Introduction
In Japan, the number of lung cancer patients is
increasing and lung cancer has become the leading and the second
largest cause of cancer-related mortality in men and women,
respectively (1). Since the
improvement in diagnostic technologies for lung cancer, an
increasing number of patients are being diagnosed in the early
stages of the disease. In cases where non-small cell lung cancer
(NSCLC) is diagnosed in the early stages, favorable prognoses have
been reported after treatment with lobectomy (2–5), and
lobectomy without any adjuvant therapy is an approved standard of
therapy for these patients (3–5).
However, we often encounter rapid tumor progression after
lobectomy, even in these patients. If, therefore, the likelihood of
this rapid progression could be predicted, it would be reasonable
to initiate adjuvant therapy in advance.
The regenerating gene (Reg) was originally
discovered in the regeneration of pancreatic β-cells (6–8). There
are currently five genes in the REG family found in humans
(REG Iα, REG Iβ, REG III, HIP/PAP and
REG IV) (9), encoding a
growth factor family of proteins involved not only in regeneration
of damaged tissues but also in the growth of various types of
cancers, including gastrointestinal cancer, cholangiocarcinoma,
pancreatic cancer, breast cancer and prostate cancer (10–27). A
correlation between REG Iα expression and poor prognosis has
also been reported in NSCLC (28).
While studies have indicated that poor prognosis in patients
expressing REG Iα appears to be due to an increased cell
number in gastric and pancreatic cancers (13,26),
the impact of REG Iα on cancer cells has not been examined
in NSCLC.
In the present study, the effects of the expression
of REG Iα and REG Iβ, which has a similar structure
to REG Iα and seems to have an identical function to REG
Iα, on adenocarcinoma (AD) and squamous cell carcinoma (SCC)
cells were examined in vitro. In addition, we investigated
the correlation between expression of REG family genes and
the prognosis of AD and SCC patients.
Materials and methods
Human lung cancer cell lines
The HLC-1 human lung adenocarcinoma cell line and
the EBC-1 human squamous cell carcinoma cell line were obtained
from Riken BioResource Center (Tsukuba, Japan). HLC-1 and EBC-1
cells were maintained in Ham’s F12 and minimum essential medium
(MEM), respectively. No expression of any of the REG family
genes was confirmed in these cells by real-time RT-PCR.
Establishment of stable transfectants for
REG Iα and REG Iβ
We established two cell lines expressing the REG
Iα or the REG Iβ gene in HLC-1 and EBC-1 cells and one
mock-transfected cell line as a control for each cell type. The
expression vectors or a control vector (without insert DNA) were
then transfected into HLC-1 or EBC-1 cells by electroporation
(17). Stable transfectants were
selected after 2 weeks of culture with 500 μg/ml
Geneticin® (Gibco, Carlsbad, CA, USA). REG Iα or
REG Iβ expression was confirmed by real-time RT-PCR and
immunoblot analysis of the culture medium, as previously described
(17). The resulting
Geneticin-resistant clones were designated as HLC-1 REG Iα-1, -2;
HLC-1 REG Iβ-1, -2; HLC-1 mock; EBC-1 REG Iα-1, -2; EBC-1 REG Iβ-1,
-2; and EBC-1 mock.
Cell number, cell invasive capacity and
anchorage-independent cell growth
For evaluation of cell growth in the HLC-1 and EBC-1
cell lines, cells were cultured in Ham’s F12 or MEM containing 1 or
0.5% FBS, respectively. The cell number for the HLC-1 cells was
determined using a Cell Counting Kit-8 (Dojindo, Mashikimachi,
Japan) on 1, 3, 5 and 7 days of culture, and that for EBC-1 was
monitored on 0, 1, 2 and 3 days of culture. Increases in the cell
number were expressed as the percentage of the cell number at
culture day 1 or 0, respectively. Cell invasive activity was
monitored using a Cultrex 96 Well BME Cell Invasion assay
(Trevigen, Gaithersburg, MD, USA). To evaluate
anchorage-independent cell growth, cells (1.75×103) were
plated into 12-well plates in culture medium containing 0.35% agar
on top of 0.5% agar, prepared in the same medium. The plates were
incubated at 37°C for 16 days. Colonies were stained with 0.005%
crystal violet for 1 h. Colonies, containing at least 50 cells,
were counted.
Patients
Fifty-one AD and 23 SCC patients, who underwent
surgery at Nara Medical University Hospital from 2004 to 2007, were
enrolled. The present study was approved by the Ethics Committee of
the Nara Medical University School of Medicine. Fifty-one were male
and 23 were female, and the mean age was 68.3±1.1 years. Forty-six
patients (AD, 32 patients; SCC, 14 patients) were in pathological
stage I, 8 patients (AD, 2; SCC, 6) were in stage II and 20
patients (AD, 17; SCC, 3) were in stage III. Sixty-eight patients
(AD, 47; SCC, 21) underwent complete resection and the remaining 6
patients (AD, 4; SCC, 2) in stage III received incomplete resection
because of the extensive invasion of the tumors into the
surrounding organs.
Real-time RT-PCR of surgical tissue
samples
Samples (tumor and normal lung tissues) were
collected immediately after lung resection (surgical sample), and
frozen in liquid nitrogen until RNA isolation. Total RNA was
isolated for real-time reverse transcription-polymerase chain
reaction (real-time RT-PCR), as previously described (27,28).
The primers and probes (Table I)
were synthesized by Nihon Gene Research Laboratories (Sendai,
Japan). Real-time RT-PCR was then carried out using
TaqMan® Universal PCR Master Mix in an ABI
PRISM® 7700 Sequence Detection system (Applied
Biosystems, Foster City, CA, USA). Expression of REG family
genes was normalized with respect to β-actin. The cut-off levels
for expression of each gene were set at the average + 3SD
expression of the normal lung tissues. The expression of each
REG family gene, which was higher or lower than the cut-off
level, was defined as high or weak, respectively, and the absence
of the expression of each gene was defined as no expression. For
analysis of the correlation between the expression of each gene and
prognosis, patients with high expression were defined as positive,
and those with weak or absence of expression were defined as
negative.
 | Table IPrimers and probes for real-time
RT-PCR. |
Table I
Primers and probes for real-time
RT-PCR.
| Gene (Accession
no.) | Sequence |
|---|
| β-actin
(NM_001101) | Forward:
5′-GCGAGAAGATGACCCAGA-3′
Reverse: 5′-CAGAGGCGTACAGGGATA-3′
Probe: 5′-FAM-ACAGCCTGGATAGCAACGTACATGGCT-TAMRA-3′ |
| REG Iα
(NM_002909) | Forward:
5′-AGGAGAGTGGCACTGATGACTT-3′
Reverse: 5′-TAGGAGACCAGGGACCCACTG-3′
Probe: 5′-FAM-TGGCCTCCATGACCCCAAAAAGAAC-TAMRA-3′ |
| REG Iβ
(NM_006507) | Forward:
5′-GCTGATCTCCTCCCTGATGTTC-3′
Reverse: 5′-GGCAGCTGATTCGGGGATTA-3′
Probe: 5′-FAM-TGTCTCTGAGCCAAGGCCAGGAGTCCCA-TAMRA-3′ |
| REG III
(AB161037) | Forward:
5′-GAATATTCTCCCCAAACTG-3′
Reverse: 5′-GAGAAAAGCCTGAAATGAAG-3′
Probe: 5′-FAM-CCTACCTGACTACCTTGTCATGATCCTCC-TAMRA-3′ |
| HIP/PAP
(NM_138937) | Forward:
5′-AGAGAATATTCGCTTAATTCC-3′
Reverse: 5′-AATGAAGAGACTGAAATGACA-3′
Probe: 5′-FAM-CCAACCTGACCACCTCATTCTTATCTTTC-TAMRA-3′ |
| REG IV
(AY007243) | Forward:
5′-ATCCTGGTCTGGCAAGTC-3′
Reverse: 5′-CGTTGCTGCTCCAAGTTA-3′
Probe: 5′-FAM-CTGTGCTGAGATGAGCTCCAATAACAACTT-TAMRA-3′ |
Real-time RT-PCR of formalin-fixed
paraffin-embedded (FFPE) samples
Total RNA was isolated from FFPE tissue specimens
(AD, 10; SCC, 8, randomly selected) using the RNeasy FFPE kit
(Qiagen, Hilden, Germany) and reverse transcribed as described
above. Real-time PCR was performed using KAPA SYBR® FAST
qPCR Master Mix (Kapa Biosystems, Boston, MA, USA) and the Thermal
Cycler Dice Real-Time System (Takara, Otsu, Japan) as previously
described (29–31).
Disease-specific survival
Patient death in the progression of lung cancer was
defined as the end point. Kaplan-Meier survival curves for
disease-specific survival (DSS) were constructed according to the
expression of REG Iα or REG IV genes.
Statistics
Data are expressed as the mean ± standard error of
the mean (SEM), and cell number, cell invasive activity and
anchorage-independent cell growth were compared by unpaired
t-tests. Comparison of clinicopathological parameters according to
the expression of the REG Iα gene was carried out by
Chi-squared analyses. Kaplan-Meier survival curves for DSS were
compared using the log-rank test. Correlations of the expression
levels of the REG Iα gene from surgical and FFPE samples
were analyzed using Pearson non-parametric tests. A P-value of
<0.05 was considered to indicate a statistically significant
result.
Results
Effects of the transfection of REG Iα and
REG Iβ on cell number, cell invasive activity and
anchorage-independent cell growth in lung cancer cells
The expression of REG Iα or REG Iβ in
HLC-1 REG Iα/Iβ and EBC-1 REG Iα/Iβ cells was confirmed by
real-time RT-PCR, whereas no expression of REG Iα or REG
Iβ was detected in the HLC-1 and EBC-1 mock control cell lines.
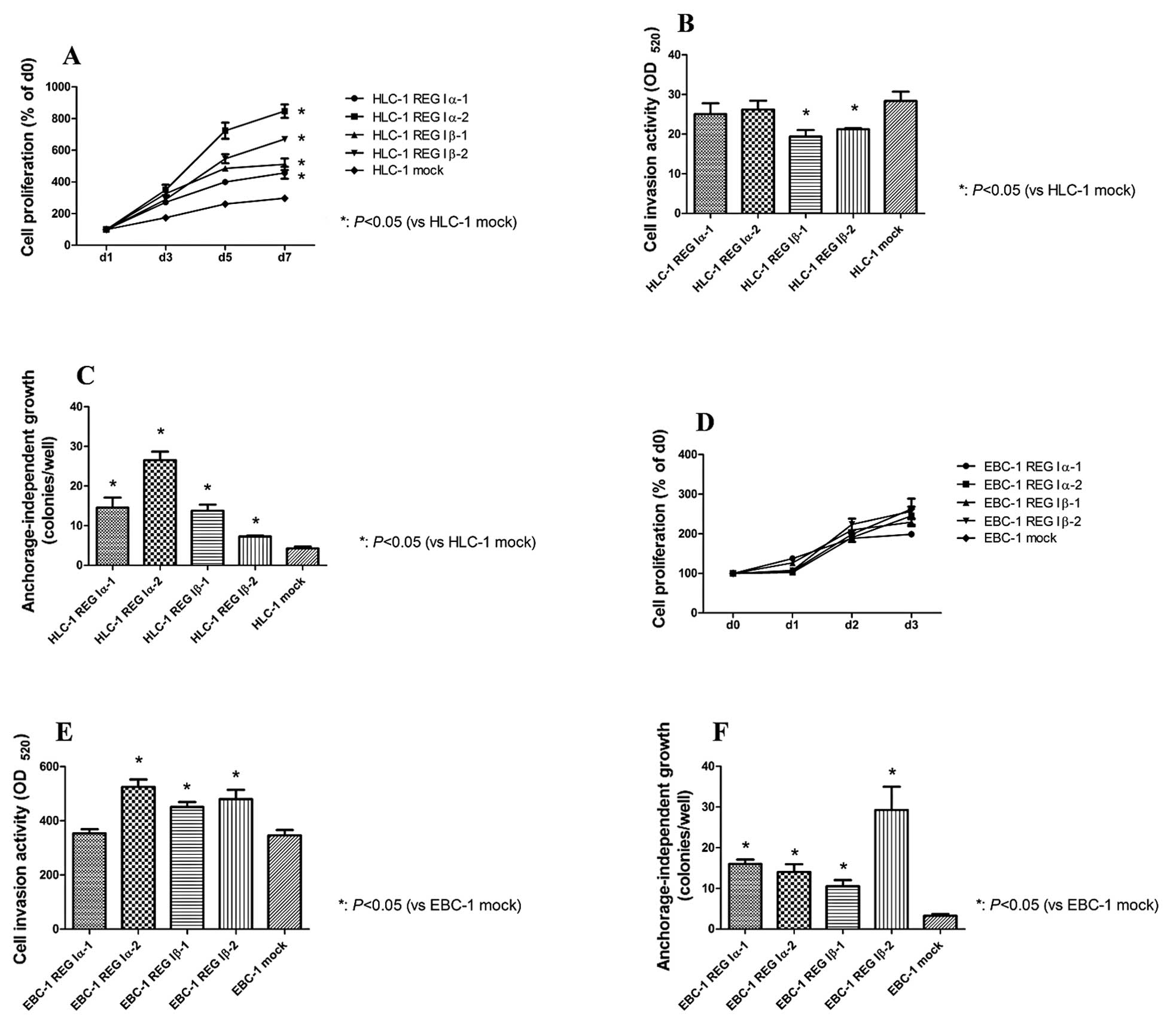
All of the HLC-1 REG Iα/Iβ-transfected cell lines showed a
significant increase in cell number when compared with the HLC-1
mock cells on culture day 7 (Fig.
1A). In contrast, HLC-1 REG Iα cells did not show increased
cell invasive activity when compared with the HLC-1 mock cells,
while HLC-1 REG Iβ cells in fact showed a decelerated invasive
potential (Fig. 1B). HLC-1 REG
Iα/Iβ cells showed significant increases in anchorage-independent
cell growth as compared with the HLC-1 mock cells (Fig. 1C).
By comparison, we observed no significant increases
in cell number for the EBC-1 REG Iα/Iβ-transfected cells as
compared with the EBC-1 mock cells after 3 days of culture
(Fig. 1D). EBC-1 REG Iα-2, EBC-1
REG Iβ-1 and -2 cells showed increased cell invasive activity as
compared with the EBC-1 mock cells (Fig. 1E), and all of the EBC-1 REG Iα/Iβ
cell lines showed a significant increase in anchorage independent
cell growth when compared with the EBC-1 mock cells (Fig. 1F).
REG family gene expression in normal lung
and tumor tissues
Expression of all the REG family genes,
except for REG Iβ, was observed in both normal lung and
tumor tissues (Fig. 2). REG
Iβ was expressed only in 3 AD patients. The expression of
REG III and HIP/PAP was noted in ~90% of both normal
lung and tumor tissues. The expression profile of these genes was
not different between the normal lung and tumor tissues.
Comparatively, REG Iα and REG IV mRNAs were observed
more frequently in tumor tissues than in normal lung tissues.
Therefore, we focused on the correlation between the expression of
REG Iα and REG VI in tumor tissues and the prognosis
of patients in the subsequent studies.
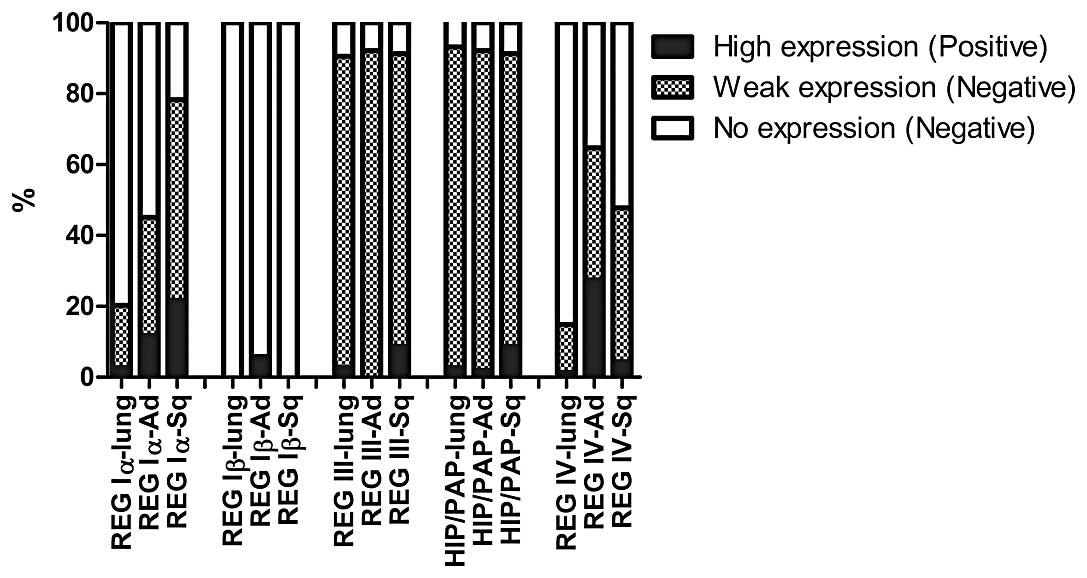 | Figure 2Expression levels of REG
family genes in normal lung and tumor tissues. The cut-off levels
for expression of each REG family gene were set at average +
3SD expression of the normal lung tissues. The expression of each
REG family gene, which was higher or lower than the cut-off
level, was defined as high expression (positive) or weak expression
(negative), respectively. The absence of expression of each gene,
was defined as no expression (negative). REG Iα-lung, REG Iα
expression in normal lung tissues; REG Iα-Ad, REG Iα
expression in AD; REG Iα-Sq, REG Iα expression in SCC; REG
Iβ-lung, REG Iβ expression in normal lung tissues; REG
Iβ-Ad, REG Iβ expression in AD; REG Iβ-Sq, REG Iβ
expression in SCC; REG III-lung, REG III expression in
normal lung tissues; REG III-Ad, REG III expression in AD;
REG III-Sq, REG III expression in SCC; HIP/PAP-lung,
HIP/PAP expression in normal lung tissues; HIP/PAP-Ad,
HIP/PAP expression in AD; HIP/PAP-Sq, HIP/PAP
expression in SCC; REG IV-lung, REG IV expression in one
normal lung tissue; REG IV-Ad, REG IV expression in AD; REG
IV-Sq, REG IV expression in SCC. |
REG Iα expression and prognosis of
patients
In the 68 patients (AD, 47; SCC, 21) who underwent
complete resection, there were no significant differences in
gender, age or pathological stage between patients who were
positive and those who were negative for REG Iα expression
(Table II). First, we evaluated
the relationship between the expression of REG Iα and
prognosis in these 68 patients. Ten patients (AD, 5; SCC, 5) showed
positive expression for REG Iα, whereas 58 patients (AD, 42;
SCC, 16) showed negative expression. Overall, there was no
significant correlation between patients with positive or negative
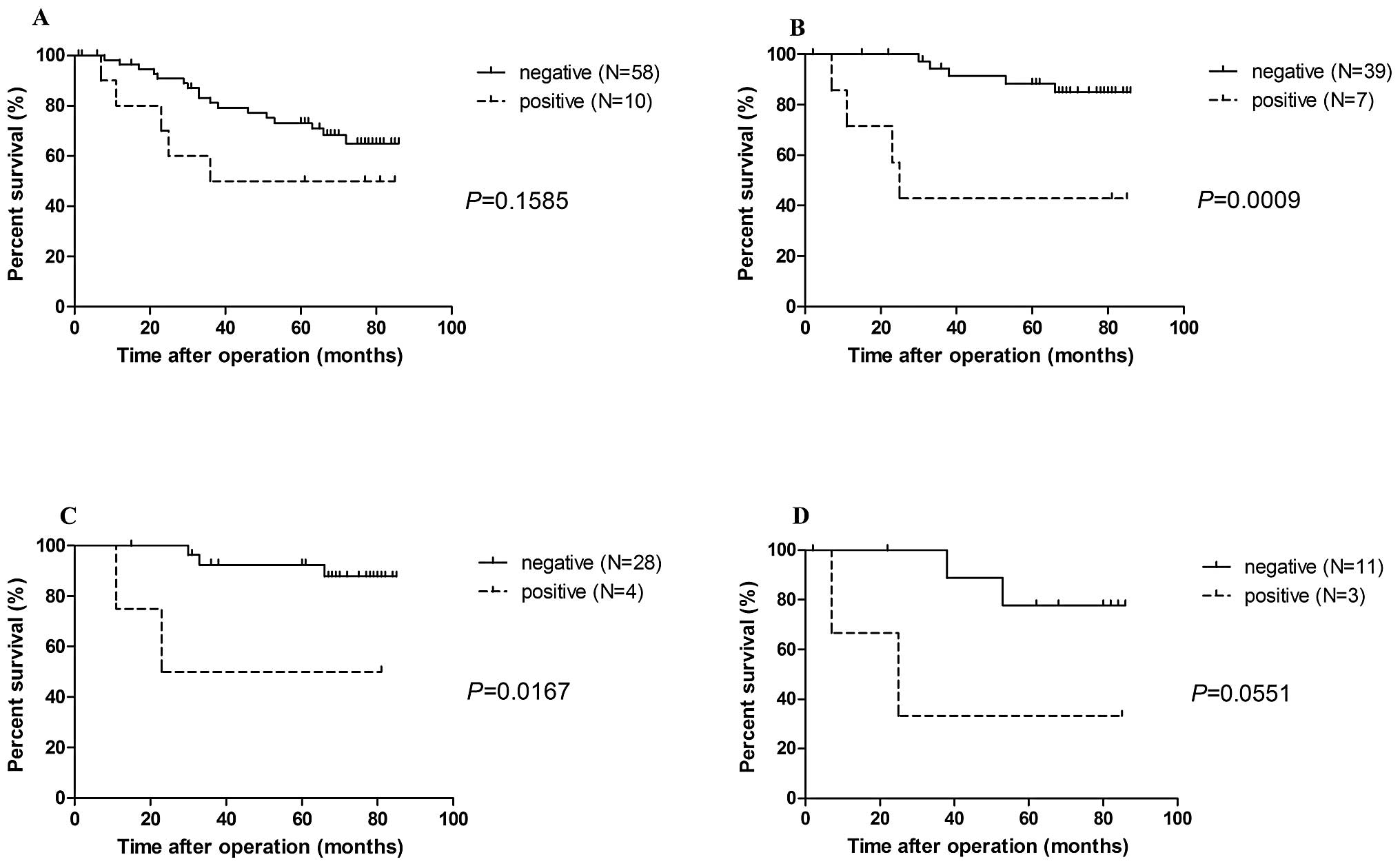
REG Iα expression and prognosis (P=0.1585; Fig. 3A). However, when we examined the 46
stage I patients separately, we observed a significantly worse
prognosis in patients with positive REG Iα expression (n=7)
than those with negative REG Iα expression (n=39) (P=0.0009;
Fig. 3B). In addition, the 5-year
survival in these patients with positive REG Iα expression
was significantly lower than that in patients with negative REG
Iα expression (42.9 vs. 84.9%; P=0.034). Next, we divided 46
stage I patients into two groups by histological types: AD (n=32)
and SCC (n=14). The prognosis of stage I AD patients positive for
REG Iα expression was significantly worse than that for
patients negative for REG Iα (P=0.0167; Fig. 3C). In stage I SCC patients, however,
there was a trend toward poor prognosis in patients with positive
REG Iα expression (P=0.0551; Fig. 3D) when compared with the negative
patients. Concerning REG IV expression and patient
prognosis, no correlation was noted for any of the subgroupings
detailed above (data not shown).
 | Table IICharacteristics of the lung cancer
patients with complete resection. |
Table II
Characteristics of the lung cancer
patients with complete resection.
| Adenocarcinoma | Squamous cell
carcinoma |
|---|
|
|
|
|---|
| REG Iα gene
expression | REG Iα gene
expression |
|---|
|
|
|
|---|
| Positive | Negative | P-value | Positive | Negative | P-value |
|---|
| Gender |
| Male | 3 | 25 | 0.64 | 4 | 15 | 0.97 |
| Female | 2 | 17 | | 1 | 1 | |
| Age (years) | 72.0±2.2 | 66.0±1.5 | 0.15 | 73.0±2.7 | 71.6±2.0 | 0.72 |
| Tumor stage |
| I | 4 | 28 | 0.92 | 3 | 11 | 0.86 |
| II and III | 1 | 14 | | 2 | 5 | |
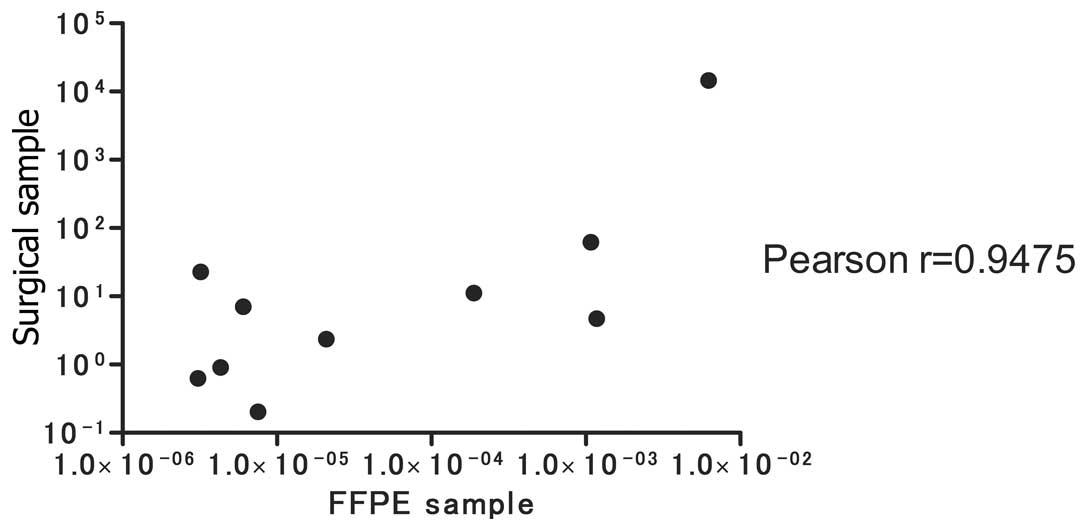
REG Iα expression in FFPE samples
Next, we tested REG Iα expression in FFPE
samples taken from a random selection of AD (n=10) and SCC (n=8)
patients, and compared the results with REG Iα expression in
surgical samples. As shown in Fig.
4, a significant correlation was noted between REG Iα
expression from the surgical samples and that from the FFPE samples
(Pearson correlation r=0.9475).
Discussion
The effect of REG family genes on
malignancies has been studied mainly in gastrointestinal cancers
(10–15,17–20).
REG Iα and REG IV was found to be correlated with
poor prognosis in gastric and colorectal cancers (12–15,27). A
correlation, however, has recently been reported to exist between
high expression of REG Iα and a more favorable prognosis in
esophageal cancer patients (18).
The authors indicated that high expression of REG Iα
enhanced the chemosensitivity and radiosensitivity of esophageal
cancer cells, which may explain the better prognosis of the
patients (17). This high
expression is contradictory to the findings of others (14–16,26–28),
but may be explained by histopathological differences between other
gastrointestinal tumors and esophageal tumors; most esophageal
cancers are SCCs, whereas other gastrointestinal cancers are ADs.
Gastric AD patients with REG Iα expression were reported to
show poor prognosis, and REG Iα-expressing cells exhibit an
increase in cell number (13). In
lung cancer, the reason why REG Iα expression leads to a
poorer prognosis has not been clarified (28). We hypothesized that discrete
mechanisms may exist in lung cancer cells due to histological
distinctions between AD and SCC cells. Thus, we performed an in
vitro study to clarify the effect of the expression of REG
Iα and REG Iβ, which has a similar structure to REG
Iα and seems to have an identical function to REG Iα, on
AD and SCC cells.
In AD cells, both REG Iα and Iβ
increased cell numbers as compared with the control cells, whereas,
in SCC cells, neither REG Iα nor Iβ influenced cell
number. In contrast, no clear effect was found in the AD cells in
regards to enhanced cell invasion in response to either gene,
whereas a positive effect was demonstrated in SCC cells.
Anchorage-independent cell growth, however, was upregulated for
both cell types expressing REG Iα and Iβ. These
results suggest that the effect of REG Iα and Iβ on
lung cancer is specific in regards to the type of tumor. From these
results, we hypothesized that patients who express the REG
Iα and Iβ genes may have poor prognosis by different
mechanisms as described above. We evaluated the relationship
between the expression of these genes and patient prognosis.
Despite recent findings that a link exists between
the REG family genes and various significant cancer subtypes
(10–27), including lung cancer (28), the expression levels of this family
of genes have not been explored. In the present study, we evaluated
the expression levels of REG family genes in lung cancer
tissues. Almost all of the REG family genes were expressed
both in normal lung and tumor tissues except for REG Iβ.
However, positive ratios of gene expression levels varied for each
REG family member. REG III and HIP/PAP were
high in both normal lung and tumor tissues. Conversely, the
expression ratios of REG Iα and REG IV in tumor
tissues were higher than those in normal lung tissues. Previous
studies have shown that prognoses are worse in patients with
stomach, pancreatic, lung, and breast cancers with high REG
Iα expression (14–16,26–28).
Likewise, high REG IV expression in colorectal and prostate
cancers is linked with a worse prognosis (12,24).
Therefore, we also tested correlations between the expression of
REG Iα and REG IV and patient prognosis in lung
cancer patients. We found that a high expression of REG Iα
was correlated with poor prognosis in stage I lung cancer patients,
suggesting that REG Iα is a reliable marker for the
prognosis of stage I lung cancer patients. The in vitro
study confirmed that the REG Iβ gene promoted an increased
cell number and anchorage-independent cell growth in AD cells, and
increased cell invasive activity and anchorage-independent cell
growth in SCC cells. However, REG Iβ was expressed only in 3
AD patients. Therefore, the expression of the REG Iβ gene
seems to have no meaning clinically. Together with the in
vitro data, we surmised that poorer prognosis in REG
Iα-expressing AD patients stems from an increase in cell number
and anchorage-independent cell growth, whereas the tendency for a
poorer prognosis in SCC patients with positive expression of REG
Iα might be due to enhanced cell invasion and
anchorage-independent cell growth. In comparison, we found no
correlation between REG IV and prognosis, suggesting a
different role for REG IV in lung cancer (12,24).
As it is not easy to obtain fresh frozen surgical
samples, we also evaluated the expression of the REG Iα gene
in FFPE samples to compare an easier, more practical and more
economical method for RNA extraction for future clinical
applications. We found a significant correlation in REG Iα
expression between the two different sampling and real-time RT-PCR
methods (Fig. 4). Clinically, the
positive effect of REG Iα in lung cancer cells implies that
REG Iα could be used as an indicator to initiate adjuvant
therapy, even in stage I lung cancer patients; alternatively, it
may become a target for therapy or a marker of chemosensitivity and
radiosensitivity (18).
One of the limitations of the present study was the
small number of participants in the SCC group, as the correlation
between REG Iα and prognosis could only be evaluated in 14
stage I SCC patients. This may explain the lack of a significant
correlation between the expression of REG Iα and SCC
prognosis.
In summary, the REG Iα gene increased the
cell number and anchorage-independent cell growth of lung
adenocarcinoma cells, and the cell invasive activity and
anchorage-independent cell growth in lung squamous cell carcinoma.
Overexpression of the REG Iα gene is a risk factor for poor
prognosis in lung cancer patients functioning via different
mechanisms in adenocarcinoma and squamous cell carcinoma.
Acknowledgements
We are grateful to Dr Maiko Takeda and Dr Takahiko
Kasai, Nara Medical University School of Medicine for their kind
assistance. The present study was for partial academic fulfillment
of the degree thesis by M.K. of Medical Science at Nara Medical
University. The study was also supported in part by grants-in-aid
for Scientific Research (Practical Application Research) from the
Japan Science and Technology Agency.
Abbreviations:
|
AD
|
adenocarcinoma
|
|
DSS
|
disease-specific survival
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
NSCLC
|
non-small cell lung cancer
|
|
Reg
|
regenerating gene
|
|
SCC
|
squamous cell carcinoma
|
References
|
1
|
Cancer statistics in Japan ’08. November
5–2008, Available at: http://ganjoho.jp/pro/statistics/en/gdb_year.html?1%1.
Accessed July 24, 2012
|
|
2
|
Whitson BA, Groth SS, Duval SJ, Swanson SJ
and Maddaus MA: Surgery for early-stage non-small cell lung cancer:
a systematic review of the video-assisted thoracoscopic surgery
versus thoracotomy approaches to lobectomy. Ann Thorac Surg.
86:2008–2016. 2008. View Article : Google Scholar
|
|
3
|
Wright G, Manser RL, Byrnes G, Hart D and
Campbell DA: Surgery for non-small cell lung cancer: systematic
review and meta-analysis of randomised controlled trials. Thorax.
61:597–603. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mountain CF: Revisions in the
International System for Staging Lung Cancer. Chest. 111:1710–1717.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ginsberg RJ and Rubinstein LV: Randomized
trial of lobectomy versus limited resection for T1 N0 non-small
cell lung cancer. Lung Cancer Study Group Ann Thorac Surg.
60:615–622. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Terazono K, Yamamoto H, Takasawa S, et al:
A novel gene activated in regenerating islets. J Biol Chem.
263:2111–2114. 1988.PubMed/NCBI
|
|
7
|
Watanabe T, Yonemura Y, Yonekura H, et al:
Pancreatic beta-cell replication and amelioration of surgical
diabetes by Reg protein. Proc Natl Acad Sci USA. 91:3589–3592.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Takasawa S, Ikeda T, Akiyama T, et al:
Cyclin D1 activation through ATF-2 in Reg-induced pancreatic β-cell
regeneration. FEBS Lett. 580:585–591. 2006.PubMed/NCBI
|
|
9
|
Nata K, Liu Y, Xu L, et al: Molecular
cloning, expression and chromosomal localization of a novel human
REG family gene, REG III. Gene. 340:161–170. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sekikawa A, Fukui H, Fujii S, et al: REG
Iα protein may function as a trophic and/or anti-apoptotic factor
in the development of gastric cancer. Gastroenterology.
128:642–653. 2005.
|
|
11
|
Mitani Y, Oue N, Matsumura S, et al: Reg
IV is a serum biomarker for gastric cancer patients and predicts
response to 5-fluorouracil-based chemotherapy. Oncogene.
26:4383–4393. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Oue N, Kuniyasu H, Noguchi T, et al: Serum
concentration of Reg IV in patients with colorectal cancer:
overexpression and high serum levels of Reg IV are associated with
liver metastasis. Oncology. 72:371–380. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fukui H, Fujii S, Takeda J, et al:
Expression of REG Iα protein in human gastric cancers. Digestion.
69:177–184. 2004.
|
|
14
|
Yamagishi H, Fukui H, Sekikawa A, et al:
Expression profile of REG family proteins REG Iα and REG IV in
advanced gastric cancer: comparison with mucin phenotype and
prognostic markers. Mod Pathol. 22:906–913. 2009.
|
|
15
|
Dhar DK, Udagawa J, Ishihara S, et al:
Expression of regenerating gene I in gastric adenocarcinomas:
correlation with tumor differentiation status and patient survival.
Cancer. 100:1130–1136. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sasaki Y, Minamiya Y, Takahashi N, et al:
REG1A expression is an independent factor predictive of poor
prognosis in patients with breast cancer. Ann Surg Oncol.
15:3244–3251. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hayashi K, Motoyama S, Koyota S, et al:
REG I enhances chemo- and radiosensitivity in squamous cell
esophageal cancer cells. Cancer Sci. 99:2491–2495. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hayashi K, Motoyama S, Sugiyama T, et al:
REG Iα is a reliable marker of chemoradiosensitivity in squamous
cell esophageal cancer patients. Ann Surg Oncol. 15:1224–1231.
2008.
|
|
19
|
Motoyama S, Sugiyama T, Ueno Y, et al: REG
I expression predicts long-term survival among locally advanced
thoracic squamous cell esophageal cancer patients treated with
neoadjuvant chemoradiotherapy followed by esophagectomy. Ann Surg
Oncol. 13:1724–1731. 2006. View Article : Google Scholar
|
|
20
|
Usami S, Motoyama S, Koyota S, et al:
Regenerating gene I regulates interleukin-6 production in squamous
esophageal cancer cells. Biochem Biophys Res Commun. 392:4–8. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kiji T, Dohi Y, Takasawa S, Okamoto H,
Nonomura A and Taniguchi S: Activation of regenerating gene
Reg in rat and human hearts in response to acute stress. Am
J Physiol Heart Circ Physiol. 289:H277–H284. 2005.PubMed/NCBI
|
|
22
|
Kiji T, Dohi Y, Nishizaki K, et al:
Enhancement of cell viability in cryopreserved rat vascular grafts
by administration of regenerating gene (REG) inducers. J Vasc Res.
40:132–139. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Harada K, Zen Y, Kanemori Y, et al: Human
REG I gene is up-regulated in intrahepatic cholangiocarcinoma and
its precursor lesions. Hepatology. 33:1036–1042. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ohara S, Oue N, Matsubara A, et al: Reg IV
is an independent prognostic factor for relapse in patients with
clinically localized prostate cancer. Cancer Sci. 99:1570–1577.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou L, Zhang R, Wang L, et al:
Upregulation of REG Iα accelerates tumor progression in pancreatic
cancer with diabetes. Int J Cancer. 127:1795–1803. 2010.
|
|
26
|
Yonemura Y, Sakurai S, Yamamoto H, et al:
REG gene expression is associated with the infiltrating
growth of gastric carcinoma. Cancer. 98:1394–1400. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zheng HC, Sugawara A, Okamoto H, et al:
Expression profile of the REG gene family in colorectal
carcinoma. J Histochem Cytochem. 59:106–115. 2011.PubMed/NCBI
|
|
28
|
Minamiya Y, Kawai H, Saito H, et al: REG1A
expression is an independent factor predictive of poor prognosis in
patients with non-small cell lung cancer. Lung Cancer. 60:98–104.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Takasawa S, Kuroki M, Nata K, et al: A
novel ryanodine receptor expressed in pancreatic islets by
alternative splicing from type 2 ryanodine receptor gene. Biochem
Biophys Res Commun. 397:140–145. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ota H, Tamaki S, Itaya-Hironaka A, et al:
Attenuation of glucose-induced insulin secretion by intermittent
hypoxia via down-regulation of CD38. Life Sci. 90:206–211. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Masui T, Ota I, Itaya-Hironaka A, et al:
Expression of REG III and prognosis in head and neck cancer. Oncol
Rep. 30:573–578. 2013.PubMed/NCBI
|