Introduction
Lung cancer was the most commonly diagnosed cancer
as well as the leading cause of cancer-related mortality in males
in 2008 worldwide (1). Similar to
other cancers, lung cancer is thought to be caused by the
deregulation of normal gene expression (2).
Fibulins are widely distributed and are localized at
basal membranes mediating cell-to-cell and cell-to-matrix
communication. They are characterized by repeated epidermal growth
factor (EGF)-like domains and a unique C-terminal structure
(3,4). The relationship of the fibulin gene
family members and many types of cancer has been reported (5–7). The
fibulin-3 gene, also known as EFEMP1 (epidermal
growth factor-containing fibulin-like extracellular matrix protein
1) located at chromosome 2p16, is one of the 7 members of the
fibulin gene family of extracellular glycoproteins (8). The level of fibulin-3 expression was
found to be decreased in many cancer types due to aberrant promoter
methylation and is correlated with poor survival of patients with
breast cancer (9) lung cancer
(10) and hepatocellular carcinoma
(11). Lecka-Czernik et
al(12) predicted that this
protein has 2 possible isoforms, a ‘long’ form of 53 to 55 kDa,
with an NH2-terminal Ca2+-binding epidermal growth
factor-like repeat, and a ‘short’ form of 40 to 43 kDa without this
domain. However, which form of fibulin-3 is expressed in lung
cancer cells remains unclear.
Pathologic and functional studies were carried out
to determine the role of fibulin-3 in suppressing lung
cancer both in vivo and in vitro. Further elucidation
of the functions of fibulin-3 may lead to a better
understanding of the pathogenesis of lung cancer. Fibulin-3 is a
potential molecular marker for lung cancer.
Patients and methods
Cell culture
The A549 cell line was obtained from ATCC
(Rockville, MD, USA). Cells were maintained at 37°C in a 5%
CO2 incubator in RPMI-1640 medium (Sigma-Aldrich,
Carlsbad, CA, USA) containing 10% heat-inactivated fetal bovine
serum (FBS) and 1% penicillin-streptomycin.
Subjects
This study was approved by China Medical University
Ethics Committee and was conducted according to regulations of the
Helsinki Declaration. All patients provided written informed
consent to participate in the present study. Fifty-six patients who
were diagnosed with primary non-small cell lung cancer (NSCLC) at
the Department of Thoracic Surgery, The First Affiliated Hospital
of China Medical University from January 2006 to December 2010 were
included in the present study. Twenty-eight patients had stage IIIB
NSCLC (locally advanced) and 28 patients had stage IV NSCLC
(metastatic) according to the International Association for the
Study of Lung Cancer (IASLC) staging committee (13). All diagnoses were based on standard
laboratory tests (cytology and histology), and confirmed by
computerized tomography of the thorax. None of the patients
underwent radiotherapy or chemotherapy prior to surgery.
Quantitative real-time PCR
Total RNA was isolated from the tissues using TRIzol
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according
to the manufacturer's protocol. First-strand cDNA was reverse
transcribed with 1 μg of total RNA, using Takara Reverse
Transcription kit and oligo(dT)15 primers (both from Takara,
Dalian, China). The resultant cDNA was then used for quantitative
PCR reactions. The fibulin-3 primers were:
5′-GAGCAGCTCCAGGGGACCGCCGCG-3′ (sense) and
5′-TCCCCGACACGCTACCTTCG-3′ (antisense). The housekeeping gene,
GAPDH, was used as an internal control for normalization of
the results. The GAPDH primers were:
5′-AGAAGGCTGGGGCTCATTTG-3′ (sense) and 5′-AGGGGCCATCCACAGTCTTC-3′
(antisense). Amplification of fibulin-3 and GAPDH was
performed with 1 cycle at 95°C for 10 min, and 40 cycles at 95°C
for 15 sec and 60°C for 60 sec. Calculation of the relative
expression of each transcript was performed using the
2−ΔΔCt method.
Methylation-specific PCR (MSP)
Genomic DNA was extracted from lung cancer specimens
using a TissueGen DNA kit (CWbiotech, Beijing, China). Genomic DNA
(2 μg) was denatured with 0.2 M NaOH. Then, 10 mM hydroquinone and
3 M sodium-bisulfite (both from Sigma) were added. The solution was
incubated at 55°C for 16 h. DNA samples were then purified using a
Wizard® DNA Purification Resin (Promega Corporation,
Madison, WI, USA). In this procedure unmethylated (but not
methylated) cytosines are converted to uracil, which is then
converted to thymidine during subsequent PCR to give sequence
differences between methylated and unmethylated DNA. The modified
DNA was used as a template both for MSP and USP. The primer
sequences for the methylated fibulin-3 gene were:
5′-GTAGTTTTAGGGGATCGTCGC-3′ (sense) and 5′-TCCCCGACACGCTACCTTCG-3′
(antisense); and for the unmethylated allele were: 5′-GAGTAGTTTTAGG
GGATTGTTGT-3′ (sense) and 5′-TCCCCAACACACTACCT TCA-3′ (antisense).
The PCR products were separated on 2% agarose gel with ethidium
bromide and visualized under UV illumination.
Western blot analysis
The tissues or cells were lysed in lysis buffer (20
mM Tris-HCl, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100) containing a
protease inhibitor cocktail (Sigma). The extracted protein amounts
were quantified using the BCA protein assay kit (CWbiotech).
Equivalent amounts of protein (40 μg) were separated using 10%
SDS-PAGE and transferred to a PVDF membrane (Millipore, Billerica,
MA, USA). Western blotting was performed using the following
primary antibodies: fibulin-3 (sc-365224), MMP-2 (sc-8835), MMP-9
(sc-12759), phospho-JNK (sc-12882), p38 (sc-7149), phospho-p38
(sc-166182), ERK 1/2 (sc-135900), phospho-ERK 1/2 (sc-16982) and
β-actin (sc-130657) (all from Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA). The binding for each specific antibody was detected
with horseradish peroxidase (HRP)-conjugated respective secondary
antibodies and ECL solutions (both from Amersham Biosciences,
Amersham, UK).
Immunohistochemical staining
Tissues were fixed with 10% buffered formalin,
embedded in paraffin, and decalcified in 10% EDTA solution.
Representative blocks were then cut into 4-μm sections,
deparaffinized with xylene and rehydrated in a series of ethanol
washes (100, 90, 80 and 70%). The sections were then incubated with
3% H2O2 and 5% serum to block endogenous
peroxidase activity and non-specific binding. For the fibulin-3
protein, sections were incubated with the anti-human fibulin-3
antibody. The sections were then incubated with biotinylated
secondary antibodies and visualized by DAB. Counterstaining was
carried out with hematoxylin. The sections were dehydrated in
alcohol and coverslipped. For the negative controls, PBS replaced
the primary antibody.
Enzyme-linked immunosorbent assay
Levels of fibulin-3 were measured in peripheral
blood of the patients with NSCLC and quantified in
nanograms/milliliter with the use of the human fibulin-3
enzyme-linked immunosorbent assay (Uscn Life Science Inc., Wuhan,
China).
Isolation and enumeration of circulating
tumor cells (CTCs)
Blood samples (7.5 ml) from the patients with NSCLC
were drawn into CellSave® tubes (Veridex LLC, Warren,
NJ, USA), which were maintained at room temperature (RT) and
processed within 72 h of collection. CTCs were defined as nucleated
EpCAM-positive cells, lacking CD45 but expressing cytoplasmic
cytokeratins 8, 18 and 19. All CTC evaluations were performed by
qualified and trained personnel.
Transient transfection
The plasmid pEGFP-C1-fibulin-3 (gifted by Li Yin,
China Medical University) was transfected into A549 cells by
Lipofectamine™ 2000 (Invitrogen Life Technologies) according to the
manufacturer's protocol. Twenty four hours post-transfection,
drug-resistant clones were isolated and expanded. A549 cells
transfected with pEGFP-C1 were used as a control. All gene
expression studies were conducted using pools of colonies (n=150)
to avoid a clonal bias.
Immunofluorescence
Cells were washed with PBS, fixed in 4%
paraformaldehyde, permeabilized in 1% Triton X-100 for 5 min, and
blocked with 5% bovine serum albumin (BSA) in PBS containing 0.5%
Triton X-100 for 1 h. Fibulin-3 expression was detected using the
anti-fibulin-3 antibody for 1 h at room temperature. Cells were
washed with PBS and incubated with the appropriate secondary
fluorophore-conjugated antibody for 1 h at RT, washed with PBS, and
mounted using SlowFade® Gold Antifade reagent
(Invitrogen Life Technologies). The secondary antibody used for
detection of fibulin-3 was Alexa Fluor® 594 rat
anti-mouse IgG (H+L) (Invitrogen Life Technologies).
MTT assay
Cells were plated in 96-well plates (1,500
cells/well). After 24 h, 0.5 mg/ml
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT;
Sigma) was added to each well. Four hours later, cells were lysed
with dimethyl sulfoxide (DMSO), and the absorbance rates were
measured at 550–560 nm using a microplate reader (Bio-Rad,
Hercules, CA, USA).
In vitro wound healing assay
Cells were grown in a 6-well dish. A confluent
monolayer of cells was scratched with a 200-μl pipette tip to
simulate a wound. Cells were washed twice with PBS and then
supplemented with medium and incubated for 4 h at 37°C. Cell
migration into the wounded area was monitored microscopically.
Images were captured at the interface of the unwounded and wounded
areas.
Transwell migration assay
Cells were plated at 2×105 cells/well in
0.5 ml of serum-free medium in 24-well Matrigel-coated Transwell
units with polycarbonate filters (8-μm pore size, Costar Inc.,
Milpitas, CA, USA). The lower chamber was loaded with 600 μl of
RPMI-1640 containing 10% FBS. After incubation for 24 h in normal
culture conditions, we did not see any cells floating in the upper
chamber, indicating that the cells had not undergone apoptosis at
this time-point. The top surface of the membrane was gently
scrubbed with a cotton bud and fixed in 4% paraformaldehyde (Sigma)
and stained with crystal violet. The cells that had invaded through
the membrane filters were counted using a light microscope. Ten
microscopic fields (x400) were randomly selected to count the
cells.
Statistical analysis
All experiments were performed in triplicate, and
the results are expressed as the means ± standard deviation (SD).
Kaplan-Meier survival plots were generated and comparisons were
made with log-rank statistics. P-values <0.05 were considered to
indicate statistically significant results. All the statistical
analyses and graphics were performed with GraphPad Prism version
5.00 for Windows (GraphPad Software, San Diego, CA, USA).
Results
Detection of fibulin-3 mRNA and protein
levels in lung tissue samples
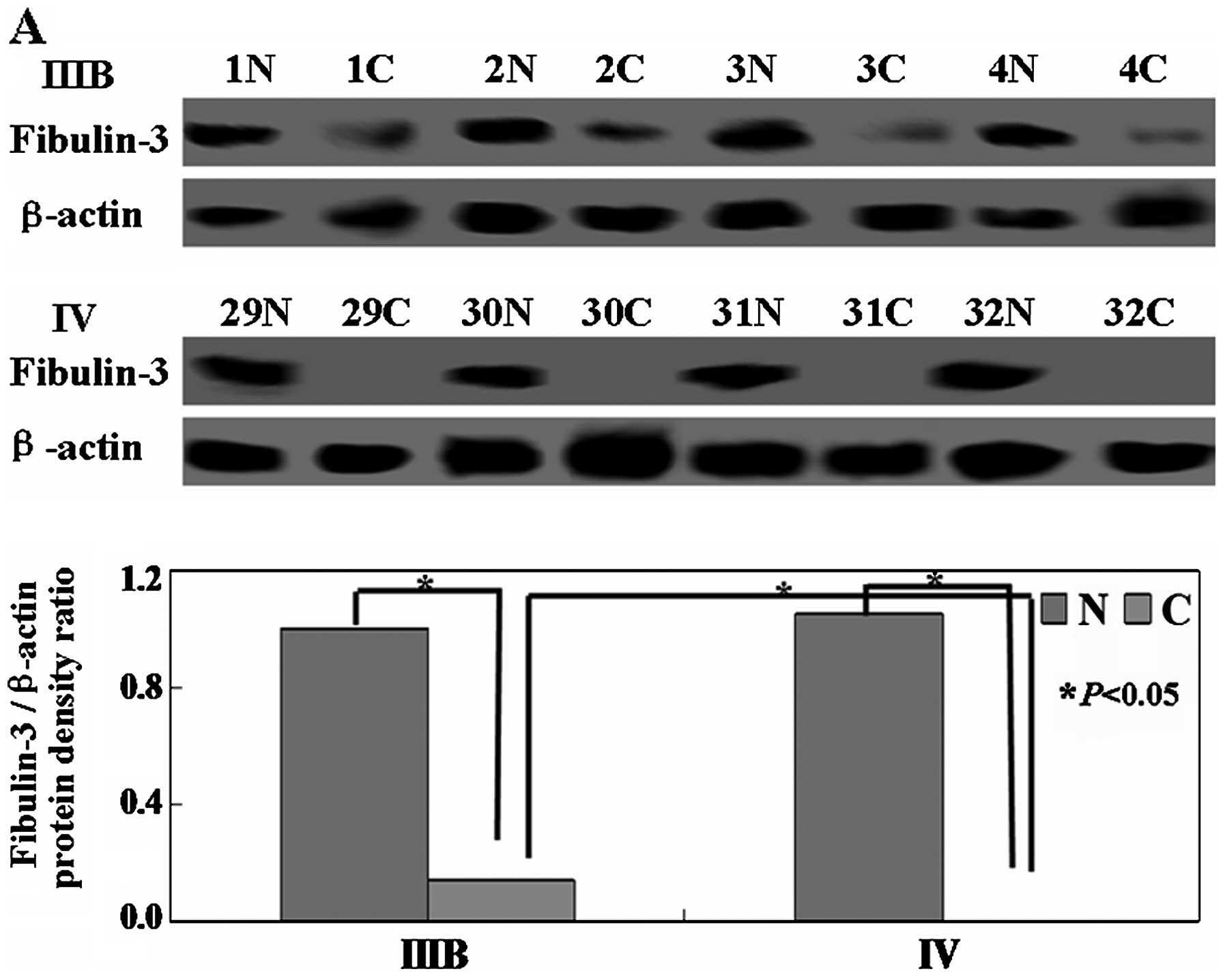
Real-time PCR and western blot assays were performed
to detect the levels of fibulin-3 mRNA and protein. As shown in
Fig. 1A and B, the levels of
fibulin-3 mRNA and protein were low in the low-stage NSCLC (IIIB)
tissues and undetectable in the advanced stage (IV) tissues
(P<0.05). Immunohistochemistry was then performed on 56 NSCLC
tissue samples and the corresponding non-tumor tissue samples.
Representative examples of fibulin-3 protein expression in NSCLC
tissue samples showed that fibulin-3 protein was weakly expressed
in the lung cancer specimens, but was highly expressed in the
normal areas of the specimens (Fig.
1C). A correlation was noted between fibulin-3 promoter
methylation and downregulation of fibulin-3 mRNA levels in
the tumor samples. Methylated tumor lung tissue samples showed loss
of fibulin-3 mRNA (Fig. 1D).
Taken together, the loss of fibulin-3 gene expression in
NSCLC may be correlated with the methylation of the
fibulin-3 gene promoter.
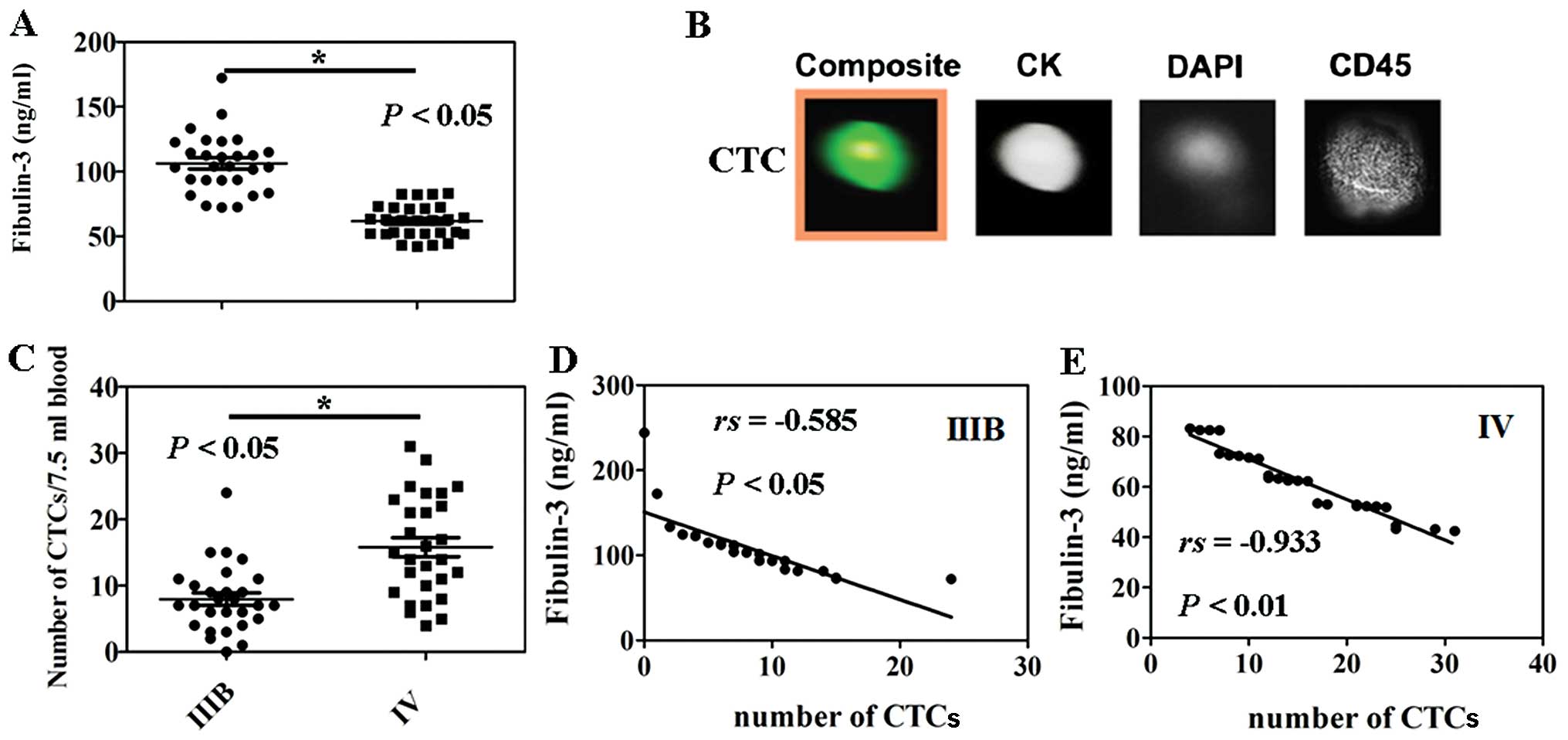
Correlation of fibulin-3 serum levels and
CTCs
A statistically significant difference was observed
between circulating fibulin-3 levels in the low stage NSCLC (IIIB)
patients and the advanced stage (IV) patients (P<0.05, Fig. 2A). In the present study, we isolated
and enumerated the CTCs in patients using the
CellSearch® system (Fig.
2B). Less CTCs were noted in the peripheral blood of patients
with low stage NSCLC (IIIB) when compared with the patients with
advanced stage NSCLC (IV) (P<0.05, Fig. 2C). The plasma levels of fibulin-3
and the number of CTCs were significantly negatively correlated
between stage IIIB and IV NSCLC patients (r=−0.585, P<0.05,
Fig. 2D or r=−0.933, P<0.05,
Fig. 2E), respectively.
Fibulin-3 and clinicopathological
variables
We analyzed the potential relationship between the
expression of fibulin-3 and the clinicopathological characteristics
of the NSCLC patients. Fibulin-3 expression was not associated with
any of the clinicopathological characteristics of the patients with
stage IIIB or IV NSCLC (Table I).
However, the serum level of fibulin-3 was associated with lymphatic
invasion (P<0.05, Table II). To
investigate the correlation of serum level of fibulin-3 with
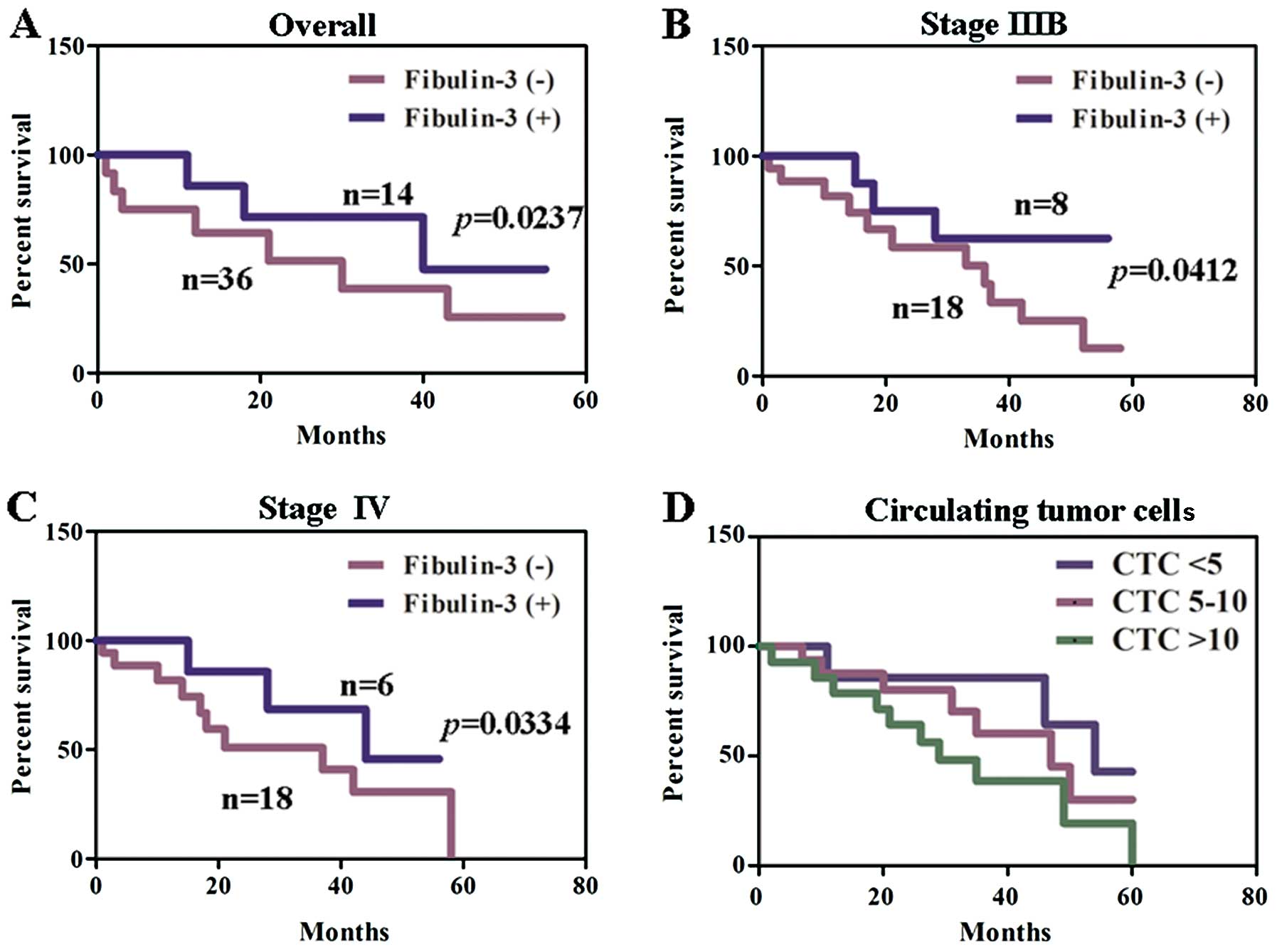
patient survival, the survival data from 50 patients with NSCLC (6
missing at follow-up) were assessed. In patients with stage IIIB
and IV NSCLC, comparison by the Kaplan-Meier method according to
low vs. high fibulin-3 expression showed a significant difference
in the 5-year survival rate (P<0.05, Fig. 3A–C). The patients with <5 CTCs
showed a higher survival rate when compared with the patients with
5–10 CTCs or with >10 CTCs. A significantly different survival
rate was confirmed between the patients with 5–10 CTCs and the
patients with >10 CTCs in the Cox model (P<0.05, Fig. 3D).
 | Table ICorrelation between fibulin-3
expression and clinicopathological parameters of the patients with
stage IIIB and IV NSCLC. |
Table I
Correlation between fibulin-3
expression and clinicopathological parameters of the patients with
stage IIIB and IV NSCLC.
| Clinicopathological
features | Fibulin-3 expression
(IIIB) | Fibulin-3 expression
(IV) |
|---|
|
|
|---|
| n | − | + | PR (%) | χ2 | P-value | n | − | + | PR (%) | χ2 | P-value |
|---|
| Gender | | | | | 0.69 | 0.406 | | | | | 1.70 | 0.192 |
| Female | 8 | 4 | 4 | 50.0 | | | 10 | 6 | 4 | 40.0 | | |
| Male | 20 | 15 | 5 | 25.0 | | | 18 | 16 | 2 | 11.1 | | |
| Age (years) | | | | | 0.00 | 0.976 | | | | | 0.05 | 0.827 |
| <50 | 11 | 8 | 3 | 27.3 | | | 8 | 6 | 2 | 25.0 | | |
| ≥50 | 17 | 11 | 6 | 35.3 | | | 20 | 16 | 4 | 20.0 | | |
|
Differentiation | | | | | 0.12 | 0.734 | | | | | 1.85 | 0.173 |
| Well or
moderate | 9 | 6 | 3 | 33.3 | | | 6 | 3 | 3 | 50.0 | | |
| Poor | 19 | 13 | 6 | 31.6 | | | 22 | 19 | 3 | 13.6 | | |
| Lymphatic
invasion | | | | | 0.00 | 1.00 | | | | | 0.18 | 0.673 |
| − | 14 | 9 | 5 | 35.7 | | | 19 | 14 | 5 | 26.3 | | |
| + | 14 | 10 | 4 | 28.6 | | | 9 | 8 | 1 | 11.1 | | |
| Venous
invasion | | | | | 1.13 | 0.287 | | | | | 0.38 | 0.536 |
| − | 12 | 6 | 6 | 50.0 | | | 18 | 13 | 5 | 27.8 | | |
| + | 16 | 13 | 3 | 18.8 | | | 10 | 9 | 1 | 10.0 | | |
| Histological
type | | | | | 0.23 | 0.891 | | | | | 0.01 | 0.905 |
| Squamous cell | 9 | 6 | 3 | 33.3 | | | 9 | 7 | 2 | 22.2 | | |
|
Adenocarcinoma | 8 | 5 | 3 | 37.5 | | | 9 | 7 | 2 | 22.2 | | |
| Small cell | 11 | 8 | 3 | 27.3 | | | 10 | 8 | 2 | 20.0 | | |
| Tumor size
(cm) | | | | | 0.07 | 0.794 | | | | | 0.02 | 0.893 |
| <3 | 15 | 11 | 4 | 26.7 | | | 17 | 14 | 3 | 17.6 | | |
| ≥3 | 13 | 8 | 5 | 38.5 | | | 11 | 8 | 3 | 27.3 | | |
| pN category | | | | | 0.19 | 0.979 | | | | | 1.38 | 0.713 |
| pN0 | 6 | 4 | 2 | 33.3 | | | 6 | 5 | 1 | 16.7 | | |
| pN1 | 7 | 5 | 2 | 28.6 | | | 7 | 5 | 2 | 28.6 | | |
| pN2 | 8 | 5 | 3 | 37.5 | | | 6 | 4 | 2 | 33.3 | | |
| pN3 | 7 | 5 | 2 | 28.6 | | | 9 | 8 | 1 | 11.1 | | |
 | Table IICorrelation of the
clinicopathological features of the NSCLC patients and serum levels
of fibulin-3 using ELISA. |
Table II
Correlation of the
clinicopathological features of the NSCLC patients and serum levels
of fibulin-3 using ELISA.
| Clinicopathological
features | Positive for
fibulin-3 (n=18) | Negative for
fibulin-3 (n=38) | P-value |
|---|
| Mean age
(years) | 61.4±5.7 | 64.3±5.8 | 0.742 |
| Gender | | | 0.294 |
| Female | 8 | 10 | |
| Male | 10 | 28 | |
|
Differentiation | | | 0.846 |
| Well or
moderate | 5 | 10 | |
| Poor | 13 | 28 | |
| Lymphatic
invasion | | | 0.003 |
| − | 5 | 28 | |
| + | 13 | 10 | |
| Venous
invasion | | | 0.512 |
| − | 8 | 22 | |
| + | 10 | 16 | |
| Histological
type | | | 0.155 |
| Squamous cell | 3 | 15 | |
|
Adenocarcinoma | 8 | 9 | |
| Small cell | 7 | 14 | |
| Tumor size
(cm) | | | 0.901 |
| <3 | 11 | 21 | |
| ≥3 | 7 | 17 | |
| pN category | | | 0.391 |
| pN0 | 4 | 8 | |
| pN1 | 2 | 12 | |
| pN2 | 6 | 8 | |
| pN3 | 6 | 10 | |
| Stage | | | 0.775 |
| IIIB | 9 | 19 | |
| IV | 9 | 19 | |
Effect of fibulin-3 expression on the
proliferation and migratory capacity in A549 cells
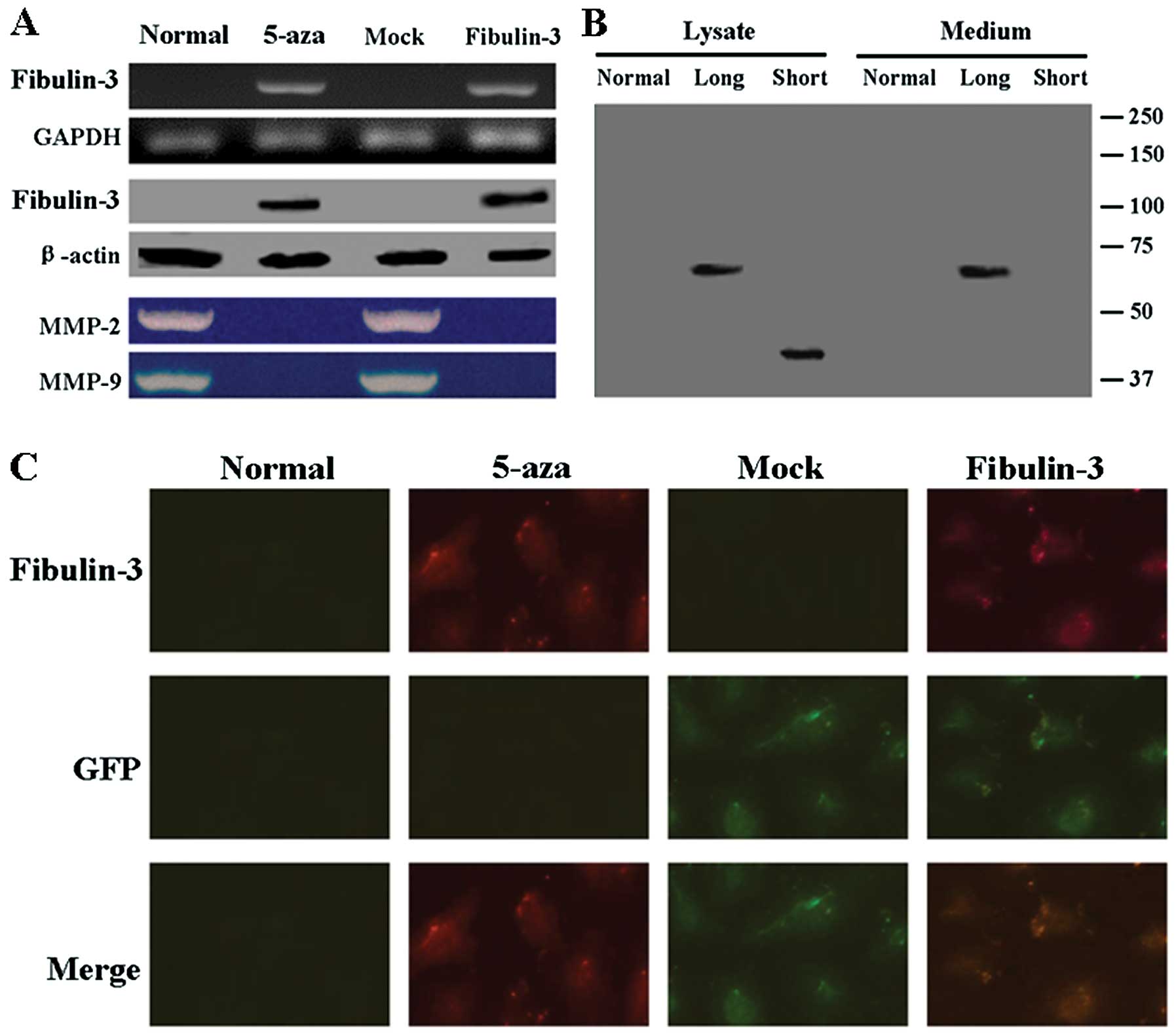
A549 cells were transfected with the
pEGFP-C1-fibulin-3 expression vector, and the levels of fibulin-3
protein and mRNA were measured by western blotting, RT-PCR and
immunofluorescence. As shown in Fig.
4A, the results of RT-PCR and western blot analysis confirmed
exogenous expression of fibulin-3 in A549 cells after transfection.
Immunofluorescence analysis showed the localization of GFP and
fibulin-3 in the fibulin-3-expressing A549 cells (Fig. 4C). We observed that the long form of
fibulin-3 was largely secreted into the culture medium, whereas the
short form was expressed at lower levels, accumulated in the total
cell lysate, and was virtually absent from the culture medium
(Fig. 4B). We next assessed the
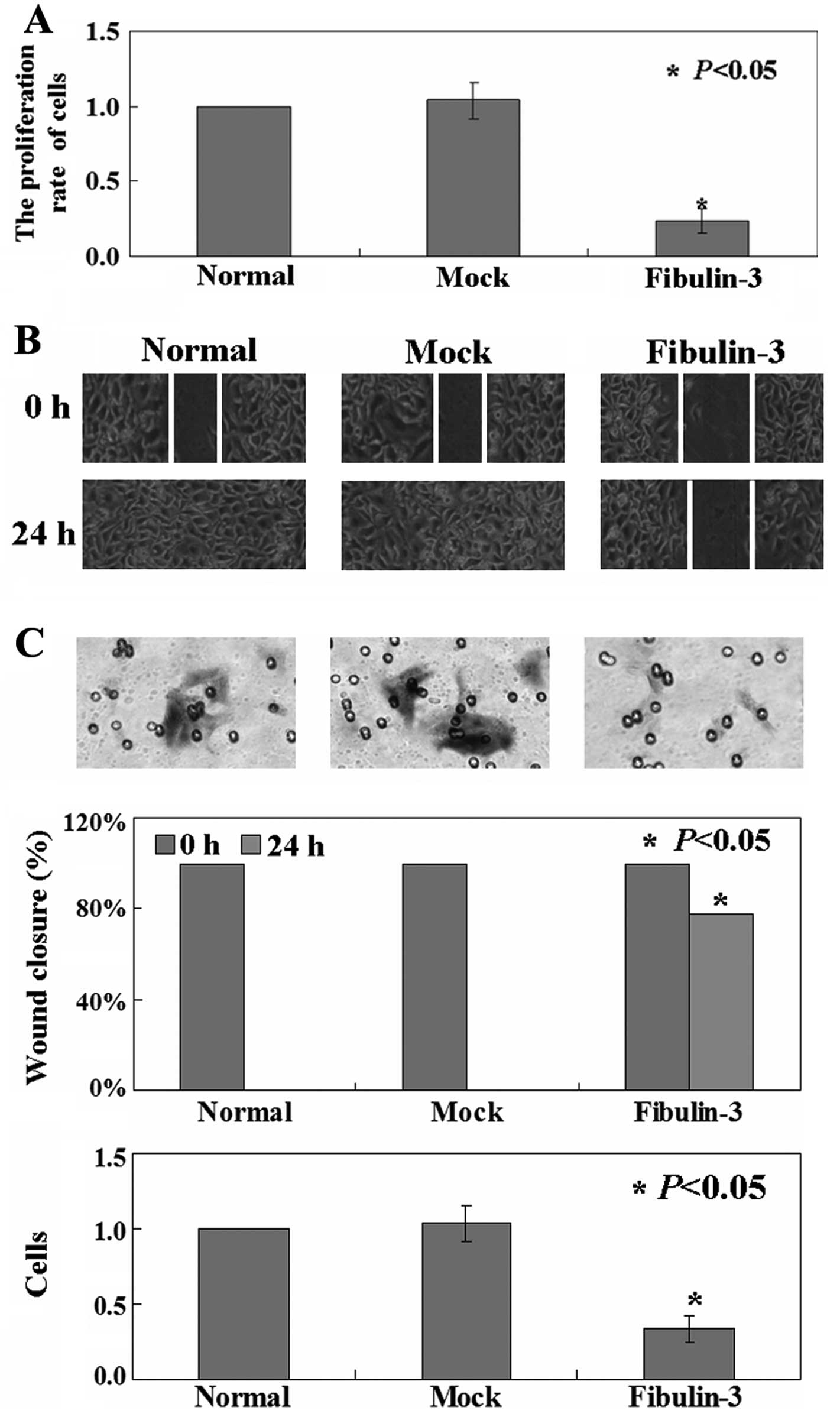
effect of fibulin-3 expression on the growth of A549 cells. The
growth curves obtained by MTT assays demonstrated that fibulin-3
inhibited the growth of A549 cells (P<0.05, Fig. 5A). Next, the mobility of
fibulin-3-expressing A549 cells was determined using wound-healing
and Transwell assays. As shown in Fig.
5B, the percentage of wound closure of A549 cells (16.7%) was
decreased when compared to the untreated (100.0%) and mock
transfected (100.0%) cells (P<0.05). Overexpression of the
fibulin-3 gene significantly inhibited the invasion of A549
cells into the Matrigel as detected using Transwell assay
(P<0.05, Fig. 5C). Furthermore,
the activities of MMP-2 and −9 were significantly decreased in
cells transfected with pEGFP-C1-fibulin-3 when compared to the
untreated cells (Fig. 4A).
Fibulin-3 suppresses the p38/MAPK
signaling pathway
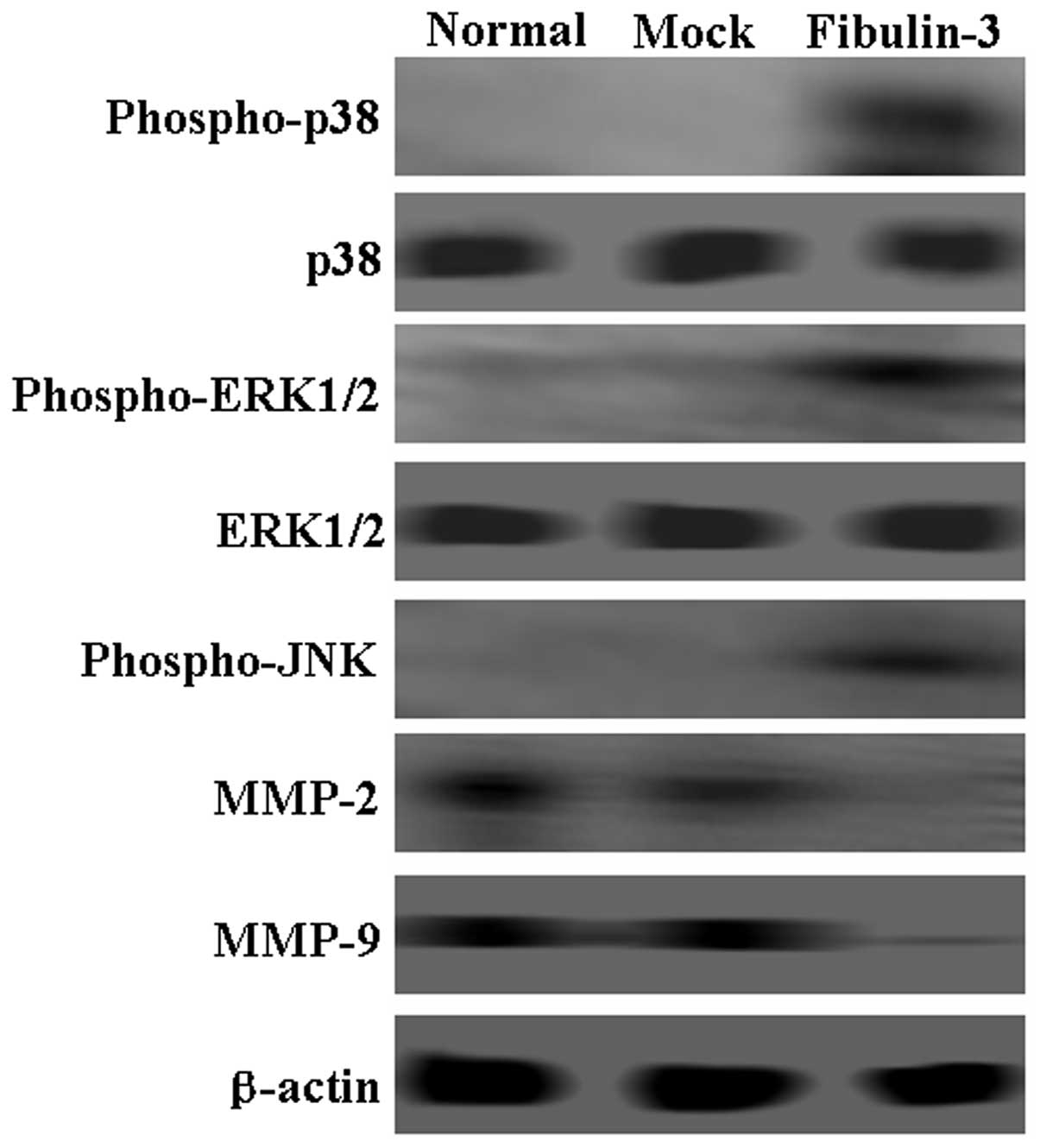
To identify the mechanism of fibulin-3, western blot
assays were performed to detect changes in possible signaling
pathway proteins. Expression of MMP-2 and −9 was downregulated in
fibulin-3-expressing A549 cells. While total levels of p38 and
ERK1/2 showed no changes, the levels of phospho-p38 and
phospho-ERK1/2 were observed to be significantly higher in the
fibulin-3-positive cells when compared to that in negative cells
(Fig. 6). Notably, phospho-JNK was
also increased in the fibulin-3-positive cells (Fig. 6). In combination, these results
suggest that fibulin-3 inhibited the mobility of A549 cells via the
p38 and ERK 1/2 proteins.
Discussion
Fibulin-3 has been found to be downregulated in
several types of solid tumors (14)
but surprisingly, is the most upregulated member of the fibulin
family in malignant gliomas (15).
Fibulin-3 is also upregulated in aggressive pancreatic
adenocarcinoma, resulting in enhanced in vivo orthotopic and
metastatic tumor growth (16).
Consistent with previous studies on lung cancer (17,18),
downregulation of fibulin-3 expression was observed in the present
study. These results suggest that fibulin-3 has dual functions as a
positive and negative regulator of cancer cell growth depending on
cell type. Wang et al(17)
found that the frequency of methylation of the fibulin-3
gene promoter was significantly higher in NSCLC tissue samples than
the frequency in the corresponding non-tumor tissue samples. Nomoto
et al(11) found that the
fibulin-3 gene was decreased in hepatocellular carcinoma
(HCC) tumor tissue, and 24 of 48 HCC samples showed promoter
hypermethylation, in the 24 methylated cases. In the study of Tong
et al(19), the
fibulin-3 gene showed a higher methylation frequency in
colorectal cancer (CRC) specimens while rare in non-cancer
colorectal tissues, and its promoter methylation state was
specifically associated with weak or absent expression of fibulin-3
protein. In the present study, we also confirmed that
downregulation of fibulin-3 was caused by promoter
hypermethylation. Notably, we did not find any association between
fibulin-3 expression and the clinicopathological characteristics of
the patients with stage IIB or IV NSCLC. However, in previous
studies, downregulation of fibulin-3 was associated with advanced
stage and lymph node metastasis in lung cancer (17) and CRC (19). We did find that circulating
fibulin-3 was associated with lymph node metastasis. The
discrepancy may be attributable to differences in the tumor
entities examined and in the microenvironment of the different
experimental settings.
Circulating tumor cells (CTCs) captured from
peripheral blood were recently used as a predictor for disease
outcome and therapy response in cancer patients (20). Cytokeratin-expressing cells can be
found in peripheral blood of advanced cancer patients but are rare
in healthy donors (21). In the
present study, we used the CellSearch® system which is
the only US Food and Drug Administration authorized test for CTC
enumeration in clinical practice to isolate CTCs (22). We found that fibulin-3 expression
decreased the number of CTCs. The results indicate that upregulated
expression of fibulin-3 can be used as a therapeutic strategy for
NSCLC. In addition, levels of fibulin-3 in plasma and effusions may
aid in determining the diagnosis and prognosis of pleural
mesothelioma (23). Our results
were lacking healthy controls, thus we could not conclude that
fibulin-3 is a marker for lung cancer. However, we confirmed that
fibulin-3 may be used to indicate the progression of lung cancer.
Circulating fibulin-3 level was correlated with favorable survival
in total, stage IIIB and IV NSCLC patients. Previous studies also
showed that reduced fibulin-3 expression was associated with worse
prognosis of breast cancer (9),
lung cancer (10) and
hepatocellular carcinoma (11).
Both in our previous study (10) and the present study, fibulin-3
expression was revealed to be associated with the regulation of
tumor cell growth and invasiveness in A549 cells by suppressing the
expression of MMP-9 and −2. Furthermore, the mechanism of fibulin-3
was identified by using western blotting. The positive regulation
of phospho-p38, phospho-ERK1/2 and phospho-JNK by fibulin-3 was
shown in the present study. p38-MAPK was previously shown to
regulate invasion by modulation of MMP-2/−9 mRNA levels and
zymographic activity in bladder cancer model (24). p38-MAPK also modulated the
inhibition of migration in 17β-estradiol-treated human colon cancer
cells by inhibition of MMP-2-/9 expression (25). Similar to the present study,
fibulin-3 inhibited invasion by upregulating p38-MAPK
phosphorylation and downregulating the MMP-2/−9 protein expression
in A549 cells.
Taken together, fibulin-3 downregulation is a common
abnormality in lung cancer tissue and its expression is regulated
by hypermethylation of the promoter region. Circulating fibulin-3
may play a role in tumor progression and the survival rate of
patients with NSCLC. Our results suggest that fibulin-3 negatively
modulates the invasiveness of lung cancer cells via regulation of
p38-MAPK and MMP-2/9. In particular, fibulin-3 negatively controls
the number of CTCs. These facts imply the diagnostic and
therapeutic potential of fibulin-3 for NSCLC. Further research is
needed prior to its clinical applications.
Acknowledgements
We are indebted to Li Yin for the plasmid
pEGFP-C1-fibulin-3 and constructive suggestions in the preparation
of this manuscript.
References
|
1
|
Jemal A, Bray F, Center MM, et al: Global
cancer statistics. CA Cancer J Clin. 61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Lam WK, White NW and Chan-Yeung MM: Lung
cancer epidemiology and risk factors in Asia and Africa. Int J
Tuberc Lung Dis. 8:1045–1057. 2004.PubMed/NCBI
|
|
3
|
Kobayashi N, Kostka G, Garbe JH, et al: A
comparative analysis of the fibulin protein family. Biochemical
characterization, binding interactions, and tissue localization. J
Biol Chem. 282:11805–11816. 2007. View Article : Google Scholar
|
|
4
|
Timpl R, Sasaki T, Kostka G and Ghu ML:
Fibulins: a versatile family of extracellular matrix proteins. Nat
Rev Mol Cell Biol. 4:479–489. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pupa SM, Giuffré S, Castiglioni F, et al:
Regulation of breast cancer response to chemotherapy by fibulin-1.
Cancer Res. 67:4271–4277. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yi CH, Smith DJ, West WW and Hollingsworth
MA: Loss of fibulin-2 expression is associated with breast cancer
progression. Am J Pathol. 170:1535–1545. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Albig AR and Schiemann WP: Fibulin-5
antagonizes vascular endothelial growth factor (VEGF) signaling and
angiogenic sprouting by endothelial cells. DNA Cell Biol.
23:367–379. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Y and Marmorstein LY: Focus on
molecules: fibulin-3 (EFEMP1). Exp Eye Res. 90:374–375. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sadr-Nabavi A, Ramser J, Volkmann J, et
al: Decreased expression of angiogenesis antagonist EFEMP1 in
sporadic breast cancer is caused by aberrant promoter methylation
and points to an impact of EFEMP1 as molecular biomarker. Int J
Cancer. 124:1727–1735. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yue W, Dacic S, Sun Q, et al: Frequent
inactivation of RAMP2, EFEMP1 and Dutt1 in lung cancer by promoter
hypermethylation. Clin Cancer Res. 13:4336–4344. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nomoto S, Kanda M, Okamura Y, et al:
Epidermal growth factor-containing fibulin-like extracellular
matrix protein 1, EFEMP1, a novel tumor-suppressor gene
detected in hepatocellular carcinoma using double combination array
analysis. Ann Surg Oncol. 17:923–932. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lecka-Czernik B, Lumpkin CK Jr and
Goldstein S: An overexpressed gene transcript in senescent and
quiescent human fibroblasts encoding a novel protein in the
epidermal growth factor-like repeat family stimulates DNA
synthesis. Mol Cell Biol. 15:120–128. 1995.PubMed/NCBI
|
|
13
|
Hata K, Dhar DK, Watanabe Y, et al:
Expression of metastin and a G-protein-coupled receptor (AXOR12) in
epithelial ovarian cancer. Eur J Cancer. 43:1452–1459. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Albig AR, Neil JR and Schiemann WP:
Fibulins 3 and 5 antagonize tumor angiogenesis in vivo. Cancer Res.
66:2621–2629. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu B, Thirtamara-Rajamani KK, Sim H and
Viapiano MS: Fibulin-3 is uniquely upregulated in malignant gliomas
and promotes tumor cell motility and invasion. Mol Cancer Res.
7:1756–1770. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Seeliger H, Camaj P, Ischenko I, et al:
EFEMP1 expression promotes in vivo tumor growth in human pancreatic
adenocarcinoma. Mol Cancer Res. 7:189–198. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang R, Zhang YW and Chen LB: Aberrant
promoter methylation of FBLN-3 gene and clinicopathological
significance in non-small cell lung carcinoma. Lung Cancer.
69:239–244. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim EJ, Lee SY, Woo MK, et al: Fibulin-3
promoter methylation alters the invasive behavior of non-small cell
lung cancer cell lines via MMP-7 and MMP-2 regulation. Int J Oncol.
40:402–408. 2012.PubMed/NCBI
|
|
19
|
Tong JD, Jiao NL, Wang YX, Zhang YW and
Han F: Downregulation of fibulin-3 gene by promoter methylation in
colorectal cancer predicts adverse prognosis. Neoplasma.
58:441–448. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cristofanilli M, Hayes DF, Budd GT, et al:
Circulating tumor cells: a novel prognostic factor for newly
diagnosed metastatic breast cancer. J Clin Oncol. 23:1420–1430.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Osta WA, Chen Y, Mikhitarian K, et al:
EpCAM is overexpressed in breast cancer and is a potential target
for breast cancer gene therapy. Cancer Res. 64:5818–5824. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Allard WJ, Matera J, Miller MC, et al:
Tumor cells circulate in the peripheral blood of all major
carcinomas but not in healthy subjects or patients with
nonmalignant diseases. Clin Cancer Res. 10:6897–6904. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pass HI, Levin SM, Harbut MR, et al:
Fibulin-3 as a blood and effusion biomarker for pleural
mesothelioma. N Engl J Med. 367:1417–1427. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hsu HH, Liu CJ, Shen CY, et al: p38α MAPK
mediates 17β-estradiol inhibition of MMP-2 and −9 expression and
cell migration in human lovo colon cancer cells. J Cell Physiol.
227:3648–3660. 2012.
|
|
25
|
Kumar B, Koul S, Petersen J, et al: p38
mitogen-activated protein kinase-driven MAPKAPK2 regulates invasion
of bladder cancer by modulation of MMP-2 and MMP-9 activity. Cancer
Res. 70:832–841. 2010. View Article : Google Scholar : PubMed/NCBI
|