by
lung adenocarcinoma.
Materials and methods
Tissue samples
The 51 formalin-fixed and paraffin-embedded (FFPE)
samples from 2 groups of cancer patients were studied, including 40
primary lung adenocarcinoma samples (20 samples with lymph node
metastasis, 20 samples without) and 11 BM samples from lung
adenocarcinoma. All samples were collected from West China
Hospital, Sichuan University. A pathologist evaluated histologic
tumor type, tumor grade, and tumor percentage using
hematoxylin-eosin (H&E)-stained samples. Histologic
classifications of the samples are summarized in Table I. Ethics approval for the study was
obtained from the Ethics Committee of the West China Hospital of
Sichuan University.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Parameter | Primary lung
adenocarcinoma samples (n=40) | Brain metastatic
lung adenocarcinoma samples (n=11) |
|---|
| Age (years) | 53.55±9.35 | 57.27±9.76 |
| Gender |
| Male | 20 | 7 |
| Female | 20 | 4 |
| LNM |
| N0 | 20 | |
| N1 | 4 | |
| N2 | 14 | |
| N3 | 2 | |
|
Differentiation |
| II | 15 | |
| II–III | 19 | |
| III | 6 | |
RNA extraction and purification
Total RNA was extracted and purified using
RecoverAII™ Total Nucleic Acid Isolation (Cat No. AM1975; Ambion,
Austin, TX, US) following the manufacturer’s instructions and
checked for an RIN number to inspect RNA integration by an Agilent
Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA).
Microarray analysis
miRNA molecular in total RNA was labeled by miRNA
Complete Labeling and Hyb kit (Cat No. 5190-0456; Agilent
Technologies) following the manufacturer’s instructions, labeling
section. Each slide was hybridized with 100 ng Cy3-labeled RNA
using miRNA Complete Labeling and Hyb Kit (Cat No. 5190-0456) in
hybridization Oven (Cat No. G2545A; Agilent Technologies) at 55°C,
20 rpm for 20 h according to the manufacturer’s instructions,
hybridization section. After hybridization, slides were washed in
staining dishes (Cat No. 121; Thermo Shandon, Waltham, MA, USA)
with Gene Expression Wash Buffer kit (Cat No. 5188-5327, Agilent
Technologies). Slides were scanned by Agilent Microarray Scanner
(Cat No. G2565BA) and Feature Extraction software 10.7 (both from
Agilent Technologies) with default settings. Raw data were
normalized by Quantile algorithm, GeneSpring Software 11.0 (Agilent
Technologies).
TaqMan real-time PCR
Real-time quantitative PCR (qPCR) was carried out to
quantify the mature miR-9*, miR-1471, miR-214 and
miR-145 by TaqMan MicroRNA Assays according to the manufacturer’s
protocol (Applied Biosystems, USA). The qPCR reaction was performed
with the following parameter values: 15 min at 50°C, 10 min at
95°C, followed by 40 cycles at 95°C for 15 sec and 60°C for 1 min.
The TaqMan probes were designed by ABI. miR-145, GUCCAGUUUU
CCCAGGAAUCCCU; miR-214, ACAGCAGGCACAGAC AGGCAGU; miR-471,
ACACCTGGCTCCACAGUGUGAC; miR-9*, UAAAGCUAGAUAACCGAAAGU;
U6B, CGCAAG GATGACACGCAAATTCGTGAAGCGTTCCATATTTTT. qPCR was
performed on ABI 7900 HT Sequence Detection System, and data were
analyzed with 7900 System SDS software. U6B snRNA was used as
normalization control. Relative expression values from 3
independent experiments were calculated following the
2−ΔΔCt method.
Cell culture
Human lung adenocarcinoma cell lines A549 and
SPC-A1, and the adenovirus-immortalized human embryonic kidney
epithelial cells HEK-293 were from the American Type Culture
Collection and were maintained in DMEM (Gibco-BRL) with 10% fetal
bovine serum. Cells were grown in a humidified atmosphere of 5%
CO2 at 37°C.
Recombinant adenoviral vectors for
overexpression of miR-145
The pri-miR-145 sequence −179 to +287 (+1 being the
first base of the mature miR-145) was amplified from HEK-293 cell
genomic DNA. PCR product was cloned into pMD18-T (Takara Bio,
Inc.), verified by sequencing, and subcloned into shuttle plasmid
pAdTrack-CMV (designated as pAdTrack-miR-145) (26). pAdTrack-miR-145 linearized with PmeI
was used to transform BJ5183-AD-1 cells harboring the adenoviral
pAdeasy-1 vector for homologous recombination. Colonies were
screened by plasmid miniprep and PacI restriction analysis to
obtain clones with recombinant miR-145 (designated as
pAdeasy-miR-145). PacI linearized pAdeasy-miR-145 was used to
transfect HEK-293 cells to obtain packaged recombinant miR-145
adenovirus (designated as AD-miR-145). AD-miR-145 was amplified by
repeated infection and verified by PCR. The pAdTrack-CMV empty
vector was used as control (designated as AD-control). The titers
and the multiplicity of infection were determined according to the
manufacturer’s protocols.
Cell proliferation assay
Cell proliferation was evaluated by Cell Counting
Kit-8 (CCK-8) (Dojindo) according to the manufacturer’s
instructions. Briefly, A549 cells (5,000 cells) and SPC-A1 cells
(3,000 cells) were plated in 96-well plates in 100 μl/well and
incubated for 24 h. Then, cells were infected with AD-miR-145 and
AD-control (multiplicity of infection, 80). After 48 h, CCK-8 was
added under sterile conditions, A549 cells were incubated for 2 h
and SPC-A1 cells for 3 h before reading absorbance at 450 nm in an
enzyme-linked immunosorbent assay plate reader (BioTeck
Instruments, Inc.). Each experiment was performed in 4 replicate
wells and independently repeated three times. Absorbance values
were normalized to media control.
Cell invasion and migration assay
The invasion assay was performed by using 24-well
Millicell hanging cell culture inserts consisting of 8-μm PET
membrane (Millipore) coated with BD Matrigel Basement Membrane
Matrix. Briefly, the infected A549 and SPC-A1 cells were
trypsinized and resuspended in DMEM containing 1% FBS and
5×104 cells were added to the upper chamber of each
well. After 24 h at 37°C, cells on the upper membrane surface were
removed by careful wiping with a cotton swab and the filters were
fixed by treatment with methanol for 20 min and stained with 0.1%
crystal violet solution for 20 min. Invasive cells adhering to the
undersurface of the filter were then counted (5 high-power fields)
using an inverted microscope. The migration assay is the same as
the invasion assay except no Matrigel was used.
Statistical analysis
The SPSS 18.0 program was used for general
statistical analysis. Data are expressed as means ± standard
deviation (SD). Statistical significance of the studies was
analyzed by Student’s t-test. To understand the relationship
between miRNAs, the significant correlations were determined using
the Kendall rank correlation test. P-value <0.05 was considered
to indicate a statistically significant difference.
Results
miRNA expression profiling between
primary and BM in lung adenocarcinoma
To investigate differentially expressed miRNAs
between primary and brain metastatic lung adenocarcinoma, 5 primary
lung adenocarcinoma samples without lymph node metastasis and 3 BMs
in lung adenocarcinoma were analyzed using Agilent Human miRNA
Microarray (8*60k) v16.0. The miRNAs that were altered
by at least 2-fold and P-value was <0.05 were considered to be
significant candidates. Heat maps depicted the relative expression
level of mature miRNAs indicated by microarray analyses of the
samples from 8 patients (Fig.
1).
Using strict criteria above, we identified 5
upregulated miRNAs and 3 downregulated miRNAs. miR-214 was the most
downregulated miRNA with an average 20.33-fold change. miR-145 and
miR-23a were also downregulated, with 6.9 and 3.62-fold change,
respectively. We also observed an apparent increased expression of
miRs-9*, -1471, -718, -3656 and -720. miR-9*
was the most upregulated miRNA with an average 64.84-fold change.
All the differential miRNAs are shown in Table II.
 | Table IIDifferentially expressed miRNAs
identified in brain metastasis from lung adenocarcinoma compared to
primary lung adenocarcinoma by Agilent miRNA Microarray. |
Table II
Differentially expressed miRNAs
identified in brain metastasis from lung adenocarcinoma compared to
primary lung adenocarcinoma by Agilent miRNA Microarray.
| microRNA | Expression | Fold-change | P-value | Location |
|---|
| miR-145 | Downregulation | 6.9 | 0.0001 | 5q32 |
| miR-214 | Downregulation | 20.33 | 0.021 | 1q24 |
| miR-23a | Downregulation | 3.62 | 0.022 | 19p13 |
|
miR-9* | Upregulation | 64.84 | 0.008 | 1q22 |
| miR-1471 | Upregulation | 26.05 | 0.017 | 2q37 |
| miR-718 | Upregulation | 11.97 | 0.048 | Xq28 |
| miR-3656 | Upregulation | 4.38 | 0.046 | 11q23 |
| miR-720 | Upregulation | 2.23 | 0.006 | 3q26 |
Verification of miRNA microarray data by
TaqMan real-time PCR analysis
To confirm our microarray data, TaqMan real-time PCR
was performed to analyze the expression of the most significantly
deregulated miRNAs, including miR-9*, miR-1471, miR-214
and miR-145, as shown in Table II.
We examined the expression of 4 miRNAs in 51 samples (including 8
samples for microarray analysis). The miRNA expression level of
each sample was quantified and normalized to U6B expression. The
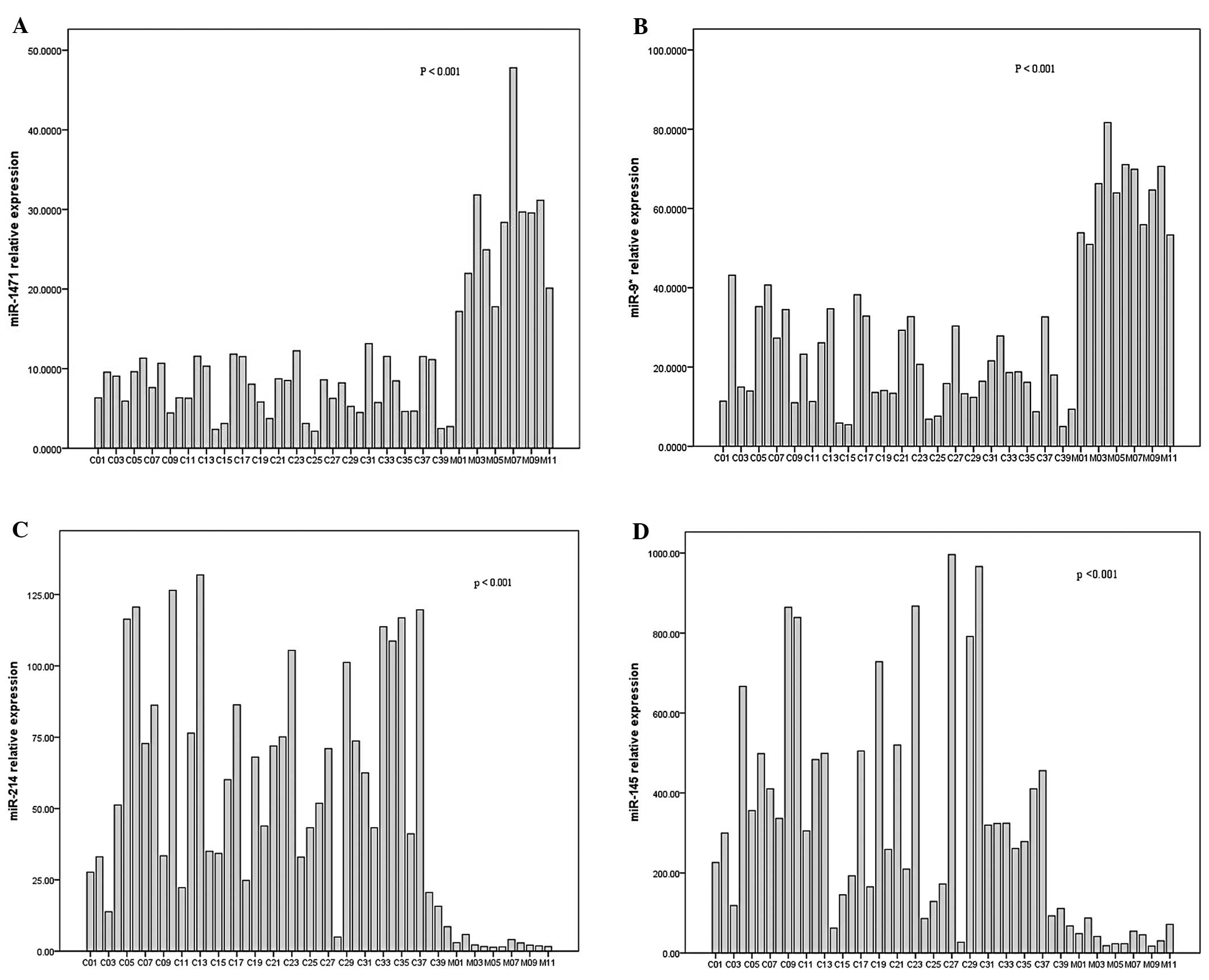
TaqMan real-time PCR data showed that the expression of miR-1471
and miR-9* was upregulated significantly in BM samples
vs. primary lung adenocarcinoma (p<0.001, p<0.001,
respectively), with the increase of 3.65 and 3.14-fold,
respectively (Fig. 2A and 2B).
Consistent with the microarray data, as shown in Fig. 2C, miR-214 was downregulated
significantly in BM samples with a decrease of 24.85-fold
(P<0.001). miR-145 was also verified to be significantly
downregulated 9.17-fold in BM samples (P<0.001, Fig. 2D).
Bioinformatics analysis
We explored the role of the dysregulated miRNAs in
primary lung adenocarcinoma progression and metastasis through
bioinformatics analysis. Of note, a literature search showed that
several of the miRNAs that we found dysregulated in BM from lung
adenocarcinoma have also been reported to be altered in other types
of tumor progression and metastasis (Table III).
 | Table IIIThe involvement of miRNAs
dysregulated in brain metastatic lung adenocarcinoma in the
progression and metastasis of other types of cancer and their
biological roles and targets. |
Table III
The involvement of miRNAs
dysregulated in brain metastatic lung adenocarcinoma in the
progression and metastasis of other types of cancer and their
biological roles and targets.
| miRNA | Deregulation | Cancer type | Biological
function | Validated target
genes |
|---|
| miR-214 | Down | Ovarian cancer | Cell survival | PTEN |
| Down | Cervical
cancer | Cell growth and
invasion | Plexin-B1 |
| Down | Breast cancer | Cell proliferation,
invasion | Polycomb Ezh2
methyltransferase |
| Down | Hepatocellular
cancer | | |
| miR-145 | Down | Gastric cancer | | |
| Down | Hepatocellular
carcinoma | | |
| Down | Lung cancer | Cell
proliferation | c-Myc, EGFR,
NUDT1 |
| Down | Ovarian
carcinoma | | |
| Down | Colon cancer | Cell growth | IRS-1, YES,
STAT1 |
| Down | Prostate
cancer | Cell proliferation
and migration, regulation of EMT | BNIP3 |
| Down | Breast cancer | Cell motility and
invasiveness | MUC1, JAM-A,
fascin |
| Down | Esophageal squamous
cell carcinoma | Cell motility and
invasiveness | |
| miR-23a | Down | Hepatocellular
carcinoma | | |
| Down | Lung cancer | Regulation of
EMT | E-cadherin |
| Up | Colon
carcinoma | Cell growth,
invasion and metastasis | MTSS1 |
| Up | Gliomas | | CREB, PTEN |
|
miR-9* | Up | Brain cancer | | |
| miR-1471 | Up | Rectal cancer | | |
| Up | Breast cancer | | |
| miR-718 | Up | Breast cancer | | |
To further evaluate the effect of these dysregulated
miRNAs on the process of BM of lung adenocarcinoma, we focused on
miR-145 and characterized its functional behavior first.
Expression of miR-145 in primary lung
adenocarcinoma with/without lymph node involvement
To investigate the relationship of the expression of
miR-145 with lymph node involvement, we analyzed the relative
expression level of miR-145 in 20 primary lung adenocarcinoma
samples with and in 20 samples without lymph node involvement
(including 5 primary lung adenocarcinoma samples used in microarray
assay). However, the miR-145 expression level in our primary lung
adenocarcinoma showed no significant difference between with and
without lymph node metastasis.
miR-145 overexpression inhibits the
proliferation of human lung adenocarcinoma cell lines
One measure of the tumorigenic nature of cells is
the ability to proliferate to form colonies in distant metastatic
sites. To determine the effect of miR-145 expression on cell
proliferation, we used AD-miR-145 and AD-control to infect human
lung adenocarcinoma cell lines A549 and SPC-A1. After infection, we
found the expression of miR-145 was increased in both cell lines
that were infected with AD-miR-145 compared to AD-control and MOCK
groups (data not shown). By CCK-8 proliferation assay, it was
demonstrated that overexpression of miR-145 markedly inhibited cell
proliferation in both cell lines. Compared to cells infected with
AD-control, the average growth rate of A549 (Fig. 3A) and SPC-A1 (Fig. 3B) cells overexpressing miR-145
decreased by ~41 and 22%, respectively, in 3 separate
experiments.
miR-145 overexpression does not alter
cell migration and invasion
The TaqMan qPCR results showed that the expression
of miR-145 in primary lung adenocarcinoma was not related to lymph
node metastasis, suggesting that miR-145 may have no effect on the
early stages of metastasis. To further determine the effect of
miR-145 overexpression on the migration and invasion ability of
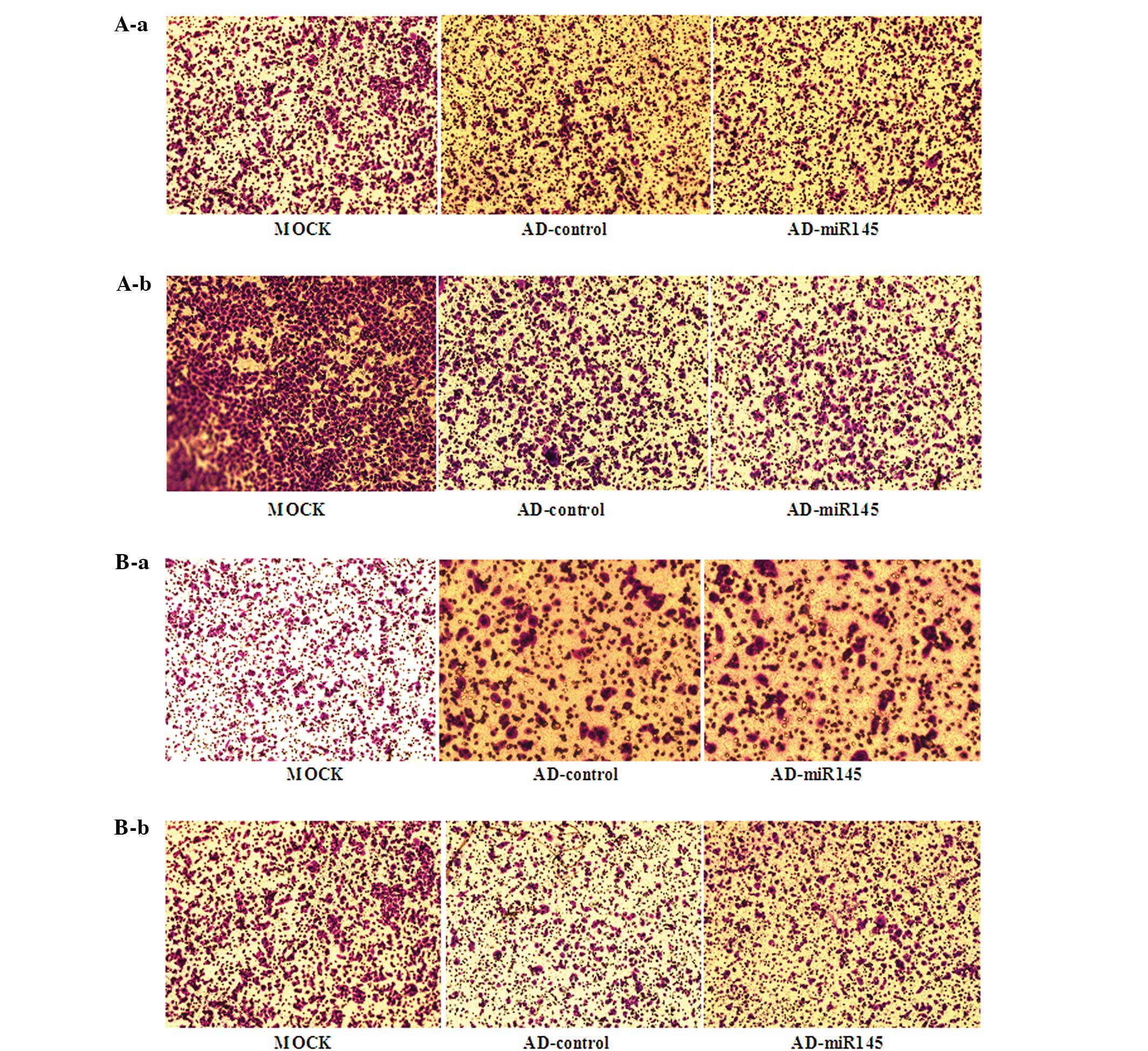
A549 and SPC-A1 cell lines, the transwell assays were performed.
Overexpression of miR-145 had no effect on migration (Fig. 4A) and invasion ability (Fig. 4B) of A549 and SPC-A1 cell lines.
Discussion
Metastatic colonization of the brain has a poor
prognosis and high mortality rates. To understand the molecular
mechanisms that regulate the process of BM from primary lung
adenocarcinoma, we initially compared the miRNA profiles between 5
primary lung adenocarcinoma samples and 3 BMs in lung
adenocarcinoma samples by using Agilent Human miRNA Microarrays. In
this study, 5 upregulated miRNAs (miRs-9*, -1471, -718,
-3656 and -720) and 3 downregulated miRNAs (miRs-214, -145 and
-23a) were significantly detected in brain metastatic tumors
compared to primary lung adenocarcinoma. The 4 most significantly
altered miRNAs from microarray were verified by using TaqMan
real-time PCR with additional samples. Consistent with the
microarray results, TaqMan real-time PCR results showed the
expression of miRs-1471 and -9* were significantly
upregulated, but miR-214 and miR-145 were significantly
downregulated in 11 BM samples compared to 40 primary lung
adenocarcinoma samples. These miRNAs may be involved in BM from
primary lung adenocarcinoma and have the potential to be
biomarkers.
Downregulation of miR-145 was found in several tumor
types including breast, gastric, lung, ovary, prostate cancer and
esophageal squamous cell carcinoma (ESCC) (27–34).
Accumulating evidence indicates that the processing of miR-145 is
also involved in cancer metastasis. In breast cancer, miR-145 was
identified to suppress breast cancer cell line invasion and
metastasis by directly targeting MUC1 (35). miR-145-dependent regulation of 3′UTR
of the JAM-A and fascin decreased motility and invasiveness of
MDA-MB-231 and MCF-7 breast cancer cells (33). Using the prostate cancer cell lines,
PC3 and DU145, miR-145 transfection can inhibit cell proliferation,
migration and invasion by targeting FSCN1 (36). miR-145 is associated with bone
metastasis of prostate cancer and may be involved in the regulation
of EMT by reducing the ability of migration and invasion in
vitro, and tumor development and bone invasion in vivo
of PC-3 cells (37). In ESCC, by
direct deregulation of FSCN1, miR-145 can inhibit cell
proliferation and cell invasion in ESCC cells; it can also inhibit
cell mobility (34,38). In addition, the high levels of
expression of mature miR-145 were associated with recurrence of
metastasis in ESCC patients (39).
In bladder cancer, miR-145 also directly targets FSCN1 and inhibits
bladder cancer cell line growth, migration and invasion
significantly (40). These studies
demonstrated miR-145 acts as a tumor suppressor in the progression
and metastasis of cancer.
Our study demonstrated that miR-145 is downregulated
in the BM compared to primary lung adenocarcinoma samples.
Furthermore, we found that upregulation of miR-145 in lung
adenocarcinoma cells suppresses proliferation of tumor cells.
Consistent with our result, other reports also confirmed miR-145
can inhibit cell proliferation of human lung adenocarcinoma by
targeting c-Myc, EGFR and NUDT1 (41,42).
However, the transwell assays showed that upregulation of miR-145
has no effect on lung adenocarcinoma cancer cell migration and
invasion. The miR-145 expression level also showed no significant
difference between our primary lung adenocarcinoma samples with and
without lymph node involvement. These findings suggest that the
functional expression of miR-145 may also be cell type-specific.
Considerable evidence indicates that molecular factors that are
present in specific organs can, indeed, influence whether or not
various types of cancer cells will grow there (43,44).
The ability of cancer cells to grow in a specific site depends on
features that are inherent to the cancer cell, features inherent to
the organ and the active interplay between these factors. To
colonize a new organ outright, disseminated tumor cells must have
the capacity to productively interact with the new microenvironment
in order to extract growth and survival advantages (45–47).
Our studies suggest that downregulation of miR-145 should mainly
contribute to growth and colonization advantages of lung
adenocarcinoma cells at the metastatic site of the brain, but does
not functionally support the involvement at early stages of
metastatic disease (migration and invasion) in brain.
The formation of metastases requires the acquisition
of genetic or epigenetic changes that allow for the detachment of
cells from the primary tumor, transport and survival in the
circulation and colonization in distant organs. We found that
miR-145 is downregulated in the BM compared to primary lung
adenocarcinoma samples. Our functional analyses showed that
overexpression of miR-145 suppresses the growth of lung
adenocarcinoma cells. These results outline an important role of
miR-145 in the development of BM of lung adenocarcinoma and
implicate its potential application in cancer therapy. To further
understand the interaction between miR-145 and its targeted gene(s)
in the BM of lung adenocarcinoma, functional characterizing of the
target gene(s) is required in future studies.
Acknowledgements
The authors thank Mrs. Xueqin Chen (the Pathology
Laboratory of West China Hospital) for her assistance and tissue
samples were collected from the Pathology Department of West China
Hospital. The Agilent miRNA Microarray experiments were performed
by Shanghai Biotechnology Corporation, China. This study was
supported by the National Major Project of China (No.
2011ZX09302-001-01).
References
|
1
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lai EC: MicroRNAs are complementary to 3′
UTR sequence motifs that mediate negative post-transcriptional
regulation. Nat Genet. 30:363–364. 2002.
|
|
3
|
Berezikov E, Guryev V, van de Belt J,
Wienholds E, Plasterk RH and Cuppen E: Phylogenetic shadowing and
computational identification of human microRNA genes. Cell.
120:21–24. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha
V, Lindblad-Toh K, Lander ES and Kellis M: Systematic discovery of
regulatory motifs in human promoters and 3′ UTRs by comparison of
several mammals. Nature. 434:338–345. 2005.
|
|
5
|
Shi XB, Tepper CG and deVere White RW:
Cancerous miRNAs and their regulation. Cell Cycle. 7:1529–1538.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mezzanzanica D, Bagnoli M, De Cecco L,
Valeri B and Canevari S: Role of microRNAs in ovarian cancer
pathogenesis and potential clinical implications. Int J Biochem
Cell Biol. 42:1262–1272. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen X, Gong J, Zeng H, et al: MicroRNA145
targets BNIP3 and suppresses prostate cancer progression. Cancer
Res. 70:2728–2738. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dong Q, Meng P, Wang T, et al: MicroRNA
let-7a inhibits proliferation of human prostate cancer cells in
vitro and in vivo by targeting E2F2 and CCND2. PLoS One.
5:e101472010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs - the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Patchell RA: The management of brain
metastases. Cancer Treat Rev. 29:533–540. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nathoo N, Chahlavi A, Barnett GH and Toms
SA: Pathobiology of brain metastases. J Clin Pathol. 58:237–242.
2005. View Article : Google Scholar
|
|
13
|
Schouten LJ, Rutten J, Huveneers HA and
Twijnstra A: Incidence of brain metastases in a cohort of patients
with carcinoma of the breast, colon, kidney, and lung and melanoma.
Cancer. 94:2698–2705. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Smedby KE, Brandt L, Bäcklund ML and
Blomqvist P: Brain metastases admissions in Sweden between 1987 and
2006. Br J Cancer. 101:1919–1924. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Penel N, Brichet A, Prevost B, Duhamel A,
Assaker R, Dubois F and Lafitte JJ: Pronostic factors of
synchronous brain metastases from lung cancer. Lung Cancer.
33:143–154. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Steeg PS: Tumor metastasis: mechanistic
insights and clinical challenges. Nat Med. 12:895–904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chambers AF, Groom AC and MacDonald IC:
Dissemination and growth of cancer cells in metastatic sites. Nat
Rev Cancer. 2:563–572. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Braun S, Vogl FD, Naume B, et al: A pooled
analysis of bone marrow micrometastasis in breast cancer. N Engl J
Med. 353:793–802. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Roodman GD: Mechanisms of bone metastasis.
Discov Med. 4:144–148. 2004.
|
|
20
|
Fidler IJ: Modulation of the organ
microenvironment for treatment of cancer metastasis. J Natl Cancer
Inst. 87:1588–1592. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Radinsky R: Molecular mechanisms for
organ-specific colon carcinoma metastasis. Eur J Cancer.
31:1091–1095. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Radinsky R and Ellis LM: Molecular
determinants in the biology of liver metastasis. Surg Oncol Clin N
Am. 5:215–229. 1996.PubMed/NCBI
|
|
23
|
Fitzgerald DP, Palmieri D, Hua E, Hargrave
E, Herring JM, Qian Y, Vega-Valle E, Weil RJ, Stark AM, Vortmeyer
AO and Steeg PS: Reactive glia are recruited by highly
proliferative brain metastases of breast cancer and promote tumor
cell colonization. Clin Exp Metastasis. 25:799–810. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dykxhoorn DM, Wu Y, Xie H, et al: miR-200
enhances mouse breast cancer cell colonization to form distant
metastases. PLoS One. 4:e71812009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang H, Ach RA and Curry B: Direct and
sensitive miRNA profiling from low-input total RNA. RNA.
13:151–159. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He TC, Zhou S, da Costa LT, et al: A
simplified system for generating recombinant adenoviruses. Proc
Natl Acad Sci USA. 95:2509–2514. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lima RT, Busacca S, Almeida GM, Gaudino G,
Fennell DA and Vasconcelos MH: MicroRNA regulation of core
apoptosis pathways in cancer. Eur J Cancer. 47:163–174. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Varnholt H, Drebber U, Schulze F,
Wedemeyer I, Schirmacher P, Dienes HP and Odenthal M: MicroRNA gene
expression profile of hepatitis C virus-associated hepatocellular
carcinoma. Hepatology. 47:1223–1232. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu X, Sempere LF, Galimberti F, et al:
Uncovering growth-suppressive MicroRNAs in lung cancer. Clin Cancer
Res. 15:1177–1183. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dyrskjøt L, Ostenfeld MS, Bramsen JB, et
al: Genomic profiling of microRNAs in bladder cancer: miR-129 is
associated with poor outcome and promotes cell death in vitro.
Cancer Res. 69:4851–4860. 2009.PubMed/NCBI
|
|
31
|
Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim
JH, Kim JW and Kim S: MicroRNA expression profiles in serous
ovarian carcinoma. Clin Cancer Res. 14:2690–2695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Arndt GM, Dossey L, Cullen LM, et al:
Characterization of global microRNA expression reveals oncogenic
potential of miR-145 in metastatic colorectal cancer. BMC Cancer.
9:3742009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Götte M, Mohr C, Koo CY, et al:
miR-145-dependent targeting of junctional adhesion molecule A and
modulation of fascin expression are associated with reduced breast
cancer cell motility and invasiveness. Oncogene. 29:6569–6580.
2010.PubMed/NCBI
|
|
34
|
Wu BL, Xu LY, Du ZP, et al: MiRNA profile
in esophageal squamous cell carcinoma: downregulation of miR-143
and miR-145. World J Gastroenterol. 17:79–88. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sachdeva M and Mo YY: MicroRNA-145
suppresses cell invasion and metastasis by directly targeting mucin
1. Cancer Res. 70:378–387. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fuse M, Nohata N, Kojima S, et al:
Restoration of miR-145 expression suppresses cell
proliferation, migration and invasion in prostate cancer by
targeting FSCN1. Int J Oncol. 38:1093–1101. 2011.
|
|
37
|
Peng X, Guo W, Liu T, et al:
Identification of miRs-143 and -145 that is associated with bone
metastasis of prostate cancer and involved in the regulation of
EMT. PLoS One. 6:e203412011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kano M, Seki N, Kikkawa N, et al:
miR-145, miR-133a and miR-133b: Tumor-suppressive
miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J
Cancer. 127:2804–2814. 2010. View Article : Google Scholar
|
|
39
|
Akagi I, Miyashita M, Ishibashi O, et al:
Relationship between altered expression levels of MIR21, MIR143,
MIR145, and MIR205 and clinicopathologic features of
esophageal squamous cell carcinoma. Dis Esophagus. 24:523–530.
2011.
|
|
40
|
Chiyomaru T, Enokida H, Tatarano S, et al:
miR-145 and miR-133a function as tumour suppressors
and directly regulate FSCN1 expression in bladder cancer. Br J
Cancer. 102:883–891. 2010. View Article : Google Scholar
|
|
41
|
Cho WC, Chow AS and Au JS: Restoration of
tumour suppressor hsa-miR-145 inhibits cancer cell growth in
lung adenocarcinoma patients with epidermal growth factor receptor
mutation. Eur J Cancer. 45:2197–2206. 2009.PubMed/NCBI
|
|
42
|
Cho WC, Chow AS and Au JS: MiR-145
inhibits cell proliferation of human lung adenocarcinoma by
targeting EGFR and NUDT1. RNA Biol. 8:125–131. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fidler IJ: Seed and soil revisited:
contribution of the organ microenvironment to cancer metastasis.
Surg Oncol Clin N Am. 10:257–269. 2001.PubMed/NCBI
|
|
44
|
Radinsky R: Modulation of tumor cell gene
expression and phenotype by the organ-specific metastatic
environment. Cancer Metastasis Rev. 14:323–338. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gupta GP and Massagué J: Cancer
metastasis: building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Poste G and Fidler IJ: The pathogenesis of
cancer metastasis. Nature. 283:139–146. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Fidler IJ: The organ microenvironment and
cancer metastasis. Differentiation. 70:498–505. 2002. View Article : Google Scholar : PubMed/NCBI
|